Abstract
The heat-shock response in Escherichia coli depends primarily on the transient increase in the cellular level of heat-shock sigma factor σ32 encoded by the rpoH gene, which results from both enhanced synthesis and transient stabilization of normally unstable σ32. Heat-induced synthesis of σ32 was previously shown to occur at the translation level by melting the mRNA secondary structure formed within the 5′ coding sequence of rpoH including the translation initiation region. The subsequent decrease in the σ32 level during the adaptation phase has been thought to involve both shutoff of synthesis (translation) and destabilization of σ32-mediated by the DnaK–DnaJ chaperones, although direct evidence for translational repression was lacking. We now show that the heat-induced synthesis of σ32 does not shut off at the translation level by using a reporter system involving translational coupling. Furthermore, the apparent shutoff was not observed when the synthesis rate was determined by a very short pulse labeling (15 s). Examination of σ32 stability at 10 min after shift from 30 to 42°C revealed more extreme instability (t1/2=20 s) than had previously been thought. Thus, the dynamic change in σ32 stability during the heat-shock response largely accounts for the apparent shutoff of σ32 synthesis observed with a longer pulse. These results suggest a mechanism for maintaining the intricate balance between the antagonistic pathways: the rpoH translation as determined primarily by ambient temperature and the turnover of σ32 as modulated by the chaperone (and presumably protease)-mediated autogenous control.
Most organisms respond to heat or other stresses by transiently inducing molecular chaperones and other heat-shock proteins (HSPs) to cope with stress-induced protein damage (1). When Escherichia coli cells are exposed to modest heat shock by a shift from 30 to 42°C, the synthesis of HSP increases for several minutes (induction phase) and gradually decreases (adaptation phase) to reach a new steady-state level within 20–30 min. The induction results from a rapid but transient increase in the cellular level of σ32 (the rpoH gene product) which directs RNA polymerase to transcribe specifically from heat-shock promoters (2–4). The increase in σ32 level results from both increased synthesis and stabilization of normally unstable σ32 (t1/2 = 1 min), whereas the subsequent decrease in σ32 has been thought to depend on shutoff of synthesis and destabilization of σ32 by the DnaK–DnaJ chaperone-mediated autogenous negative control (5–9). The free pool of DnaK and/or DnaJ was thought to act as a cellular thermometer that modulates expression of all HSPs by monitoring the state of protein folding (9–11).
Heat-induced stabilization of σ32 occurs rapidly although transiently. The half-life of σ32 increases from 1 to 8 min for the first several minutes, and returns to about 1 min by 10 min after shift (7–9). The initial stabilization probably results from sequestering σ32 away from the DnaK–DnaJ chaperones because of heat-induced accumulation of unfolded or misfolded proteins and facilitating σ32 to bind core RNA polymerase, whereas subsequent destabilization results from accumulation of DnaK–DnaJ chaperones and proteases caused by the increase in σ32 (9–11). Accumulation of abnormal proteins without temperature upshift induces HSP synthesis (12) through stabilization but not induced synthesis of σ32 (13, 14). Although the membrane-bound ATP-dependent metalloprotease FtsH (HflB) was first shown to be responsible for rapid turnover of σ32 (15, 16), a set of cytosolic proteases including HslVU (ClpYQ) also participate in degradation of σ32 in vivo and in vitro (17, 18). The bulk of heat-shock proteases may therefore collectively serve to modulate the heat-shock response as well as degrade much of misfolded or abnormal proteins to cope with heat and other stresses.
Heat-induced synthesis of σ32 occurs primarily at the level of translation and is independent of the DnaK–DnaJ chaperone functions (8, 9, 19, 20). Mutational analyses of expression of rpoH–lacZ gene fusion combined with in vitro structural probing of rpoH mRNA established the importance of secondary structure (with appropriate stability) of the 5′ portion (+1 to 230 nt) of mRNA formed between the translation initiation region and part of the internal region for thermoregulation (20–22). Temperature-melting profiles of RNA segments (−60 to + 247 nt) with or without mutation(s) revealed an inverse correlation between thermostability and expression in vivo. Moreover, toeprint analyses with a synthetic mRNA fragment, purified 30S ribosome, and tRNAfMet revealed a strong correlation between the formation of mRNA-30S ribosome-tRNAfMet ternary complex in vitro and expression in vivo at different temperatures (23). These results led us to propose that the rpoH mRNA alone, with no additional regulatory factors, acts as a thermosensor in the translational control of σ32. However, a major question remained concerning the nature of shutoff of σ32 synthesis during the adaptation phase.
Early observation of shutoff of σ32 synthesis even under control of the λPL promoter suggested that it occurs posttranscriptionally (6). E. coli mutants deficient in the dnaK–dnaJ chaperones showed both defective shutoff of synthesis and degradation of σ32, suggesting negative control mediated by the chaperones (8, 9). Analyses of the rpoH–lacZ gene fusion suggested involvement of an internal region of σ32 (region C; 122–144 aa) in the control of both DnaK–DnaJ-mediated shutoff of synthesis and stability of the σ32–β-galactosidase fusion protein (24), although core RNA polymerase binding rather than DnaK binding or σ32 stability was recently shown to be affected by mutations in this region (25, 26). In any event, shutoff of σ32 synthesis could not be separated from the control of σ32 stability, despite the clear distinction between controls of heat-induced synthesis and stability of σ32 (3, 4, 27). In addition, the changing stability of σ32 at different phases prevented accurate determination of synthesis rates during the heat-shock response.
We now show that the heat-induced synthesis of σ32 does not shut off at the translation level by using a reporter system without the complication of σ32 stability change. In addition, σ32 was shown to be destabilized to an extent much greater than had previously been thought. The control of the σ32 level during the heat-shock response appears to rest on an intricate balance between the efficiency of rpoH mRNA translation primarily determined by ambient temperature and the rate of σ32 turnover modulated by the chaperone (and protease)-mediated negative control.
Materials and Methods
Bacteria, Phage, and Growth Media.
E. coli K-12, strains MC4100 [araD Δ(argF-lac)U169 rpsL relA flbB deoC ptsF rbsR] (28) and MG1655 (prototroph) were used for most experiments. Strain KY1603 (ΔrpoH30∷kan zhf50∷Tn10 suhX401), which lacks σ32 and overproduces GroEL–GroES chaperones (29), was used to examine σ32-like function encoded by λrpoHBAZ and its derivatives. λTLF97–3 vector (30) was used to construct rpoH–lacZ gene fusions. Synthetic medium M9 (31) supplemented with 0.2% glucose, thiamine (2 μg/ml), and all amino acids except for Met (20 μg/ml each) was used for pulse-labeling experiments. MacConkey lactose agar (Difco) and L agar containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) (30 μg/ml) were used for isolation of lysogens carrying λrpoHBAZ prophage.
Construction of a rpoH–lacZ Fusion with the trpBA Junction.
The entire rpoH gene including promoters and 3′ untranslated region was inserted into the pACYC177 vector between the XhoI and BamHI sites. The C-terminal (Ala) and termination (UAA) codons of rpoH were replaced by a SphI site without changing the Ala residue by using PCR site-directed mutagenesis (Fig. 1). A DNA fragment containing the trpBA junction was obtained by PCR with pMS436S (32) as a template and 5′ and 3′ primers that correspond to the respective ends of the fragment with some protruding extra nucleotides to create the SphI or BamHI site, respectively. The resulting SphI–BamHI fragment was then substituted for the SphI–BamHI segment on pACYC177. The XhoI–BamHI fragment cut out from the latter plasmid was inserted into λTLF97–3 vector to yield an in-frame fusion with lacZ (λrpoHBAZ). A derivative of rpoHBAZ carrying the G123A mutation was obtained by replacing the rpoH gene on pACYC177 by the corresponding region of pFRP103 containing the mutation (21). To construct rpoHΔC17BAZ and 123ΔC17BAZ, the 17th codon from the C terminus of rpoH was converted to UGA by PCR mutagenesis and substituted for rpoH on pACYC177. Nucleotide sequences of all pertinent regions were confirmed by DNA sequencing. The resulting gene fusions were transferred to λTLF97–3 by in vitro packaging, introduced into strain MC4100, and confirmed for monolysogeny by PCR (33).
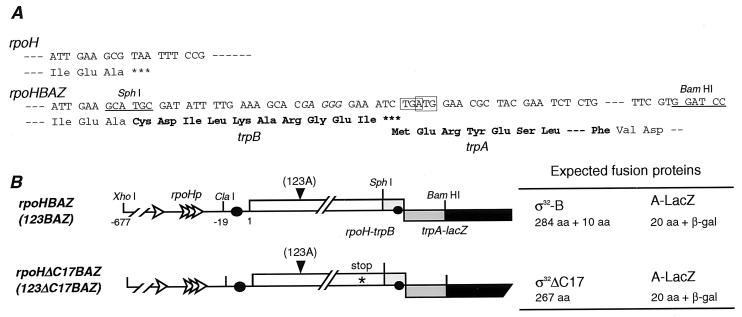
Figure 1.
(A) Sequences of the 3′ end of rpoH (Upper) and the trpBA junction of rpoHBAZ (Lower). In the rpoHBAZ construct, the C-terminal Ala codon of rpoH was fused in-frame with the 5′ end of trpB at the SphI site, whereas the 5′ end of trpA is fused in-frame to codon 9 of lacZ at the BamHI site. The stop codon (TGA) of trpB and the initiation codon (ATG) of trpA are boxed. Amino acids in bold letters originate from the trpBA junction. (B) Schematic diagrams of rpoHBAZ and its derivatives constructed as described in Materials and Methods. Arrowheads indicate the position of the G123A mutation that enhances translation, and parentheses indicate the mutation or constructs containing the mutation. The expected fusion protein products for each construct are indicated to the right.
Determination of Synthesis Rates and Stability of σ32 and Fusion Proteins.
Mid-logarithmic-phase cells were labeled with l-[35S]Met (600 or 1,200 Ci/mmol; 100 or 200 μCi/ml) for 15–60 s with or without chase with unlabeled Met (200 μg/ml) to determine synthesis rates or stability as indicated for each set of experiments. Portions of labeled cells were treated with 5% trichloroacetic acid, and the resulting precipitates were washed with acetone and suspended in buffer containing SDS. Samples with equal radioactivities were mixed with a fixed amount of CAG11033 cell extract containing a truncated form of σ32 (7) or JM103 cell extract containing β-galactosidase ω protein (both labeled with [35S]Met), and treated with antibody against σ32 or β-galactosidase (Organon Teknika–Cappel) for determining synthesis rates of σ32 or LacZ fusion proteins, respectively. Immunoprecipitates were subjected to SDS/PAGE essentially as described (22). Intensities of radioactive protein bands were quantified with a FUJIX (Tokyo) BAS2000 Imaging Analyzer to determine synthesis rates or stability after correction for their recovery by using truncated σ32 or ω as a reference.
β-Galactosidase Activity.
Cells were grown at 30°C in M9 medium and assayed for β-galactosidase activity by the standard procedure (34).
Recombinant DNA and Other General Techniques.
These were performed essentially as described by Sambrook et al. (35) and Miller (34).
Results
Translational Coupling Between σ32 and LacZ: Analysis of Shutoff of Synthesis Independent of Degradation.
To analyze the possible translational shutoff of σ32 without complications arising from change in σ32 stability during the heat-shock response, we constructed a reporter system by using translational coupling that occurs at the trpB–trpA junction, in which translation of the downstream gene (trpA) depends largely on complete translation of upstream trpB (36). The entire rpoH gene including promoters but omitting the stop codon (UAA) was fused with the 92-bp trpBA junction and with lacZ (Fig. 1A; rpoHBAZ). This generated two fusion genes encoding σ32 with the C-terminal 10 aa of TrpB (σ32-B) and LacZ with the N-terminal 20 aa of TrpA (A-LacZ) (Fig. 1B). The trpBA junction containing a direct overlap between the trpB stop codon (UGA) and the trpA initiation codon (AUG) was expected to couple translation of upstream σ32-B with that of downstream A-LacZ. The fusion was constructed on a single copy vector λTLF97–3 (30) and was inserted into the chromosome of strain MC4100, resulting in a lysogen that carries the λrpoHBAZ prophage.
To test the validity of this system, three mutant derivatives were constructed: the 123BAZ fusion exhibiting high-translation efficiency caused by the G123A mutation (of rpoH) which disrupts the mRNA secondary structure (21), and a pair of fusions that carry an extra stop codon (UGA) at the 17th codon from the C terminus of rpoH with or without G123A (rpoHΔC17BAZ and 123ΔC17BAZ) (Fig. 1B). When expression of A-LacZ from these constructs was determined by measuring β-galactosidase activity, expression from 123BAZ was 3.4-fold higher than that from the control (rpoHBAZ), as expected (Table 1). Moreover, expression from the two constructs carrying an extra stop codon was equally low, indicating that A-LacZ expression was markedly reduced independent of the efficiency of upstream translation. The low but significant activities observed with the latter constructs represent intrinsic trpA–lacZ translation independent of translation from upstream. These results showed effective translational coupling between the two fusion genes.
Table 1.
Expression of A-LacZ fusion protein from MC4100 (λrpoHBAZ) and its derivatives
| Construct | β-gal activity of A-LacZ, Miller U |
|---|---|
| rpoHBAZ | 222.93 ± 10.10 |
| rpoHΔC17BAZ | 82.36 ± 2.40 |
| 123BAZ | 762.25 ± 25.78 |
| 123ΔC17BAZ | 81.77 ± 2.21 |
Cells were grown in M9 medium at 30°C to midlog phase and assayed for β-galactosidase activity.
We also confirmed that the activity and expression pattern of σ32-B is similar to those of authentic σ32. When λrpoHBAZ was introduced into strain KY1603 (ΔrpoH strain that overproduces GroE proteins) unable to grow at above 40°C (29), the resulting lysogen regained the ability to grow at 42°C (data not shown). In contrast, λrpoHΔC17BAZ was inactive in this respect, indicating that σ32-B but not the C-terminal truncated derivative is functionally active and promotes transcription from the heat-shock promoters. Furthermore, when the λrpoHBAZ lysogen of wild-type MC4100 grown at 30°C was shifted to 42°C, synthesis of σ32-B as well as authentic σ32 encoded by the chromosomal rpoH was appreciably enhanced and shut off after about 3 min, as expected (Fig. 2A). The stability of σ32-B and σ32ΔC17 was also tested and found to be similar to that of authentic σ32 (Fig. 4 C and D).
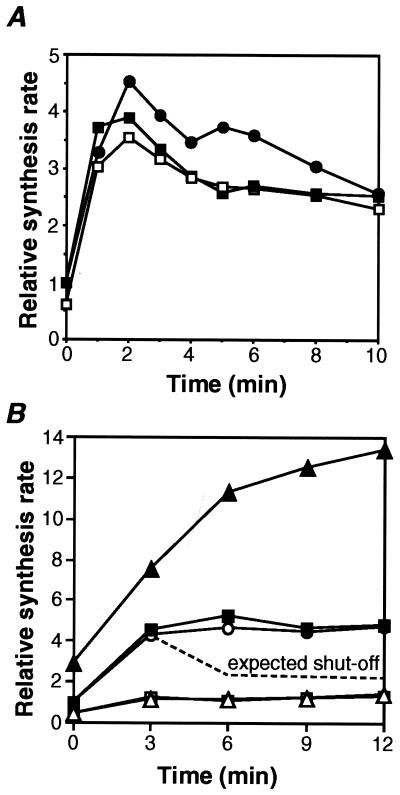
Figure 2.
Heat-induced synthesis of σ32, σ32-B, and A-LacZ in strain MC4100 carrying λrpoHBAZ or its derivative. (A) Cells were grown in minimal medium at 30°C and shifted to 42°C at t = 0. Samples were taken at the times indicated and pulse labeled with [35S]Met (1,200 Ci/mmol; 100 μCi/ml) for 30 s. The labeled σ32 and σ32-B were precipitated by σ32-specific antiserum and analyzed by SDS/PAGE followed by quantification as described in Materials and Methods. Values were normalized to the t = 0 value for each protein and then to σ32 in MC4100. (●) σ32 in MC4100; (■) σ32 in MC4100 (λrpoHBAZ); and (□) σ32-B in MC4100 (λrpoHBAZ). (B) Cells were grown and treated essentially as in A, except that the pulse labeling with [35S]Met was done at 600 Ci/mmol for 60 s followed by chase with excess unlabeled Met for 60 s. The labeled A-LacZ was precipitated with anti-β-galactosidase antiserum and analyzed by SDS/PAGE. Quantification was done as in A and normalized to t = 0 for each protein and then to rpoHBAZ. The dotted line indicates the expected curve if the heat-induced synthesis from rpoHBAZ was shut off. (○) TLF247; (■) rpoHBAZ; (▴) 123BAZ; (□) rpoHΔC17BAZ; and (▵) 123ΔC17BAZ.
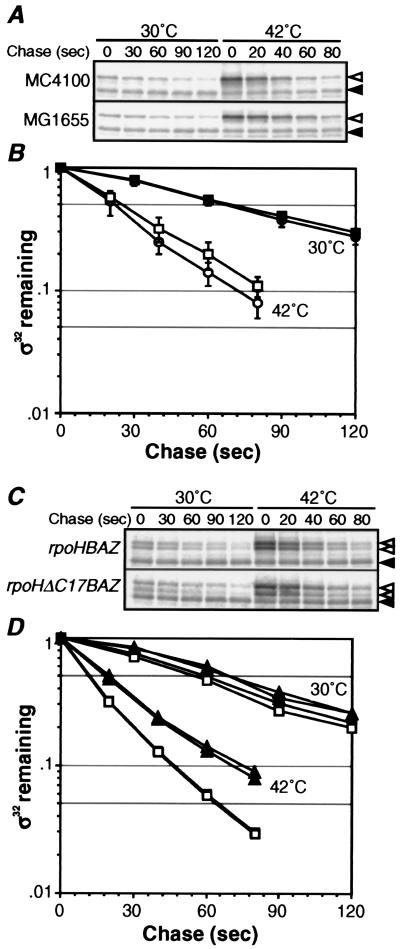
Figure 4.
Differential stability of σ32 at 30°C and 10 min after shift to 42°C. (A and C) SDS/PAGE patterns of σ32 remaining in pulse-chase experiments. Cells were grown at 30°C, shifted to 42°C, and portions taken at t = 0 and 10 min were pulse labeled with [35S]Met (1,200 Ci/mmol, 200 μCi/ml) for 30 s, and chased with excess unlabeled Met for 30 or 20 s at 30 or 42°C, respectively, and set as t = 0. Aliquots were then taken at the times indicated, and σ32 (◃) and σ32-B or σ32ΔC17 (hatched arrowheads) were immunoprecipitated and analyzed by SDS/PAGE as in Fig. 2A. (◂) Reference as in Fig. 2. (B and D) Quantification of protein stability. (B) ●, MC4100, 30°C; ○, MC4100, 42°C; ■, MG1655, 30°C; and □, MG1655, 42°C. (D) □, σ32-B in MC4100 (λrpoHBAZ); ■, σ32 in MC4100 (λrpoHBAZ); ▵, σ32ΔC17 in MC4100 (λrpoHΔC17BAZ); and ▴, chromosomally encoded σ32 in MC4100 (λrpoHΔC17BAZ).
Heat-Induced Translation of σ32 Does Not Shut Off During the Adaptation Phase.
By using the above reporter system, we asked whether heat-induced synthesis of σ32 is shut off at the level of translation. If shutoff occurs at the translational level, expression of A-LacZ from rpoHBAZ should be heat induced and shut off, like that observed with σ32-B or authentic σ32 (Fig. 2A). Thus, synthesis rates of A-LacZ from rpoHBAZ and its mutant derivatives were determined after temperature upshift. Contrary to the expectation, synthesis of A-LacZ was normally heat induced but did not shut off (Fig. 2B), like the rpoH–lacZ fusion TLF247 constructed (22) and thought to be unable to shut off because of the lack of region C (24). Moreover, the expression of A-LacZ from 123BAZ was about threefold higher than that from the control at 30°C and further enhanced on shift to 42°C as expected, but again failed to shut off.
In contrast, A-LacZ expression from rpoHΔC17BAZ or 123ΔC17BAZ carrying an extra stop codon was very low at 30°C and only slightly enhanced on shift to 42°C, consistent with the lack of translational coupling (Fig. 2B). This also showed that heat induction of A-LacZ observed with rpoHBAZ occurs as the result of translational coupling and not independent of upstream translation. These results strongly suggested that shutoff does not take place at the translational level. The apparent shutoff of synthesis of σ32 or σ32-LacZ fusion observed (7, 20, 24) therefore most probably reflects posttranslational events occurring during the adaptation phase.
Absence of Apparent Shutoff with Shorter Pulse Labeling.
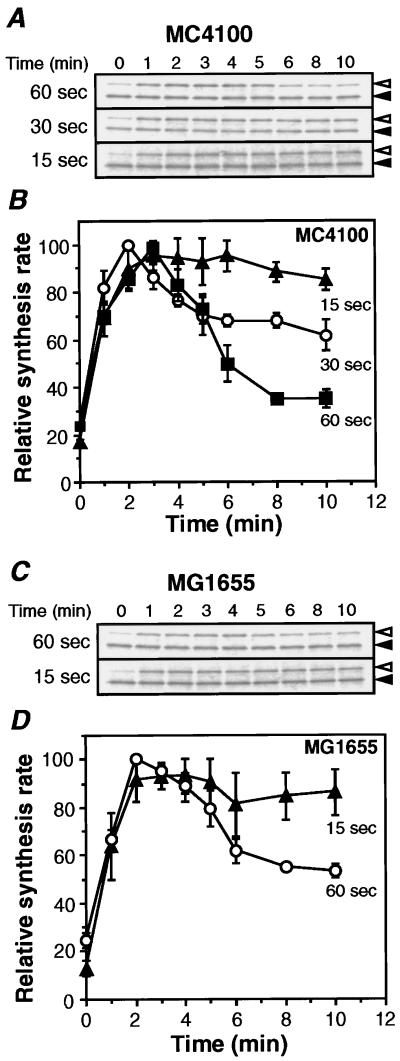
Because of the very low content of σ32, most previous work used 1-min pulse labeling with [35S]Met to measure the synthesis rates. However, it was difficult to follow the change in synthesis rates accurately because of the intrinsic instability. We therefore reexamined σ32 synthesis by using shorter pulse-labeling protocols with strain MC4100. It was anticipated that apparent synthesis rates obtained even with 30-s pulse can represent only approximations. In agreement with the results presented above, apparent shutoff was observed with a 60-s pulse but less clearly with a 30-s pulse (Fig. 3 A and B). Most significantly, when the 15-s pulse was used, little or no apparent shutoff occurred even after a 10-min incubation.
Figure 3.
Effects of varying length of pulse labeling on σ32 synthesis on heat shock. (A and C) SDS/PAGE patterns of σ32 synthesized in MC4100 or MG1655. Cells were grown in minimal medium at 30°C, shifted to 42°C, and samples taken at various points were pulse labeled for the indicated period with [35S]Met (600 Ci/mmol, 100 μCi/ml for 60-s pulse; 1,200 Ci/mmol, 100 μCi/ml for 30- or 15-s pulse). (◃) The labeled σ32 was precipitated and analyzed by SDS/PAGE as in Fig. 2A. (◂) Reference protein to correct for sample loss. (B and D) Quantitation of relative synthesis rates. The band intensities were quantified and normalized to the maximum value (set as 100) for each experiment and shown as relative synthesis rates. Average values of at least three independent experiments are presented with standard errors.
It should be noted that the initial heat induction seen for 2–3 min was essentially identical in all cases, and differential responses were found only after 4–5 min during the adaptation phase, by which time σ32 was known to be destabilized. Experiments with another prototrophic strain (MG1655) gave similar results, although the apparent shutoff with a 60-s pulse was slightly less than that with MC4100 (Fig. 3 C and D). These results unambiguously demonstrated the lack of shutoff of heat-induced σ32 synthesis and suggested that the apparent shutoff observed with longer pulse may result from much severer destabilization of σ32 than had previously been suspected.
Extreme Instability of σ32 During Adaptation Phase May Account for Apparent Shutoff of Synthesis.
Consistent with the above findings, the apparent synthesis rate of σ32 in cells pulse labeled at 10 min after temperature upshift was only about twofold higher than that at 30°C (t = 0) when the results with 60-s pulse were compared (Fig. 3), in contrast to the four- to fivefold heat induction of stable σ32-β-galactosidase fusion observed with TLF247 (22). This suggested that stability of σ32 during the adaptation phase may be lower than at 30°C. Indeed, recent results revealed that the half-life of σ32 in cells steadily growing at 42°C is much shorter (≈15 s) than at 30°C (1 min) with strain MG1655 (18). We therefore examined whether such a drastic destabilization of σ32 occurs within 10 min after temperature upshift.
Cells of MC4100 were pulse labeled for 30 s before (30°C) and 10 min after shift to 42°C, and chased with excess unlabeled Met at the respective temperatures. Based on these experiments, the half-life of σ32 at 30°C and 10 min after shift to 42°C was estimated to be 65 and 20 s, respectively (Fig. 4 A and B). Quite similar results were obtained with strain MG1655 (data not shown). These results indicated that stability of σ32 changes drastically and dynamically during the heat-shock response, as predicted from the above experiments on σ32 synthesis. Thus, normally unstable σ32 at 30°C (t1/2 = 1 min) is stabilized almost immediately for 4–5 min (t1/2 = 8 min) on shift to 42°C (8), followed by rapid destabilization leading to an extreme instability (t1/2 = 20 s) within 10 min. Apparent shutoff of synthesis of σ32 observed with a 60-s pulse may therefore be explained primarily by extreme instability of σ32 that presumably counterbalances the excessive synthesis of σ32 that would arise from continuously enhanced translation.
Discussion
By using the reporter system involving translational coupling and the very short pulse-labeling protocols, we demonstrated that the apparent shutoff of heat-induced σ32 synthesis does not result from translational repression, contrary to what had previously been believed. Instead, σ32 was found to become extremely unstable (t1/2 = 20 s) within 10 min after temperature shift (Fig. 4), consistent with the instability observed with strain MG1655 under steady-state growth at 42°C (t1/2=10–15 s; ref. 18). The apparent shutoff of σ32 synthesis during the adaptation phase can therefore be explained primarily by severe instability of σ32 at high temperature. Whereas the early work established the basic regulatory features including initial stabilization of σ32 followed by destabilization, the extent of destabilization was underestimated and thought to be comparable to that at 30°C (t1/2 = 1 min; ref. 7). The unusual instability together with the changing stability precluded accurate measurement of synthesis rates of σ32 by 60-s pulse labeling.
The above conclusion is consistent with the previous failure to separate shutoff of synthesis and degradation when involvement of specific segments of σ32 or transacting factors in the two processes were assessed (9, 24). However, some results seemed to be an apparent contradiction. First, heat-induced synthesis of σ32 in the dnaJ259 mutant failed to shut off (60-s pulse) despite the fact that σ32 synthesized 3 min after shift to 42°C was as unstable as the wild type (9). When we examined σ32 stability during the adaptation phase with the dnaJ259 mutant of MC4100, the half-life of σ32 was about 1 min, significantly longer than that for the wild type, indicating marked stabilization (data not shown). However, this difference in half-life may not be sufficient to explain the difference in the shutoff profile reported (9). Second, heat-induced synthesis of σ32 in the ΔftsH mutant containing a suppressor (ΔshfC) seemed to shut off normally (60-s pulse) even though σ32 was markedly stabilized (37). We confirmed that σ32 synthesis with the same ΔftsH ΔshfC mutant exhibits gradual apparent shutoff, although not as striking as that reported previously (data not shown). It should also be noted that this mutant shows very slow growth (by about threefold). Besides, we found that apparent shutoff hardly occurs with the ΔshfC control, as also seen in the reported results (37). Thus, significance of the apparent shutoff observed specifically with the ΔftsH mutant remains unclear.
The inability of σ32ΔC17 to complement the temperature-sensitive growth of ΔrpoH strain (KY1603) agrees with the recent report on σ32 lacking the C-terminal 15 residues (38). When we measured the stability of σ32-B and σ32ΔC17, they both exhibited stability very similar to that of authentic σ32, the half-life in the λrpoHBAZ lysogen of MC4100 before or after temperature upshift being similar to, or slightly shorter than, that of σ32 in MC4100 (Fig. 4 C and D). The results with σ32ΔC17 was unexpected, because it was recently reported that σ32 with the C-terminal 15 aa replaced by 6 unrelated residues (σ32CΔ) expressed in strain KY1603 (ΔrpoH suhX401) was quite stable at 30°C (38). The apparent discrepancy could be caused by the extra amino acids added during construction of the expression plasmid or the different host bacteria used (ΔrpoH vs. rpoH+).
To sum up, HSP synthesis in E. coli is primarily regulated by the dynamic interplay between two antagonistic pathways affecting synthesis and degradation of σ32. One pathway controls rpoH translation by temperature-directed melting of the mRNA secondary structure (22, 23) in which the mRNA serves not only as a messenger but as a thermosensor and regulator of translation. Such a multifunctional mRNA must provide a unique and sensitive means of responding very rapidly to sudden changes in ambient temperature. However, this is a steady state rather than transient response to high temperature. In contrast, the other pathway controls degradation of σ32 as mediated by the DnaK–DnaJ chaperones and ATP-dependent heat-shock proteases which should serve to monitor the cellular state of protein folding, thereby fine-tuning the level of σ32 to cope with constant changes in cellular requirements. The DnaK–DnaJ chaperones can also modulate HSP synthesis by inhibiting σ32 activity (37, 39). Recent work suggested that σ32 itself directly responds to high temperature by changing its susceptibility to proteases (18). The combined results therefore indicate that regulation of the σ32 level during the heat-shock response rests on intricate balance between elevated synthesis and elevated turnover of σ32 that are controlled by distinct but interconnected pathways. The dynamic role played by the chaperones and proteases in modulating the stress response in this and other systems remains an outstanding issue for future investigation.
Acknowledgments
We thank M. Nakayama for technical assistance and M. Tanaka for assistance in preparing figures.
Abbreviation
- HSP
heat shock protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.080495197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.080495197
References
- 1.Morimoto R I, Tissieres A, Georgopoulos C. The Biology of Heat Shock Proteins and Molecular Chaperones. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 2.Gross C A, Straus D B, Erickson J W, Yura C. In: Stress Proteins in Biology and Medicine. Morimoto R I, Tissieres A, Georgopoulos C, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. pp. 167–189. [Google Scholar]
- 3.Gross C A. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: ASM Press; 1996. pp. 1389–1399. [Google Scholar]
- 4.Yura T, Nagai H, Mori H. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]
- 5.Tilly K, McKittrick N, Zylicz M, Georgopoulos C. Cell. 1983;34:641–646. doi: 10.1016/0092-8674(83)90396-3. [DOI] [PubMed] [Google Scholar]
- 6.Grossman A D, Straus D B, Walter W A, Gross C A. Genes Dev. 1987;1:179–184. doi: 10.1101/gad.1.2.179. [DOI] [PubMed] [Google Scholar]
- 7.Straus D, Walter W, Gross C A. Nature (London) 1987;329:348–351. doi: 10.1038/329348a0. [DOI] [PubMed] [Google Scholar]
- 8.Tilly K, Spence J, Georgopoulos C. J Bacteriol. 1989;171:1585–1589. doi: 10.1128/jb.171.3.1585-1589.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Straus D, Walter W, Gross C A. Genes Dev. 1990;4:2202–2209. doi: 10.1101/gad.4.12a.2202. [DOI] [PubMed] [Google Scholar]
- 10.Craig E A, Gross C A. Trends Biochem Sci. 1991;16:135–140. doi: 10.1016/0968-0004(91)90055-z. [DOI] [PubMed] [Google Scholar]
- 11.Bukau B. Mol Microbiol. 1993;9:671–680. doi: 10.1111/j.1365-2958.1993.tb01727.x. [DOI] [PubMed] [Google Scholar]
- 12.Goff S A, Goldberg A L. Cell. 1985;41:587–595. doi: 10.1016/s0092-8674(85)80031-3. [DOI] [PubMed] [Google Scholar]
- 13.Wild J, Walter W A, Gross C A, Altman E. J Bacteriol. 1993;175:3992–3997. doi: 10.1128/jb.175.13.3992-3997.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanemori M, Mori H, Yura T. J Bacteriol. 1994;176:5648–5653. doi: 10.1128/jb.176.18.5648-5653.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herman C, Thevenet D, D'Ari R, Bouloc P. Proc Natl Acad Sci USA. 1995;92:3516–3520. doi: 10.1073/pnas.92.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomoyasu T, Gamer J, Bukau B, Kanemori M, Mori H, Rutman A J, Oppenheim A B, Yura T, Yamanaka K, Niki H, et al. EMBO J. 1995;14:2551–2560. doi: 10.1002/j.1460-2075.1995.tb07253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanemori M, Nishihara K, Yanagi H, Yura T. J Bacteriol. 1997;179:7219–7225. doi: 10.1128/jb.179.23.7219-7225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanemori M, Yanagi H, Yura T. J Biol Chem. 1999;274:22002–22007. doi: 10.1074/jbc.274.31.22002. [DOI] [PubMed] [Google Scholar]
- 19.Kamath-Loeb A S, Gross C A. J Bacteriol. 1991;173:3904–3906. doi: 10.1128/jb.173.12.3904-3906.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagai H, Yuzawa H, Yura T. Proc Natl Acad Sci USA. 1991;88:10515–10519. doi: 10.1073/pnas.88.23.10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuzawa H, Nagai H, Mori H, Yura T. Nucleic Acids Res. 1993;21:5449–5455. doi: 10.1093/nar/21.23.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morita M, Kanemori M, Yanagi H, Yura T. J Bacteriol. 1999;181:401–410. doi: 10.1128/jb.181.2.401-410.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morita T M, Tanaka Y, Kodama T, Kyogoku Y, Yanagi H, Yura T. Genes Dev. 1999;13:655–665. doi: 10.1101/gad.13.6.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagai H, Yuzawa H, Kanemori M, Yura T. Proc Natl Acad Sci USA. 1994;91:10280–10284. doi: 10.1073/pnas.91.22.10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joo D M, Nolte A, Calendar R, Zhou Y-N, Jin D J. J Bacteriol. 1998;180:1095–1102. doi: 10.1128/jb.180.5.1095-1102.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arsene F, Tomoyasu T, Mogk A, Schirra C, Schulze-Specking A, Bukau B. J Bacteriol. 1999;181:3552–3561. doi: 10.1128/jb.181.11.3552-3561.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yura T. Genes Cells. 1996;1:277–284. doi: 10.1046/j.1365-2443.1996.28028.x. [DOI] [PubMed] [Google Scholar]
- 28.Casadaban M. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 29.Kusukawa N, Yura T. Genes Dev. 1988;2:874–882. doi: 10.1101/gad.2.7.874. [DOI] [PubMed] [Google Scholar]
- 30.St. Pierre R, Linn T. Gene. 1996;169:65–68. doi: 10.1016/0378-1119(95)00787-3. [DOI] [PubMed] [Google Scholar]
- 31.Nagai H, Yano R, Erickson J W, Yura T. J Bacteriol. 1990;172:2710–2715. doi: 10.1128/jb.172.5.2710-2715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirano M, Shigesada K, Imai M. Gene. 1987;57:89–99. doi: 10.1016/0378-1119(87)90180-6. [DOI] [PubMed] [Google Scholar]
- 33.Powel B S, Court D L, Nakamura Y, Rivas M P, Turnbough C L., Jr Nucleic Acids Res. 1994;22:5765–5766. doi: 10.1093/nar/22.25.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller J. A Short Course in Bacterial Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 36.Das A, Yanofsky C. Nucleic Acids Res. 1984;12:4757–4768. doi: 10.1093/nar/12.11.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tatsuta T, Tomoyasu T, Bukau B, Kitagawa M, Mori H, Karata K, Ogura T. Mol Microbiol. 1998;30:583–593. doi: 10.1046/j.1365-2958.1998.01091.x. [DOI] [PubMed] [Google Scholar]
- 38.Blaszczak A, Georgopoulos C, Liberek K. Mol Microbiol. 1999;31:157–166. doi: 10.1046/j.1365-2958.1999.01155.x. [DOI] [PubMed] [Google Scholar]
- 39.Tomoyasu T, Ogura T, Tatsuta T, Bukau B. Mol Microbiol. 1998;30:567–581. doi: 10.1046/j.1365-2958.1998.01090.x. [DOI] [PubMed] [Google Scholar]