Abstract
Retinoschisin (RS) is a 24 kDa secreted protein expressed in retina and is required for the structural and functional integrity of the retina. RS has been predicted to serve as an adhesive protein but the precise molecular mechanism by which it functions in retina is not yet known. During investigations on structural and functional aspects of RS in murine retina using proteomic tools, we identified two isoforms of RS that differed in mass by 200 Da with no apparent change in charge. Mass spectra and amino acid sequence analysis of the tryptic peptides revealed that these isoforms differed by two amino acids at the N-terminus which suggested processing of RS signal sequence at two cleavage sites by signal peptidase as the basic mechanism underlying the occurrence of two mature RS isoforms in retina. Bioinformatic analysis identified two potential cleavage sites (between amino acids 21-22 and 23-24) in RS signal sequence. The flexibility of the signal peptidase to cleave at two sites is correlated to the amino acid composition of the RS signal sequence. This finding represents a rare example of a naturally occurring signal sequence cleavage at more than one site in vivo.
Keywords: Retina, Retinoschisin, Post-Translational Modifications, Isoforms, Signal Sequence, Signal peptidase, Processing, X-linked retinoschisis, Missense mutations
Retinoschisin (RS) is a 24 kDa secreted protein expressed in retina and pineal* and is required for the structural and functional integrity of the retina [1]. Based on its conserved discoidin domain sequence, RS is predicted to serve as an adhesion protein in maintaining the organization of the retina [1] but a precise molecular description of RS function is not yet known. Mutations in the RS1 gene that encodes the RS protein cause monogenic X-linked retinoschisis (XLRS) in young males [2-4]. Retinoschisis is a form of juvenile macular degeneration in which schisis or splitting within the nuclear and plexiform layers of the retina [5] leads to progressive loss of vision beginning at a young age [6]. Gene therapy is being considered as a therapeutic approach to treat XLRS disease which otherwise has no known cure [7].
RS is synthesized in retinal photoreceptor, bipolar amacrine and ganglion cells [8, 9]. The 224 amino acid RS polypeptide consists of a 23 amino acid leader sequence followed by a 39 amino acid RS domain, a highly conserved 157 amino acid discoidin domain and a 5 amino acid C-terminal flanking region. The 23 amino acid leader sequence of RS (Fig-1A) possesses all the structural and functional features associated with eukaryotic signal sequences [10], including a polar (n) region with a net positive charge on its N-terminal side, a central hydrophobic core (h) region and a polar (c) region at C-terminal side with a signal peptidase cleavage site. These structural features of signal sequence serve as a platform on which the signal peptidase and signal recognition particles interact and cleave the 23 amino acid leader peptide from the nascent protein during the translocation of the polypeptide chain into the lumen of the endoplasmic reticulum [11, 12]. The folding of the RS subunit with the formation of intramolecular disulfide bonds and the assembly of RS subunits into RS dimer and octamer take place at this stage [13]. Following this, the protein is sorted at the trans-Golgi network into discrete carriers for delivery to the plasma membrane.
Figure-1.
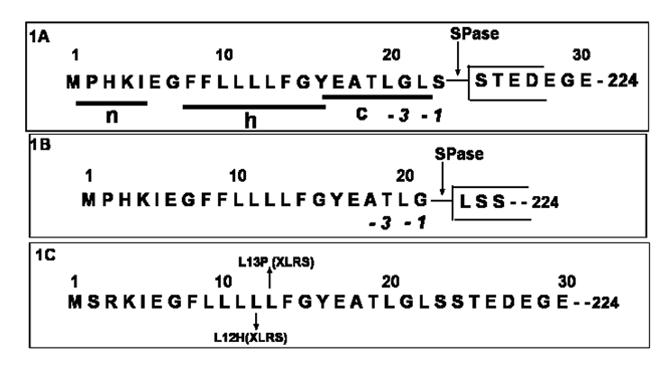
NH2-terminal sequence of pre-Retinoschisin: (A) Schematic drawing of tripartite structure of mouse RS signal sequence. Processing of signal sequence at two cleavage sites 21 and 23 generate two isoforms of RS: (A) RS: Ser 24-224 (B) RS: Leu 22-224. Signal peptidase (SPase) cleavage sites are indicated by arrows. (C) NH2-terminal sequence of human pre-retinoschisin. Mutations within the signal sequence that are associated with human X-linked retinoschisis disease are marked. Protein Database accession numbers to access entire RS sequences: NP_000321, X-linked juvenile retinoschisis protein [Homo sapiens] and NP_035432, retinoschisis 1 homolog [Mus musculus].
In an effort towards understanding RS structure-function relationship, we carried out a comprehensive analysis of RS in murine retina using a proteomic approach. Immunoprecipitation followed by 2D gel electrophoresis analysis of murine retinal cell lysates revealed several low abundant isoforms of RS that differed in charge. However, the two most abundant isoforms differed in mass by 200 Da with no apparent change in charge. Mass spectra and amino acid sequence analysis of the tryptic peptides established that both isoforms are mature forms of RS with different NH2 terminal sequences which suggest that alternate processing of the signal sequence might be the basic mechanism underlying the occurrence of two mature RS isoforms in retina. Bioinformatic analysis identified two potential cleavage sites (between amino acids 21-22 and 23-24) in RS signal sequence. The flexibility of the signal peptidase to cleave at two sites is correlated to the amino acid composition of the RS signal sequence.
Materials and Methods
Animals and Reagents
C57BL/6J (wild type) mice were used in this study. Animal experimental procedures were performed in accordance with the institutional animal experimentation ethics in the National Institutes of Health (NIH). All reagents of analytical grade were obtained either from Sigma-Aldrich or Bio-Rad Laboratories, USA. A rabbit polyclonal RS antibody raised against a synthetic peptide corresponding to the amino acid residues 24-37 of retinoschisin was used in this study [7].
Immunoprecipitation
Whole cell lysates from adult mouse retina were prepared by freeze thaw method in a buffer containing 10 mM Tris-HCl, pH 7.4, 150mM NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM EGTA, and protease inhibitor cocktail. Immunoprecipitation was carried out by standard protocols in the presence of protease inhibitors. About 500 μg of retinal cell lysates and 2 μg anti-RS antibody or rabbit IgG were used for each reaction.
Isoelectricfocusing (IEF)
Following the extraction of the immunocomplexes into SDS boiling buffer, the contents of 4-6 reactions were combined, dialyzed and lyophilized. The sample was redissolved in 50 μl of 1:1 SDS-boiling buffer, heated and loaded on isoelectric focusing gel (IEF). Two-dimensional gel electrophoresis was performed by O’Farrell method [14] by Kendrick Labs Inc (Madison, WI) in a glass tube of inner diameter 2 mm using 2% pH 3.5-10 ampholines (GE Health Care) for 9600 volt-hrs (700 volts for 13 h 45 min), with tropomyosin (1μg) as an internal standard. Following 1 dimension electrophoresis the gels were equilibrated for 10 min in buffer (10% glycerol, 50 mM DTT, 2.3% SDS, 62.5 mM Tris-HCl ph 6.8). The second dimension SDS-PAGE was carried out by standards methods on 10% gels.
In-gel digestion
Following electrophoresis, the gels were stained with Coomassie Brilliant Blue R-250. Coomassiestained spots were excised from the gel, washed first with 0.05M Tris, pH 8.5/25% acetonitrile, then 100% acetonitrile, and dried in a vacuum centrifuge. The gel pieces were rehydrated with 0.06μg trypsin in 15 μL 0.025 M Tris, pH 8.5, and digested overnight at 32° C. Peptides were extracted with 50% acetonitrile/2% TFA and subjected to either MALDI analysis or LC-MS/MS.
MALDI-TOF analysis
The extracted peptides were dried and redissolved in matrix solution (10mg/mL 4-hydroxy-α-cyanocinnamic acid in 50% acetonitrile/0.1% TFA) and analyzed on an Applied Biosystems Voyager DE Pro MALDI-TOF mass spectrometer in the linear mode.
LC-MS/MS
LC-MS/MS analysis of the digest mixture was performed on a Micromass Q-tof mass spectrometer using in-line reversed-phase separation with a linear acetonitrile gradient at 200nL/min flow rate. MS/MS spectra were scanned in the mass range 350-1500 amu. Additional runs were performed in the range 800-900 amu to focus on the N-terminal peptide ion. Raw data were processed with the Micromass MaxEnt 3 software and spectra were evaluated manually.
Data Presentation
All experiments were performed in triplicate, and representative results are presented.
Results
RS exists in several isoforms that differ in charge or mass
To determine whether isoforms of RS exist in retina, whole cell lysates from retina were immunoprecipitated with anti-RS antibody and subjected to 2D-gel electrophoresis. The position of RS on 2D gel immunoblot (Fig-2A) is consistent with the 26-28 kDa known molecular mass of RS. In this gel, RS occurs as a trail of closely migrating spots along the pH gradient, suggesting multiple protein isoforms that differ in pI and/or apparent mass. The pI of these RS isoforms ranged between 4.6-5.6, with a calculated value at 5.51. A similar profile of RS isoforms was obtained when IEF was performed on a narrow range IPG strips of linear pH 5-8 gradient. A trail of RS antibody specific spots was not seen in the negative control when cell lysates immunoprecipitated with rabbit IgG were probed with RS antibody (results not shown).
Figure-2.
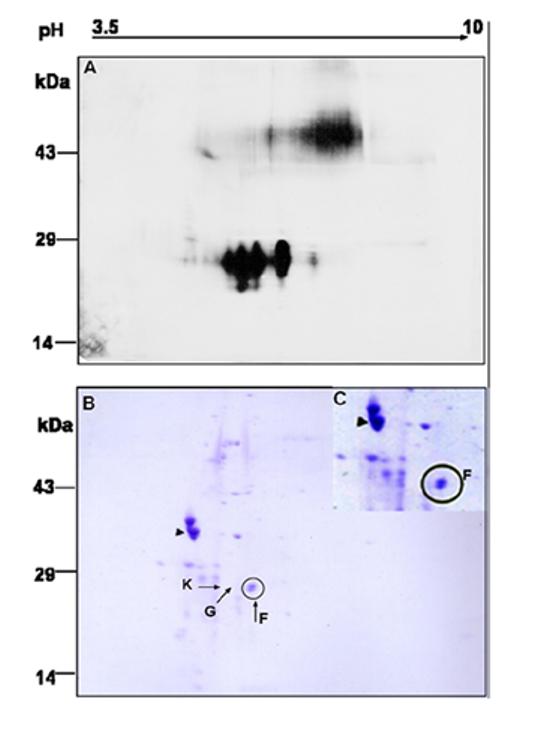
Identification of multiple isoforms of RS by 2D gel electrophoresis: Whole cell lysates from mouse retina were immunopreicipited with anti-RS antibody, and the immunocomplexes were subjected to 2D gel electrophoresis. The gel was either (A) electoblotted for immunoblot analysis or subjected to (B &C) staining with Coomassie blue. The short arrow indicates IEF marker, tropomyosin (33kDa; pI 5.2). The protein spots labeled F, G and K were excised from the gel for peptide mass analysis. (C) Enlarged image of the F doublet from a duplicate gel.
Mass spectra and amino acid sequence analysis of RS isoform specific tryptic peptides by MALDI/MS and LC/MS/MS
To characterize the isoforms of the proteins pulled down by RS antibody, a duplicate 2D gel electrophoressed under identical conditions was stained with Coomassie blue (Fig-2B). The pI marker, tropomyosin (33kDa), migrates as a doublet with a pI 5.2 (short arrow, Fig-2B). The darkest visible protein (spot F) specific to RS antibody appeared to be a doublet of higher and lower molecular weight (enlarged image-Fig-2C). The two components of the doublet were excised, digested with trypsin, and analyzed by MALDI-MS. The faint spots K and G, corresponding to apparently separate RS isoforms, were also analyzed.
On peptide mass mapping by MALDI-TOF mass spectrometry, the lower weight spot F (Fig-2B) corresponded to RS with high confidence, with 10 peptides matched and 38% sequence coverage (Table-1). Peptide masses consistent with RS protein fragments were also detected in less basic spots, G & K, although spectra from these barely visible spots were far less intense than from F. Differences in charge without significant changes in mass suggested that small molecules such as phosphates or monosaccharides might be involved consequent to post-translational modifications. However, a preliminary search of the mass spectra indicated no phosphorylated peptides in the RS protein spots.
Table-1.
MALDI-TOF analysis of tryptic peptides of retinoschisin found in F upper and lower spots
| Peptide | Upper spot. MH+ Observed | Lower spot. MH+ Observed | MH+ Calculated (Average) | Peptide sequence | Residues |
|---|---|---|---|---|---|
| 1 | 958.07 | 957.94 | 958.017 | VFYGNSDR | 175-182 |
| 2 | 1000.18 | 1000.16 | 1000.184 | LNWIYYK | 161-167 |
| 3 | 1044.27 | 1044.27 | 1044.238 | VISGILTQGR | 132-141 |
| 4 | 1091.41 | 1091.35 | 1091.343 | LIPLGWHVR | 201-209 |
| 5 | 1180.34 | 1180.24 | 1180.3174 | CDIDEWVTK | 142-150 |
| 6 | 1482.70 | 1482.62 | 1482.6965 | LNSQGFGCAWLSK | 103-115 |
| 7 | 1584.61 | 1584.60 | 1584.592 | STEDEGEDPWYQK | 24- 36 |
| 8 | 1624.78 | 1624.74 | 1624.789 | YQDSSQWLQIDLK | 116-128 |
| 9 | 1681.99 | 1681.94 | 1681.975 | SSTVQNLLRPPIISR | 183-197 |
| 10 | 1784.98 | 1785.18 | 1784.8310 | LSSTEDEGEDPWYQK | 22-36 |
| 11 | 3088.48* | ----- | 3088.4637 | LNSQGFGCAWLSKYQ-DSSQWLQIDLK | 103-208 |
Cysteine residues are modified by acrylamide.
Peptide 103-208, containing one missed cleavage appeared only in the upper spot.
MALDI-MS analysis confirmed that the upper part of spot F was RS (Fig-2B&C), and the F upper and lower spots represented two RS isoforms that differed in mass but with no difference in charge. MALDI-MS/MS analysis of tryptic digests showed that these two isoforms had different fragmentation patterns (Table-1). The N-terminus tryptic fragment with the mass 1584.60 in the F-lower spot (Fig-3B, peak-7) was shifted to 1784.98 mass in the F-upper spot (Fig-3A, peak-10 & Table-1), which suggested a post-translational modification involving the N-terminus of RS. The identification of fragment ion peak-10 (Fig-3B) in lower spot is most likely due to upper spot cross contamination.
Figure-3.
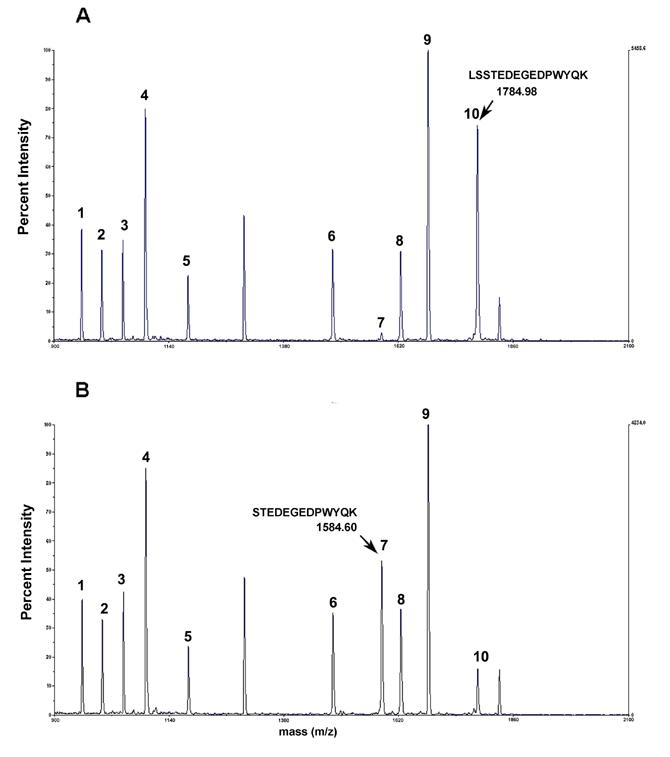
MALDI-TOF mass spectra of RS tryptic peptides from upper (A) and lower (B) portions of Coomassie-stained spot F shown in Fig. 2B. The arrow indicates the mass of the N-terminal RS peptide in each spectrum. The addition of Leu-Ser in the upper isoform shown in Fig. 3A adds 200.38 Da to the observed mass. The experimental conditions for in gel digestion and MALDI-TOF analysis are described in Materials & Methods.
Amino acid sequencing of the N-terminus tryptic peptides by LC/MS/MS demonstrated that the F-upper spot had two additional amino acids at the N-terminus (Fig-4 Upper), and the addition of Leu and Ser in the F-upper N-terminus tryptic fragment accounted for the 200Da difference in peptide mass compared with the F-lower tryptic fragment (Fig-4 Lower). These results indicate that two mature forms of RS exist in intact mouse retina. One isoform starts from amino acid 24 (RS: Ser 24-224/Fig-1A) and the second isoform starts from amino acid 22 (RS: Leu 22-224/Fig-1B).
Figure-4.
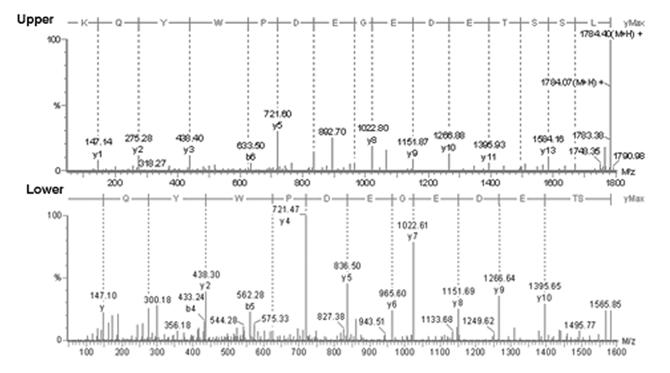
Amino Acid sequence of N-terminus RS tryptic peptides: Tryptic digests from F upper and lower spots were subjected to LC-MS/MS analysis on a Micromass Q-tof mass spectrometer as described in Materials & Methods.
Discussion
In this study we observed multiple RS isoforms in intact retina. Post-translational modifications that result in amino acid charge or mass change are the most likely cause of their occurrence. Because of their extremely low abundance, the nature of the modifications that resulted in changes in charge could not be characterized. Further, the relevance of such low abundance RS isoforms in the context of the RS function as a structural protein cannot easily be interpreted. The two most abundant isoforms differed in mass with no apparent change in charge. Based on the results of this study we propose that processing of the RS signal sequence at two cleavage sites is the basis for their occurrence in retina. RS represents a rare example of a secretory protein being found in two mature isoforms.
Blobel in 1971 formulated the “signal hypothesis” and postulated that protein secretion out of a cell requires an intrinsic signal (N-terminal signal sequence) that governs translocation across membranes [11, 12]. Several functional interactions of the signal sequence with the eukaryotic translocation apparatus have been defined. During translocation the signal sequence cleavage site is recognized by a signal peptidase, and co-translational proteolytic processing occurs. The membrane spanning properties of the signal peptidase, as well as the pre-protein substrate (the h-region in the signal sequence/Fig-1), are known to limit the enzyme substrate interactions such that the cleavage normally occurs at only one site [15]. The presence of two mature isoforms of RS in retina indicates that RS signal sequence is probably processed at two cleavage sites. Alternatively, the RS precursor may undergo a two step processing similar to that of pre-prohormones, in which cleavage of the signal sequence from pre-prohormone is the first step, followed by endoproteolytic processing of the cleaved prohormone product into active hormone, a process mediated by a tissue and hormone specific prohormone convertase [16]. However, the structural properties of the RS signal sequence would indicate the two mature isoforms of RS in intact retina occur because of two cleavage sites.
Structural features of RS signal sequence
A number of features known to be important for signal sequences have been defined in multiple ways, including amino acid sequence comparisons of known eukaryotic signal peptide sequences. Substrate specificities of eukaryotic and prokaryotic signal peptidases are conserved in evolution [17]. The relative contributions to cleavage site selection by specific amino acids at different positions within the signal peptide are known from site-directed mutagenesis of eukaryotic preproteins followed by their expression in bacterial systems [18, 19]. The resulting rules for predicting cleavage sites within a signal sequence were further improved by incorporating neural network and hidden Markov model algorithms [20] (SignalP3.0-publicly available web server: http://www.cbs.dtu.dk/services/SignalP).
Analysis of RS signal sequence identified two putative cleavage sites. The amino acid sequence and composition of these sites are in agreement with the -3, -1 N-terminal rule of the cleavage site proposed by von Heijne [10, 21, 22]. For the Leu 22-224 isoform of RS, cleavage occurs with the neutral short side chain Gly at -1 and the uncharged Thr at -3 (Fig-1B). For the Ser 24-224 isoform of RS, the -1 and -3 amino acids are Ser and Gly respectively (Fig-1A). Normally the helix breaker (Gly/Pro) occurs between -4 to -6 positions of the cleavage site. However, in the RS signal sequence the Gly lies at -3 for the Ser 24-224 isoform (Fig-1A) and at -7 for the Leu 22-224 isoform (Fig-1B). Hence the -4 to-6 constraint for helix breaker might be a less stringent condition.
SignalP 3.0 predicted RS cleavage at both 21 (0.425 probability) and 23 (0.350 probability) and reported the most likely cleavage site to be between positions 21 and 22, based on the assumption that cleavage at one site inhibits cleavage at a second site [20]. Currently there are only five examples of signal sequence cleavage at alternate or multiple sites [reviewed in 18]. Cleavage of the signal sequence at multiple sites was studied in vitro by Nothwehr & Gordon [19] who demonstrated that cleavage efficiency depended on the property of the -1 amino acid. For a signal sequence with multiple cleavage sites, the efficiency of cleavage was much higher after Gly compared to Ser. The RS signal sequence shows several of the structural features which can influence the efficiency and site of cleavage in Nothwehr & Gordon’s in vitro model. According to cleavage efficiencies calculated by Nothwehr & Gordon, the Leu 22-224 isoform (cleaved after Gly, Fig-1B) would occur at higher concentration than the RS: Ser 24-224 isoform (cleaved after Ser, Fig-1A), and this conforms to our observation on the relative concentrations of RS isoforms (Fig-2B&C). The amino acid composition at cleavage sites Gly 21 and Ser 23 (Fig-1A&B) confirm that that the physical and chemical properties of the amino acid at the -1 position of the cleavage site are important [18, 19].
Missense mutations in signal peptide and XLRS
The human RS signal sequence differs from the mouse sequence in having two positive charges in n region Arg-3, Lys-4 as compared to one Lys-4 in mouse (Fig-1C). The positively charged residues in n-region contribute to the stability and efficiency of the secretion process. Since human and mouse RS signal sequences share well conserved h- and c- regions (Fig-1), we anticipate that signal sequence cleavage occurs at both 21 and 23 positions and leads to two mature isoforms of RS in the human retina. Predictions (SignalP 3.0) showed identical patterns of cleavage both for human and mouse RS signal sequences.
X-linked retinoschisis disease was reported in several families with missense mutations in signal sequence of RS1 gene, a 38T→ C which changes an amino acid residue Leu13Pro and 35T→ A which changes an amino acid residue Leu12H is in the signal sequence (Fig-1C) [23, 24]. Replacing the hydrophobic Leu by the highly hydrophilic and neutral/positively charged His in the Leu12His mutant caused intracellular retention and failure to secrete the mutant protein [24]. The prediction of secondary structure of mutant signal sequence (amino acids 1-30 in Leu13Pro) by GOR method [25] revealed significant reduction in helical content when the helix breaker Pro replaced Leu.
However, our analysis of the Leu13Pro and Leu12H is mutations by SignalP 3.0 indicated no alterations in cleavage pattern or cleavage site positions in these mutants. Hence, disease mechanism in XLRS with mutations in h-region of signal peptide might involve loss of signal peptidase binding function due to the change in secondary structure of the pre-protein substrate. The h-region extrinsic to the cleavage site is a critical determinant of signal sequence recognition by signal peptidase. These results demonstrate how several features of signal sequence influence the overall signal processing and cleavage.
In summary, the cleavage of RS signal sequence, as observed in intact retina, demonstrates that cleavage site position and amino acid composition of the signal sequence are correlated and two or more potential sites of cleavage compete for recognition by signal peptidase. This finding represents a rare example of a naturally occurring signal sequence cleavage at more than one site. Further studies are needed to determine whether these isoforms have functional significance or accidental in nature because of the flexibility of the signal peptidase.
Acknowledgments
We thank Dr. Donita Garland, Dr. Barbara Wiggert for useful discussions and Maria Santos-Muffley, Juanita Marner for technical assistance. This study was supported by the Intramural Research Program of NIH, NIDCD and NEI.
Footnotes
Takada et. al.(2006).Retinoschisin expression and localization in rodent pineal and consequences of mouse RS1 gene knockout.(Unpublished)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Weber BH, Schrewe H, Molday LL, Gehrig A, White KL, Seeliger MW, Jaissle GB, Friedburg C, Tamm E, Molday RS. Inactivation of the murine X-linked juvenile retinoschisis gene, Rs1h, suggests a role of retinoschisin in retinal cell layer organization and synaptic structure. Proc Natl Acad Sci U S A. 2002;99:6222–7. doi: 10.1073/pnas.092528599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Sauer CG, Gehrig A, Warneke-Wittstock R, Marquardt A, Ewing CC, Gibson A, Lorenz B, Jurklies B, Weber BH. Positional cloning of the gene associated with X linked juvenile retinoschisis. Nat Genet. 1997;17:164–70. doi: 10.1038/ng1097-164. [DOI] [PubMed] [Google Scholar]
- [3].Gehrig AE, Warneke-Wittstock R, Sauer CG, Weber BH. Isolation and characterization of the murine X-linked juvenile retinoschisis (Rs1h) gene. Mamm Genome. 1999;10:303–7. doi: 10.1007/s003359900991. [DOI] [PubMed] [Google Scholar]
- [4].Consortium TR. Functional implications of the spectrum of mutations found in 234 cases with X-linked juvenile retinoschisis. The Retinoschisis Consortium. Hum Mol Genet. 1998;7:1185–92. doi: 10.1093/hmg/7.7.1185. [DOI] [PubMed] [Google Scholar]
- [5].Mooy CM, Van Den Born LI, Baarsma S, Paridaens DA, Kraaijenbrink T, Bergen A, Weber BH. Hereditary X-linked juvenile retinoschisis: a review of the role of Muller cells. Arch Ophthalmol. 2002;120:979–84. [PubMed] [Google Scholar]
- [6].Sieving PA. Juvenile Retinoschisis. In: Traboulsi E, editor. Genetic Diseases of the Eye. Oxford University Press; New York: 1999. pp. 347–356. [Google Scholar]
- [7].Zeng Y, Takada Y, Kjellstrom S, Hiriyanna K, Tanikawa A, Wawrousek E, Smaoui N, Caruso R, Bush RA, Sieving PA. RS-1 Gene Delivery to an Adult Rs1h Knockout Mouse Model Restores ERG b-Wave with Reversal of the Electronegative Waveform of X-Linked Retinoschisis. Invest Ophthalmol Vis Sci. 2004;45:3279–85. doi: 10.1167/iovs.04-0576. [DOI] [PubMed] [Google Scholar]
- [8].Molday RS. Focus on Molecules: Retinoschisin (RS1) Exp Eye Res. 2006 doi: 10.1016/j.exer.2005.12.013. [DOI] [PubMed] [Google Scholar]
- [9].Takada Y, Fariss RN, Tanikawa A, Zeng Y, Carper D, Bush R, Sieving PA. A retinal neuronal developmental wave of retinoschisin expression begins in ganglion cells during layer formation. Invest Ophthalmol Vis Sci. 2004;45:3302–12. doi: 10.1167/iovs.04-0156. [DOI] [PubMed] [Google Scholar]
- [10].Martoglio B, Dobberstein B. Signal sequences: more than just greasy peptides. Trends Cell Biol. 1998;8:410–5. doi: 10.1016/s0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- [11].Blobel G, Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975;67:835–51. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Blobel G, Dobberstein B. Transfer to proteins across membranes. II. Reconstitution of functional rough microsomes from heterologous components. J Cell Biol. 1975;67:852–62. doi: 10.1083/jcb.67.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wu WW, Wong JP, Kast J, Molday RS. RS1, a discoidin domain-containing retinal cell adhesion protein associated with X-linked retinoschisis, exists as a novel disulfide-linked octamer. J Biol Chem. 2005;280:10721–30. doi: 10.1074/jbc.M413117200. [DOI] [PubMed] [Google Scholar]
- [14].O’Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–21. [PMC free article] [PubMed] [Google Scholar]
- [15].Carlos JL, Paetzel M, Brubaker G, Karla A, Ashwell CM, Lively MO, Cao G, Bullinger P, Dalbey RE. The role of the membrane-spanning domain of type I signal peptidases in substrate cleavage site selection. J Biol Chem. 2000;275:38813–22. doi: 10.1074/jbc.M007093200. [DOI] [PubMed] [Google Scholar]
- [16].von Eggelkraut-Gottanka R, Beck-Sickinger AG. Biosynthesis of peptide hormones derived from precursor sequences. Curr Med Chem. 2004;11:2651–65. doi: 10.2174/0929867043364405. [DOI] [PubMed] [Google Scholar]
- [17].Garcia PD, Ghrayeb J, Inouye M, Walter P. Wild type and mutant signal peptides of Escherichia coli outer membrane lipoprotein interact with equal efficiency with mammalian signal recognition particle. J Biol Chem. 1987;262:9463–8. [PubMed] [Google Scholar]
- [18].Folz RJ, Nothwehr SF, Gordon JI. Substrate specificity of eukaryotic signal peptidase. Site-saturation mutagenesis at position -1 regulates cleavage between multiple sites in human pre (delta pro) apolipoprotein A-II. J Biol Chem. 1988;263:2070–8. [PubMed] [Google Scholar]
- [19].Nothwehr SF, Gordon JI. Eukaryotic signal peptide structure/function relationships. Identification of conformational features which influence the site and efficiency of co-translational proteolytic processing by site-directed mutagenesis of human pre(delta pro)apolipoprotein A-II. J Biol Chem. 1989;264:3979–87. [PubMed] [Google Scholar]
- [20].Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–95. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- [21].von Heijne G. How signal sequences maintain cleavage specificity. J Mol Biol. 1984;173:243–51. doi: 10.1016/0022-2836(84)90192-x. [DOI] [PubMed] [Google Scholar]
- [22].von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–90. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hiriyanna KT, Bingham EL, Yashar BM, Ayyagari R, Fishman G, Small KW, Weinberg DV, Weleber RG, Lewis RA, Andreasson S, Richards JE, Sieving PA. Novel mutations in XLRS1 causing retinoschisis, including first evidence of putative leader sequence change. Hum Mutat. 1999;14:423–7. doi: 10.1002/(SICI)1098-1004(199911)14:5<423::AID-HUMU8>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- [24].Wang T, Waters CT, Rothman AM, Jakins TJ, Romisch K, Trump D. Intracellular retention of mutant retinoschisin is the pathological mechanism underlying X-linked retinoschisis. Hum Mol Genet. 2002;11:3097–105. doi: 10.1093/hmg/11.24.3097. [DOI] [PubMed] [Google Scholar]
- [25].Garnier J, Gibrat JF, Robson B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996;266:540–53. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]


