Abstract
Recent evidence using GT1-7 cells indicates that GnRH pulsatility depends on exocytotic-release and gene transcription events. To determine whether calcium or DREAM may play a role in linking these processes, we used an L-type Ca2+-blocker (nimodipine) and found that not only GnRH gene expression (GnRH-GE) pulse activity was abolished but also that binding of proteins to OCT1BS-a (essential site for GnRH-GE pulses) was reduced. We further found that only EF-hand forms of DREAM were expressed in GT1-7 and that DREAM was part of the complex binding to OCT1BS-a. Finally, microinjection of DREAM antibody into cells abolished GnRH-GE pulses demonstrating its importance in pulsatility. These results reveal that calcium and DREAM may bridge cytoplasmic and nuclear events enabling temporal coordination of intermittent activity. Expression of DREAM in various cell types coupled with the universal role of calcium raise the possibility that these factors may play similar role in other secretory cells.
Keywords: rhythmicity, single-cell imaging, GT1-7 cells, luciferase reporter gene, microinjection, pulsatility, nimodipine, calcium binding protein, DREAM
1. Introduction
The pulsatile release of GnRH is critical for proper reproductive function and acts by governing the intermittent release of gonadotropic hormones from the anterior pituitary (Terasawa, 1995). These gonadotropin episodes are needed for the continued function of various reproductive tissues (Crowley et al., 1985; Spratt et al., 1987). The importance of this intermittent process has stimulated a great deal of interest in determining the cellular mechanisms that underlie this intermittent release. Because neurons that produce GnRH are scattered in the preoptic and mediobasal areas of the hypothalamus (Silverman and Krey, 1978) and are very difficult to study effectively, many of the investigative efforts directed toward this problem have utilized immortalized GT1-7 GnRH neurons, as a cell model system (Krsmanovic et al., 1999; Martinez de la Escalera et al., 1992; Martinez de la Escalera et al., 1995; Mellon et al., 1990; Weiner et al., 1992). It has been established that cultures of these cells release GnRH in an episodic manner with an interpulse interval comparable to that observed for primary cultures from castrated rodents (Masotto and Negro-Vilar, 1988). In fact using single cell techniques in our laboratory, we found that pulsatile GnRH release is an intrinsic characteristic of individual GT1-7 neurons (Vazquez-Martinez et al., 2001a).
Intermittent activity is not restricted to just GnRH release, but also is associated with GnRH gene expression (GnRH-GE). Recent evidence revealed that GnRH promoter activity occurs in a pulsatile manner (Nunez et al., 1998; Vazquez-Martinez et al., 2002) and that a region termed the neuron specific enhancer (NSE) (Whyte et al., 1995) was necessary for this pulse activity. Within this region, the interactions of the homeoprotein OCT1 with specific sites (OCT1BS-a and OCT1BS-c) were identified as being critical for generation of these pulses (Leclerc and Boockfor, 2005; Vazquez-Martinez et al., 2002). Interestingly, simultaneous measurement of exocytosis and GnRH-GE in individual GT1-7 cells revealed a strong temporal association between exocytotic pulses and GnRH-GE episodes (Vazquez-Martinez et al., 2001b). In fact, blockage of exocytosis using a treatment that obstructs secretory granule fusion abolished GnRH-GE pulse activity indicating that certain steps in the exocytotic process are responsible for triggering episodes of GnRH gene transcription. Quantitative estimates of these processes in GT1-7 cells revealed that a careful balance was maintained between release and biosynthesis suggesting that GnRH neurons were kept in a constant state of readiness to respond to functional demands such as those occurring in different reproductive states. When taken together, it was clear not only that episodic activity was central to both the gene expression and secretory processes, but also that there was a functional link between the events occurring at the cell membrane and those involving gene expression in the nucleus.
A growing body of evidence suggests that calcium is a critical component in the initiation and propagation of exocytotic pulse activity in GT1-7 cells (Spergel et al., 1996). The most compelling results obtained revealed that blockage of calcium channels by pharmacological means or removal of calcium abolished GnRH release in cultures of these cells (Martinez de la Escalera et al., 1992; Martinez de la Escalera et al., 1995; Spergel et al., 1996). Studies from our laboratory performed on single GT1-7 cells showed that intracellular calcium concentration ([Ca2+]i) shifted from quiescence to high oscillatory behavior and that these shifts were frequently associated with exocytotic events (Nunez et al., 2000). Also, we found that treatment of individual GT1-7 cells with the L-type voltage gated Ca2+ channel blocker, nimodipine, markedly reduced the frequency of secretory pulse activity (Vazquez-Martinez et al., 2001a). These results demonstrate clearly that [Ca2+]i plays a central role in the exocytotic pulse process and suggests that this element alone or in conjunction with other factors may be an important functional link between cytoplasmic and nuclear pulse processes. In many cases, the ability of Ca2+ to initiate a cellular response depends on proteins that first bind calcium and then interact with other regulatory components of the transcriptional machinery. Among these proteins, calretinin, calcineurin and calmodulin received a great deal of attention (for review see Ikura et al., 2002). Recently, a novel calcium binding protein capable of interacting with components in both the cytoplasmic and nuclear compartments of the cell was identified in GT1-7 neurons (Leclerc et al., 2002). This protein termed DREAM (Downstream Regulatory Element Antagonist Modulator) contains four EF-hands capable of binding calcium with high affinity (Carrion et al., 1999; Craig et al., 2002). These characteristics of DREAM combined with the importance of Ca2+ in GnRH pulse activity raises the exciting possibility that these two components may play an important role in coupling intermittent gene expression and exocytosis. In the following study, our goal was to determine first whether [Ca2+]i influx is needed for pulsatile GnRH-GE to occur and next to identify whether DREAM protein plays a role in this process. To accomplish this, we used real-time monitoring of luciferase reporter activity driven by a GnRH promoter construct in individual GT1-7 cells to evaluate both the impact of a reduction in Ca2+ influx (nimodipine treatment) and the action of DREAM protein (immunological neutralization) on GnRH-GE pulse elaboration.
2. Materials and methods
2.1 Cell Culture
GT1-7 cells (provided by Dr. Richard I. Weiner, University of California, San Francisco, CA) were cultured as described previously (Vazquez-Martinez et al., 2001a). Briefly, cells were maintained in high glucose DMEM supplemented with antibiotics (100 U/ml of penicillin, 100 μg/ml of streptomycin, and 2.5 μg/ml fungizone), and 10% fetal bovine serum (FBS) in an atmosphere of 5% CO2/95% air at 37°C. Unless indicated otherwise, all tissue culture supplies were obtained from Invitrogen (Carlsbad, CA). Cells were detached from the plates by treatment with 0.05% trypsin and 0.53 mM EDTA for 5 min at 37°C, and replated onto Matrigel (1:25) (BD-Biosciences, Bedford, MA) coated, grid-etched 25 mm coverslips (Bellco Glass, Inc., Vineland, NJ) at a density of 12,500 cells/cm2. Cells were kept in culture for 2–7 d before experimental manipulation. For drug treatment experiments, cells were seeded in 75 cm2 flasks (8x105 cells per flask) and cultured for 48 h at 37°C. Then, drug (ethanol (0.08%, control vehicle) or nimodipine (1 μM, diluted in ethanol)) (Calbiochem-Novabiochem, San Diego, CA) was added to the cell culture and incubated for an additional 24 h before manipulation.
2.2 Bioluminescent Imaging of GnRH Gene Expression in Individual GnRH Cells
For real-time bioluminescent measurement of GnRH gene promoter activity, individual GT1-7 cells were microinjected (Eppendorf 5242) with Endofree-purified (Qiagen, Valencia, CA) pA3GnRH-LUC plasmids using a concentration of 2 μg/μl as described previously (Vazquez-Martinez et al., 2002). This plasmid contains 3026 bp of the rat GnRH promoter (region −3026/+116) fused upstream of the coding sequence of the firefly luciferase gene (Kepa et al., 1992; Maxwell et al., 1989; Wood et al., 1989). Cells were incubated under normal culture conditions for 24–48 h prior to bioluminescent imaging. Then, coverslips containing the cells were assembled in Sykes-Moore chambers and perifused (10 μl/min) with high glucose DMEM (phenol-red free) supplemented with 10% FBS, 10mM HEPES, 1mM sodium pyruvate, 4 mM L-glutamine, antibiotics (penicillin, streptomycin, and fungizone), 0.1 mM luciferin (Sigma, St. Louis, MO), and drugs as indicated (ethanol (0.08%, control vehicle) or nimodipine (1 μM)). The chamber was placed on the heated stage of a photon-capture microscope (Axioskop, Carl Zeiss, Jena, Germany) and photon images were visualized using a 20X objective (Neofluar, Carl Zeiss). Photonic signals from single cells were accumulated and quantified continuously in 30-min bins for at least 12 h using a photon-counting camera system (Argus 50, Hamamatsu, Bridgewater, NJ). Photonic images were analyzed off-line and the presence of GnRH promoter pulses were identified as previously described (Leclerc and Boockfor, 2005; Vazquez-Martinez et al., 2002). A pulse was defined as a significant rise in photonic activity corresponding to a value greater than 5 % of the photonic signal count at the beginning of the pulse plus two times the total signal-noise variation (SNV). SNV was calculated by the square root of each value plus the standard deviation (SD) of the corresponding background values squared (SNV = √[(signal count) + (SD of detected background2)]) (Takasuka et al., 1998). All the cells analyzed expressed a detectable level of luciferase activity (generating photonic patterns with or without pulses) that was significantly higher than the background level indicating that the cells remained viable during the course of the experiment. Specific photonic signal emitted from each cell was background subtracted and plotted as a function of time. Pulse frequencies of cells microinjected with pA3GnRH-LUC resulting from different treatments were compared to control values using one-tailed, unpaired t tests. All data are expressed as mean ± SEM.
2.3 RNA Extraction and RT-PCR
Total RNA was isolated from GT1-7 cells using the RNeasy Kit (Qiagen, Chatsworth, CA) according to the manufacturer’s specification. First-strand cDNA synthesis and RT-reactions were carried out as described elsewhere (Leclerc et al., 2002). For PCR amplification of the mouse DREAM gene, we used the PCR primers DAMF1 (5′-GGG AAG ATT AGT GAC GG) and DAMR2 (5′-CTC TGG GTT GAG AAG CAA TGA) which were based on the mouse DREAM cDNA sequence (GeneBank Accession Number: AF184624) and predicted to amplify a fragment of 852 bp. These primers has been used successfully to amplify specifically the whole coding sequence of the rat DREAM gene from the pituitary (Leclerc et al., 2002). The amplifications were performed in a 25 μl reaction volume containing 1X Qiagen PCR buffer, 1X Qiagen Q-solution, 0.5 μM primers, 0.2 mM each dNTPs (Invitrogen), and 0.6 U HotStar Taq DNA Polymerase (Qiagen). The PCR conditions were as follows: initial incubation at 95°C for 15 min and then 35 cycles of denaturation at 95°C for 60 sec, annealing at 50°C for 60 sec, and elongation at 72°C for 80 sec. The PCR amplified products were separated by 1.0% TAE agarose gel electropheresis and visualized by ethidium bromide staining. The amplified RT-PCR fragment DAM12 (852 bp) was cloned into pCR2.1-TOPO vector using the TOPO TA Cloning Kit (Invitrogen). The resulting plasmids were purified using the QIAprep Spin Miniprep Kits (Qiagen Inc., Valencia, CA). Integrity and identity of six independent cloned RT-PCR DNA fragments were confirmed by nucleotide DNA sequencing which was performed by the Biotechnology Resource Laboratory of the Medical University of South Carolina (Charleston, SC).
2.4 Nuclear Protein Extraction and Electrophoretic Mobility Shift Assays (EMSAs)
Nuclear protein extracts from GT1-7 cells were prepared using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce, Rockford, IL) and supplemented with 2X final concentration of the Halt Protease Inhibitor Cocktail Kit (Pierce). Then, protein lysates were purified, concentrated 3-fold, and protein concentration determined as described previously (Leclerc and Boockfor, 2005). Protein-DNA interaction between nuclear proteins with duplex oligonucleotides containing the wild-type OCT1BS-a site (−1774 to −1781, sequence underlined below) of the GnRH NSE region (Leclerc and Boockfor, 2005; Vazquez-Martinez et al., 2002) was analyzed by EMSAs using the LightShift Chemiluminescent EMSA Kit (Pierce, Rockford, IL). The DNA duplex consisted of the following annealed pair of 5′-end biotinylated oligonucleotides: OCT1AF-W (5′-Bio/GCT GAG ATT TTA CAT TAG GGC AA) and OCT1AR-W (5′-Bio/TTG CCC TAA TGT AAA ATC TCA GC). The binding reactions consisted of incubation of 6 μg of nuclear protein extracts from GT1-7 with a reaction buffer composed of 10 mM Tris-HCl (pH 7.5), 50 mM KCl, 1 mM DTT, 50 ng/μl Poly (dI-dC)·Poly (dI-dC) (GE Healthcare, Piscataway, NJ), 2.5% glycerol, and 0.7X concentrated Complete Mini Protease Inhibitor Cocktail (Roche Applied Science, Indianapolis, IN) for 10 min at room temperature. This was followed by addition of 5 nM of the 5′-Biotinylated OCT1BS-a probe to the reaction and incubated for an additional 30 min at room temperature. Then, either 4 μg of specific antibody or 1 mM phosphate buffered saline pH 7.4 was added to the reaction mixture and incubated for 50 min at room temperature. For competition experiments, a 100-fold molar excess of unlabeled OCT1BS-a DNA duplex was added to the reaction mixture before addition of the labeled probe. For supershift experiments, 4 μg of IgG affinity-purified goat polyclonal antibody raised against either DREAM (sc-9309), GATA-4 (sc-1237), GATA-5 (sc-7280), or GATA-6 (sc-7244) was used. The effect of calcium on protein-DNA complex formation was evaluated by addition of EGTA (12.5 mM) (Sigma) to the binding reaction. The mixtures were resolved in non-denaturing 6% polyacrylamide gel electrophoresis (Invitrogen) at 4°C with 0.25X TBE (90 mM Tris-Borate, 2 mM EDTA, pH 8.3) for 125 min at 120 volts. Then, the protein-DNA complexes were transferred to Biodyne B Nylon Membranes (Pierce). The biotin-labeled DNA probes were detected by chemiluminescence according to manufacturer’s instructions. The density of each retarded band was determined using a densitometry ChemiImager digital imaging system (Alpha Innotech, San Leandro, CA). Statistical differences in relative integrated density values (IDV) were assessed using one-tailed, unpaired t tests according to the Graph Pad PRISM software version 2 (GraphPad Software, Inc., San Diego, CA). The data are expressed as mean ± SEM.
2.5 Immunological neutralization
Immunological neutralization (immunoneutralization) of DREAM protein in GT1-7 cells was performed as described elsewhere (Vazquez-Martinez et al., 2002). Briefly, GT1-7 cells were first microinjected with the pA3GnRH-LUC plasmid and 48 h later monitored for photonic activity. Active luciferase expressing cells were again microinjected with a specific antibody solution (1 mg/ml). For DREAM, we used an affinity-purified goat polyclonal antibody (sc-9309) that was shown to react specifically with the DREAM protein in Western blottings (Leclerc et al., 2002). For control experiments, cells first microinjected with pA3GnRH-LUC and found to be photonically active were reinjected with a normal rabbit polyclonal IgG (sc-2027). Immediately after microinjection of the antibody, photonic emissions from individual cells were recorded.
3. Results
3.1 Influence of L-type Ca2+ channel blocker on GnRH gene expression pulse activity
As shown in examples presented in Figure 1A, GT1-7 cells microinjected with a construct containing 3026 bp of the rat GnRH promoter fused to the luciferase gene reporter (pA3GnRH-LUC, see Materials and methods) and treated with vehicle (0.08% ethanol) exhibited “pulses” or episodes of photonic activity (0.123 ± 0.011 pulse/h, n = 24 cells). Treatment of injected cells with 1 μM nimodipine, a specific L-type voltage gated Ca2+ channel blocker (McCarthy and TanPiengco, 1992), resulted in a complete abolition of these pulses (Fig. 1B, p < 0.001) (0.026 pulse/h ± 0.012, n = 13 cells vs. 0.123 ± 0.011 pulse/h, n = 24 cells; nimodipine- vs. control/ethanol-treated cells, respectively), though basal photonic activity was still present. These results suggest that calcium influx plays an important role in the generation of GnRH promoter pulses.
Figure 1. Effect of nimodipine on GnRH promoter pulse activity.
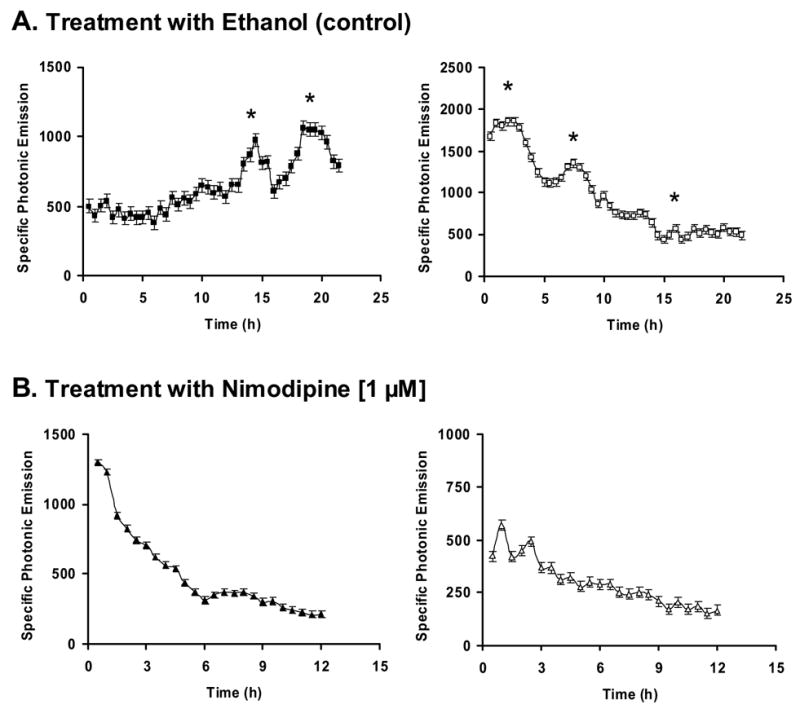
Representative photonic profiles measured from single GT1-7 cells microinjected with pA3GnRH-LUC and treated with either (A) 0.08% ethanol (control vehicle, n = 24 cells) or (B) 1 μM nimodipine (n = 13 cells). Two examples of expression profiles are presented for each treatment. Every point represents photonic signal accumulated for 30 min and is expressed as a function of the SNV (see Materials and methods). Vertical bars indicate SEM. Asterisks represent significant GnRH-GE pulses.
3.2 Effect of nimodipine on the formation of the OCT1BS-a/protein complex
In order to begin to elucidate the mechanism underlying these nimodipine-induced effects in gene expression, we evaluated whether reduced calcium levels interfered with protein binding at a site found to be critical for GnRH pulse activity (Leclerc and Boockfor, 2005; Vazquez-Martinez et al., 2002). The OCT1BS-a site was found by our laboratory to be essential for gene expression pulse activity (Vazquez-Martinez et al., 2002). For our experiments, EMSA analysis was used to compare quantitatively the manner in which nuclear proteins obtained from control/ethanol- and nimodipine-treated GT1-7 cells bound to a DNA duplex containing the OCT1BS-a site. As presented in Figure 2A, incubation of protein lysate from control/ethanol-treated cells with the OCT1BS-a probe resulted in the formation of a very distinct specific binding complex (NE, lane 1) as previously reported (Clark and Mellon, 1995). Interestingly, addition of 12.5 mM EGTA (NE + EGTA, lane 2) to the binding reaction diminished the amount of this complex suggesting that calcium was needed for optimal binding of proteins at the OCT1BS-a site. When nuclear protein extract obtained from nimodipine-treated cells was used in these reactions, we observed a band forming that migrated at the same rate as the control extract, but appeared to be less intense (Nimodipine, lane 3). As expected, addition of 100-fold molar excess of the specific DNA competitor OCT1BS-a completely eliminated the formation of the protein/OCT1BS-a complex (NE + OCT1BS-a, lane 4). Quantification of this band formation in four independent EMSA experiments using densiometric scan analysis revealed that inclusion of EGTA in the binding reaction or use of lysates from nimodipine-treated cells resulted in a significant decrease in band intensity indicating that the amount of protein binding to the OCT1BS-a probe was calcium dependent (Fig. 2B) (21640 ± 4522 for NE + EGTA and 23690 ± 5037 for Nimodipine vs. 51930 ± 6711 for ethanol-treated cells (NE), respectively). Taken together, these results reveal that changes in cellular calcium influx can influence the degree of protein binding to at least one site of the GnRH promoter (OCT1BS-a) found to be essential for the generation of GnRH-GE pulses.
Figure 2. Effect of calcium on proteins binding to the OCT1BS-a site located in the GnRH NSE region.
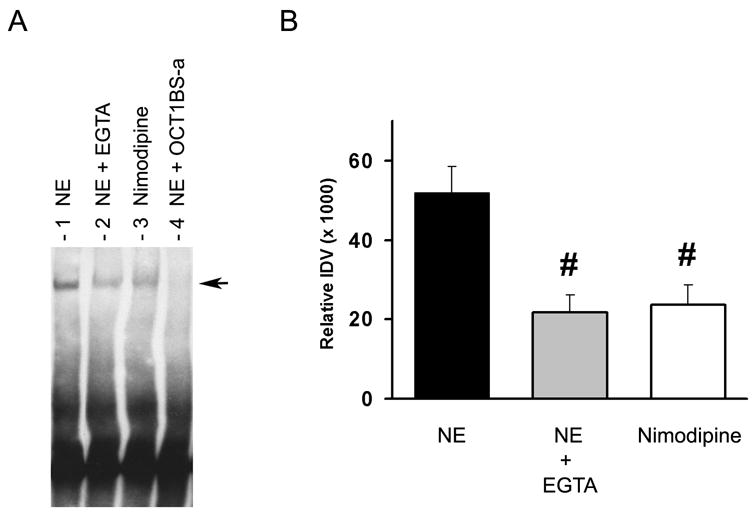
A, EMSAs analysis of 5′-biotinylated oligonucleotides containing the OCT1BS-a site (−1774 to −1781) and nuclear protein extracts from GT1-7 cells treated with either 0.08% ethanol (control, NE, lane 1) or 1 μM nimodipine (lane 3) which generated the formation of a specific band (arrow) (see Materials and methods). Incubation of the ethanol-treated extract/OCT1BS-a mixture with 12.5 mM EGTA resulted in significant reduction in the formation of the protein/DNA complex (NE + EGTA, lane 2). Addition of 100-fold molar excess of unlabeled OCT1BS-a DNA to the reaction mixture resulted in elimination of the specific band (NE + OCT1BS-a, lane 4). B, Densitometry scan analysis of the resulting protein-OCT1BS-a DNA complex formed in (A). The integrated density values (IDV) for each band were obtained from four independent EMSA experiments. Comparison of relative IDV was assessed by one-way ANOVA followed by a Newman-Keuls multiple comparison test (#, p < 0.05 for NE + EGTA (21640 ± 4522) and Nimodipine (23690 ± 5037) vs. NE (51930 ± 6711)). All data are expressed as mean ± SEM.
3.3 Expression of the Ca2+-binding protein DREAM in GT1-7 cells
As mentioned above, treatment of cells with nimodipine has been shown to reduce the influx of Ca2+ through blockage of L-type voltage gated channels in the cell membrane resulting in a decrease in intracellular calcium levels ([Ca2+]i) (McCarthy and TanPiengco, 1992). This reduction in [Ca2+]i is believed to affect specific cellular events such as protein phosphorylation, and activity of second messengers (cyclic adenosine monophosphate (cAMP) and inositol 1,4,5-triphosphate) (Berridge et al., 2003; Bootman et al., 2001; Carafoli, 2004). Our identification that nimodipine induced changes in protein binding activity at the OCT1BS-a site in the GnRH promoter suggested that calcium may be active in this nuclear transcription process. Certain Ca2+-binding proteins may be capable of linking cytoplasmic and nuclear calcium and have the potential to impact transcriptional regulation. One of these proteins, identified as DREAM, was shown to be expressed in GT1-7 cells, but little information was available concerning its function (Leclerc et al., 2002). In order to explore the possibility that DREAM may be involved in this process, we assessed first whether or not the DREAM protein that was expressed in GT1-7 cells contained all the elements needed for proper calcium binding activity. Using known primers previously used to clone the rat DREAM cDNA, we performed RT-PCR using GT1-7 RNA as the template. As shown in Figure 3A, bands of predicted size (852 bp, lanes 2 and 3) were identified using these primers suggesting that the full coding region corresponding to DREAM was expressed in these cells. As expected, RT-PCR reactions performed with these primers, but in the absence of RNA, did not yield an 852 bp band. The resulting RT-PCR amplified products were cloned into pCR2.1-TOPO vector as described in Materials and methods. Six of the clones obtained were sequenced and analyzed (Fig. 3B). From this we were able to determine that the DREAM molecules expressed in GT1-7 cells contained each of the four putative EF hands believed to be important for interaction with calcium (Ikura, 1996; Osawa et al., 2001). Although a splice variant containing an extra aspartic acid residue (position nucleotide 216) was identified (Fig. 3B), it was clear from our analysis that both forms of DREAM expressed in GT1-7 cells contained intact EF-hand Ca2+-binding motifs.
Figure 3. RT-PCR analysis and nucleotide DNA sequence of the mouse DREAM gene cDNA from GT1-7 cells.
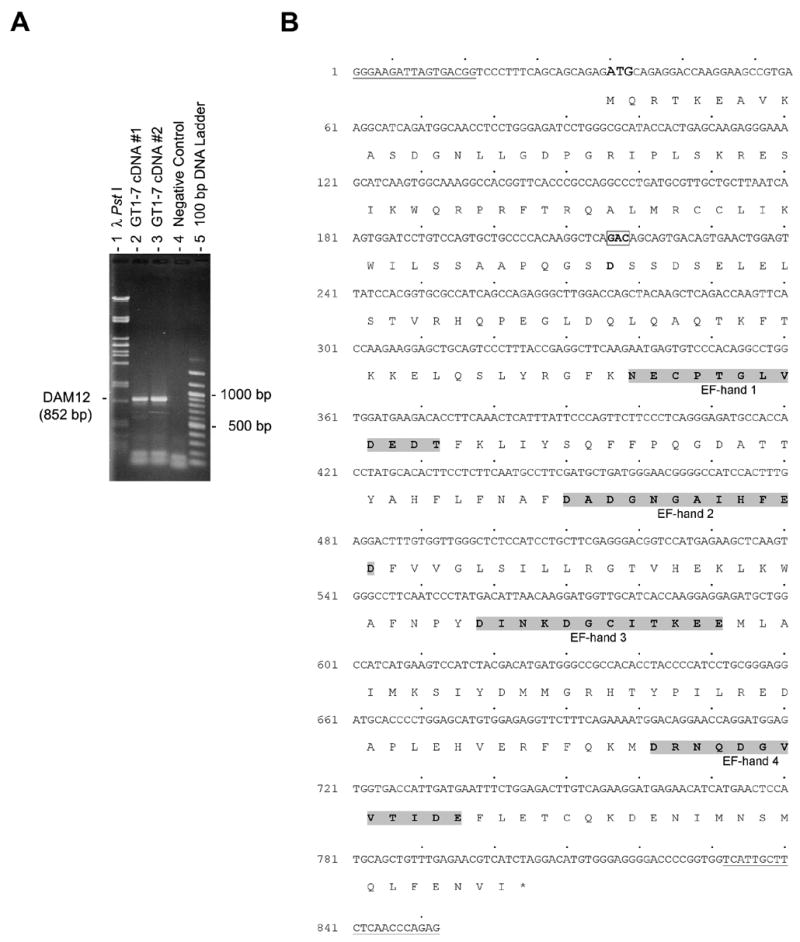
A, RT-PCR analysis of DREAM gene expression in GT1-7 cells. One microgram of total RNA was reversed transcribed and amplified with mouse specific primers for the DREAM gene cDNA yielding an expected product of 852 bp (DAM12), as described in Materials and methods are shown (lanes 2 and 3). The negative control (lane 4) contained water instead of cDNA template. One third of the PCR mixture was resolved on 1.0 % TAE-agarose gel stained with ethidium bromide. Two cDNA templates generated with total RNA isolated from GT1-7 cells were employed in the PCR amplifications (GT1-7 cDNA #1, lane 2 and GT1-7 cDNA #2, lane 3). A lambda PstI digest (lane 1) and a 100 bp DNA ladder (lane 5) were used as molecular weight markers. B, Nucleotide DNA sequence resulting from six-independent cloned RT-PCR amplified fragments. The DAM12 fragments were cloned in pCR2.1 TOPO vector as outlined in Materials and methods. The deduced amino acid sequence is shown directly below the first base of each codon. Regions corresponding to primers DAM-F1 and DAM-R2 are underlined. The splice variant GAC (coding for an extra aspartic acid residue) is indicated in bold and enclosed in a box.
3.4 EMSA analysis of DREAM binding to the OCT1BS-a site
Recent studies revealed that in addition to direct binding to DNA through downstream regulatory elements (DRE), DREAM can also interact and modulate the action of transcriptional factors through protein-protein interaction (Ledo et al., 2002; Rivas et al., 2004; Zaidi et al., 2006). In the NSE region of the GnRH promoter, there are no sequences that offer a high degree of homology to consensus DRE sequence. From this, we hypothesized that any DREAM influence on gene expression pulse activity would be mediated by interaction with other transcriptional proteins. Our observation that nimodipine treatment reduced protein binding at the OCT1BS-a site revealed that not only this site was essential for the generation of GnRH promoter pulses (Vazquez-Martinez et al., 2002) but also can act as active sensitive site for calcium changes. Therefore, we reasoned that if DREAM mediated any of these calcium-related changes, then it should exert some or all of its action at this site. To address this, EMSA experiments were performed with oligonucleotides containing the OCT1BS-a site as probe and nuclear proteins from GT1-7. As shown in Figure 4 (NE, lane 1), this reaction yielded a distinct specific band as described previously (Clark and Mellon, 1995). In supershift reactions, addition of an antibody against DREAM resulted in the elimination of the specific band demonstrating that DREAM protein is part of the complex binding to OCT1BS-a (NE + αDREAM, lane 5). Our use of several other polyclonal antibodies as controls did not influence the formation of the specific band (lanes 2–4). As expected, addition 100-fold molar excess of the OCT1BS-a specific competitor completely abolished the formation of the protein/OCT1BS-a complex (NE + OCT1BS-a, lane 6). Taken together, these results indicate that DREAM is part of the protein complex that binds to the OCT1BS-a site and may interact with OCT1 directly or indirectly through association with other proteins of the complex.
Figure 4. EMSA analysis of DREAM-binding to the OCT1BS-a site in the GnRH NSE promoter region.
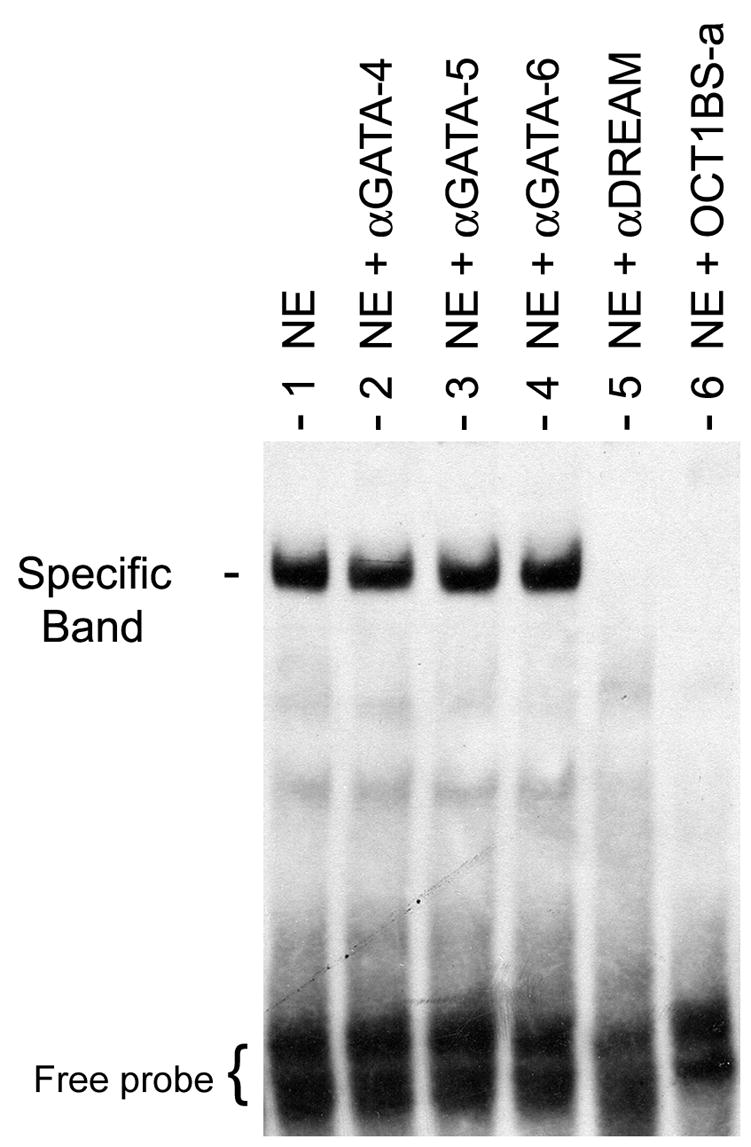
EMSAs were performed with nuclear protein extracts from GT1-7 cells and 5′-biotinylated OCT1BS-a duplex DNA as a probe (see Materials and methods). Incubations of the labeled OCT1BS-a probe with GT1-7 nuclear extracts alone (NE, lane 1) or combined with various antisera or specific DNA competitor were conducted. Band intensity remained unchanged with addition of either antibody against GATA-4 (lane 2) GATA-5 (lane 3) or GATA-6 (lane 4). The presence of DREAM protein within the protein-DNA complex was identified by disappearance of the specific band in the supershift reaction when an antiserum specific for DREAM was used (NE + αDREAM, lane 5). In competition experiment, results of 100-fold molar excess of unlabeled specific OCT1BS-a duplex DNA (NE + OCT1BS-a, lane 6) is shown. These results are representative of those obtained in 4 independent EMSA experiments.
3.5 Immunoneutralization of DREAM
In our final studies, we evaluated the role of DREAM in the generation of GnRH promoter pulse activity. This was accomplished by first injecting an anti-DREAM antibody into individual GT1-7 cells and then evaluating the photonic pattern of GnRH-GE that was generated. As shown in representative examples presented in Figure 5A, injection of control IgG into individual GT1-7 cells did not influence the generation of pulses of GnRH-GE (0.092 pulse/h ± 0.020, n = 6 cells). In contrast, GT1-7 cells injected with antibody against DREAM generated photonic patterns with virtually no pulse activity (Fig. 5B, p < 0.001) (0.024 pulse/h ± 0.010, n = 11 cells vs. 0.092 ± 0.0.020 pulse/h, n = 6 cells; αDREAM vs. control IgG, respectively). This demonstrates clearly that the DREAM protein is needed for the elaboration of GnRH-GE pulses.
Figure 5. Impact of immunoneutralization of DREAM protein on GnRH gene promoter pulse activity.
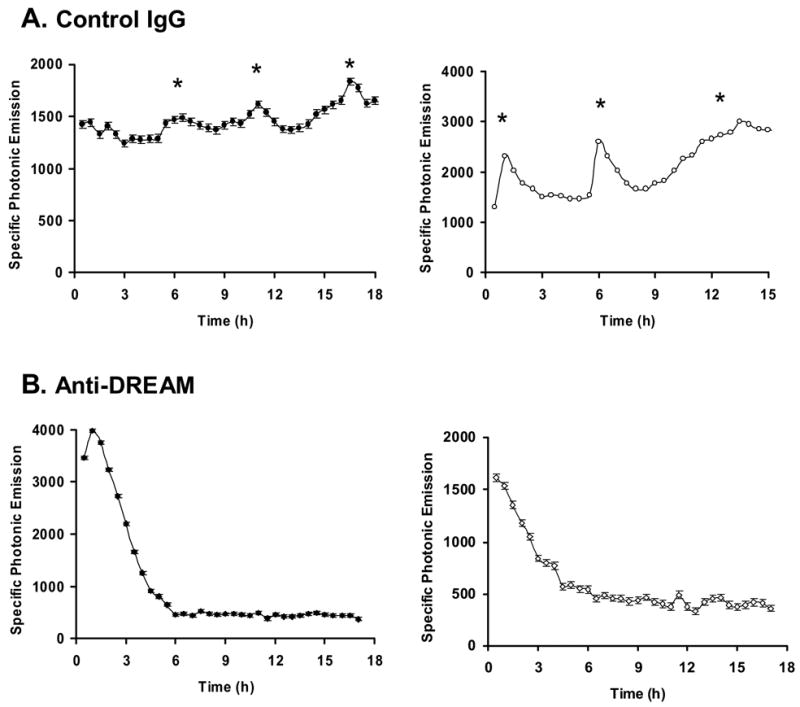
Representative examples of photonic profiles generated by single GT1-7 cells first microinjected with the reporter plasmid pA3GnRH-LUC, monitored to identify photonic active cells, and then microinjected with either (A) control polyclonal IgG antibody solution (n = 6 cells), or (B) specific antibody against DREAM (n = 11 cells). Two photonic profiles for each treatment are presented. Asterisks represent significant GnRH gene promoter pulses as described in Materials and methods.
4. Discussion
Previously, we have reported that GnRH-GE occurs in episodes and that these episodes or bursts of activity maintain a close functional relationship with GnRH secretory pulses (Nunez et al., 1998; Vazquez-Martinez et al., 2001b). This functional relationship is further evidenced in the present study by the demonstration that nimodipine, an L-type voltage gated Ca2+ channel blocker which markedly reduces secretory pulse activity (Vazquez-Martinez et al., 2001a), also abolishes GnRH-GE pulses. During attempts to elucidate the mechanism(s) that may link secretory and gene expression events, we identified DREAM expression in GT1-7 cells (Leclerc et al., 2002) and found in this investigation that DREAM complexed with factors binding at the OCT1BS-a site which is an essential element for the generation of GnRH gene promoter pulses. Moreover, immunoneutralization of DREAM in individual GT1-7 cells eliminated episodic gene expression. In light of the calcium binding properties of DREAM, our results raise the possibility that by responding to changes in calcium levels, this protein may serve as a functional link bridging episodic secretory events in the cytoplasm with gene expression pulses in the nucleus.
The release of GnRH in discrete pulses or bursts is critical for proper reproductive function (Terasawa, 1995). Despite its importance, little information is available on the mechanism underlying this phenomenon. A growing body of evidence suggests that spontaneous intracellular free calcium oscillations are involved in the regulation of pulsatile hormone secretion (Martinez de la Escalera et al., 1992; Martinez de la Escalera et al., 1995; Spergel et al., 1996; Terasawa et al., 1999). Nunez et al. (2000) demonstated that [Ca2+]i underwent shifts from a quiescent to a high oscillatory state in individual GT1-7 cells and that these shifts were frequently associated with exocytotic events. The possibility that L-type calcium channels may be involved in this process was suggested by the observation that nimodipine caused a striking reduction in the frequency of secretory pulses in individual GT1-7 cells (Vazquez-Martinez et al., 2001a). Later studies from our laboratory revealed that exocytotic pulses are not isolated cytoplasmic events in these cells, but are functionally linked to nuclear gene expression pulses with the number and size of secretory episodes dictating the initiation of GnRH gene expression events (Vazquez-Martinez et al., 2001b). Although the process of communication between these two cellular compartments remains obscure, it would not be unreasonable to suggest that a factor (Ca2+) that is central to the secretory pulse process may be also important in the generation of gene expression episodic events as well. Indeed, our results demonstrating that nimodipine treatment markedly decreased hormone secretion as well as abolished gene expression pulse activity suggest that a nimodipine-induced response signal (e.g., altered influence of Ca2+ at the cell membrane) is transduced to the nucleus to regulate gene expression.
In order for a communicating molecule such as Ca2+ to be able to elicit a transcriptional change, it would have to act in some manner with nuclear components and at specific regulatory sites. Recent studies demonstrate that the NSE region of the GnRH promoter is important in dictating the basal level of GnRH-GE (Lawson et al., 2002; Nelson et al., 2000; Whyte et al., 1995). Within this region, we have established that the OCT1BS-a is critical for the elaboration of gene promoter pulses (Vazquez-Martinez et al., 2002). Indeed, mutation of this promoter element abolishes GnRH gene pulse activity. In addition, this site, as with other sites binding OCT1, is capable of complexing with several transcriptional elements that contribute to GnRH-GE (Clark and Mellon, 1995; Givens et al., 2004). In this regard, experimental approaches, such as two-hybrid system assays directed toward identifying elements that associate with OCT1 in GT1-7 cells, have revealed a number of nuclear transcription factors belonging to major families of co-regulators including the TALE homeodomain proteins (PBX1) (Rave-Harel et al., 2004), and the Groucho-Related Gene proteins (GRG5) (Rave-Harel et al., 2005). In light of the importance of OCT1BS-a in GnRH-GE, it would be reasonable to suggest that an element that is capable of recognizing changes in calcium may have a profound influence on this intermittent transcriptional process, if it interacted with this site. A likely candidate to function in this capacity is the Ca2+-binding DREAM protein. DREAM was one of the first nuclear transcription factors observed to be directly regulated by calcium binding (Carrion et al., 1999). This protein was found to have four binding motifs termed EF hands that interact directly with Ca2+ molecules. It was further discovered that binding of calcium to these EF regions resulted in a change in configuration that impairs interaction of the DREAM molecule with DNA (Osawa et al., 2001). Recent studies utilizing yeast two-hybrid screening reveals that DREAM also has the capacity to interact with a number of proteins in different cellular compartments (Savignac et al., 2005; Zaidi et al., 2006). Our demonstration that DREAM is expressed in GT1-7 cells and that it is a part of the OCT1BS-a binding complex supports its involvement in the process of NSE-related GnRH promoter activity. Moreover, our observation that gene expression pulses are abolished when DREAM is immunoneutralized in individual GT1-7 cells indicates that this protein is critical for the elaboration of gene expression pulse events.
In summary, our findings that the Ca2+-binding DREAM protein binds to the OCT1BS-a site in the NSE region of the GnRH promoter and that immunoneutralization of this protein in individual cells abolishes GnRH-GE pulses clearly identifies a role for this factor in gene expression pulse activity. These observations, when coupled with its calcium binding properties and our findings of calcium requirements for pulses to occur, suggest that DREAM via calcium may serve as the basis for the cytoplasm/nuclear communication that links secretion and gene expression pulse events, and provides a mechanism that may well maintain the cell in a state of readiness for responding to critically timed demands for release of GnRH during changing reproductive events. In fact, the importance of DREAM in gene expression pulse elaboration and its potential to respond to secretory-related changes raises the intriguing possibility that this protein may form the basis of a molecular on/off switch that dictates the elaboration of a gene expression pulse in GnRH neurons. Moreover, considering that DREAM is widely expressed in a variety of tissues and that Ca2+ is an universal intracellular messenger, it is possible that these two factors may play a role coupling nuclear and cytoplasmic events in other secretory cells. Further studies are in progress to identify additional components that are involved in this calcium-regulatory process.
Acknowledgments
The authors would like to thank Dr. Guy J. Leclerc for helpful comments and discussion, and members of the Medical University of South Carolina Biotechnology Resource Laboratory for their help with DNA sequence analysis. This investigation was supported by NIH grant HD-37657 to F.R.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- Bootman MD, Collins TJ, Peppiatt CM, Prothero LS, MacKenzie L, De Smet P, Travers M, Tovey SC, Seo JT, Berridge MJ, Ciccolini F, Lipp P. Calcium signalling--an overview. Semin Cell Dev Biol. 2001;12:3–10. doi: 10.1006/scdb.2000.0211. [DOI] [PubMed] [Google Scholar]
- Carafoli E. The ambivalent nature of the calcium signal. J Endocrinol Invest. 2004;27:134–136. [PubMed] [Google Scholar]
- Carrion AM, Link WA, Ledo F, Mellstrom B, Naranjo JR. DREAM is a Ca2+-regulated transcriptional repressor. Nature. 1999;398:80–84. doi: 10.1038/18044. [DOI] [PubMed] [Google Scholar]
- Clark ME, Mellon PL. The POU homeodomain transcription factor Oct-1 is essential for activity of the gonadotropin-releasing hormone neuron-specific enhancer. Mol Cell Biol. 1995;15:6169–6177. doi: 10.1128/mcb.15.11.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig TA, Benson LM, Venyaminov SY, Klimtchuk ES, Bajzer Z, Prendergast FG, Naylor S, Kumar R. The metal-binding properties of DREAM: evidence for calcium-mediated changes in DREAM structure. J Biol Chem. 2002;277:10955–10966. doi: 10.1074/jbc.M109660200. [DOI] [PubMed] [Google Scholar]
- Crowley WF, Jr, Filicori M, Spratt DI, Santoro NF. The physiology of gonadotropin-releasing hormone (GnRH) secretion in men and women. Recent Prog Horm Res. 1985;41:473–531. doi: 10.1016/b978-0-12-571141-8.50015-9. [DOI] [PubMed] [Google Scholar]
- Givens ML, Kurotani R, Rave-Harel N, Miller NL, Mellon PL. Phylogenetic footprinting reveals evolutionarily conserved regions of the gonadotropin-releasing hormone gene that enhance cell-specific expression. Mol Endocrinol. 2004;18:2950–2966. doi: 10.1210/me.2003-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura M. Calcium binding and conformational response in EF-hand proteins. Trends Biochem Sci. 1996;21:14–17. [PubMed] [Google Scholar]
- Ikura M, Osawa M, Ames JB. The role of calcium-binding proteins in the control of transcription: structure to function. Bioessays. 2002;24:625–636. doi: 10.1002/bies.10105. [DOI] [PubMed] [Google Scholar]
- Kepa JK, Wang C, Neeley CI, Raynolds MV, Gordon DF, Wood WM, Wierman ME. Structure of the rat gonadotropin releasing hormone (rGnRH) gene promoter and functional analysis in hypothalamic cells. Nucleic Acids Res. 1992;20:1393–1399. doi: 10.1093/nar/20.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krsmanovic LZ, Martinez-Fuentes AJ, Arora KK, Mores N, Navarro CE, Chen HC, Stojilkovic SS, Catt KJ. Autocrine regulation of gonadotropin-releasing hormone secretion in cultured hypothalamic neurons. Endocrinology. 1999;140:1423–1431. doi: 10.1210/endo.140.3.6588. [DOI] [PubMed] [Google Scholar]
- Lawson MA, Macconell LA, Kim J, Powl BT, Nelson SB, Mellon PL. Neuron-specific expression in vivo by defined transcription regulatory elements of the GnRH gene. Endocrinology. 2002;143:1404–1412. doi: 10.1210/endo.143.4.8751. [DOI] [PubMed] [Google Scholar]
- Leclerc GM, Boockfor FR. Identification of a novel OCT1 binding site that is necessary for the elaboration of pulses of rat GnRH promoter activity. Mol Cell Endocrinol. 2005;245:86–92. doi: 10.1016/j.mce.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Leclerc GM, Leclerc GJ, Shorte SL, Stephen Frawley L, Boockfor FR. Cloning and mRNA expression of the Ca2+-binding DREAM protein in the pituitary. Gen Comp Endocrinol. 2002;129:45–55. doi: 10.1016/s0016-6480(02)00509-9. [DOI] [PubMed] [Google Scholar]
- Ledo F, Kremer L, Mellstrom B, Naranjo JR. Ca2+-dependent block of CREB-CBP transcription by repressor DREAM. Embo J. 2002;21:4583–4592. doi: 10.1093/emboj/cdf440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Escalera G Martinez, Choi AL, Weiner RI. Generation and synchronization of gonadotropin-releasing hormone (GnRH) pulses: intrinsic properties of the GT1-1 GnRH neuronal cell line. Proc Natl Acad Sci U S A. 1992;89:1852–1855. doi: 10.1073/pnas.89.5.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Escalera G Martinez, Choi AL, Weiner RI. Signaling pathways involved in GnRH secretion in GT1 cells. Neuroendocrinology. 1995;61:310–317. doi: 10.1159/000126853. [DOI] [PubMed] [Google Scholar]
- Masotto C, Negro-Vilar A. Gonadectomy influences the inhibitory effect of the endogenous opiate system on pulsatile gonadotropin secretion. Endocrinology. 1988;123:747–752. doi: 10.1210/endo-123-2-747. [DOI] [PubMed] [Google Scholar]
- Maxwell IH, Harrison GS, Wood WM, Maxwell F. A DNA cassette containing a trimerized SV40 polyadenylation signal which efficiently blocks spurious plasmid-initiated transcription. Biotechniques. 1989;7:276–280. [PubMed] [Google Scholar]
- McCarthy RT, TanPiengco PE. Multiple types of high-threshold calcium channels in rabbit sensory neurons: high-affinity block of neuronal L-type by nimodipine. J Neurosci. 1992;12:2225–2234. doi: 10.1523/JNEUROSCI.12-06-02225.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon PL, Windle JJ, Goldsmith PC, Padula CA, Roberts JL, Weiner RI. Immortalization of hypothalamic GnRH neurons by genetically targeted tumorigenesis. Neuron. 1990;5:1–10. doi: 10.1016/0896-6273(90)90028-e. [DOI] [PubMed] [Google Scholar]
- Nelson SB, Lawson MA, Kelley CG, Mellon PL. Neuron-specific expression of the rat gonadotropin-releasing hormone gene is conferred by interactions of a defined promoter element with the enhancer in GT1-7 cells. Mol Endocrinol. 2000;14:1509–1522. doi: 10.1210/mend.14.9.0521. [DOI] [PubMed] [Google Scholar]
- Nunez L, Faught WJ, Frawley LS. Episodic gonadotropin-releasing hormone gene expression revealed by dynamic monitoring of luciferase reporter activity in single, living neurons. Proc Natl Acad Sci U S A. 1998;95:9648–9653. doi: 10.1073/pnas.95.16.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez L, Villalobos C, Boockfor FR, Frawley LS. The relationship between pulsatile secretion and calcium dynamics in single, living gonadotropin-releasing hormone neurons. Endocrinology. 2000;141:2012–2017. doi: 10.1210/endo.141.6.7491. [DOI] [PubMed] [Google Scholar]
- Osawa M, Tong KI, Lilliehook C, Wasco W, Buxbaum JD, Cheng HY, Penninger JM, Ikura M, Ames JB. Calcium-regulated DNA binding and oligomerization of the neuronal calcium-sensing protein, calsenilin/DREAM/KChIP3. J Biol Chem. 2001;276:41005–41013. doi: 10.1074/jbc.M105842200. [DOI] [PubMed] [Google Scholar]
- Rave-Harel N, Givens ML, Nelson SB, Duong HA, Coss D, Clark ME, Hall SB, Kamps MP, Mellon PL. TALE homeodomain proteins regulate gonadotropin-releasing hormone gene expression independently and via interactions with Oct-1. J Biol Chem. 2004;279:30287–30297. doi: 10.1074/jbc.M402960200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rave-Harel N, Miller NL, Givens ML, Mellon PL. The Groucho-related gene family regulates the gonadotropin-releasing hormone gene through interaction with the homeodomain proteins MSX1 and OCT1. J Biol Chem. 2005;280:30975–30983. doi: 10.1074/jbc.M502315200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas M, Mellstrom B, Naranjo JR, Santisteban P. Transcriptional repressor DREAM interacts with thyroid transcription factor-1 and regulates thyroglobulin gene expression. J Biol Chem. 2004;279:33114–33122. doi: 10.1074/jbc.M403526200. [DOI] [PubMed] [Google Scholar]
- Savignac M, Pintado B, Gutierrez-Adan A, Palczewska M, Mellstrom B, Naranjo JR. Transcriptional repressor DREAM regulates T-lymphocyte proliferation and cytokine gene expression. Embo J. 2005;24:3555–3564. doi: 10.1038/sj.emboj.7600810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman AJ, Krey LC. The luteinizing hormone-releasing hormone (LH-RH) neuronal networks of the guinea pig brain. I. Intra- and extra-hypothalamic projections. Brain Res. 1978;157:233–246. doi: 10.1016/0006-8993(78)90026-4. [DOI] [PubMed] [Google Scholar]
- Spergel DJ, Catt KJ, Rojas E. Immortalized GnRH neurons express large-conductance calcium-activated potassium channels. Neuroendocrinology. 1996;63:101–111. doi: 10.1159/000126946. [DOI] [PubMed] [Google Scholar]
- Spratt DI, Carr DB, Merriam GR, Scully RE, Rao PN, Crowley WF., Jr The spectrum of abnormal patterns of gonadotropin-releasing hormone secretion in men with idiopathic hypogonadotropic hypogonadism: clinical and laboratory correlations. J Clin Endocrinol Metab. 1987;64:283–291. doi: 10.1210/jcem-64-2-283. [DOI] [PubMed] [Google Scholar]
- Takasuka N, White MR, Wood CD, Robertson WR, Davis JR. Dynamic changes in prolactin promoter activation in individual living lactotrophic cells. Endocrinology. 1998;139:1361–1368. doi: 10.1210/endo.139.3.5826. [DOI] [PubMed] [Google Scholar]
- Terasawa E. Control of luteinizing hormone-releasing hormone pulse generation in nonhuman primates. Cell Mol Neurobiol. 1995;15:141–164. doi: 10.1007/BF02069563. [DOI] [PubMed] [Google Scholar]
- Terasawa E, Keen KL, Mogi K, Claude P. Pulsatile release of luteinizing hormone-releasing hormone (LHRH) in cultured LHRH neurons derived from the embryonic olfactory placode of the rhesus monkey. Endocrinology. 1999;140:1432–1441. doi: 10.1210/endo.140.3.6559. [DOI] [PubMed] [Google Scholar]
- Vazquez-Martinez R, Leclerc GM, Wierman ME, Boockfor FR. Episodic activation of the rat GnRH promoter: role of the homeoprotein Oct-1. Mol Endocrinol. 2002;16:2093–2100. doi: 10.1210/me.2002-0139. [DOI] [PubMed] [Google Scholar]
- Vazquez-Martinez R, Shorte SL, Boockfor FR, Frawley LS. Synchronized exocytotic bursts from gonadotropin-releasing hormone- expressing cells: dual control by intrinsic cellular pulsatility and gap junctional communication. Endocrinology. 2001a;142:2095–2101. doi: 10.1210/endo.142.5.8123. [DOI] [PubMed] [Google Scholar]
- Vazquez-Martinez R, Shorte SL, Faught WJ, Leaumont DC, Frawley LS, Boockfor FR. Pulsatile exocytosis is functionally associated with GnRH gene expression in immortalized GnRH-expressing cells. Endocrinology. 2001b;142:5364–5370. doi: 10.1210/endo.142.12.8551. [DOI] [PubMed] [Google Scholar]
- Weiner RI, Wetsel W, Goldsmith P, de la Escalera G Martinez, Windle J, Padula C, Choi A, Negro-Vilar A, Mellon P. Gonadotropin-releasing hormone neuronal cell lines. Front Neuroendocrinol. 1992;13:95–119. [PubMed] [Google Scholar]
- Whyte DB, Lawson MA, Belsham DD, Eraly SA, Bond CT, Adelman JP, Mellon PL. A neuron-specific enhancer targets expression of the gonadotropin- releasing hormone gene to hypothalamic neurosecretory neurons. Mol Endocrinol. 1995;9:467–477. doi: 10.1210/mend.9.4.7659090. [DOI] [PubMed] [Google Scholar]
- Wood SF, Szuts EZ, Fein A. Inositol trisphosphate production in squid photoreceptors. Activation by light, aluminum fluoride, and guanine nucleotides. J Biol Chem. 1989;264:12970–12976. [PubMed] [Google Scholar]
- Zaidi NF, Kuplast KG, Washicosky KJ, Kajiwara Y, Buxbaum JD, Wasco W. Calsenilin interacts with transcriptional co-repressor C-terminal binding protein(s) J Neurochem. 2006;98:1290–1301. doi: 10.1111/j.1471-4159.2006.03972.x. [DOI] [PubMed] [Google Scholar]


