Summary
Clp ATPases are protein machines involved in protein degradation and disaggregation. The common structural feature of Clp ATPases is the formation of ring-shaped oligomers. Recent work has shown that the function of all Clp ATPases is based on an energy-dependent threading of substrates through the narrow pore at the center of the ring. This review gives an outline of known mechanistic principles of threading machines that unfold protein substrates either before their degradation (ClpA, ClpX, HslU) or during their reactivation from aggregates (ClpB). The place of Clp ATPases within a broad AAA+ superfamily of ATPases associated with various cellular activities suggests that similar mechanisms can be used by other protein machines to induce conformational rearrangements in a wide variety of substrates.
Keywords: Clp ATPases, AAA+ ATPases, protein unfolding, protein degradation, protein aggregation, molecular chaperone
Living cells maintain an elaborate network of molecular processes that depend on the function of large protein complexes. The principles of assembly and mechanism of those “protein machines” that carry out so many essential cellular functions provide a challenging new frontier to be explored in the post-genomic era (Alberts, 1998). Research on Clp ATPases is an ongoing inquiry into the inner workings of machinery designed to maintain the quality of cellular proteins by performing the functions of molecular chaperones and energy-dependent proteases. Although many unanswered questions remain, major progress in understanding the mechanism of protein quality control by Clp ATPases has been achieved in recent years. The place of Clp ATPases within the AAA+ superfamily of ATPases associated with various cellular activities (Neuwald et al., 1999) suggests that analogous mechanisms might by applicable to a substantially broader range of biological processes.
ClpA from Escherichia coli, the ATP-binding component of an energy-dependent “caseinolytic protease” was the first Clp ATPase discovered (Hwang et al., 1987; Katayama-Fujimura et al., 1987). It was followed by identification of a number of closely related ATPases: ClpB, ClpC, ClpE, ClpL, ClpV, ClpX, and HslU (ClpY) (reviewed in (Butler et al., 2006; Dougan et al., 2002a; Gottesman, 2003; Schlieker et al., 2005)). Four members of the family: ClpA, ClpB, ClpX, and HslU will be the focus of the present review, as these are the most thoroughly characterized Clp ATPases.
Many Clp ATPases, with the notable exception of ClpB, form complexes with peptidase subunits (ClpP or HslV, see Fig. 1). The ClpAP, ClpXP, and HslUV proteases are similar in design to the eukaryotic 26S proteasome, with the ATPase subunits guarding the entrances to the proteolytic chambers (Kessel et al., 1995; Rohrwild et al., 1997). Clp proteases are found in all prokaryotes as well as in mitochondria and chloroplasts of eukaryotic cells (Gottesman, 2003).
Fig. 1.
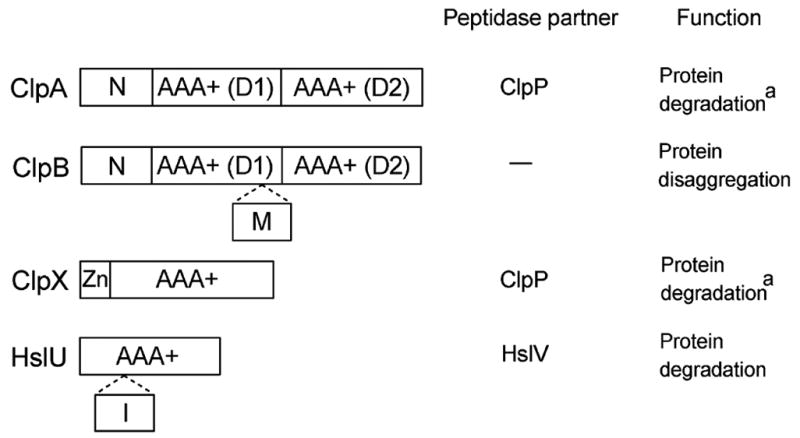
Domain organization and function of selected Clp ATPases. AAA+ sequence modules are shown along with the N-terminal domains of ClpA and ClpB (N), the middle domain of ClpB (M), the Zn-binding domain of ClpX (Zn), and the intermediate domain of HslU (I). a In addition to their ClpP-dependent protease activity, ClpA and ClpX remodel the conformation of the replication initiation protein RepA and MuA transposase, respectively (reviewed in (Burton and Baker, 2005).
ClpB is different from ClpA, ClpX, and HslU because it does not associate with peptidase subunits (see Fig. 1) and is found not only in prokaryotes and in the eukaryotic organelles, but also in the cytosol of yeast and plants. Indeed, the function of ClpB is distinct from that of other Clp ATPases: ClpB is not involved in protein degradation (Woo et al., 1992), instead, it disaggregates and reactivates strongly aggregated proteins (Goloubinoff et al., 1999; Motohashi et al., 1999; Zolkiewski, 1999). The aggregation-reversing activity of ClpB requires cooperation with the Hsp70/Hsp40 chaperone machinery (bacterial DnaK/DnaJ).
Presently, Clp ATPases are considered to be a subgroup of the AAA+ protein superfamily (Lupas and Martin, 2002; Neuwald et al., 1999). Thus, their protein degradation and disaggregation activities are just two of a wide variety of AAA+ functions that include membrane fusion and vesicular trafficking, cytoskeletal regulation and intracellular transport, organelle biogenesis, DNA replication and repair, and transcription regulation (Hanson and Whiteheart, 2005; Ogura and Wilkinson, 2001; Vale, 2000). The functional diversity of AAA+ ATPases is based on the common protein architecture: all AAA+ proteins contain conserved sequence modules (one or two, see Fig. 1) with ATP-binding motifs (Walker A and B, sensor-1 and -2) (reviewed in (Hanson and Whiteheart, 2005)). The AAA+ ATPases form ring- or cylinder-shaped oligomers (usually hexamers) with a narrow channel located at the center (see Fig. 2) and the nucleotide binding sites at the inter-subunit interfaces. In addition to the AAA+ modules, the ATPases contain other domains that often specify their cellular localization, substrate identity, and/or their interacting partners (Iyer et al., 2004). Many AAA+ ATPases, including ClpA and ClpX, interact with partner proteins that modulate their function (Dougan et al., 2002b).
Fig. 2.

Substrate threading mechanism of ClpA. (Left), ClpA contains two AAA+ modules (D1, D2) and forms a cylinder-shaped hexamer with a narrow central channel. The degradation tag (red) at the terminus of a protein substrate (blue) is inserted into the ClpA channel. (Right), Flexible loops located inside the channel (orange) mediate the ATP-hydrolysis-driven translocation of the substrate along the channel and its forced unfolding. Two out of the six ClpA subunits are shown.
A channel of destruction: protein unfolding induced by ClpA, ClpX, and ClpY (HslU)
A common theme in the function of AAA+ ATPases is their ability to remodel the conformation of macromolecules. Researchers in the field found it attractive to postulate a common mechanism of action of AAA+ ATPases linked to their conserved design and, in particular, their characteristic assembly into oligomeric rings. The mechanistic breakthrough came when several groups showed that ClpA and ClpX fully unfold their substrates by translocating them through the channel at the center of the oligomeric ring (see Fig. 2) (Kim et al., 2000; Reid et al., 2001; Singh et al., 2000; Weber-Ban et al., 1999). In the reconstruction of hexameric ClpA obtained from the monomer crystal structure, the channel is ~24 Å in diameter at its entrance and only ~9 Å-wide at its narrowest point (Guo et al., 2002), which implies that folded proteins cannot pass through the pore, unless transient increases in the channel size occur during the work cycle of the ATPase. Thus, the main function of the ATPase appears to be induction of substrate unfolding by energy-driven threading through the narrow “eye of the needle” at the center of the oligomeric ring. The channel might be wide enough to accommodate two unstructured polypeptides at the same time, as demonstrated by successful degradation of disulfide-crosslinked substrates by ClpXP (Burton et al., 2001). The unfolded substrates are directly transferred from the ATPase rings into the associated peptidases for degradation. An analogous mechanism applies to the proteasome, whose six AAA+ subunits (Rpt1 through Rpt6) form a ring that gradually unfolds and translocates substrates into the proteolytic chamber (Lee et al., 2001).
How does the threading machine work? Major progress in our understanding of the translocation process came from studies on ClpA and ClpX that recognize the SsrA degradation tag (Keiler et al., 1996) (reviewed in (Sauer et al., 2004)). Tagging the C-termini of many substrate variants with the SsrA sequence allowed observation of their unfolding and translocation. Several groups have focused on the process of substrate engagement by the structural parts of a Clp ATPase that form the narrow channel. In each monomer of ClpA, which contains two AAA+ modules (D1, D2), there are three channel motifs that are involved in substrate binding: two in D1 and one in D2 (Hinnerwisch et al., 2005). Two of those are unstructured loops (one in each AAA+ module, marked in orange in Fig. 2) containing tyrosine residues flanked by a pair of glycines (see Fig. 3, below). In the hexameric ClpA, the width of the channel is restricted by two diaphragms (in D1 and D2), each containing six loops (Guo et al., 2002). Mutational analysis showed that the loop sequence motifs are essential for substrate binding and translocation (Hinnerwisch et al., 2005). In HslU, the loop tyrosine changes its position in the channel as the ATPase switches between different conformational states (Wang et al., 2001), which suggests that the channel loop movement could directly drive the translocation of an inserted substrate. It is not yet known what part(s) of a threaded polypeptide would directly interact with the channel loop or how the loop tyrosine could mediate such interactions. Replacement of the flanking glycines, which might reduce loop flexibility, inhibits not only substrate translocation but also the ATPase activity of HslU (Park et al., 2005). This demonstrates a tight conformational coupling between the “engine” (ATP-binding sites located between the subunits of the oligomeric ring) and the “drive” (flexible loops inside the central channel).
Fig. 3.
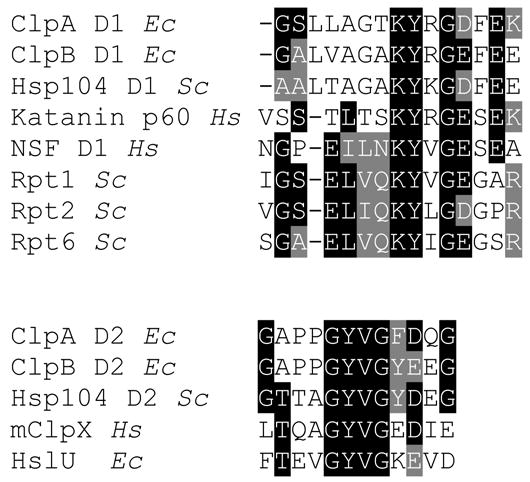
Sequence alignment of the channel loop regions in Clp ATPases and selected other AAA+ ATPases. The sequences shown are from ClpA, ClpB, and HslU from Escherichia coli (Ec), a ClpB homolog Hsp104 and the proteasomal ATPases Rpt1, Rpt2, and Rpt6 from Saccharomyces cerevisiae (Sc), katanin p60, N-ethylmaleimide-sensitive factor (NSF), and mitochondrial ClpX (mClpX) from Homo sapiens (Hs).
Elegant studies on ClpX performed by Sauer and Baker groups have given us the closest view so far on the operation of a polypeptide-threading ATPase. ClpX contains a single AAA+ module and works like a machine with a two-speed transmission (Kenniston et al., 2003). The rate of ATP hydrolysis by ClpX is high during translocation of unfolded substrates that do not resist translocation. About one molecule of ATP is hydrolyzed per one translocated residue. Folded substrates offer significant resistance to translocation, directly related to the local stability of structures near the degradation tag (Lee et al., 2001). During unfolding and translocation of folded proteins, ClpX switches to the lower gear and lower rates of ATP hydrolysis. However, the amount of ATP consumed during unfolding and translocation of folded substrates is higher than that needed for the unfolded ones (Kenniston et al., 2003).
A hexameric Clp ATPase is a “six-cylinder” ATP-driven polypeptide-threading engine. The diaphragms restricting the size of the central channel and possibly mediating substrate translocation consist of six flexible loops, each provided by one of the six protein subunits. Are the movements of the six loops in each diaphragm coordinated? Do all cylinders of the threading machine fire in concert? Extensive studies on ClpX show strong asymmetry in interactions of the six subunits with nucleotides, which implies that the subunits of the hexamer work quasi-independently without concerted hydrolysis of ATP (Hersch et al., 2005). Furthermore, hexamers of ClpX with different configurations of active and inactive subunits support translocation of polypeptides (Martin et al., 2005). Indeed, even a ClpX hexamer containing only one active subunit is capable of substrate threading, albeit at a low speed. Thus, substrate translocation through the ClpX channel does not require simultaneous movements of the six channel loops, although some coordination of loop strokes in a fully-active hexameric ATPase might occur to stimulate the directional translocation of a substrate. During translocation, different amino acids of the substrate will appear in proximity of the channel loops and a particular conformation and orientation of the substrate might offer the most favorable interaction to a particular loop within the ring of six. The random mechanism of ATP hydrolysis and channel loop movements within the hexameric ATPase could enable the residues of the loops to search favorable contacts with the substrate and induce substrate translocation independently of the contacts maintained by the neighboring loops.
A channel of life: protein disaggregation mediated by ClpB
While progress continues to be made in deciphering the mechanism of ClpA, ClpX, and HslU, the principles of function of their cousin ClpB remain less clear. Whereas both ClpA and ClpB contain two AAA+ modules (D1, D2, see Fig. 1), a ~120 amino-acid-long segment is inserted into D1 of ClpB. The presence of this inserted middle domain is the most conspicuous difference between ClpA, a protein-unfolding component of a protease, and ClpB, a protein-disaggregating chaperone. The middle domain forms a propeller-shaped coiled-coil at the outer surface of the oligomeric ring of ClpB and does not have any apparent connection with the central channel (Lee et al., 2003). Restricting the mobility of the coiled-coil by engineering inter-domain disulfide bonds causes significant inhibition of the chaperone activity of ClpB. Thus, the disaggregation activity of ClpB might be linked to the motions of the coiled-coil domains in the hexameric ClpB. The coiled-coils could interact with protein aggregates and work as a “crowbar”, breaking apart aggregated particles (Glover and Tkach, 2001; Lee et al., 2003). However, direct experimental evidence for the crowbar model has not been obtained and the role of the middle domain in ClpB remains a mystery.
By mutating surface residues within the D2 module of ClpB, Bukau and coworkers produced a ClpA-like ClpB variant (BAP), which formed complex with the peptidase ClpP (Weibezahn et al., 2004). Instead of reactivating aggregated substrates, BAP guides them to degradation by ClpP, which demonstrates that the substrates are threaded through the central channel in the oligomeric ring of BAP. Importantly, the channel-loop residues in ClpB can be crosslinked to various substrates (Schlieker et al., 2004; Weibezahn et al., 2004). Thus, the mechanism of protein disaggregation mediated by ClpB is based on the same principle as the one used by other Clp ATPases: energy-dependent translocation of polypeptides through the central pore in the oligomeric ring and their forced unfolding. Whereas ClpA, ClpX, and HslU unfold mostly soluble substrates, ClpB extracts single polypeptides from aggregated particles by unfolding whichever misfolded structures that stabilize the aggregates. Unlike in ClpA, ClpX, and HslU, where the channel exit leads to the attached peptidase subunit, the ClpB channel exit is open and releases its substrates into solution, where their refolding might be assisted by other chaperones.
Discovery of the threading-mediated protein disaggregation has shown that ClpB is mechanistically related to its better understood peers involved in protein degradation. The channel loop motifs in ClpB are very similar to those in ClpA (see Fig. 3, below) and it is likely that the operating principles of the ATP-driven translocation engine, as described in the previous section, apply to ClpB as well as to ClpA, ClpX, and HslU. However, the mechanism of protein disaggregation mediated by ClpB still offers a number of puzzles.
Although the channel loops are similar in ClpA and ClpB, the latter protein does not interact with the SsrA tag (Hinnerwisch et al., 2005). This result makes sense because aggregated proteins, unlike those destined for degradation, are not tagged by any known cellular mechanism. If ClpB interacts with polypeptide termini that are exposed at the aggregate surface and extracts those polypeptides from aggregates by inserting the termini into its channel, it needs to possess unique aggregate-recognition capabilities. A machine that is capable of unfolding any untagged protein whose termini are available for binding would be harmful rather than beneficial in vivo. Indeed, native glucose-6-phosphate dehydrogenase, whose N- and C-terminal peptides are exposed, is not recognized by ClpB, whereas aggregates of the same protein become substrates of ClpB (M. Nagy and M. Zolkiewski, unpublished results).
Peptide-library binding data suggest that electrostatic attraction between positive charges in a protein substrate and conserved negatively charged residues at the entrance of the ClpB channel plays a role in the ClpB-substrate recognition (Schlieker et al., 2004). In addition, two acidic residues in the N-terminal domain of ClpB support binding of an aggregated substrate (Barnett et al., 2005). However, whether aggregated proteins preferentially expose positively charged groups at their surface while hiding negative charges has not been determined. The general mechanism of protein aggregation implies that the aggregates might expose hydrophobic surfaces that could be recognized by a chaperone or, if left alone, could promote further aggregation (Fink, 1998). Surprisingly, hydrophobic peptides do not preferentially interact with ClpB (Schlieker et al., 2004). How ClpB can distinguish between a properly folded and an aggregated protein remains a mystery. Aggregated proteins might expose a specific blend of hydrophobic and charged areas at their surface, which could be recognized by the chaperone. Further studies on the mechanisms of protein aggregation should help in solving this problem.
Perhaps the most puzzling aspect of ClpB is its cooperation with DnaK (Hsp70). The threading of aggregated substrates by BAP is strictly dependent on the presence of the DnaK chaperone system (Weibezahn et al., 2004). Since the substrates of BAP are degraded and are never released to solution in intact form, some action of the DnaK system must occur up-stream of ClpB, in addition to a possible down-stream function in refolding the extracted polypeptides. ClpB does interact with aggregates even in the absence of DnaK (Barnett et al., 2005) and, in fact, it competes with DnaK for substrate binding sites (Weibezahn et al., 2003). Since the peptide-binding preferences of both chaperones overlap (Schlieker et al., 2004), ClpB and DnaK might independently scan the surface of an aggregate in search of an appropriate initiation site. DnaK might also prepare the aggregates for disaggregation, since the pre-treatment of aggregated proteins with DnaK/DnaJ accelerates their subsequent reactivation by ClpB (Zietkiewicz et al., 2004). Evidence for a direct transfer of a substrate from DnaK to ClpB has not been found (Kedzierska et al., 2005).
Finally, the presence of the unique propeller-like coiled-coil domain in ClpB suggests that this chaperone might use another mechanism, in addition to substrate threading, to disaggregate proteins. Perhaps, ClpB can work either as a crowbar or as an unfoldase, depending on conditions or the nature of a substrate. Attaching the ClpB ring to ClpP in the studies of Weibezahn et al. (Weibezahn et al., 2004) might have modified the properties of ClpB and placed an emphasis on substrate threading, while inhibiting other potential mechanisms. Indeed, ClpP rescues the substrate translocation and unfolding defects of several ClpX mutants, which suggests that substrate threading by an ATPase ring can be enhanced in the presence of the peptidase (Joshi et al., 2004). It has been also suggested that ClpB might use two different sites during disaggregation: one for immobilizing itself at the surface of a substrate and one for initiation of the threading (Maurizi and Xia, 2004). Thus, the coiled-coil domain of ClpB might be involved in interactions with substrates even if it does not directly support their insertion into the channel.
Other channels and threading machines
The analysis of Clp ATPases demonstrates that forced unfolding of polypeptides by threading them through a narrow channel can be applied to achieve destruction as well as resurrection of proteins. It is tempting to speculate that the function of other AAA+ ATPases that form ring-shaped oligomers might be similarly linked to threading and unfolding of their substrates. It should be noted that a number of helicases belong to the AAA+ superfamily (Ogura and Wilkinson, 2001) and use their central channel to enclose and translocate either single-stranded or double-stranded DNA.
In search of polypeptide-threading machines, one can assume that the channel loop motifs would identify an ATPase capable of translocating a polypeptide substrate through the channel. In the AAA+ module, the loop motif is located between Walker A and B. As shown in Fig. 3, the channel loop motifs contain a conserved tyrosine and flanking glycine residues that convey flexibility to the loop structure. In the first variant of the loop motif (Fig. 3, top), the tyrosine is preceded by a conserved lysine and the flanking glycine is followed by an acidic residue. These motifs can be found at the entrance of the channel of ClpA, ClpB and its yeast homolog Hsp104, as well as the proteasomal ATPases (Rpt1, Rpt2, Rpt6). The other loop variant (Fig. 3, bottom) contains a conserved GYVG motif followed by an acidic residue and is found in the bottom half the channel in ClpA, ClpB, and Hsp104 as well as in the channels of ClpX and HslU. The loop motifs are not present in the wide sequence alignment that defined the AAA+ family (Neuwald et al., 1999).
Fig. 3 lists two examples of AAA+ ATPases that contain the ClpA- and ClpB-like loop motifs: katanin p60 and NSF. Katanin p60 is the AAA+ subunit of a protein complex that disassembles microtubules (Hartman and Vale, 1999). It has been proposed that microtubule disruption is linked to dissociation of the oligomeric rings of katanin. However, based on the presence of the loop sequence in katanin, it can be also hypothesized that katanin could insert a substrate (tubulin) into the channel, induce its unfolding by translocation, and cause microtubule severing by removing one of its building blocks. This prediction could be tested experimentally.
NSF (N-ethylmaleimide-sensitive factor) disassembles SNARE complexes after membrane fusion in cooperation with α-SNAP adaptor proteins (Sollner et al., 1993). Like ClpA and ClpB, NSF contains two AAA+ modules. However, unlike in ClpA and ClpB, only the “top” D1 module of NSF contains the loop motif (see Fig. 3), which suggests that NSF could pull a substrate into the channel and then perhaps release it without achieving complete translocation. This interesting mechanism would imply that it is possible to destabilize a protein complex by mechanically tugging on one of its components. As described above, advances in research on the mechanism of Clp ATPases not only gave us a close insight into the processes of destruction and reactivation of proteins, but also provided a novel general perspective on possible ways of using energy-driven protein machines to remodel, disrupt, and disassemble the structure of macromolecules.
Acknowledgments
I thank Dr. Larry Davis and the anonymous reviewers for comments and suggestions. MZ has been supported by the NIH (R01GM58626) and by the Kansas Agricultural Experiment Station (contribution 06-293-J).
References
- Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- Barnett ME, Nagy M, Kedzierska S, Zolkiewski M. The amino-terminal domain of ClpB supports binding to strongly aggregated proteins. J Biol Chem. 2005;280:34940–34945. doi: 10.1074/jbc.M505653200. [DOI] [PubMed] [Google Scholar]
- Burton BM, Baker TA. Remodeling protein complexes: insights from the AAA+ unfoldase ClpX and Mu transposase. Protein Sci. 2005;14:1945–1954. doi: 10.1110/ps.051417505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RE, Siddiqui SM, Kim YI, Baker TA, Sauer RT. Effects of protein stability and structure on substrate processing by the ClpXP unfolding and degradation machine. EMBO J. 2001;20:3092–3100. doi: 10.1093/emboj/20.12.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler SM, Festa RA, Pearce MJ, Darwin KH. Self-compartmentalized bacterial proteases and pathogenesis. Mol Microbiol. 2006;60:553–562. doi: 10.1111/j.1365-2958.2006.05128.x. [DOI] [PubMed] [Google Scholar]
- Dougan DA, Mogk A, Bukau B. Protein folding and degradation in bacteria: to degrade or not to degrade? That is the question. Cell Mol Life Sci. 2002a;59:1607–1616. doi: 10.1007/PL00012487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan DA, Mogk A, Zeth K, Turgay K, Bukau B. AAA+ proteins and substrate recognition, it all depends on their partner in crime. FEBS Lett. 2002b;529:6–10. doi: 10.1016/s0014-5793(02)03179-4. [DOI] [PubMed] [Google Scholar]
- Fink AL. Protein aggregation: folding aggregates, inclusion bodies and amyloid. Fold Des. 1998;3:R9–23. doi: 10.1016/S1359-0278(98)00002-9. [DOI] [PubMed] [Google Scholar]
- Glover JR, Tkach JM. Crowbars and ratchets: hsp100 chaperones as tools in reversing protein aggregation. Biochem Cell Biol. 2001;79:557–568. [PubMed] [Google Scholar]
- Goloubinoff P, Mogk A, Zvi AP, Tomoyasu T, Bukau B. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc Natl Acad Sci U S A. 1999;96:13732–13737. doi: 10.1073/pnas.96.24.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. Proteolysis in bacterial regulatory circuits. Annu Rev Cell Dev Biol. 2003;19:565–587. doi: 10.1146/annurev.cellbio.19.110701.153228. [DOI] [PubMed] [Google Scholar]
- Guo F, Maurizi MR, Esser L, Xia D. Crystal Structure of ClpA, an Hsp100 Chaperone and Regulator of ClpAP Protease. J Biol Chem. 2002;277:46743–46752. doi: 10.1074/jbc.M207796200. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- Hartman JJ, Vale RD. Microtubule disassembly by ATP-dependent oligomerization of the AAA enzyme katanin. Science. 1999;286:782–785. doi: 10.1126/science.286.5440.782. [DOI] [PubMed] [Google Scholar]
- Hersch GL, Burton RE, Bolon DN, Baker TA, Sauer RT. Asymmetric interactions of ATP with the AAA+ ClpX6 unfoldase: allosteric control of a protein machine. Cell. 2005;121:1017–1027. doi: 10.1016/j.cell.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell. 2005;121:1029–1041. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Hwang BJ, Park WJ, Chung CH, Goldberg AL. Escherichia coli contains a soluble ATP-dependent protease (Ti) distinct from protease La. Proc Natl Acad Sci U S A. 1987;84:5550–5554. doi: 10.1073/pnas.84.16.5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Joshi SA, Hersch GL, Baker TA, Sauer RT. Communication between ClpX and ClpP during substrate processing and degradation. Nat Struct Mol Biol. 2004;11:404–411. doi: 10.1038/nsmb752. [DOI] [PubMed] [Google Scholar]
- Katayama-Fujimura Y, Gottesman S, Maurizi MR. A multiple-component, ATP-dependent protease from Escherichia coli. J Biol Chem. 1987;262:4477–4485. [PubMed] [Google Scholar]
- Kedzierska S, Chesnokova LS, Witt SN, Zolkiewski M. Interactions within the ClpB/DnaK bi-chaperone system from Escherichia coli. Arch Biochem Biophys. 2005;444:61–65. doi: 10.1016/j.abb.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Kenniston JA, Baker TA, Fernandez JM, Sauer RT. Linkage between ATP consumption and mechanical unfolding during the protein processing reactions of an AAA+ degradation machine. Cell. 2003;114:511–520. doi: 10.1016/s0092-8674(03)00612-3. [DOI] [PubMed] [Google Scholar]
- Kessel M, Maurizi MR, Kim B, Kocsis E, Trus BL, Singh SK, Steven AC. Homology in structural organization between E. coli ClpAP protease and the eukaryotic 26 S proteasome. J Mol Biol. 1995;250:587–594. doi: 10.1006/jmbi.1995.0400. [DOI] [PubMed] [Google Scholar]
- Kim YI, Burton RE, Burton BM, Sauer RT, Baker TA. Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol Cell. 2000;5:639–648. doi: 10.1016/s1097-2765(00)80243-9. [DOI] [PubMed] [Google Scholar]
- Lee C, Schwartz MP, Prakash S, Iwakura M, Matouschek A. ATP-dependent proteases degrade their substrates by processively unraveling them from the degradation signal. Mol Cell. 2001;7:627–637. doi: 10.1016/s1097-2765(01)00209-x. [DOI] [PubMed] [Google Scholar]
- Lee S, Sowa ME, Watanabe YH, Sigler PB, Chiu W, Yoshida M, Tsai FT. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- Lupas AN, Martin J. AAA proteins. Curr Opin Struct Biol. 2002;12:746–753. doi: 10.1016/s0959-440x(02)00388-3. [DOI] [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Rebuilt AAA + motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- Maurizi MR, Xia D. Protein binding and disruption by Clp/Hsp100 chaperones. Structure. 2004;12:175–183. doi: 10.1016/j.str.2004.01.021. [DOI] [PubMed] [Google Scholar]
- Motohashi K, Watanabe Y, Yohda M, Yoshida M. Heat-inactivated proteins are rescued by the DnaK.J-GrpE set and ClpB chaperones. Proc Natl Acad Sci U S A. 1999;96:7184–7189. doi: 10.1073/pnas.96.13.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: common structure--diverse function. Genes Cells. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- Park E, Rho YM, Koh OJ, Ahn SW, Seong IS, Song JJ, Bang O, Seol JH, Wang J, Eom SH, Chung CH. Role of the GYVG pore motif of HslU ATPase in protein unfolding and translocation for degradation by HslV peptidase. J Biol Chem. 2005;280:22892–22898. doi: 10.1074/jbc.M500035200. [DOI] [PubMed] [Google Scholar]
- Reid BG, Fenton WA, Horwich AL, Weber-Ban EU. ClpA mediates directional translocation of substrate proteins into the ClpP protease. Proc Natl Acad Sci U S A. 2001;98:3768–3772. doi: 10.1073/pnas.071043698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohrwild M, Pfeifer G, Santarius U, Muller SA, Huang HC, Engel A, Baumeister W, Goldberg AL. The ATP-dependent HslVU protease from Escherichia coli is a four-ring structure resembling the proteasome. Nat Struct Biol. 1997;4:133–139. doi: 10.1038/nsb0297-133. [DOI] [PubMed] [Google Scholar]
- Sauer RT, Bolon DN, Burton BM, Burton RE, Flynn JM, Grant RA, Hersch GL, Joshi SA, Kenniston JA, Levchenko I, Neher SB, Oakes ES, Siddiqui SM, Wah DA, Baker TA. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlieker C, Weibezahn J, Patzelt H, Tessarz P, Strub C, Zeth K, Erbse A, Schneider-Mergener J, Chin JW, Schultz PG, Bukau B, Mogk A. Substrate recognition by the AAA+ chaperone ClpB. Nat Struct Mol Biol. 2004;11:607–615. doi: 10.1038/nsmb787. [DOI] [PubMed] [Google Scholar]
- Schlieker C, Zentgraf H, Dersch P, Mogk A. ClpV, a unique Hsp100/Clp member of pathogenic proteobacteria. Biol Chem. 2005;386:1115–1127. doi: 10.1515/BC.2005.128. [DOI] [PubMed] [Google Scholar]
- Singh SK, Grimaud R, Hoskins JR, Wickner S, Maurizi MR. Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proc Natl Acad Sci U S A. 2000;97:8898–8903. doi: 10.1073/pnas.97.16.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- Vale RD. AAA proteins. Lords of the ring. J Cell Biol. 2000;150:F13–F19. doi: 10.1083/jcb.150.1.f13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Song JJ, Seong IS, Franklin MC, Kamtekar S, Eom SH, Chung CH. Nucleotide-dependent conformational changes in a protease-associated ATPase HsIU. Structure (Camb ) 2001;9:1107–1116. doi: 10.1016/s0969-2126(01)00670-0. [DOI] [PubMed] [Google Scholar]
- Weber-Ban EU, Reid BG, Miranker AD, Horwich AL. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature. 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- Weibezahn J, Schlieker C, Bukau B, Mogk A. Characterization of a trap mutant of the AAA+ chaperone ClpB. J Biol Chem. 2003;278:32608–32617. doi: 10.1074/jbc.M303653200. [DOI] [PubMed] [Google Scholar]
- Weibezahn J, Tessarz P, Schlieker C, Zahn R, Maglica Z, Lee S, Zentgraf H, Weber-Ban EU, Dougan DA, Tsai FT, Mogk A, Bukau B. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Woo KM, Kim KI, Goldberg AL, Ha DB, Chung CH. The heat-shock protein ClpB in Escherichia coli is a protein-activated ATPase. J Biol Chem. 1992;267:20429–20434. [PubMed] [Google Scholar]
- Zietkiewicz S, Krzewska J, Liberek K. Successive and synergistic action of the Hsp70 and Hsp100 chaperones in protein disaggregation. J Biol Chem. 2004;279:44376–44383. doi: 10.1074/jbc.M402405200. [DOI] [PubMed] [Google Scholar]
- Zolkiewski M. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. A novel multi-chaperone system from Escherichia coli. J Biol Chem. 1999;274:28083–28086. doi: 10.1074/jbc.274.40.28083. [DOI] [PubMed] [Google Scholar]


