SUMMARY
During organogenesis, pluripotent precursor cells acquire a defined identity such as muscle or nerve. The transition from naïve precursor towards the differentiated state is characterized by sequential waves of gene expression that are determined by regulatory transcription factors. A key question is how transcriptional circuitry dictates the succession of events that accompanies developmental competence, cell fate specification and differentiation. To address this question, we have examined how anterior muscles are established within the C. elegans foregut (pharynx). We find that the T-box transcription factor tbx-2 is essential to form anterior pharyngeal muscles from the ABa blastomere. In the absence of tbx-2 function, ABa-derived cells initiate development normally: they receive glp-1/Notch signaling cues, activate the T-box gene TBX-38 and express the organ selector gene PHA-4/FoxA. However, these cells subsequently arrest development, extinguish PHA-4 and fail to activate PHA-4 target genes. tbx-2 mutant cells do not undergo apoptosis and there is no evidence for adoption of an alternative fate. TBX-2 is expressed in ABa descendants and depends on activation by pha-4 and repression by glp-1/Notch signaling. Our analysis suggests that a positive feedback loop between tbx-2 and pha-4 is required for ABa-derived precursors to commit to pharyngeal muscle fate.
Keywords: Tbx2, Tbx3, Tbx4, Tbx5, omb, FoxA, pharynx, pha-4, Notch, glp-1, tbx-38
INTRODUCTION
During the orderly progression of development, multipotent progenitors adopt specific cell fates. For example, in the vertebrate retina, a pool of retinal precursors produces seven distinct cell types in response to proneural transcription factors and intercellular signaling pathways (Ahmad et al., 2004; Kim et al., 2005). During C. elegans embryogenesis, a progenitor population generates the seven cell types of the pharynx (Albertson and Thomson, 1976). Many genes have been identified that are expressed in different pharyngeal cell types, but none is essential to specify a particular cell fate (Ao et al., 2004; Gaudet and Mango, 2002; Okkema and Fire, 1994; Thatcher et al., 2001). Thus, a critical question is how the pharyngeal precursors are patterned to produce the seven cell types of the mature pharynx. Here we explore how one cell type, anterior pharyngeal muscle, is established during embryogenesis.
Studies over the past ten years have lead to an outline of how the pharyngeal precursors are formed during embryogenesis. At the 4-cell stage, two blastomeres, ABa and EMS, are destined to generate all of the pharyngeal cells, as well as additional, non-pharyngeal cells (Sulston et al., 1983). ABa will give rise to many anterior pharyngeal cells, including anterior muscles. The appropriate development of ABa depends on intercellular signaling by the Notch receptor orthologue GLP-1 at the 12-15 cell stage (Hutter and Schnabel, 1994; Mango et al., 1994b; Moskowitz et al., 1994; Priess et al., 1987; Priess and Thomson, 1987). At the 24-cell stage, the ABa descendants express the T-box genes TBX-37 and TBX-38, and these proteins function in combination with glp-1 signaling to activate the FoxA factor PHA-4 at the 44-cell stage (Good et al., 2004). pha-4 is a selector gene that specifies pharyngeal identity for both ABa and EMS descendants. In the absence of pha-4 activity, cells destined to become part of the pharynx are transformed into ectoderm (Horner et al., 1998; Mango et al., 1994a). Conversely ectopic pha-4 is sufficient to drive non-pharyngeal cells towards a pharyngeal fate (Horner et al., 1998). These effects are global, affecting many different cell types within the pharynx. How, then, are particular pharyngeal cell fates established? The transcription factor ceh-22/Nkx2.5 is expressed in a subset of pharyngeal muscles and its loss results in feeding abnormalities (Okkema et al., 1997). Nevertheless, pharyngeal muscles are still present in ceh-22 mutants (Okkema and Fire, 1994), indicating that additional factors are required.
Here we identify the T-box factor tbx-2 and demonstrate that it is critical to establish pharyngeal muscles derived from ABa. The T-box family of transcription factors are defined by the T-box DNA binding domain, and can function as either activators or repressors (Wilson and Conlon, 2002). T-box genes are essential for developmental processes such as patterning the germ layers (Brachyury), vertebrate limb bud development (Tbx4, Tbx5), and cardiac development (Tbx2, Tbx20) (Harrelson et al., 2004; Singh et al., 2005). The most similar vertebrate homologues of C. elegans tbx-2 are H. Sapiens TBX2/3 (pBlast E value 4.4e-87) and TBX/4/5 (E value 6.2e-67). Tbx2/3/4/5 represent a subgroup of closely related T-box genes that were likely generated by two duplication events (Agulnik et al., 1996). TBX2/3 are involved anterior/posterior digit identity (Harrelson et al., 2004; Suzuki et al., 2004), and TBX4/5 are responsible for hindlimb/forelimb development. However, the function of these genes in these processes is still mysterious. For example, misexpression of Tbx4 and Tbx5 can alter limb identity in chick (Logan and Tabin, 1999; Rodriguez-Esteban et al., 1999; Takeuchi et al., 1999), but in mouse, loss of Tbx5 affects limb outgrowth not patterning (Agarwal et al., 2003; Minguillon et al., 2005; Rallis et al., 2003). Similarly, misexpression of Tbx2 in chick embryos leads to transformation of digit III to IV (Suzuki et al., 2004), whereas in mouse, loss of Tbx2 leads to digit IV duplications (Harrelson et al., 2004). Thus, the roles of Tbx2 and Tbx5 in cell fate specification remain murky.
There are at least 20 T-box transcription factors in the C. elegans genome but tbx-2 is the only clear member of the Tbx 2/3/4/5 subfamily. A handful of T-box proteins are necessary for worm development, including tbx-8 and tbx-9, tbx-30, tbx-37 and tbx-38, and mab-9 (Andachi, 2004; Baugh et al., 2005; Chisholm and Hodgkin, 1989; Good et al., 2004; Pocock et al., 2004; Woollard and Hodgkin, 2000). For example, tbx-8 and tbx-9 are important for epidermal, intestinal and body wall muscle development, while mab-9 affects the fates of two cells in the hindgut. Here, we find that tbx-2 is critical for ABa-derived pharyngeal precursors to differentiate into pharyngeal muscles. Based on the expression and phenotypes, we suggest that a positive feedback loop between pha-4 and tbx-2 is required for commitment to anterior muscle fate during pharyngeal development.
MATERIAL AND METHODS
Nematode strains
Caenorhabditis elegans was Bristol N2, maintained as described (Brenner, 1974). The following strains were used: ceh-22::GFP (Okkema et al., 1997), fog-2(q71) pha-4(q490)/ stu-3(q265) V (Mango et al., 1994a), tbx-37(zu467) tbx-38(zu460) /qT1 III; lin-2(e1309) X (Good et al., 2004), ced-10(n1993) IV (Ellis et al., 1991), and a pha-4::GFP transcriptional fusion (Alder et al., 2003). tbx-2(ok529) was provided by the Gene Knockout Consortium (http://www.celeganskoconsortium.omrf.org), outcrossed 8x and balanced with dpy-17(e164) unc-32(e189).
GFP reporter strains
tbx-2(ok529)/dpy-17(e164) unc-32(e189) containing ceh-22::GFP, ajm-1::GFP, pha-4::GFP or avr-15::GFP were constructed with integrated GFP reporters. itr-1::GFP was created by injecting 2 ng/μl itr-1::GFP (Dal Santo et al., 1999), 40 ng/μl pRF4 (rol-6, Mello et al., 1991), and 60 ng/μl herring sperm DNA into tbx-2(ok529)/ dpy-17(e164) unc-32(e189). tbx-2(ok529)/dpy-17(e164) unc-32(e189); myo-2::GFP:his2B transgenic worms were created by microinjection of 2 ng/μl 920bp myo-2 promoter::GFP::his2B DNA (PCR-amplified with 5′-GACATCTAGACAAGTAGAGACATTGAGGGA-3′ and 5′-GCATCCGCTTACAGACAA GCTGTGAC-3′ (Gaudet and Mango, 2002)), 40 ng/μl pRF4 and 60 ng/μl herring sperm DNA. avr-15::GFP (Dent et al., 1997) transgenic worms were created by microinjection of 89 ng/μl avr-15::GFP, 39 ng/μl pRF4, 62 ng/μl herring sperm DNA and 1.0 ng/μl 1 Kb DNA ladder (Gibco/BRL) into wild-type worms. avr-15::GFP was integrated by gamma irradiation (Mello and Fire, 1995).
The 5.2kb promoter tbx-2::GFP transgenic strain was produced by PCR amplification with forward primer 5′-GATTTATTCTCGTGCCTCGGTTCTCCTTTGAAG-3′ and reverse primer 5′-AGGCTTTGACAACAAATCGAGAACATCAAAACC-3′ (penultimate codon from Y54H5). The product was re-amplified with nested primers 5′-TTAAGAAGCATTTGAAAGCA-3′ and 5′-AGTCGACCTGCAGGCATGCAAGCTAGGCTTTGACAACAAATCGAG-3′, which placed GFP at the 3′ end of the tbx-2 coding region. Independently, the entire GFP and unc-54 terminator sequence of pPD95.75 (ftp://www.ciwemb.edu/pub/FireLabInfo/) was amplified using forward primer 5′-AGCTTGCATGCCTGCAGGTCGACT-3′ and reverse primer 5′-AAGGGCCCGTACGGCCGACTAGTAGG-3′. The tbx-2 and pPD95.75 PCR products were used to generate the 5.2, 4.6, 4.1, 2.4, 2.3, 1.2, and 1.0 kb tbx-2::GFP::unc-54 reporter DNAs (protocol described in Hobert, 2002) (primer sequences available upon request). Wild-type, tbx-2(ok529) / dpy-17(e164) unc-32(e189), and fog-2(q71) pha-4(q490)/ stu-3(q265) hermaphrodites were injected with 0.2 ng/μl tbx-2::GFP fragment, herring sperm DNA (60 ng/μl) and EcoRI-linearized pRF4 (rol-6; 40 ng/μl). A 1.0 kb transcriptional tbx-2::GFP reporter was constructed by cloning the PCR product from F21H11 with primers 5′-ACGCGTCGACTTGTTCTCTTTTGAATTTCACATTTCATTTTG-3′ and 5′-GCAAAAGCTTCATCGGAATGTAAAG-3′ into pPD95.77 using SalI and HindIII digestion. The resulting plasmid was used to amplify the 1kb promoter tbx-2::GFP insert using the same forward primer and reverse primer 5′-GCATCCGCTTACAGACAAGCTGTGAC-3. The microinjection mixture contained 1 ng/μl 1.0 kb promoter tbx-2::GFP PCR product, 40 ng/μl pRF4, and 50 ng/μl Herring sperm genomic DNA. The 11.4 kb tbx-2 genomic PCR product was amplified from the YAC Y54H5 using forward primer 5′-GATTTATTCTCGTGCCTCGGTTCTCCTTTGAAG-3 and reverse primer 5′-CTCTTTCCAGACAATTACAGTAATCCGGGCGAG - 3′, then injected as a mixture of 0.5 ng/μl tbx-2, 50 ng/μl pRF4, and 130 ng/μl herring sperm DNA.
RNA interference
DNA templates for F21H11.3 (tbx-2) and C48D1.2 (ced-3) were obtained in the pL4440 dsT7 RNAi plasmid from the ORFeome library (Reboul et al., 2003). C35D10.9 (ced-4), W02D3.9 (unc-37), and REF genes T01E8.2 (ref-1), C17C3.7 (hlh-25), C17C3.8 (hlh-26), F31A3.2 (hlh-28), and F31A3.4 (hlh-29) were obtained from the MRC library (Kamath et al., 2003). tbx-7 double-stranded RNA (dsRNA) was generated from a PCR product using ZK328 template (primer sequences available upon request) and cloned into pDONRdT7 (protocol describe in Reddien et al., 2005).
dsRNA was transcribed using the AmpliScribe T7 polymerase kit (Epicenter) with linear PCR products amplified using T7 promoters added to each end (tbx-2, ced-3, ced-4) or the T7 terminator primer 5′-TCAAGACCCGTTTAGAGGC-3′ (tbx-7) as DNA templates. dsRNA was purified with RNEasy Mini spin columns (Qiagen), and annealed in 50mM NaCl, 10mM Tris pH 7.5. =10 young hermaphrodites were microinjected with 0.5 - 1.0 mg/ml dsRNA (Fire et al., 1998). After 12-24 hours recovery at 24°C, each injected worm was moved to a separate plate for 12 hours, removed, and progeny scored 12 hours later. Feeding RNAi was performed using HT115 bacteria transformed with the ced-3, ced-4, or tbx-7-containing plasmids using standard procedures (Kamath et al., 2003).
Heat Shock
HS::tbx-2 was PCR amplified from ORFeome clone 10052@F12 using primers 5′-AAGCTAGCGGATCCCCATGGCATTCAATCCATTTGC-3′ and 5′-TTGGTACCGTCGACTTAAGGCTTTGACAACAAATCG-3′ and cloned into Topo TA pCR2.1 (Invitrogen). The resulting plasmid was digested with NheI and KpnI and tbx-2 moved into pPD49.78 and pPD49.83 (Firelab vectors ftp://www.ciwemb.edu/pub/FireLabInfo, Fire et al., 1990). 0.02 ng/μl HS::tbx-2 (pPD49.78), 0.02ng/μl HS::tbx-2 (pPD49.83), 0.05 ng/μl myo-2::GFP::His2B, 40 ng/μl pRF4, and 60 ng/μl herring sperm DNA were injected into wild-type worms to obtain stable lines.
Transgenic HS::tbx-2, myo-2::GFP::His2B and control myo-2::GFP::His2B embryos were isolated and treated in 0.2ml PCR tubes for 30 minutes at 33°C, then recovered at 20°C. Embryos were examined for reporter gene activity 11.5 hours post heat-shock. No heat shock controls were incubated at 20°C for 12 hours.
Staining and Image analysis
Staining with 3NB12, anti-intermediate filament, or 9.2.1 antibodies, was performed using standard procedures except slides were incubated in methanol followed by acetone, for 3 minutes each (modification of Albertson, 1984; modification of Mango et al., 1994a). Larvae were incubated with TNB [100 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5% blocking reagent] containing 10% normal goat serum (NGS) for 30 minutes. Slides were incubated 12-18 hours at 15°C using primary antibodies diluted in TNB/NGS: 3NB12 (1:5) (Okamoto and Thomson, 1985; Priess and Thomson, 1987), αIF (1:3) (Priess and Thomson, 1987; Pruss et al., 1981), 9.2.1 (1:5000) (Miller et al., 1983), MH27 (1:100) (Francis and Waterston, 1991), αPHA-4 N-terminal (1:2000), αTBX-38 (1:2) (Good et al., 2004), αLIN-26 (1:1000) (Labouesse et al., 1996), and αGFP mAB3580 (1:250) (Chemicon). Slides were washed three times in TBS + 0.5% Tween20, before addition of donkey anti-mouse or anti-rabbit, Cy-2 or Cy-3-conjugated antibodies (1:200 dilution, Jackson ImmunoReseach) for two hours. Slides were washed 3x in TBS + Tween20 and mounted in 50.0% glycerol, 0.006 M Na Citrate, 0.05 M NaH2PO4, 0.5 mg/ml 4′,6-diamidino-2-phenylindole (DAPI), 25mg/ml 1,4-diazobicyclo-[2.2.2]-octane (DABCO) and a pinch of p-phenylenediamine.
Confocal microscope images were captured on an Olympus FluoView confocal microscope with FluoView 2.1 software. Z-series were computed by ImageJ software using maximum intensity. PHA-4::GFP::Histone nuclei were counted using live embryos and a Zeiss axioskop, fitted with a Ludl focus drive (0.4 μm step setting), Roper Scientific Micromax digital camera, Zeiss Attoarc fluorescent burner (5% power setting for time-lapse), and Improvision Openlab 3.1.5 software.
4D movies were acquired using a Applied Precision Deltavision RT with 0.2 – 0.4 μm step sections taken every 15 minutes. Images were deconvolved using Applied Precision RTView suite software with default settings.
RESULTS
Discovery of the tbx-2 locus
Our interest in tbx-2/(F21H11.3) derived from a global RNAi screen of genes with expression enriched within pharyngeal cells (P.A.S., C. Armstrong, M. Vidal and S.E.M., unpublished; (Ao et al., 2004; Gaudet and Mango, 2002). RNAi of tbx-2 led to defects in the anterior pharynx, an Uncoordinated phenotype and a first larval stage arrest (described below). To confirm the phenotype, we obtained the deletion allele ok529 from the Gene Knockout Consortium (http://celeganskoconsortium.omrf.org/). The tbx-2(ok529) deletion eliminated bases 5,132,217 to 5,133,298 of chromosome III (1082bp), resulting in the in-frame removal of exon four (Fig. 1A, B). Exon 4 contains part of the DNA binding domain (Miyahara et al., 2004), which in the homologous T-box factor Brachyury directly contacts DNA (Muller and Herrmann, 1997).
Fig. 1. The tbx-2 locus and its predicted product.
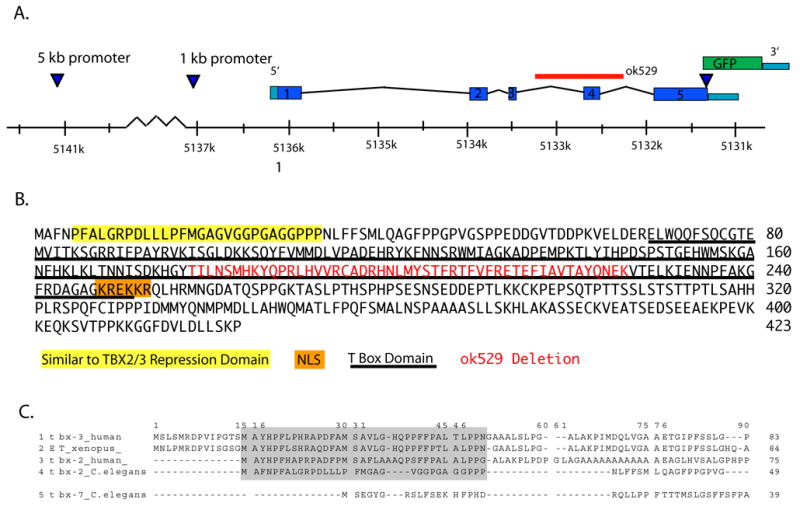
(A) tbx-2 consists of five exons based on EST alignments, Genefinder, and ORFeome 3.1 (Lamesch et al., 2004). We constructed a GFP reporter with 5.2 kb of sequence upstream of the ATG and the open reading frame fused to GFP at the predicted carboxyl terminus. This construct uses the unc-54 3′UTR and polyadenylation site (Fire et al., 1990). The ok529 deletion spans exon four (red). (B) TBX-2 has sequences similar to the amino-terminal repressor domain of vertebrate Tbx2 and Tbx3 (yellow) (He et al., 1999), the conserved T-box (underlined, Wilson and Conlon, 2002), and nuclear localization signal (orange; (Carlson et al., 2001; Miyahara et al., 2004). The ok529 mutation results in an in-frame deletion of exon 4 (red), compromising the conserved T-box DNA binding domain. (C) Alignment of the amino terminus of C. elegans TBX-2 with sequences from the amino terminal repression domain of vertebrate Tbx 2/3 (He et al., 1999).
Hypothetically, tbx-2(ok529) could give rise to a protein with either partial activity or an interfering dominant-negative function. To determine if either of these possibilities was likely to be true, we examined the phenotype of tbx-2(ok529) mutants in which tbx-2 was further inactivated by RNAi. Heterozygous tbx-2(ok529)/+ mothers were injected with dsRNA and their progeny scored for the terminal phenotype. 100% of progeny from either wild-type or tbx-2(ok529)/+ mothers arrested as embryos or larvae, indicating that RNAi was effective. For tbx-2/+ mothers, 27% of their progeny arrested with a pharynx phenotype identical to tbx-2(ok529) homozygotes (n=44, Table 1, Fig 2D), suggesting that reduction of TBX-2(ok529) protein by RNAi did not lead to a more severe phenotype for tbx-2(ok529) homozygotes. The remaining 73% of progeny arrested with a slightly weaker phenotype that resembled tbx-2(RNAi) of wild-type worms (Fig 2B). These animals were likely of the genotype tbx-2/+ and +/+. We conclude tbx-2(ok529) is a strong loss-of-function, possibly null, allele. This is in agreement with data from Roy Chowdhuri et al. examining this allele and a second, null allele (S. Roy Chowdhuri and P. Okkema, pers. comm.).
Table 1.
Lack of interactions for tbx-2 with tbx-7 or cell death genes
| Strain | WT progeny | L1 arrest | Embryonic | Total arrest | n |
|---|---|---|---|---|---|
| (ok529)/+ | 75.0% (422) | 19.9% (112) | 5.2% (29) | 25.0% (131) | 563 |
| N2 + tbx-2 (RNAi) | 77.7% (14) | 22.2% (4) | 0% (0) | 22.2% (4) | 18 |
| (ok529)/+ tbx-2(RNAi) | 0 % (0) | n.d | n.d | 100% (44) | 44 |
| (ok529)+, tbx-7(RNAi) | 78.8% (256) | 18.8% (61) | 2.5% (8) | 21.2% (69) | 325 |
| (ok529)/+, ced-3(RNAi) | 74.4% (734) | n.d. | n.d. | 25.5% (252) | 986 |
| (ok529)/+, ced-4(RNAi) | 73.6% (234) | n.d. | n.d. | 26.4% (84) | 318 |
Progeny counts of heterozygous tbx-2(ok529)hermaphrodites vs. wild type(N2) tbx-2(RNAi), (ok529) tbx-2(RNAi), (ok529) tbx-7(RNAi), (ok529) ced-3(RNAi) and (ok529) ced-4(RNAi).
Fig. 2. Loss of anterior muscles in tbx-2 mutants.

(A). Diagram of a pharynx showing the eight groups of pharyngeal muscles (pm; adapted from Albertson and Thomson, 1976). Cells affected by tbx-2 mutations are shaded. (B,C,D) Differential interference contrast micrographs of first stage (L1) larvae. (B) The pharynx of a tbx-2(RNAi) L1 with a wild-type posterior bulb (asterisk), but abnormal anterior bulb (arrow). (C, E, G, I, K, M, O) wild-type larvae, (D, F, H, J, L, N, P) tbx-2(ok529) larvae. (C) Wild-type larva with anterior and posterior bulbs (arrow, asterisk). (D) tbx-2(ok529) L1 with a relatively normal posterior bulb (asterisk), but morphologically aberrant procorpus, anterior bulb, and isthmus (arrows). Muscle marker 3NB12 is lost from anterior cells in tbx-2 mutants compared to wild type (E,F; (Okamoto and Thomson, 1985; Priess and Thomson, 1987). Pharyngeal myosin is absent from anterior muscles in tbx-2(ok529) larvae (Miller et al., 1983). CEH-22::GFP (Okkema et al., 1997) is expressed in fewer pm3, 4, and 5 cells (arrows) while pm7 expression appears WT (asterisk) in tbx-2(ok529) (I, J). ITR-1::GFP, (Baylis et al., 1999) is lost from pm4 and pm5 (arrows) while expression is still detected in pm6 (asterisk) and the intestine of tbx-2(ok529) worms (K, L). AVR-15::GFP expression in wild type (M) and tbx-2(ok529) (N) larvae (Dent et al., 1997) reveals only a subset of pm4 and pm5 cells express in mutant larvae. Wild-type (O) and tbx-2(ok529) (P) larvae stained with an antibody that recognizes intermediate filaments to reveal pharyngeal marginal cells (Pruss et al., 1981). All three groups of marginal cells are present and morphologically wild-type (arrows). Wild-type L1 pharynx is ~45 μm long.
Many C. elegans T-box factors function in pairs, including tbx-37 and tbx-38 (Good et al., 2004), and tbx-8 and tbx-9 (Andachi, 2004; Pocock et al., 2004). tbx-7 has the greatest sequence similarity to tbx-2 (62% similar, 2e−55), and is located 0.63 map units from tbx-2, suggesting the two share a common ancestor gene. We investigated if loss of tbx-7 (ZK328.8) would enhance the pharynx phenotype associated with tbx-2. Neither the tbx-7 allele tm478 nor tbx-7(RNAi) resulted in a phenotype when examined alone. Nor did we observe a stronger phenotype associated with tbx-2(ok529) tbx-7(RNAi) double inactivation compared to tbx-2(ok529) single mutations, as monitored under the light microscope (n=61, Table 1). These data, as well as the lack of a tbx-7 homolog in C. briggsae, suggest tbx-7 does not contribute significantly to pharynx development, and therefore we focus on tbx-2 alone.
tbx-2 is required for anterior pharynx muscle development
The most striking phenotype associated with tbx-2 mutations was a loss of anterior pharyngeal muscles. Wild-type C. elegans posses eight rings of pharyngeal muscles, (Fig. 2A, Table 2) (Albertson and Thomson, 1976). tbx-2 mutants lacked expression of muscle markers 3NB12 (Okamoto and Thomson, 1985; Priess and Thomson, 1987) and 9.2.1 (Miller et al., 1983) in 2/3 pm3 cells, 1/3 pm4 cells, and 1/3 pm5 cells (Table 2, Fig. 2E-H). By contrast, posterior muscles pm6, pm7 and pm8 looked wildtype, indicating that these cells were not affected. CEH-22::GFP expression showed a similar pattern, with one pm3, two pm4, and one pm5 cell expressing the reporter (Fig 2I,J). These numbers suggested that muscles derived from ABa were altered or missing, whereas those from EMS were not (compare Table 2 and Table 3). Three additional markers supported this idea. First, we counted pharyngeal muscle nuclei with the nuclear muscle marker myo-2::GFP (Miller et al., 1986). Wild-type embryos had an average of 35.5 GFP+ nuclei (n=9 embryos), close to the published number of pharyngeal nuclei (37; (Albertson and Thomson, 1976)). By contrast, tbx-2 mutants had an average of 19 GFP+ nuclei, all of which were located in the posterior of the head (n=7). This number agreed well with the number of pharyngeal muscles produced by EMS in wild-type embryos (18, Albertson and Thomson, 1976)), consistent with an absence of ABa-derived muscles . Second, we examined ITR-1::GFP, which is normally expressed in nine muscle cells of the central pharynx (pm4, pm5, pm6, and weakly in pm3; (Dal Santo et al., 1999)). In tbx-2 mutants, ITR-1::GFP was detected in three cells with pm6-like characteristics, based on location and morphology, but rarely in any other pharynx muscle cell (Fig. 2K,L; n=7 mutant vs. n=9 wild-type larvae). Mutant animals also expressed ITR-1::GFP in other cell types such as midgut and pharyngeal valves, confirming the presence of the extrachromosomal array. Third, we saw a reduction in the anterior expression of AVR-15::GFP, which is normally expressed in pm4/pm5 and extra-pharyngeal neurons (Fig. 2M-N; Dent et al., 1997). We conclude that tbx-2 activity is required for anterior muscle development, specifically those muscles generated by the ABa blastomere.
Table 2.
Pharynx muscle nuclei lineage and cell fusion
| Pharynx Muscle Nuclei | Embryonic Cell Lineage | Mature Pharynx Cell |
|---|---|---|
| pm1DL | AB.araapaaap | pm1 |
| pm1DR | AB.araappaap | |
| pm1L | AB.araaaaaap | |
| pm1R | AB.araaaaapp | |
| pm1VL | AB.alpaaaapa | |
| pm1VR | AB.araaaaapa | |
| pm2DL | AB.araapaapa | pm2 |
| pm2DR | AB.araappapa | |
| pm2L | AB.alpaaapaa | pm2 |
| pm2R | AB.arapaapaa | |
| pm2VL | AB.alpaaaaap | pm2 |
| pm2VR | AB.arapaaaap | |
| pm3DL | MS.aaapaaa | pm3 |
| pm3DR | MS.paaaapa | |
| pm3L | AB.alpaapapp | pm3 |
| pm3R | AB.arapaappa | |
| pm3VL | AB.alpappppp | pm3 |
| pm3VR | AB.arapapppp | |
| pm4DL | MS.aaaaapp | pm4 |
| pm4DR | MS.paaaapp | |
| pm4L | MS.aaapaap | pm4 |
| pm4R | AB.araaapapp | |
| pm4VL | MS.aapaaaa | pm4 |
| pm4VR | MS.papappp | |
| pm5DL | MS.aaaapap | pm5 |
| pm5DR | MS.paaappa | |
| pm5L | AB.araapapap | pm5 |
| pm5R | AB.araapppap | |
| pm5VL | MS.aapaaap | pm5 |
| pm5VR | MS.papaaap | |
| pm6D | MS.paaappp | pm6 |
| pm6VL | MS.aapappa | pm6 |
| pm6VR | MS.papappa | pm6 |
| pm7D | MS.aaaappp | pm7 |
| pm7VL | MS.aapaapp | pm7 |
| pm7VR | MS.papaapp | pm7 |
| pm8 | MS.aaapapp | pm8 |
ABa lineage pharynx muscle cells are shown in bold type.
Table 3.
tbx-2 is required to produce pharyngeal muscles
| Antibody | Strain | Cells expressing epitope at L1 stage | n | |||
|---|---|---|---|---|---|---|
| pm3 | pm4 | pm5 | pm7 | |||
| 9.2.1 | WT | 3 | 3 | 3 | 3 | 4 |
| tbx-2(ok529) | 1 | 2 | 2 | 3 | 6 | |
| 3NB12 | WT | 3 | 3 | 3 | 3 | 4 |
| tbx-2(ok529) | 1 | 2 | 2 | 3 | 6 | |
9.2.1 recognizes pharyngeal myosin (Miller et al., 1983); 3NB12 recognizes pharyngeal muscle (Okamoto and Thomson, 1985; Priess and Thomson, 1987). Cell designations based on position and morphology.
We examined whether other ABa-derived pharyngeal cells were affected by tbx-2 mutations. ABa normally produces 7/9 pharyngeal marginal cells (Albertson and Thomson, 1976; Sulston et al., 1983), which express pax-1::GFP early in development and intermediate filaments late (Bartnik et al., 1986; Portereiko et al., 2004). In tbx-2 mutants, PAX-1::GFP was expressed in eight nuclei, like the wild type (n=12), and intermediate filament staining appeared normal (Fig. 2O, P). Thus, marginal cells are made in tbx-2 mutant embryos, and tbx-2 is not required for all ABa-derived pharyngeal cells.
The fate of anterior pharyngeal muscles in mutants
When does tbx-2 affect anterior muscle development? To address this question, we examined early markers of anterior pharyngeal muscle development. We also tested for possible alternative fates in mutant embryos. This analysis suggested that pharyngeal cells initiate muscle development but then arrest or de-differentiate in the absence of tbx-2 activity.
We surveyed markers of anterior pharyngeal muscles to determine the earliest defects associated with tbx-2 mutations. First we examined TBX-38, which is normally activated at the 24-cell stage in ABa descendants (Good et al., 2004). All embryos from tbx-2/+ mothers expressed TBX-38 between the 16-cell stage and the 100-cell stage (n=17, Fig. 3A, B). Second, we examined PHA-4 using an integrated pha-4::GFP reporter (Alder et al., 2003). This reporter normally expresses GFP beginning at the 28-cell stage and continuing throughout the life of the worm (Alder et al., 2003; Horner et al., 1998). Activation of PHA-4::GFP was normal in EMS descendants at the 28-cell stage and in ABa descendents at the 44-cell stage in mutant embryos (Table 4). This result suggested that glp-1-mediated signaling and tbx-37 and/or tbx-38 were functional in tbx-2 mutants, since both signaling and TBX activity are required for PHA-4 activation in ABa descendants (Good et al., 2004; Kalb et al., 1998; Mango et al., 1994a). We conclude that the earliest stages of ABa development are normal in tbx-2 mutants.
Fig. 3. tbx-2 is required for progression of pharyngeal muscle development.

(A,B) TBX-38 (red) is expressed normally in all embryos from tbx-2(ok529)/+ mothers (DAPI, blue). (C) Wild-type comma-stage embryo with 94 PHA-4::GFP+ cells in the pharynx resembles (D) a tbx-2(ok529) comma-stage embryo with 93 PHA-4::GFP+ cells. (E) Embryo C viewed 30 minutes later with 94 PHA-4::GFP+ cells while embryo D had only 81 PHA-4::GFP+ cells (F). (G) WT and (H) tbx-2(ok529) embryos had approximately the same number of ceh-22::GFP expressing cells at the bean stage (arrows), when CEH-22::GFP is first detectable. Following the same embryos in panels G and H until the two-fold stage, wild-type embryos consistently have more ceh-22::GFP expressing cells (I, arrows) compared to tbx-2(ok529) embryos (J, arrows). (K) tbx-2::GFP WT 8E embryo and (L, arrows) tbx-2::GFP pha-4(RNAi) embryo with TBX-2::GFP expressing cells (M) Differential interference contrast micrograph (DIC) of a pha-4(q490) larva lacking a pharynx and (N) the same larva showing no TBX-2::GFP in the head other than in two non-pharyngeal cells (arrows). Each embryo is ~50 μm long.
Table 4.
tbx-2 is required to maintain PHA-4 expression.
| Strain | Stage | ||||
|---|---|---|---|---|---|
| 28 cell | 44 cell | Comma | 1.5-fold - two-fold | L1 | |
| tbx-2(ok529) | n.d. | 14 (n = 5) | 82 (n = 4) | 78 (n = 8) | 67 (n=3) |
| Progeny of (ok529)/+ | 4 (n= 12) | n.d. | n.d. | n.d. | n.d. |
| WT | 4 (n = 3) | 14 (N=6) | 95 (n = 2) | 94 (n = 9) | 93 (n = 1) |
Numbers of PHA-4::GFP:his2B+ cells in tbx-2(ok529) homozygotes. n.d. not determined.
Next we analyzed markers of later development and differentiation. There was an average of 13 fewer PHA-4::GFP+ cells at the comma stage of embryogenesis compared to the wild type, 16 fewer at the 1.5 - 2 fold stage and 18 fewer by the L1 stage (Fig. 3C-F, Table 4). Staining for endogenous PHA-4 confirmed this dramatic decrease in mutant animals (data not shown). These data were corroborated by examining the PHA-4 target gene ceh-22, which is normally activated in pm3 - pm5 and pm7 at the pre-bean stage (Okkema and Fire, 1994). We observed 4-6 CEH-22::GFP+ cells at the pre-bean stage in both mutant and wild-type embryos (Table 5). Subsequently, the number of CEH-22::GFP+ cells increased to 18 in wild-type embryos, but only to 13 cells in mutant animals (Fig. 3G-J, Fig. 4, Table 5). The expressing cells appeared to be part of pm3, pm4, pm5 and all of pm7, based on position and morphology. Normally, ABa generates 19 embryonic pharyngeal muscle cells, of which seven belong to pm3-5; the remaining twelve being the pm1 and pm2 cells (Table 2) (Sulston et al., 1983). Thus, the number and location of lost CEH-22::GFP+ cells support the notion that tbx-2 affects ABa-derived muscles (Fig. 2 I,J). We suggest that progression of muscle development is aborted in tbx-2 mutants since they cannot maintain PHA-4 expression and fail to activate CEH-22.
Table 5.
tbx-2 is required for activation of ceh-22::GFP expression
| Phenotype | Stage | n | ||
|---|---|---|---|---|
| Bean | 1.5-fold | Two-fold | ||
| WT | 5.5 | 16.0 | 17.8 | 12 |
| tbx-2(ok529) | 5.7 | 12.8 | 13.0 | 7 |
Number of ceh-22::GFP+ cells in WT and tbx-2 embryos over time by 4D microscopy.
Fig. 4. tbx-2 is required to activate ceh-22::GFP.
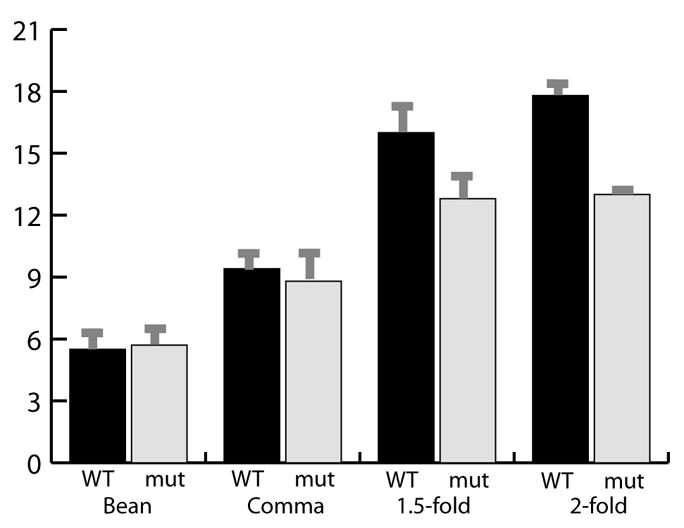
Bar graph showing the number of CEH-22::GFP+ cells in WT and mutant embryos at the bean stage, comma stage, 1.5-fold stage, and 2-fold stage. WT n = 12, mutant n = 7. error bars are s.d.
We considered three developmental options for anterior muscles in tbx-2 mutants. First, they could undergo apoptosis; second, they could adopt an alternative cell fate; third they could remain as undifferentiated or de-differentiated pharyngeal precursors. To explore apoptosis, we inactivated the cell death pathway using ced-3(RNAi) or ced-4(RNAi) (Joshi and Eisenmann, 2004; Yuan and Horvitz, 1990). We looked at the number of arrested tbx-2(ok529) larvae after ced-3 or ced-4 were inactivated by RNAi and found it identical to control (Table 1). Moreover, ten mutant larvae fed ced-3 dsRNA were phenotypically indistinguishable from non-RNAi controls (data not shown), nor did we observe extra cell corpses in tbx-2 homozygous embryos. The effectiveness of RNAi for ced-3 or ced-4 was examined in the cell corpse engulfment mutant ced-10(n1993) (Kinchen et al., 2005). We found ced-3(RNAi) or ced-4(RNAi) essentially eliminated cell corpses in terminal ced-10(n1993) embryos, demonstrating apoptosis had been reduced by RNAi (data not shown). From these data, we conclude that tbx-2 mutant cells do not undergo extra apoptosis.
Next we tested whether the pharyngeal muscles adopted an alternative fate in the absence of tbx-2. In embryos bearing null alleles of pha-4, pharyngeal cells are converted into ectoderm, which expresses LIN-26 (Horner et al., 1998; Labouesse et al., 1996). LIN-26 expression was examined in 26 tbx-2(ok529) homozygotes, confirmed by the morphology of the anterior pharynx; none activated ectopic LIN-26 or AJM-1 in the anterior pharyngeal region (Fig. 5A-D). We conclude that anterior muscles do not undergo a cell-fate transformation to ectoderm in tbx-2 mutants. We also examined the pharynges of tbx-2 embryos and larvae for markers of intestinal cells, pharyngeal glands (J126), body-wall muscles (MYO-3) and neurons (UNC-86). Wild-type staining was observed for each of these (data not shown). These data suggest that ABa-derived muscle precursors do not adopt a new fate or undergo apoptosis in tbx-2 mutants. We favor the notion that these cells arrest their development or de-differentiate.
Fig. 5. Loss of tbx-2 does not result in pharyngeal muscle transfating to ectoderm.
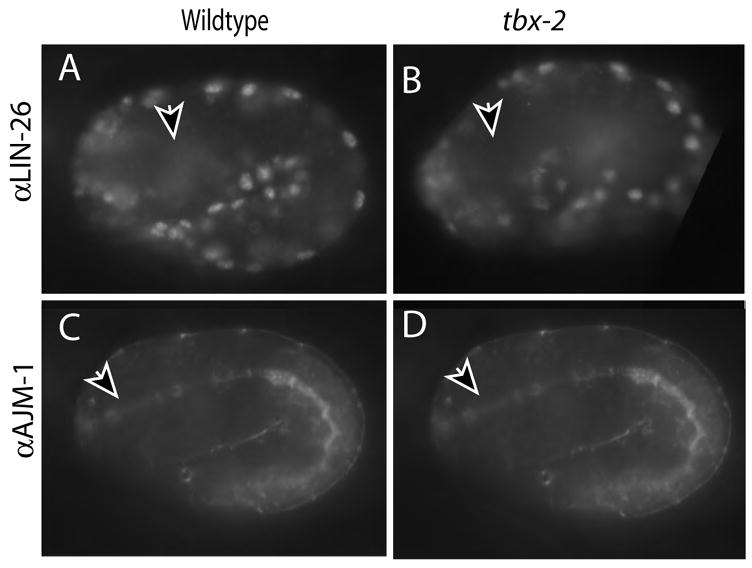
LIN-26 is not expressed in the pharynx of wild-type (A) or mutant (B) embryos, as detected by αLIN-26 antibody, but the epidermis stains (edges). WT and mutant embryos were discerned by pharyngeal adherens junction morphology with MH27 (C, D) Each embryo is ~50 μm long.
TBX-2::GFP is expressed in pharyngeal muscles
Given the effects on anterior muscle development, we wished to determine when and where TBX-2 was expressed. We relied on a tbx-2::GFP translational fusion that included 5.2kb upstream sequences and positioned GFP at the carboxyl terminus (Fig. 1A). We were unable to obtain TBX-2 antibodies, either from published sources or our own production, however our tbx-2::GFP reporter result agrees well with data from a global transcription profile that detected tbx-2 transcripts starting at the 4E stage (~102 cell stage, 143 minutes after the four cell stage; (Baugh et al., 2003)), and with mid-gastrula expression observed by in situ hybridization (Shin-i and Kohara, http://nematode.lab.nig.ac.jp/).
The Uncoordinated phenotype was rescued for all tbx-2(ok529) worms in transgenic strains, and the pharynx developmental phenotype was sufficiently rescued to allow embryos to hatch and initiate larval development. However, mutant larvae carrying either tbx-2 or tbx-2::GFP failed to reach adulthood (e.g. 4/72 F1 tbx-2::GFP progeny examined). The pharynx appeared fully formed in arrested tbx-2(ok529); tbx-2::GFP larvae, and TBX-2::GFP expression was present in the both MS- and ABa-derived pharynx cells. These data indicate that the tbx-2::GFP reporter contains sequences appropriate for expression within ABa-derived pharyngeal cells, but may lack sequences required for larval growth.
TBX-2::GFP expression initiated at the 8E stage (staging by the number of endodermal, or E, cells), in 11–12 anteriorly localized pharyngeal cells (Fig. 6C, D). Based on position, these cells are likely to be the ABa descendants that will give rise to pharyngeal muscle cells (i.e. ABalpaaa a/p, ABalpapp a/p, ABaraaaaa, ABaraapa a/p, ABarapaa a/p, ARarapapp and ABaraapp a/p; (Sulston et al., 1983). By the 1.5 fold stage, TBX-2::GFP was expressed in pm3, pm5 and variably pm4, with expression persisting throughout the larval and adult stages (Fig. 6E). Surprisingly, expression extended beyond the ABa lineage after the 1.5 fold stage. For example, only 2/6 pm5 nuclei derive from ABa but we often observed all pm5 cells expressing TBX-2::GFP in larvae (Fig. 5E). By the three-fold stage we also observed expression in pharyngeal neurons, in agreement with previous reports (Miyahara et al., 2004), occasionally in the posterior muscle pm8 and in cells outside of the pharynx such as body wall muscles (data not shown). Thus, TBX-2::GFP expression initiated within the ABa lineage but ultimately appeared in both ABa and MS-derived muscle cells.
Fig. 6. Expression of TBX-2::GFP initiates after PHA-4.
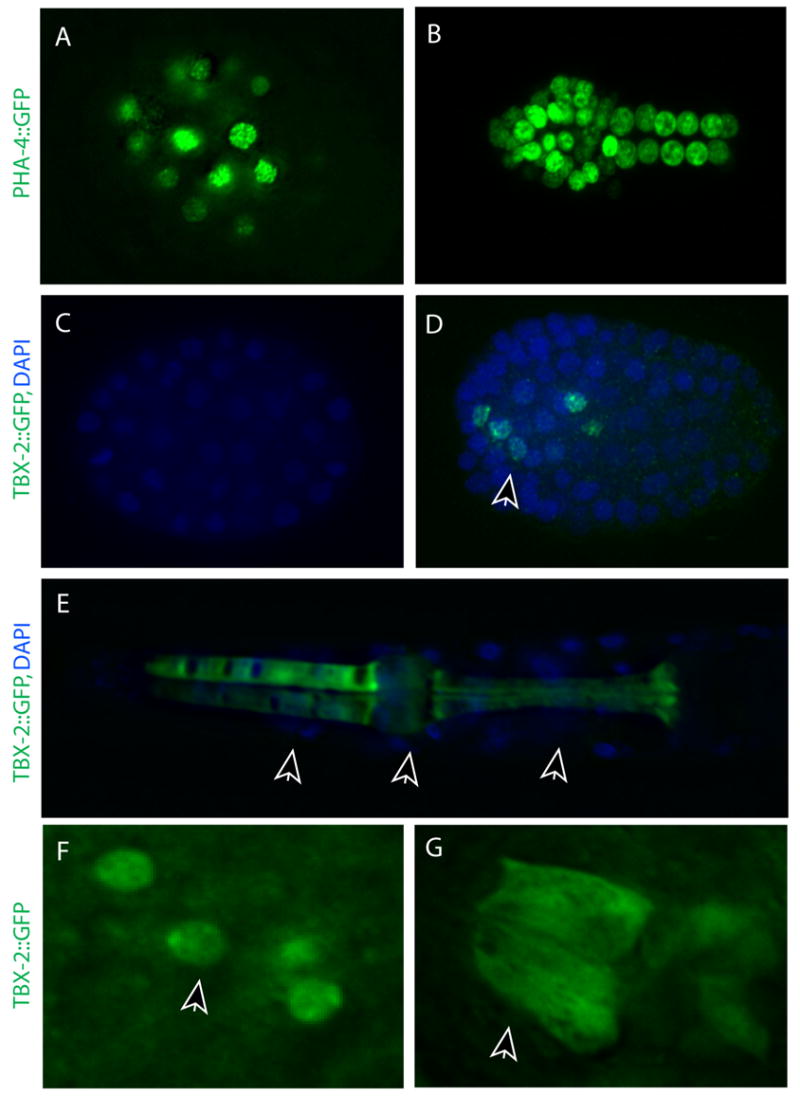
PHA-4::GFP (A, green) but not TBX-2::GFP (C, green) in both ABa and MS cell descendants at the 4E stage (nuclei, blue). (B) PHA-4::GFP in cells of the pharynx and intestine at the 8-10 E stage. (D) TBX-2::GFP is first detected in 11–12 posteriorly-located nuclei at the 8-10 E stage. (E) TBX-2::GFP in an L1 pharynx is present in pm3, pm4, and pm5 muscle groups (arrows). (F) At the 8-10E stage, TBX-2 is nuclear. (G) At the 1.5-fold stage, TBX-2::GFP is both cytoplasmic and nuclear in pm3 and pm5. Note the filamentous appearance in G. Each embryo is ~50 μm long.
Strikingly, while TBX-2::GFP was initially localized to nuclei, it was detected in the cytoplasm as well as the nucleus in pm4 and pm5 cells by the 1.5 fold stage (Fig. 6F, G). Cytoplasmic localization of T-box proteins, including TBX-2 homologues, has been noted previously in humans (Collavoli et al., 2003), zebrafish (Bruce et al., 2003), sponge (Adell and Muller, 2005), and C. elegans (Miyahara et al., 2004). Cytoplasmic expression was not homogeneous. Rather, TBX-2::GFP appeared filamentous, as though it was associated with the cytoskeleton (Fig. 6G). Cytoplasmic expression was observed even in lines expressing very low levels of TBX-2::GFP, suggesting that cytoplasmic TBX-2::GFP did not reflect over-expression from transgenes. The same pattern was observed in tbx-2(ok529); tbx-2::GFP embryos (data not shown). This localization pattern suggests that the timing of tbx-2 transcriptional function likely initiates before the 1.5 fold stage, when TBX-2 appears completely nuclear.
Regulation of tbx-2 expression
How is tbx-2 expression regulated? We generated translational GFP reporters containing 1.0, 1.2, 2.3, 2.4, 3.2, 4.1, 4.7, and 5.2 kb of putative promoter DNA to identify the minimal upstream sequences necessary for regulation (Fig. 7A). Only the 5.2 kb promoter construct conferred expression at the 8E stage, indicating that sequences between 4.7 and 5.2 kb were necessary to initiate expression appropriately (Fig. 7 B-E). All TBX-2::GFP reporter arrays between 4.7 and 2.3 kb were similar, with expression initiating at the comma stage, but weakly and in fewer cells than the 5.2 kb reporter (Fig. 7C, G, K). For the shortest promoters of 1.0 kb and 1.2 kb, we observed TBX-2::GFP only in presumed pharyngeal neurons (Fig. 7D, H, L).
Fig. 7. Complexity of tbx-2::GFP regulation.
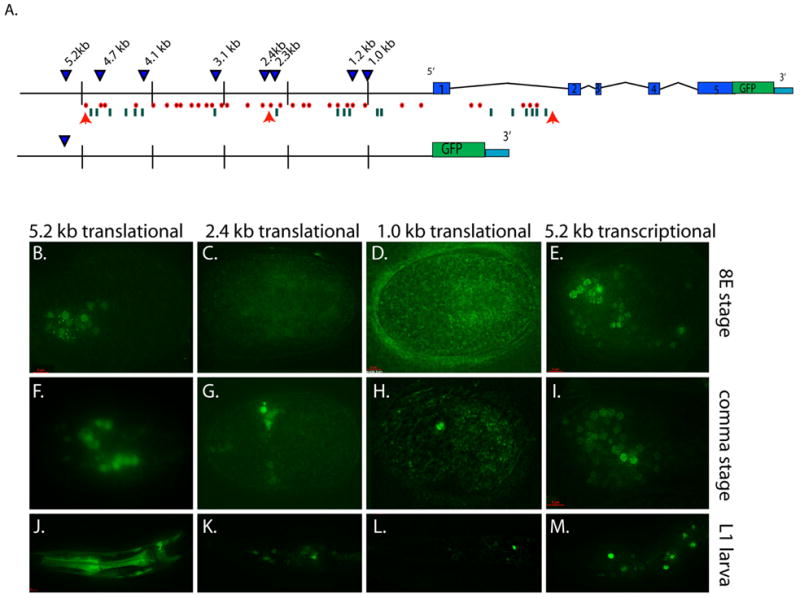
(A) Eight tbx-2::GFP translational reporter transgenes (top) and one transcriptional reporter (bottom) in wild-type worms (blue triangles). Orange arrows depict LAG-1 consensus binding sequences, red dots show potential bHLH (REF-1) binding sites, and green lines indicate PHA-4 consensus binding sequences. (B-E) TBX-2::GFP at the 8E stage for 5.2 kb (B), 2.4 kb (C), 1.0 kb (D) translational and 5.2 kb tbx-2::GFP transcriptional (E) reporters. (F-G) GFP at the comma stage for 5.2 kb (F), 2.4 kb (G), 1.0 kb (H) translational and 5.2 kb (I) transcriptional reporters. (J-M) TBX-2::GFP in an L1 larva for 5.2 kb (J), 2.4 kb (K), 1.0 kb (L) translational and 5.2 kb (M) transcriptional reporters. Embryos are ~50 μm long; an L1 pharynx is ~45 μm long.
We removed tbx-2 intronic sequences by generating a 5.2 kb transcriptional reporter that fused GFP to the ATG start site within the first exon (tbx-2p::GFP). GFP was activated at the 100-cell stage, like the translational reporter, but expression soon spread to additional, extra-pharyngeal cells and persisted throughout development (Fig. 7E, I, M). These data indicate that there are likely to be negative regulatory elements within one or more tbx-2 introns, which confine expression to the pharynx and body wall muscles. These data suggest that tbx-2 regulatory sequences are widely distributed, with both positive and negative cis-regulatory sequences that function at different developmental stages and in different cellular contexts.
Analysis of the sequences between the start codon and 5.2 kb upstream revealed two potential LAG-1 consensus sequences 2383 and 4862 bp upstream of the tbx-2 translation start (RTGGGAA), and thirty bHLH consensus sequences (CANNTG), which could be bound by members of the REF-1 family (Fig. 7A; Neves and Priess, 2005). Five additional bHLH binding sites are present in the first intron (Fig 7). The REF-1 proteins constitute a family of transcriptional repressors that are activated by Notch signaling (Neves and Priess, 2005). Based on these sequence motifs, we examined if tbx-2 was regulated positively or negatively by the Notch pathway.
TBX-2::GFP was activated normally in embryos lacking either glp-1 or lag-1 (Fig. 8A). Expression was initially observed in 8.4 cells in lag-1(RNAi) embryos (n=10/10) and in 8.4 cells in glp-1(RNAi) embryos at the 8E stage (n=10/10). This result complements our tbx-2::GFP reporter analysis, in which constructs with one or no putative LAG-1 binding site activated expression identically (2.3 kb vs. 2.4 kb of upstream sequence; data not shown). These data reveal that glp-1 signaling is not required to activate tbx-2 expression.
Fig. 8. tbx-2::GFP expression is repressed by Notch signaling.
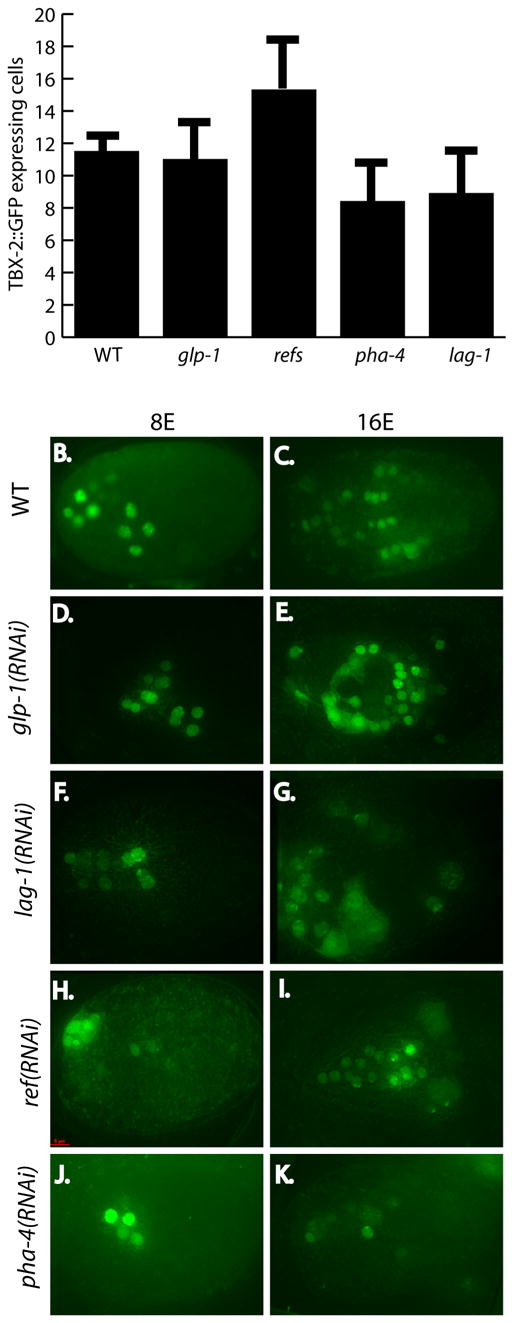
(A) tbx-2::GFP 5.2kb translational reporter is de-repressed at the 8E stage when the ref-1 family is inactivated by feeding RNAi, but activated normally with glp-1 and lag-1 inactivation. TBX-2::GFP expression in 8E cell embryos (B, D, F H, J) and 16E cell embryos (C, E, G, I, K). WT embryos (B, C), compared to feeding RNAi of glp-1 (D,E), lag-1 (F,G), and ref-1members (H,I) shows de-repression of tbx-2::GFP at the 16E cell stage. pha-4(RNAi) reduces the number of TBX-2::GFP expressing cells (J,K). Error bars indicate SD. P values of tbx-2::GFP cell counts of RNAi embryos compared to wild type are glp-1 p= 0 .32, refs p = 0.001, lag-1 p = 0.0003, pha-4 p = 0.0000003.
After the 8E stage, we observed extra TBX-2::GFP in embryos lacking lag-1. These cells were located outside of the pharynx. Similarly, we observed de-repression of TBX-2::GFP when the REF-1 family of repressors were inactivated by RNAi (Fig. 8). 15.3 cells expressed the transgene at the 8E stage when REF-1 factors ref-1, hlh-25, hlh-26, hlh-28, and hlh-29 were simultaneously inactivated instead of the usual 8 cells (n = 13) (Neves and Priess, 2005). As embryogenesis progressed, additional TBX-2::GFP+ cells were observed (Fig. 8). Thus, glp-1 signaling is required to repress tbx-2 expression, possibly by activating the REF-1 family of repressors.
Previous experiments have shown that REF-1 factors repress the T-box genes TBX-37 and TBX-38 in many cells at the 28 cell stage (Good et al., 2004). Therefore, it was possible that the activation of TBX-2::GFP in embryos lacking lag-1 or REF-1 proteins reflected activation by TBX-37 or TBX-38 rather than direct repression by REF-1 proteins. Unfortunately, it was impossible to test this idea since we could not generate a tbx-37 tbx-38 double mutant strain that carried the TBX-2::GFP reporter stably. We did observe transient TBX-2::GFP in the posterior pharynx of homozygous tbx-37/38 larvae. Thus, while we cannot distinguish direct vs. indirect repression effects of REF-1 proteins, our data demonstrate that Notch signaling restricts TBX-2 expression to a defined set of cells and is not required to activate TBX-2 within the ABa lineage.
tbx-2 and pha-4 synergize to promote pharyngeal muscle fate
Our findings suggest that TBX-2 functions in combination with PHA-4, and that a positive feedback loop between these factors is required for commitment to pharynx muscle fate. As a first test of this idea, we examined TBX-2::GFP expression in pha-4 mutant embryos. Strikingly, TBX-2::GFP was activated in 6.9 cells in pha-4(RNAi) embryos (n=14), but expression was extinguished by the L1 stage (Fig. 3K-N). As expected, TBX-2::GFP expression was unaffected in body wall muscles and non-pharyngeal cells of head. Thus, pha-4 and tbx-2 are each required to maintain expression of the other.
As a second test, we reasoned that if pharyngeal muscle development relies on a combination of tbx-2 and pha-4, then exogenous expression of tbx-2 in the presence of pha-4 might be sufficient to induce cells to adopt a pharyngeal muscle fate. To investigate this idea, we expressed tbx-2 ubiquitously in early embryos (=8E) under control of the heat-shock promoter (Jones et al., 1986). We observed an average of 40.3 myo-2::GFP+ pharyngeal muscles after heat treatment (n=7), compared to 35.7 in wild-type embryos (n=11). These myo-2::GFP+ cells were located within the pharynges of L1 larvae, as expected if TBX-2 synergizes with PHA-4, typically in the anterior pharynx and VPI region (data not shown). No excess MYO-2::GFP expressing cells were seen in worms lacking the HS::tbx-2 construct or in transgenic worms incubated at 20°C. We conclude tbx-2 can induce cells to adopt a pharyngeal muscle fate but only in the context of PHA-4+ pharyngeal cells. We favor this explanation rather than the alternative, that tbx-2 promotes division of anterior pharyngeal muscles, since the ectopic cells were the same size as non-heat-shocked pharyngeal muscles and had not become smaller by extra rounds of cell division. It is likely that additional factors are involved in these cell fate decisions since not all PHA-4+ cells adopted a pharyngeal muscle fate, despite widespread TBX-2.
DISCUSSION
Our analysis has made three contributions towards understanding the circuitry that governs cell fate specification in the C. elegans foregut. First, we identified tbx-2, which is required for ABa-derived precursors to adopt a pharyngeal muscle fate (Fig. 9). This is the first locus identified that is essential to produce a particular cell type within the pharynx. Second, tbx-2 is repressed by Notch signaling and the REF family of transcription factors. This regulation confines TBX-2 to ABa descendants, which may be important for limiting the number of pharyngeal muscles. Third, TBX-2 and PHA-4 are mutually dependent on each other to maintain expression, suggesting a direct or indirect positive regulatory loop. We propose that this regulatory loop is essential for commitment to pharyngeal muscle fate since pharyngeal muscles are lost in either pha-4 or tbx-2 mutant embryos.
Fig. 9. A model of pharyngeal muscle specification in the ABa lineage.

Notch signaling and lag-1 are required to activate the organ selector gene pha-4 in cells destined to become pharynx.. lag-1 also activates the ref-1 family of transcriptional repressors, and these limit tbx-2 expression to ABa descendants around the 8E stage, directly or indirectly. TBX-2 and PHA-4 enter a positive feedback loop, which is required for commitment and developmental progression of anterior pharyngeal muscles by the 1.5 fold/comma stage.
PHA-4 expression normally initiates at the 2-4E stage whereas TBX-2 initiates at the 8E stage. These time-points correspond precisely to the period when embryonic blastomeres lose their developmental plasticity (Fukushige and Krause, 2005; Gilleard and McGhee, 2001; Horner et al., 1998; Zhu et al., 1998). After this stage, blastomeres are not able to transform to a different cell-type identity (reviewed in Labouesse and Mango, 1999). Conversely, the loss of pha-4 or elt-1 eliminates pharynx or epidermis, respectively, demonstrating that these factors are necessary for early patterning (Mango et al., 1994a; Page et al., 1997). An intriguing notion is that the positive regulatory loop between PHA-4 and TBX-2 contributes to the cell-fate restriction that occurs in ABa descendants by the 8E stage. This model could explain the lack of a transformation in MS-derived cells that express TBX-2. TBX-2 expression initiates after the 8E stage in MS descendants, which may be too late to contribute to cell fate commitment.
pha-4 directly regulates most pharyngeally-expressed genes via a TRTTKRY consensus sequence (Gaudet and Mango, 2002). The C. elegans tbx-2 5.2 kb promoter includes 19 PHA-4 consensus sites, six of which are located within the 4.7 – 5.2 kb upstream sequence. While not conserved in position, PHA-4 binding sites are also found in tbx-2 from C. ramanei (13) and C. briggsae (8). Thus, PHA-4 may activate tbx-2 transcription directly during the maintenance phase. We note that the large number of consensus sites and the lack of direct alignment of sites complicate the analysis of direct vs. indirect regulation.
Previous studies of the Tbx2/3/4/5 subfamily suggested a role in anteroposterior patterning. For example, misexpression of Tbx2 in the chick limb can convert digit III into digit IV, and Tbx3 can convert digit II to digit III (Suzuki et al., 2004). Similarly, Tbx4 is required for hindlimb development whereas Tbx5 is required for the forelimb bud (Rodriguez-Esteban et al., 1999). These activities, in addition to the contribution of C. elegans tbx-2 for anterior pharyngeal muscles, suggest that an ancient role for the Tbx2/3/4/5 subfamily involves anterior/posterior patterning. However, the phenotypes associated with these genes are not entirely clear-cut. For example, inactivation of Tbx5 in mouse leads to loss of FGF expression and forelimb outgrowth, but patterning of the limb field appears normal (Takeuchi et al., 2003). Therefore, the effects of Tbx5 on anterior cells may reflect proliferation rather than anterior vs. posterior fate. In C. elegans, we do not observe alterations in cell division of ABa descendants in tbx-2 mutants. Moreover, FGF, which is an important Tbx target for growth and morphogenesis in vertebrates, is probably not downstream of tbx-2 in C. elegans. FGF ligands and receptors have no described phenotypes in the anterior pharynx (Birnbaum et al., 2005). Rather, our phenotypes are best explained as alterations of cell fate progression.
When glp-1 signaling is abrogated (or its downstream effectors lag-1/su(H) and the REF proteins), we observe extra cells expressing TBX-2::GFP. These data suggest that Notch signaling represses tbx-2 and ensures only some cells progress towards anterior muscle fate. Studies with Drosophila have suggested that one role for Notch is to block the progression of development (Cagan and Ready, 1989). Inhibition of tbx-2 by Notch could serve this purpose, by repressing a gene required for developmental progression.
How does TBX-2 function? In other organisms, members of the Tbx2/3/4/5 subfamily of T-box factors can be either activators (e.g. Tbx4, Tbx5) (Takeuchi et al., 2003; Zaragoza et al., 2004) or repressors (e.g. Tbx2, Tbx3) (Carreira et al., 1998; He et al., 1999). Examination of C. elegans tbx-2 reveals that it has sequences similar to the amino terminal repression domain of vertebrate Tbx2 and Tbx3 (Fig. 1C) (He et al., 1999). On the other hand, the carboxyl terminal sequence of TBX-2 is rich in serine, proline and glutamate, suggesting it could function as an activation domain, similar to some other transcription factors (Matsuzaki et al., 1995; Nagy et al., 2002; Stepchenko and Nirenberg, 2004). Thus, a speculative possibility is that in C. elegans, the sole Tbx2 family member functions as both an activator and a repressor, whereas in vertebrates, with four Tbx2 members, activation and repression activities are segregated into different proteins.
TBX-2 is both nuclear and cytoplasmic
We observe TBX-2 in the cytoplasm beginning at the 1.5 fold stage. This location agrees with previous studies where endogenous TBX-2 was observed in the cytoplasm of post-embryonic neurons (Miyahara et al., 2004). Tbx factors from other organisms also accumulate in the cytoplasm, suggesting this may be a conserved mechanism to regulate Tbx proteins (Bruce et al., 2003; Fuchikami et al., 2002; Krause et al., 2004). Our cytoplasmic expression appears filamentous, as though TBX-2 were associated with the cytoskeleton. The pattern resembles that of microtubules, although this idea remains to be tested. The cytoplasmic expression pattern was detected in 12/12 independent transgenic lines, and the cytoplasmic transition was consistent even in the weakest expressing lines; arguing that this is not an over expression phenotype. Recent studies with chicken Tbx5 suggested this protein could bind to LMP-4 in vitro, and was targeted to actin filaments in COS cells (Krause et al., 2004). Thus, members of the Tbx2 subfamily may be regulated by cytoskeletal association.
Acknowledgments
We thank Marc Vidal, Erik Jorgenson, Leon Avery, Jim Priess, and Bruce Bowerman for reagents. Some nematode strains were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). The Core facilities at the University of Utah were supported by the National Institutes of Health 2P30CA42014. This work was funded by the National Institutes of Health T32 CA93247-02 to P.A.S. and GM056264 to S.E.M. S.E.M. is an associate investigator of the Huntsman Cancer Institute and an Associate Professor of the Department of Oncological Sciences at the University of Utah.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adell T, Muller WE. Expression pattern of the Brachyury and Tbx2 homologs from the sponge Suberites domuncula. Biol Cell. 2005 doi: 10.1042/BC20040135. [DOI] [PubMed] [Google Scholar]
- Agarwal P, Wylie JN, Galceran J, Arkhitko O, Li C, Deng C, Grosschedl R, Bruneau BG. Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development. 2003;130:623–33. doi: 10.1242/dev.00191. [DOI] [PubMed] [Google Scholar]
- Agulnik SI, Garvey N, Hancock S, Ruvinsky I, Chapman DL, Agulnik I, Bollag R, Papaioannou V, Silver LM. Evolution of mouse T-box genes by tandem duplication and cluster dispersion. Genetics. 1996;144:249–54. doi: 10.1093/genetics/144.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad I, Das AV, James J, Bhattacharya S, Zhao X. Neural stem cells in the mammalian eye: types and regulation. Semin Cell Dev Biol. 2004;15:53–62. doi: 10.1016/j.semcdb.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Albertson DG. Formation of the first cleavage spindle in nematode embryos. Devel Biol. 1984;101:61–72. doi: 10.1016/0012-1606(84)90117-9. [DOI] [PubMed] [Google Scholar]
- Albertson DG, Thomson JN. The pharynx of Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1976;275:299–325. doi: 10.1098/rstb.1976.0085. [DOI] [PubMed] [Google Scholar]
- Alder MN, Dames S, Gaudet J, Mango SE. Gene silencing in Caenorhabditis elegans by transitive RNA interference. Rna. 2003;9:25–32. doi: 10.1261/rna.2650903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andachi Y. Caenorhabditis elegans T-box genes tbx-9 and tbx-8 are required for formation of hypodermis and body-wall muscle in embryogenesis. Genes Cells. 2004;9:331–44. doi: 10.1111/j.1356-9597.2004.00725.x. [DOI] [PubMed] [Google Scholar]
- Ao W, Gaudet J, Kent WJ, Muttumu S, Mango SE. Environmentally induced foregut remodeling by PHA-4/FoxA and DAF-12/NHR. Science. 2004;305:1743–6. doi: 10.1126/science.1102216. [DOI] [PubMed] [Google Scholar]
- Bartnik E, Osborn M, Weber K. Intermediate filaments in muscle and epithelial cells of nematodes. J Cell Biol. 1986;102:2033–41. doi: 10.1083/jcb.102.6.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Hill AA, Slonim DK, Brown EL, Hunter CP. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development. 2003;130:889–900. doi: 10.1242/dev.00302. [DOI] [PubMed] [Google Scholar]
- Baugh LR, Wen JC, Hill AA, Slonim DK, Brown EL, Hunter CP. Synthetic lethal analysis of Caenorhabditis elegans posterior embryonic patterning genes identifies conserved genetic interactions. Genome Biol. 2005;6:R45. doi: 10.1186/gb-2005-6-5-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylis HA, Furuichi T, Yoshikawa F, Mikoshiba K, Sattelle DB. Inositol 1,4,5-trisphosphate receptors are strongly expressed in the nervous system, pharynx, intestine, gonad and excretory cell of Caenorhabditis elegans and are encoded by a single gene (itr-1) J Mol Biol. 1999;294:467–76. doi: 10.1006/jmbi.1999.3229. [DOI] [PubMed] [Google Scholar]
- Birnbaum D, Popovici C, Roubin R. A pair as a minimum: the two fibroblast growth factors of the nematode Caenorhabditis elegans. Dev Dyn. 2005;232:247–55. doi: 10.1002/dvdy.20219. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce AE, Howley C, Zhou Y, Vickers SL, Silver LM, King ML, Ho RK. The maternally expressed zebrafish T-box gene eomesodermin regulates organizer formation. Development. 2003;130:5503–17. doi: 10.1242/dev.00763. [DOI] [PubMed] [Google Scholar]
- Cagan RL, Ready DF. Notch is required for successive cell decisions in the developing Drosophila retina. Genes Dev. 1989;3:1099–112. doi: 10.1101/gad.3.8.1099. [DOI] [PubMed] [Google Scholar]
- Carlson H, Ota S, Campbell CE, Hurlin PJ. A dominant repression domain in Tbx3 mediates transcriptional repression and cell immortalization: relevance to mutations in Tbx3 that cause ulnar-mammary syndrome. Hum Mol Genet. 2001;10:2403–13. doi: 10.1093/hmg/10.21.2403. [DOI] [PubMed] [Google Scholar]
- Carreira S, Dexter TJ, Yavuzer U, Easty DJ, Goding CR. Brachyury-related transcription factor Tbx2 and repression of the melanocyte-specific TRP-1 promoter. Mol Cell Biol. 1998;18:5099–108. doi: 10.1128/mcb.18.9.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm AD, Hodgkin J. The mab-9 gene controls the fate of B, the major male-specific balst cell in the tail region of Caenorhabditis elegans. Genes & Devel. 1989;3:1413–1423. doi: 10.1101/gad.3.9.1413. [DOI] [PubMed] [Google Scholar]
- Collavoli A, Hatcher CJ, He J, Okin D, Deo R, Basson CT. TBX5 nuclear localization is mediated by dual cooperative intramolecular signals. J Mol Cell Cardiol. 2003;35:1191–5. doi: 10.1016/s0022-2828(03)00231-1. [DOI] [PubMed] [Google Scholar]
- Dal Santo P, Logan MA, Chisholm AD, Jorgensen EM. The inositol trisphosphate receptor regulates a 50-second behavioral rhythm in C. elegans. Cell. 1999;98:757–67. doi: 10.1016/s0092-8674(00)81510-x. [DOI] [PubMed] [Google Scholar]
- Dent JA, Davis MW, Avery L. avr-15 encodes a chloride channel subunit that mediates inhibitory glutamatergic neurotransmission and ivermectin sensitivity in Caenorhabditis elegans. Embo J. 1997;16:5867–79. doi: 10.1093/emboj/16.19.5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RE, Jacobson DM, Horvitz HR. Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics. 1991;129:79–94. doi: 10.1093/genetics/129.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Harrison SW, Dixon D. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene. 1990;93:189–98. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans [see comments] Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Francis R, Waterston RH. Muscle cell attachment in Caenorhabditis elegans. J Cell Biol. 1991;114:465–79. doi: 10.1083/jcb.114.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchikami T, Mitsunaga-Nakatsubo K, Amemiya S, Hosomi T, Watanabe T, Kurokawa D, Kataoka M, Harada Y, Satoh N, Kusunoki S, et al. T-brain homologue (HpTb) is involved in the archenteron induction signals of micromere descendant cells in the sea urchin embryo. Development. 2002;129:5205–16. doi: 10.1242/dev.129.22.5205. [DOI] [PubMed] [Google Scholar]
- Fukushige T, Krause M. The myogenic potency of HLH-1 reveals widespread developmental plasticity in early C. elegans embryos. Development. 2005;132:1795–805. doi: 10.1242/dev.01774. [DOI] [PubMed] [Google Scholar]
- Gaudet J, Mango SE. Regulation of Organogenesis by the Caenorhabditis elegans FoxA Protein PHA-4. Science. 2002;295:821–5. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
- Gilleard JS, McGhee JD. Activation of hypodermal differentiation in the Caenorhabditis elegans embryo by GATA transcription factors ELT-1 and ELT-3. Mol Cell Biol. 2001;21:2533–44. doi: 10.1128/MCB.21.7.2533-2544.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good K, Ciosk R, Nance J, Neves A, Hill RJ, Priess JR. The T-box transcription factors TBX-37 and TBX-38 link GLP-1/Notch signaling to mesoderm induction in C. elegans embryos. Development. 2004;131:1967–78. doi: 10.1242/dev.01088. [DOI] [PubMed] [Google Scholar]
- Harrelson Z, Kelly RG, Goldin SN, Gibson-Brown JJ, Bollag RJ, Silver LM, Papaioannou VE. Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development. 2004;131:5041–52. doi: 10.1242/dev.01378. [DOI] [PubMed] [Google Scholar]
- He M, Wen L, Campbell CE, Wu JY, Rao Y. Transcription repression by Xenopus ET and its human ortholog TBX3, a gene involved in ulnar-mammary syndrome. Proc Natl Acad Sci U S A. 1999;96:10212–7. doi: 10.1073/pnas.96.18.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–30. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Horner MA, Quintin S, Domeier ME, Kimble J, Labouesse M, Mango SE. pha-4, an HNF-3 homolog, specifies pharyngeal organ identity in Caenorhabditis elegans. Genes Dev. 1998;12:1947–52. doi: 10.1101/gad.12.13.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter H, Schnabel R. glp-1 and inductions establishing embryonic axes in C. elegans. Development. 1994;120:2051–64. doi: 10.1242/dev.120.7.2051. [DOI] [PubMed] [Google Scholar]
- Jones D, Russnak RH, Kay RJ, Candido EP. Structure, expression, and evolution of a heat shock gene locus in Caenorhabditis elegans that is flanked by repetitive elements. J Biol Chem. 1986;261:12006–15. [PubMed] [Google Scholar]
- Joshi P, Eisenmann DM. The Caenorhabditis elegans pvl-5 gene protects hypodermal cells from ced-3-dependent, ced-4-independent cell death. Genetics. 2004;167:673–85. doi: 10.1534/genetics.103.020503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb JM, Lau KK, Goszczynski B, Fukushige T, Moons D, Okkema PG, McGhee JD. pha-4 is Ce-fkh-1, a fork head/HNF-3alpha,beta,gamma homolog that functions in organogenesis of the C. elegans pharynx. Development. 1998;125:2171–80. doi: 10.1242/dev.125.12.2171. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–7. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kim J, Wu HH, Lander AD, Lyons KM, Matzuk MM, Calof AL. GDF11 controls the timing of progenitor cell competence in developing retina. Science. 2005;308:1927–30. doi: 10.1126/science.1110175. [DOI] [PubMed] [Google Scholar]
- Kinchen JM, Cabello J, Klingele D, Wong K, Feichtinger R, Schnabel H, Schnabel R, Hengartner MO. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature. 2005;434:93–9. doi: 10.1038/nature03263. [DOI] [PubMed] [Google Scholar]
- Krause A, Zacharias W, Camarata T, Linkhart B, Law E, Lischke A, Miljan E, Simon HG. Tbx5 and Tbx4 transcription factors interact with a new chicken PDZ-LIM protein in limb and heart development. Dev Biol. 2004;273:106–20. doi: 10.1016/j.ydbio.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Labouesse M, Hartwieg E, Horvitz HR. The Caenorhabditis elegans LIN-26 protein is required to specify and/or maintain all non-neuronal ectodermal cell fates. Development. 1996;122:2579–88. doi: 10.1242/dev.122.9.2579. [DOI] [PubMed] [Google Scholar]
- Labouesse M, Mango SE. Patterning the C. elegans embryo: moving beyond the cell lineage. Trends Genet. 1999;15:307–13. doi: 10.1016/s0168-9525(99)01750-3. [DOI] [PubMed] [Google Scholar]
- Lamesch P, Milstein S, Hao T, Rosenberg J, Li N, Sequerra R, Bosak S, Doucette-Stamm L, Vandenhaute J, Hill DE, et al. C. elegans ORFeome version 3.1: increasing the coverage of ORFeome resources with improved gene predictions. Genome Res. 2004;14:2064–9. doi: 10.1101/gr.2496804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M, Tabin CJ. Role of Pitx1 upstream of Tbx4 in specification of hindlimb identity. Science. 1999;283:1736–9. doi: 10.1126/science.283.5408.1736. [DOI] [PubMed] [Google Scholar]
- Mango SE, Lambie EJ, Kimble J. The pha-4 gene is required to generate the pharyngeal primordium of Caenorhabditis elegans. Development. 1994a;120:3019–31. doi: 10.1242/dev.120.10.3019. [DOI] [PubMed] [Google Scholar]
- Mango SE, Thorpe CJ, Martin PR, Chamberlain SH, Bowerman B. Two maternal genes, apx-1 and pie-1, are required to distinguish the fates of equivalent blastomeres in the early Caenorhabditis elegans embryo. Development. 1994b;120:2305–15. doi: 10.1242/dev.120.8.2305. [DOI] [PubMed] [Google Scholar]
- Matsuzaki Y, Fujisawa J, Yoshida M. Identification of transcriptional activation domain of TREB5, a CREB/ATF family protein that binds to HTLV-1 enhancer. J Biochem (Tokyo) 1995;117:303–8. doi: 10.1093/jb/117.2.303. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–82. [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. Embo J. 1991;10:3959–70. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM, Ortiz I, Berliner GC, Epstein HF. Differential localization of two myosins within nematode thick filaments. Cell. 1983;34:477–490. doi: 10.1016/0092-8674(83)90381-1. [DOI] [PubMed] [Google Scholar]
- Miller DM, Stockdale FE, Karn J. Immunological identification of the genes encoding the four myosin heavy chain isoforms of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1986;83:2305–9. doi: 10.1073/pnas.83.8.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minguillon C, Del Buono J, Logan MP. Tbx5 and Tbx4 are not sufficient to determine limb-specific morphologies but have common roles in initiating limb outgrowth. Dev Cell. 2005;8:75–84. doi: 10.1016/j.devcel.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Miyahara K, Suzuki N, Ishihara T, Tsuchiya E, Katsura I. TBX2/TBX3 transcriptional factor homologue controls olfactory adaptation in Caenorhabditis elegans. J Neurobiol. 2004;58:392–402. doi: 10.1002/neu.10299. [DOI] [PubMed] [Google Scholar]
- Moskowitz IP, Gendreau SB, Rothman JH. Combinatorial specification of blastomere identity by glp-1-dependent cellular interactions in the nematode Caenorhabditis elegans. Development. 1994;120:3325–38. doi: 10.1242/dev.120.11.3325. [DOI] [PubMed] [Google Scholar]
- Muller CW, Herrmann BG. Crystallographic structure of the T domain-DNA complex of the Brachyury transcription factor. Nature. 1997;389:884–8. doi: 10.1038/39929. [DOI] [PubMed] [Google Scholar]
- Nagy ZS, Wang Y, Erwin-Cohen RA, Aradi J, Monia B, Wang LH, Stepkowski SM, Rui H, Kirken RA. Interleukin-2 family cytokines stimulate phosphorylation of the Pro-Ser-Pro motif of Stat5 transcription factors in human T cells: resistance to suppression of multiple serine kinase pathways. J Leukoc Biol. 2002;72:819–28. [PubMed] [Google Scholar]
- Neves A, Priess JR. The REF-1 family of bHLH transcription factors pattern C. elegans embryos through Notch-dependent and Notch-independent pathways. Dev Cell. 2005;8:867–79. doi: 10.1016/j.devcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Okamoto H, Thomson JN. Monoclonal antibodies which distinguish certain classes of neuronal and supporting cells in the nervous tissue of the nematode Caenorhabditis elegans. Journal of Neuroscience. 1985;5:643–653. doi: 10.1523/JNEUROSCI.05-03-00643.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema PG, Fire A. The Caenorhabditis elegans NK-2 class homeoprotein CEH-22 is involved in combinatorial activation of gene expression in pharyngeal muscle. Development. 1994;120:2175–86. doi: 10.1242/dev.120.8.2175. [DOI] [PubMed] [Google Scholar]
- Okkema PG, Ha E, Haun C, Chen W, Fire A. The Caenorhabditis elegans NK-2 homeobox gene ceh-22 activates pharyngeal muscle gene expression in combination with pha-1 and is required for normal pharyngeal development. Development. 1997;124:3965–73. doi: 10.1242/dev.124.20.3965. [DOI] [PubMed] [Google Scholar]
- Page BD, Zhang W, Steward K, Blumenthal T, Priess JR. ELT-1, a GATA-like transcription factor, is required for epidermal cell fates in Caenorhabditis elegans embryos. Genes Dev. 1997;11:1651–61. doi: 10.1101/gad.11.13.1651. [DOI] [PubMed] [Google Scholar]
- Pocock R, Ahringer J, Mitsch M, Maxwell S, Woollard A. A regulatory network of T-box genes and the even-skipped homologue vab-7 controls patterning and morphogenesis in C. elegans. Development. 2004;131:2373–85. doi: 10.1242/dev.01110. [DOI] [PubMed] [Google Scholar]
- Portereiko MF, Saam J, Mango SE. ZEN-4/MKLP1 is required to polarize the foregut epithelium. Curr Biol. 2004;14:932–41. doi: 10.1016/j.cub.2004.05.052. [DOI] [PubMed] [Google Scholar]
- Priess JR, Schnabel H, Schnabel R. The glp-1 locus and cellular interactions in early C. elegans embryos. Cell. 1987;51:601–611. doi: 10.1016/0092-8674(87)90129-2. [DOI] [PubMed] [Google Scholar]
- Priess JR, Thomson JN. Cellular interactions in early C. elegans embryos. Cell. 1987;48:241–250. doi: 10.1016/0092-8674(87)90427-2. [DOI] [PubMed] [Google Scholar]
- Pruss RM, Mirsky R, Raff MC, Thorpe R, Dowding AJ, Anderton BH. All classes of intermediate filaments share a common antigenic determinant defined by a monoclonal antibody. Cell. 1981;27:419–28. doi: 10.1016/0092-8674(81)90383-4. [DOI] [PubMed] [Google Scholar]
- Rallis C, Bruneau BG, Del Buono J, Seidman CE, Seidman JG, Nissim S, Tabin CJ, Logan MP. Tbx5 is required for forelimb bud formation and continued outgrowth. Development. 2003;130:2741–51. doi: 10.1242/dev.00473. [DOI] [PubMed] [Google Scholar]
- Reboul J, Vaglio P, Rual JF, Lamesch P, Martinez M, Armstrong CM, Li S, Jacotot L, Bertin N, Janky R, et al. C. elegans ORFeome version 1.1: experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat Genet. 2003;34:35–41. doi: 10.1038/ng1140. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sanchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell. 2005;8:635–49. doi: 10.1016/j.devcel.2005.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Esteban C, Tsukui T, Yonei S, Magallon J, Tamura K, Izpisua Belmonte JC. The T-box genes Tbx4 and Tbx5 regulate limb outgrowth and identity. Nature. 1999;398:814–8. doi: 10.1038/19769. [DOI] [PubMed] [Google Scholar]
- Shin-i T, Kohara Y. ( http://nematode.lab.nig.ac.jp/). NEXTDB (The Nematode Expression Pattern Database), (ed.: National Institite of Genetics.
- Singh MK, Christoffels VM, Dias JM, Trowe MO, Petry M, Schuster-Gossler K, Burger A, Ericson J, Kispert A. Tbx20 is essential for cardiac chamber differentiation and repression of Tbx2. Development. 2005 doi: 10.1242/dev.01854. [DOI] [PubMed] [Google Scholar]
- Stepchenko A, Nirenberg M. Mapping activation and repression domains of the vnd/NK-2 homeodomain protein. Proc Natl Acad Sci U S A. 2004;101:13180–5. doi: 10.1073/pnas.0404775101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Takeuchi J, Koshiba-Takeuchi K, Ogura T. Tbx Genes Specify Posterior Digit Identity through Shh and BMP Signaling. Dev Cell. 2004;6:43–53. doi: 10.1016/s1534-5807(03)00401-5. [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Koshiba-Takeuchi K, Matsumoto K, Vogel-Hopker A, Naitoh-Matsuo M, Ogura K, Takahashi N, Yasuda K, Ogura T. Tbx5 and Tbx4 genes determine the wing/leg identity of limb buds. Nature. 1999;398:810–4. doi: 10.1038/19762. [DOI] [PubMed] [Google Scholar]
- Takeuchi JK, Koshiba-Takeuchi K, Suzuki T, Kamimura M, Ogura K, Ogura T. Tbx5 and Tbx4 trigger limb initiation through activation of the Wnt/Fgf signaling cascade. Development. 2003;130:2729–39. doi: 10.1242/dev.00474. [DOI] [PubMed] [Google Scholar]
- Thatcher JD, Fernandez AP, Beaster-Jones L, Haun C, Okkema PG. The Caenorhabditis elegans peb-1 gene encodes a novel DNA-binding protein involved in morphogenesis of the pharynx, vulva, and hindgut. Dev Biol. 2001;229:480–93. doi: 10.1006/dbio.2000.9978. [DOI] [PubMed] [Google Scholar]
- Wilson V, Conlon FL. The T-box family. Genome Biol. 2002;3:REVIEWS3008. doi: 10.1186/gb-2002-3-6-reviews3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woollard A, Hodgkin J. The caenorhabditis elegans fate-determining gene mab-9 encodes a T-box protein required to pattern the posterior hindgut. Genes Dev. 2000;14:596–603. [PMC free article] [PubMed] [Google Scholar]
- Yuan JY, Horvitz HR. The Caenorhabditis elegans genes ced-3 and ced-4 act cell autonomously to cause programmed cell death. Dev Biol. 1990;138:33–41. doi: 10.1016/0012-1606(90)90174-h. [DOI] [PubMed] [Google Scholar]
- Zaragoza MV, Lewis LE, Sun G, Wang E, Li L, Said-Salman I, Feucht L, Huang T. Identification of the TBX5 transactivating domain and the nuclear localization signal. Gene. 2004;330:9–18. doi: 10.1016/j.gene.2004.01.017. [DOI] [PubMed] [Google Scholar]
- Zhu J, Fukushige T, McGhee JD, Rothman JH. Reprogramming of early embryonic blastomeres into endodermal progenitors by a Caenorhabditis elegans GATA factor. Genes Dev. 1998;12:3809–14. doi: 10.1101/gad.12.24.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]


