Summary
Successful colonization of a eukaryotic host by a microbe involves complex microbe-microbe and microbe-host interactions. Previously, we identified in Vibrio fischeri a putative sensor kinase, RscS, required for initiating symbiotic colonization of its squid host Euprymna scolopes. Here, we analyzed the role of rscS by isolating an allele, rscS1, with increased activity. Multi-copy rscS1 activated transcription of genes within the recently identified symbiosis polysaccharide (syp) cluster. Wild-type cells carrying rscS1 induced aggregation phenotypes in culture, including the formation of pellicles and wrinkled colonies, in a syp-dependent manner. Colonies formed by rscS1-expressing cells produced a matrix not found in control colonies and largely lost in an rscS1-expressing sypN mutant. Finally, multi-copy rscS1 provided a colonization advantage over control cells and substantially enhanced the ability of wild-type cells to aggregate on the surface of the symbiotic organ of E. scolopes; this latter phenotype similarly depended upon an intact syp locus. These results suggest that transcription induced by RscS-mediated signal transduction plays a key role in colonization at the aggregation stage by modifying the cell surface and increasing the ability of the cells to adhere to one another and/or to squid-secreted mucus.
Introduction
Communication between bacteria and their plant or animal hosts is an essential component of both beneficial symbioses and pathogenic associations. Perhaps equally important are interactions within the community of bacterial cells as they colonize their host. For example, during Vibrio cholerae infection of the mouse intestine, interactions mediated by the TCP pilus allow the bacteria to aggregate into microcolonies, possibly affording protection from antimicrobial agents within the intestine (Kirn et al., 2000). Notably, biofilm formation by V. cholerae increases its resistance to the toxicity of the bile acids found in the intestine (Hung et al., 2006). Similarly, biofilm formation by Pseudomonas aeruginosa in the cystic fibrosis lung is thought to contribute to chronic infection by increasing resistance of the bacterial community to antibiotics (Whiteley et al., 2001). In addition to pathogenic functions, biofilm formation is implicated in the maintenance of the complex microbial community found within the mammalian gut (Sonnenburg et al., 2004); therefore, bacterial cell-cell interactions also appear to be a component of beneficial symbioses.
An elegant model system for studying bacteria-host and inter-bacterial interactions during colonization of a host is the symbiosis between the Hawaiian bobtail squid Euprymna scolopes and the marine bioluminescent bacterium Vibrio fischeri [reviewed in (Nyholm and McFall-Ngai, 2004)]. The specialized symbiotic organ (the light organ) of E. scolopes is exclusively colonized by its bacterial symbiont, V. fischeri. Newly hatched squid lack symbionts but rapidly become colonized upon exposure to V. fischeri in the seawater. In the squid, symbiotic initiation is facilitated by ciliated cells located on the surface of the light organ that direct the flow of bacteria-containing seawater toward pores that lead into the organ. The squid also promotes colonization by secreting mucus on the surface of the light organ (Nyholm et al., 2000). Early in the association, V. fischeri cells form a tight, dense aggregate in this mucus (Nyholm et al., 2000). The cells then migrate from the aggregate into the pores and through ducts that lead to crypt spaces where finally they multiply to high cell density and luminesce (Nyholm and McFall-Ngai, 2004).
The specificity of the Vibrio-squid symbiosis is first manifested at the aggregation stage. Several species of Gram-negative bacteria, including the closely related Vibrio parahaemolyticus, can aggregate on the light organ surface (Nyholm et al., 2000). However, when both V. fischeri and V. parahaemolyticus are present, the symbiotic V. fischeri cells dominate in the aggregate (Nyholm and McFall-Ngai, 2003). These data suggest that V. fischeri actively promotes aggregation, however, the bacterial factors that contribute to this activity are unknown (Nyholm and McFall-Ngai, 2003).
Studies investigating the genetic requirements for the symbiosis have revealed a number of mutants defective for initiating colonization, some of which have been characterized with respect to aggregation (Nyholm et al., 2000). A mutant defective for the flagellar master regulator, FlrA, forms smaller aggregates than those produced by wild-type V. fischeri (Millikan and Ruby, 2003). Hyper-motile mutants, which are delayed in initiating colonization, also form smaller aggregates, typically after a lag period (Millikan and Ruby, 2002). While appropriate levels of motility appear to be required for optimal aggregation, it is unclear what other bacterial factors promote aggregation in the squid-secreted mucus. Furthermore, characteristics associated with bacterial aggregation in other bacteria – such as biofilm formation, wrinkled colony morphology and clumping of cells (Del Re et al., 2000; Romling et al., 1998) – have not been reported to date for V. fischeri cells grown in culture. This lack of culture phenotypes has hampered genetic investigation of the basis for aggregation within the squid mucus.
In bacteria, two-component regulatory systems detect environmental signals and respond by auto-phosphorylation of a sensor kinase and subsequent transfer of the phosphate to a response regulator. Signaling through these systems, in many cases, regulates transcription of a target gene(s) [reviewed in (Stock et al., 2000)]. Previously, we reported that a mutant defective for the putative sensor kinase RscS exhibited a severe defect in colonization (Visick and Skoufos, 2001). However, known symbiosis traits, including motility and bioluminescence, were unaltered under laboratory conditions. Subsequently, we reported the identification of the symbiosis polysaccharide (syp) gene locus (Yip et al., 2005) involved in symbiotic initiation. Mutation of syp genes causes over a 1000-fold decrease in symbiotic colonization. The 18 genes (sypA-R) in this cluster encode proteins with predicted roles as glycosyltransferases and other polysaccharide metabolism and export functions. In addition, four genes encode putative regulatory proteins such as response regulators. Consistent with a possible role of syp in glycosylation of a surface molecule(s), we found that multi-copy expression of a syp-encoded putative response regulator, SypG, caused a substantial increase in biofilm formation by cells grown in both static and shaking liquid cultures.
Here, we demonstrate that one role of RscS is regulation of the syp gene cluster. Multi-copy expression of an allele of rscS with increased activity induced syp transcription and dramatic syp-dependent cell-cell aggregation phenotypes, including the formation of wrinkled colonies on solid media and pellicles in static liquid culture. We then show the relevance of these phenotypes to symbiosis by identifying a role for rscS in promoting aggregation of V. fischeri cells in squid-secreted mucus on the light organ surface. Our results reveal central roles for RscS and syp in cell-cell aggregation by this bacterium, both in culture and during symbiotic initiation, and provide a basis for understanding one of the earliest known stages of symbiotic colonization of squid by V. fischeri.
Results
RscS controls the symbiosis polysaccharide gene cluster
Our preliminary data showed that the putative sensor kinase, RscS, could autophosphorylate, suggesting it did indeed function as a sensor kinase (Yip and Visick, unpublished). However, this gene was not linked to a response regulator, and our attempts to identify rscS-controlled genes through global screens with a wild-type copy of rscS on the multi-copy plasmid pLMS33 (Visick and Skoufos, 2001) were unsuccessful. We therefore hypothesized that RscS might exhibit only a low basal level of activity under laboratory culture conditions and that isolation of an rscS allele with increased activity could make it possible to detect rscS-dependent phenotypes and gene regulation. We further speculated that one potential target of RscS could be the syp cluster, which we recently identified as essential for symbiotic colonization (Yip et al., 2005). Like disruption of rscS, syp mutations caused severe initiation defects but no discernible alterations in traits required for symbiosis. Furthermore, the syp genes were not detectably transcribed under laboratory conditions unless a syp-encoded regulator, SypG, was expressed from a multi-copy plasmid (Yip et al., 2005). Therefore, we asked whether we could identify an allele of rscS capable of inducing syp transcription.
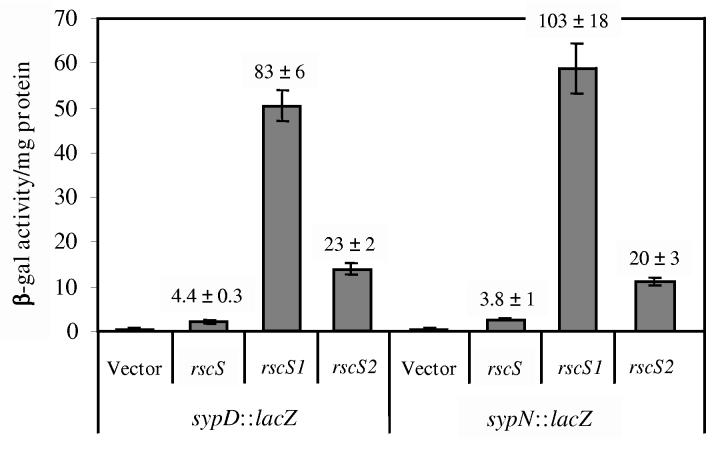
To isolate mutations in rscS, we passaged pLMS33 through an E. coli mutator strain and we introduced the resulting library of randomly mutagenized plasmids into reporter strains. To monitor the induction of syp transcription, we employed strains containing a promoterless lacZ gene carried on Tn10 inserted in either sypD or sypN (KV1635 and KV1601, respectively; Table 1) which are located within two different putative operons of the syp cluster (Yip et al., 2005). We then screened for blue colonies, and found them at a frequency of about 1 in 1000. We confirmed that the plasmid was responsible for the elevated syp transcription for six such plasmids, and following subcloning experiments (see Materials and Methods), we identified two alleles, rscS1 and rscS2, for which the mutation mapped within the subcloned rscS portion of the plasmid (carried by pKG11 and pKG13, respectively). The effect of these two alleles on syp transcription was quantified by β-galactosidase assays. The presence of multi-copy rscS1 or rscS2 increased the amount of β-galactosidase activity generated from the sypN::lacZ fusion strain by 103 ± 18-fold and 20 ± 3-fold, respectively, relative to the vector control (Fig. 1). Similar levels of induction were seen for the sypD::lacZ fusion strain (Fig. 1).
Table 1.
Strains and plasmids
| Strains | Genotype or characteristics | Reference |
|---|---|---|
| E. coli | ||
| CC118λpir | Δ(are-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA1λpir | (Herrero et al., 1990) |
| CC130 | F-, araD139, Δ(ara-leu)7697 Δlac-74 galU galK rpsL Δ(phoA-proC) phoR tsx::Tn5 mutD5 | (Manoil and Beckwith, 1985) |
| DH5α | endA1 hsdR17 (rK− mK+) glnV44 thi-1 recA1 gyrA (Nalr) relA Δ(lacIZYA-argF)U169 deoR [ϕ80dlacΔ(lacZ)M15] | (Woodcock et al., 1989) |
| TOP10F' | F' [lacIq Tn10 (TetR)] mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZ ΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (StrR) endA1 nupG | Invitrogen |
| V. fischeri | ||
| ES114 | Wild type | (Boettcher and Ruby, 1990) |
| ESR1 | RifR | (Graf et al., 1994) |
| KV905 | pKV69/ESR1 | (Visick and Skoufos, 2001) |
| KV1066 | pKV111/ES114 | This study |
| KV1223 | RifR rscS::erm + pKV111 | This study |
| KV1228 | pKV111/RifR | This study |
| KV1339 | pKV111/ES114 rscS::erm | This study |
| KV1421 | ES114 att Tn7::erm | (DeLoney et al., 2002) |
| KV1601 | RifR sypN::Tn10lacZ | (Yip et al., 2005) |
| KV1635 | RifR sypD::Tn10lacZ | (Yip et al., 2005) |
| KV1838 | ES114 sypN::pTMB54 | (Yip et al., 2005) |
| KV1844 | pKV69/ES114 | (Visick and Skoufos, 2001) |
| KV1845 | pLMS33/ES114 | This study |
| KV1956 | pKG11/ES114 | This study |
| KV1958 | pKV69/KV1601 | (Yip et al., 2005) |
| KV1959 | pLMS33/KV1601 | This study |
| KV1960 | pKG11/KV1601 | This study |
| KV1962 | pKV69/KV1635 | (Yip et al., 2005) |
| KV1963 | pLMS33/KV1635 | This study |
| KV1964 | pKG11/KV1635 | This study |
| KV1980 | pKG11/ESR1 | This study |
| KV1992 | pKG11/KV1838 | This study |
| KV2273 | pKG13/KV1601 | This study |
| KV2274 | pKG13/KV1635 | This study |
| KV2435 | pKV69/KV1421 | This study |
| KV2437 | pKG11/KV1421 | This study |
| KV2608 | pVSV208/ES114 | This study |
| KV2613 | pKG11/KV2608 | This study |
| KV2617 | pKV69/KV2608 | This study |
| KV2682 | pESY37/ES114 | This study |
| KV2683 | pESY37/KV1838 | This study |
| KV2685 | pKV69/KV2682 | This study |
| KV2686 | pKV69/KV2683 | This study |
| KV2688 | pKG11/KV2682 | This study |
| KV2689 | pKG11/KV2683 | This study |
| KV2888 | pESY48/KV1838 | This study |
| KV2976 | pKV69/KV2888 | This study |
| KV2977 | pKG11/KV2888 | This study |
| Plasmids | Characteristics or construction | |
| pCR2.1-TOPO | Commercial cloning vector; AmpR, KanR | Invitrogen |
| pESY36 | pCR2.1-TOPO + 4.6 Kb fragment of sypMNO; AmpR, KanR | This study |
| pESY37 | pVSV105 (KpnI) + 1.3 bp BamHI/XmnI fragment from pKV111 containing gfp; CmR | This study |
| pESY47 | pEVS107 (KpnI) + 4.6 Kb KpnI fragment from pESY36 encoding sypMNO; KanR, EmR | This study |
| pESY48 | pESY47 (EcoRV) + 1.2 Kb fragment encoding CmR; KanR, CmR | This study |
| pEVS107 | Mini-Tn7 delivery plasmid; mob; KanR, EmR | (McCann et al., 2003) |
| pKG11 | pKV69 (SalI) + 3 Kb SalI fragment from mutagenized pLMS33 containing rscS1 allele; CmR, TetR | This study |
| pKG13 | pKV69 (SalI) + 3 Kb SalI fragment from mutagenized pLMS33 containing rscS2 allele; CmR, TetR | This study |
| pKV69 | Mobilizable vector; CmR, TetR | (Visick and Skoufos, 2001) |
| pKV111 | Mobilizable vector containing gfp; CmR | (Stabb and Ruby, 2002) |
| pLMS33 | pKV69 containing wild-type rscS; CmR, TetR | (Visick and Skoufos, 2001) |
| pVSV105 | Mobilizable vector; CmR | (Dunn et al., 2006) |
| pVSV208 | Mobilizable vector containing rfp; CmR | (Dunn et al., 2006) |
Figure 1.
Transcription of syp genes by rscS. After 24 h growth at 22°C in HMM, β-galactosidase activity levels were determined for syp::lacZ reporter strains carrying vector (pKV69), rscS (pLMS33), rscS1 (pKG11) or rscS2 (pKG13) (see Materials and Methods). Error bars represent the standard deviation of three replicate cultures. Numbers above the bars indicate the fold induction relative to the β-galactosidase activity produced by the vector-control strain.
Sequence analysis of rscS1 and rscS2 revealed mutations within or near the putative rscS ribosome binding site, while rscS1 contained a second, silent mutation at codon Leu25 (see Materials and Methods). Because the rscS1 allele exerted the greatest effect on syp transcription, we used the plasmid (pKG11) carrying this allele in all of our subsequent investigations. Semi-quantitative RT-PCR revealed no difference in the amount of rscS transcripts produced by pKG11 relative to pLMS33 (data not shown); therefore the mutations present in rscS1 do not appear to affect transcription of the gene and thus we hypothesize that they affect translation. We verified that syp induction and the resulting syp-dependent phenotypes induced by multi-copy rscS1 in culture (described below) did indeed depend upon the rscS gene carried by pKG11 (data not shown). Furthermore, because the rscS1 allele encodes wild-type protein, we anticipate that any observed effect on syp transcription is likely to represent a normal activity of RscS. Indeed, when we introduced multi-copy wild-type rscS into the syp reporter strains, we observed a small but reproducible increase in β-galactosidase activity (∼4-fold) relative to vector control (Fig. 1). Semi-quantitative RT-PCR experiments verified rscS1-induced expression of sypD and sypN in a wild-type background and demonstrated rscS1-dependent induction of at least two other genes within the cluster (sypB and sypE; Geszvain and Visick, unpublished data). Therefore, rscS1-mediated induction of the syp cluster is neither an artifact of the particular mutations present within rscS1 nor a consequence of the Tn10lacZ insertion present in the reporter strains.
From these data, we conclude that the sensor kinase RscS regulates expression of the syp cluster. Furthermore, these experiments have generated mutagenized alleles of rscS that can be employed to increase RscS expression, allowing us to investigate the role of the regulator in culture and during symbiosis.
RscS induces novel aggregation phenotypes in culture
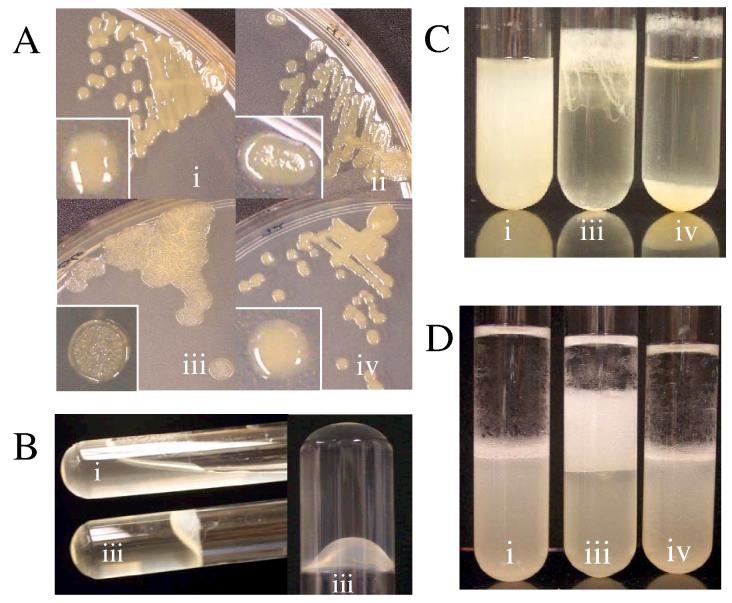
Given the large increase in syp transcription induced by rscS1 relative to wild-type rscS, we anticipated that phenotypic effects of RscS activity would be more readily observed in strains carrying rscS1. Indeed, we found that rscS1 induced a novel phenotype following prolonged incubation on complex solid medium. Normally, V. fischeri colonies are round, smooth and wet-appearing, as are colonies carrying the vector (Fig. 2Ai). Colonies containing rscS1, however, appeared dry, flat and wrinkled (Fig. 2Aiii). These wrinkled colonies formed a film that could be peeled off the plate in a continuous sheet (data not shown). This phenotype depended upon rscS1-mediated induction of the syp cluster. When a representative syp gene, sypN, which encodes one of six putative syp glycosyltransferases (Yip et al., 2005), was disrupted, rscS1 failed to induce wrinkling (Fig. 2Aiv). Wrinkling of the sypN mutant was restored upon complementation with the sypMNO operon (data not shown), further demonstrating the dependence on syp of this phenotype. Finally, wrinkling could be weakly induced by pLMS33 (Fig. 2Aii), indicating that this phenotype reflects the natural activity of RscS.
Figure 2.
Culture phenotypes of V. fischeri expressing rscS. (i) vector (pKV69) in ES114 (KV1844), (ii) rscS (pLMS33) in ES114 (KV1845), (iii) rscS1 (pKG11) in ES114 (KV1956), (iv) rscS1 (pKG11) in sypN (KV1992). (A) Growth phenotypes on solid media. Strains were streaked onto LBS with Tet and incubated at RT for 48 h. Inserts show a close-up view of a representative colony for each strain. (B) Pellicle formation in static cultures. Cultures were grown in HMM at room temperature (RT) for seven days, then photographed after inverting the tubes 90 (left panel) and 180 degrees (right panel) (C) Growth phenotypes in shaking cultures. Cultures were grown in HMM with shaking at 22 °C for 24 h. The experiment was performed in triplicate; representative results are shown. (D) Hydrophobicity of V. fischeri cultures. The organic solvent hexadecane was added to cultures that had been grown in HMM at 28 °C for 24 h. After vortexing, cells were allowed to partition into the lower, aqueous phase or the upper, hydrophobic phase. The experiment was performed in triplicate; representative results are shown.
Wrinkled colony morphology is frequently associated with increased cell aggregation phenotypes (Yildiz and Schoolnik, 1999); therefore, we screened cells for such phenotypes. First, we observed that after prolonged static growth in liquid Hepes minimal media (HMM), wild-type cells containing rscS1 formed a pellicle at the air/liquid interface that exhibited high tensile strength. In fact, the pellicle was sufficiently strong to retain the culture when the tube was completely inverted (Fig. 2B). Disruption of sypN resulted in the loss of this thick pellicle while complementation with sypMNO restored it (data not shown), demonstrating that this phenotype similarly depended upon syp. Second, during growth with shaking in HMM, we observed that rscS1 cultures formed stringy aggregates of cells (Fig. 2Ciii), which were difficult to disrupt even with vigorous vortexing. This phenotype suggested that RscS greatly increases the “stickiness” of the cell surface, allowing the cells to form aggregates even during agitation. In a sypN mutant, rscS1 did not induce the strings of aggregated cells seen in the wild-type background; however, the cells settled on the bottom of the tube (Fig. 2Civ) rather than remaining suspended as in the vector control (Fig. 2Ci). Together, these data demonstrate that RscS induces syp-dependent cell-cell aggregation.
In some organisms, such as Lactobacilli and Bifidobacterium longum, auto-aggregation and adhesion have been correlated with hydrophobic cell surfaces (Del Re et al., 2000; Wadstrom et al., 1987). Given the aggregation phenotypes induced by rscS1, we hypothesized that rscS1 might increase hydrophobicity of V. fischeri cells. When cultures of wild-type V. fischeri carrying vector were vortexed with an equal volume of hexadecane (an organic solvent), the bulk of the cells remained in the aqueous phase (Fig. 2Di; bottom layer). However, for cells carrying rscS1, a substantial proportion of the cells partitioned to the organic, hexadecane layer (Fig. 2Diii; top, “creamy” layer). The shift to the organic phase induced by rscS1 was largely lost in the sypN mutant (Fig. 2Div). These data suggest that RscS-induced expression of the syp locus results in an altered, hydrophobic cell surface that might contribute to the above aggregation phenotypes.
RscS promotes the formation of thick biofilm on a glass surface
To further understand the nature of the RscS-mediated aggregation phenotypes, we asked whether multi-copy rscS1 enhanced the ability of V. fischeri to form biofilms. We used confocal microscopy to quantitate the biomass of biofilms produced by GFP-labeled strains carrying either rscS1 or the vector control. These strains were grown statically in HMM medium in microtiter wells. To facilitate microscopy, glass cover slips were submerged vertically into medium immediately following inoculation, then removed either 6 or 30 h later. At 6 h, both vector control and rscS1-containing cells showed adherence to the glass surface. Even at this early time point, however, z-sections showed that the thickness of the adherent cells was substantially greater for rscS1-containing cells (∼18 μm) relative to the control (∼2 μm) (Fig. 3A and C). At 30 h, the difference between the two cell types was even more dramatic (Fig. 3B and D). Whereas the biofilm of vector control cells was approximately 7 μm thick, that of the rscs1-containing cells was approximately 45 μm. Furthermore, at this time point, an even thicker region (about 56 μm) could be seen for the rscS1 cells present at the air-liquid interface, where the pellicle eventually forms. These data suggest that multi-copy rscS1, while not necessary for V. fischeri cells to adhere to a solid surface, substantially promotes the events that lead to the formation of a thick biofilm.
Figure 3.
Confocal microscopy of biofilm formation on a glass surface. Development of V. fischeri biofilms formed on glass cover slips was visualized by confocal microscopy. Representative views of the xy plane and z sections were shown for GFP-labeled (A and B) vector-containing (KV2685) and (C and D) rscS1-expressing wild-type V. fischeri cells (KV2688) at 6 h and 30 h post inoculation. Arrows indicate the air-liquid interface.
RscS induces a polysaccharide extracellular matrix
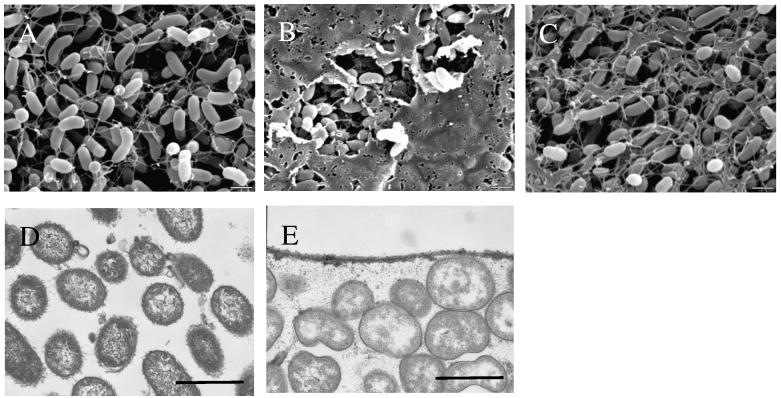
In nature, many bacteria reside in biofilms that are composed mainly of polysaccharides (Enos-Berlage and McCarter, 2000; Friedman and Kolter, 2004a, b; Jackson et al., 2004; Matsukawa and Greenberg, 2004; Solano et al., 2002; Yildiz and Schoolnik, 1999; Zogaj et al., 2001). To investigate the nature of rscS1-dependent biofilms, we first performed both scanning electron microscopy (SEM) and transmission electron microscopy (TEM) on colonies derived from rscS1-containing or vector control cells. SEM images revealed the existence of a smooth, sheet-like matrix produced by rscS1-expressing wild-type (Fig. 4B) but not vector control cells (Fig. 4A). Consistent with other culture phenotypes, the dense matrix induced by rscS1 in the wild type was largely absent in the sypN mutant strain (Fig. 4C), indicating that matrix formation required an intact syp cluster.
Figure 4.
SEM and TEM analysis of V. fischeri cells expressing rscS1. The production of an extracellular matrix was examined by SEM from (A) vector-control wild-type cells (KV1844), (B) rscS1-expressing wild-type cells (KV1956) and (C) rscS1-expressing sypN mutant cells (KV1992) and by TEM analysis of (D) vector control and (E) rscS1-containing wild type cells stained with ruthenium red. Bars represent 1 μm.
We therefore further characterized the nature of rscS1-dependent matrix by transmission electron microscopy (TEM) using ruthenium red, which is commonly used to detect acidic polysaccharides (Luft, 1971). Colonies were fixed and stained with ruthenium red and thin sections examined by TEM. The rscS1-containing wild type cells exhibited two types of ruthenium red-stained materials that were absent in vector control (Fig. 4D). First, an electron-dense layer was located at the colony surface which probably corresponds to sheet-like matrix detected by SEM (Fig. 4E. Second, electron-dense material was also observed between cells, indicative of an extracellular matrix. Together, these data suggest that rscS1 promotes the production of an extracellular matrix composed at least in part of acidic polysaccharides.
The observations by SEM and TEM of a matrix produced by syp-expressing cells supported our hypothesis that the syp cluster functions to modify the cell surface. To begin characterization of this matrix, we extracted surface-associated polysaccharide from smooth colonies and wrinkled colonies formed by vector-containing cells and rscS1-containing cells, respectively, using a capsule extraction protocol established for V. parahaemolyticus (Enos-Berlage and McCarter, 2000). Extractions from rscS1-containing wild-type cells yielded a high-molecular weight polysaccharide (Fig. 5A). This polysaccharide was absent from vector control and largely missing from rscS1-expressing sypN mutant extracts. These data support the hypothesis that the alteration induced by rscS1 involves increased surface polysaccharide.
Figure 5.
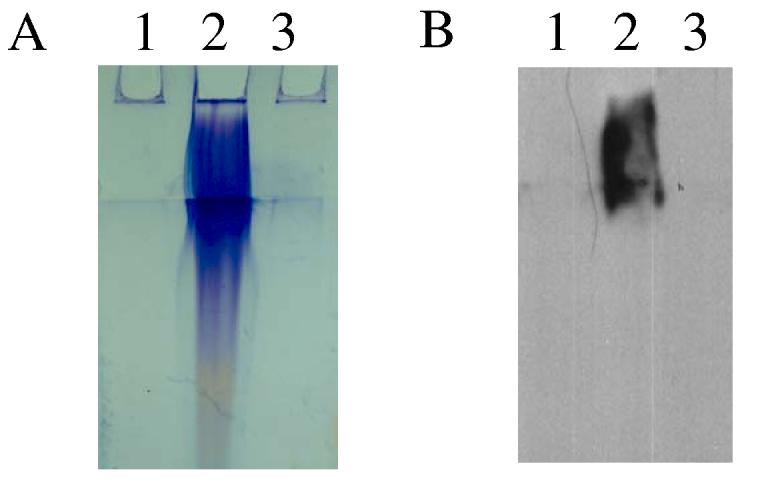
Polysaccharide extraction of V. fischeri expressing rscS1. Polysaccharide extracts were resolved on SDS-PAGE gel and (A) stained with Stains-All or (B) transferred onto membrane and probed with the lectin ConA. Lane 1: vector control (KV905), 2: rscS1-expressing wild type (KV1980) and 3: rscS1-containing sypN mutant (KV1960).
Purification of the large polysaccharide has proven difficult to date due to its insolubility. Therefore, to further characterize its nature, we performed lectin binding assays with a panel of seven biotinylated lectins. A single lectin, Concanavalin A (ConA), reacted strongly with the rscS1-induced polysaccharide (Fig. 5B), while the other six lectins tested exhibited no binding (data not shown). ConA is specific for glucose and α-linked mannose residues, indicating that one (or more) of these sugars is present in the surface-associated polysaccharide produced by rscS1-containing wild-type V. fischeri.
RscS enhances colonization efficiency by promoting aggregation outside the light organ
V. fischeri cells lacking RscS exhibit a significant colonization defect (Visick and Skoufos, 2001). We therefore wanted to determine the relevance to symbiosis, if any, of phenotypes associated with multi-copy expression of rscS1. Given the dramatic aggregation phenotypes induced in culture, we hypothesized that rscS1 could either enhance or impede colonization through its role in cell-cell aggregation. Preliminary experiments, in which juvenile squid were incubated with cells containing either vector or rscS1, indicated that the rscS1-containing cells initiated the symbiosis earlier than vector-containing cells (data not shown). These data suggested that rscS1 provided an advantage; however, the effect was difficult to quantify. As a more sensitive assay for colonization phenotypes, we performed competition assays. In these experiments, juvenile squid were inoculated with a mixture of two strains, one of which was marked with an erythromycin resistance (EmR) gene on the chromosome. As a control, we competed vector-containing cells that were either erythromycin sensitive (EmS) or resistant (EmR) against each other at a ratio of about 1:1 and screened bacteria recovered from the light organ after colonization for antibiotic resistance. The majority of these animals (8 of 11) were colonized with a roughly equal number of each strain (Fig. 6, white circles), demonstrating that the EmR cassette did not severely impact colonization fitness. Next, we inoculated squid with a mixture of vector-containing (EmR) cells and rscS1-containing (EmS) cells present at a ratio of about 2:1. All of the bacteria recovered from the light organ were EmS and therefore were derived from the rscS1-containing cells (Fig. 6, black circles). Reciprocal experiments in which the EmR marker was present in the rscS1-containing strain produced similar results: the rscS1-containing strain dominated in the light organ (data not shown). We conclude that multi-copy expression of rscS1 conferred a significant advantage to the cells during colonization.
Figure 6.
Colonization by strains carrying multi-copy rscS 1. The log of the relative competitive index (log RCI) for each juvenile squid in the experiment is plotted on the x-axis. Log RCI is calculated as the log of the fraction of EmR bacteria in the homogenate divided by the fraction of EmR bacteria in the inoculum. Each circle represents a single animal. Circles with arrows represent animals in which no EmR bacteria were present in the 100 to 200 colonies screened from the homogenate; therefore, the log RCI is below the limit of detection and the values plotted for these animals are approximate. In the competition between vector- and rscS1-containing cells, all of the colonies screened from the homogenate were the EmS rscS1-containing strain (black circles). The position of the circles on the y-axis is merely for spacing.
To understand the mechanism by which RscS controls colonization proficiency, we asked at which stage of colonization RscS exerts its influence. Specifically, we asked whether the rscS gene was required for the ability of V. fischeri cells to aggregate in squid-secreted mucus, one of the earliest known stages of symbiotic colonization. We hypothesized that disruption of rscS would result in a loss of aggregates, while multi-copy rscS1 would enhance aggregate formation.
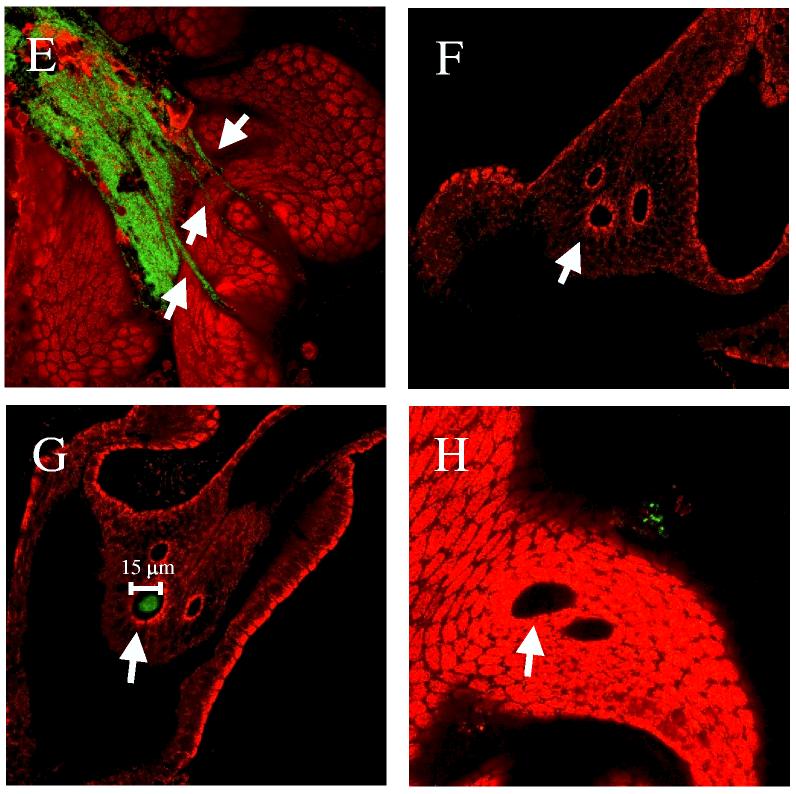
To determine if rscS mutants exhibit a defect in aggregation, we inoculated newly hatched, aposymbiotic squid with either wild-type or rscS mutant strains constitutively expressing GFP. At times ranging from 2 to 6 h post-inoculation, we dissected the animals and examined light organs for the presence of bacterial aggregates. Within 3 to 5 h of inoculation, animals exposed to GFP-labeled wild-type cells (KV1066 or KV1228) contained tightly-packed aggregates of bacteria (∼20 μm in diameter; Fig. 7A), either at or above a light organ pore, as observed previously (Nyholm et al., 2000). In contrast, animals inoculated with similar numbers of rscS mutant cells (KV1339 or KV1223) did not contain such aggregates (Fig. 7B); in fact, of 49 animals, 46 contained no visible GFP-labeled cells, while the remaining 3 animals showed only small clusters of about 3-5 cells. These data suggest that the defect of the rscS mutant in initiating symbiotic colonization stems from an inability of the cells to interact with one another and/or with mucus on the surface of the light organ.
Figure 7.
Aggregate formation on the light organ by V. fischeri. Newly hatched juvenile squid were inoculated with GFP-labeled bacteria. After 2 to 6 h, animals were stained with Cell Tracker Orange (red color) and the light organs examined by confocal microscopy. Representative images of aggregated V. fischeri cells at or near a light organ pore are shown. Animals were inoculated with (A) ES114 (KV1066), (B) rscS mutant cells (KV1339), (C) wild-type cells carrying the vector control (KV2685), (D and E) wild-type cells carrying rscS1 (KV2688), (F and G) rscS1-expressing sypN mutant cells (KV2689), and (H) sypN mutant cells carrying vector (KV2686). Arrows indicate pores of the light organ.
Next, we evaluated the effect of rscS1 on the ability of V. fischeri to aggregate in squid mucus. Whereas vector-control cells formed aggregates of approximately 10-20 μm in diameter, similar to what we and others have seen for wild-type cells (Fig. 7C; Nyholm et al., 2000), rscS1-containing wild-type cells formed substantially larger aggregates (50-200 μm in diameter; Fig. 7D and E). These data strongly suggest that the aggregation phenotypes induced by rscS1 in culture indeed reflect the role of rscS in symbiosis. In further support of this hypothesis, we found that multi-copy expression of rscS1 in a sypN mutant largely eliminated the production of large aggregates: 4 of 28 animals contained no detectable V. fischeri (Fig. 7F), 12 contained only a few cells and the remaining 12 animals had aggregates that were substantially smaller than those produced by wild-type cells expressing rscS1 (Fig. 7G; about wild type in size, 5-10 μm in diameter). These data demonstrate a role for syp in symbiotic aggregation. Furthermore, they are consistent with what we observed in culture: disruption of sypN reduces but does not eliminate aggregation induced by rscS1. Intriguingly, the sypN mutant carrying the vector control exhibited a more severe defect: of 18 animals examined, 15 contained no bacterial aggregates, while 3 contained only a few bacterial cells (Fig. 7H). These data further support a role for sypN in squid aggregation and suggest that rscS1 might partially compensate for the loss of sypN function.
Discussion
The process by which V. fischeri establishes itself as the sole species within the symbiotic organ of the squid E. scolopes is poorly understood. Here, we demonstrate that a key step, aggregation on the surface of the squid light organ, is facilitated by the action of the bacterial regulator, RscS. Our experiments show that RscS promotes aggregation in culture and initiation of symbiotic colonization in the squid light organ by regulating expression of genes that alter the cell surface. Our data thus provide an important correlation between laboratory-based biofilm studies and natural interactions that occur during the course of colonization and as well as a basis for further genetic analysis of the regulatory pathway.
Through the isolation of an allele (rscS1) with increased activity, we demonstrated that RscS regulates expression of the recently discovered syp gene cluster (Yip et al., 2005). Consistent with the predicted role of the syp genes in the production of an extracellular polysaccharide, induction of syp transcription by multi-copy rscS1 resulted in phenotypes associated with cell-cell aggregation in culture: wrinkled colony morphology, biofilm and pellicle formation, clumping in liquid culture, and increased hydrophobicity. In further support of this role, cells expressing rscS1 produce an extractable extracellular matrix visible by SEM and TEM and recognized by the lectin ConA. With this RscS-induced cell surface alteration, the ability of V. fischeri to colonize its host was greatly enhanced, as demonstrated by (i) competition experiments in which rscS1-containing cells dramatically out-competed vector-control cells and (ii) microscopy studies that uncovered the role of rscS in promoting bacterial aggregation on the surface of the light organ.
To determine the role of the syp cluster in the production of rscS1-dependent phenotypes, we disrupted a representative syp gene, sypN, and found that most of the phenotypes depended on sypN. We chose sypN because it is a structural gene (encoding a putative glycosyltransferase), the loss of which should impact primarily the product of the syp cluster, rather than producing pleiotropic effects as would deletion of a regulatory gene such as sypG. Indeed, mutations in other syp genes similarly disrupt rscS1-dependent phenotypes. For example, disruption of most of the syp genes either eliminated or reduced the formation of wrinkled colonies and pellicles (Yip and Visick, unpublished data). However, disruption of sypN (or other genes in the cluster) did not always render rscS1-containing cells indistinguishable from vector control cells. For example, shaking liquid cultures of rscS1-containing sypN mutants exhibited a cell settling phenotype unlike vector control cells (Fig. 2C). This phenotype may result either from the activities of the remaining intact syp genes or from RscS-dependent regulation of genes outside of the syp cluster.
It is not yet clear how RscS exerts its impact on syp. As a sensor kinase, it is predicted to function by signaling to a response regulator, and the syp-encoded response regulator SypG seems a prime candidate for this target regulator. Both regulators control syp transcription (Yip et al., 2005; Fig. 1) and rscS1-dependent syp induction requires SypG (Geszvain and Visick, unpublished results). However, induction of the syp cluster through multi-copy expression of sypG produces a different array of culture phenotypes than rscS1 produces. First, multi-copy sypG induces substantial adherence to the test tube when cells are grown with shaking (Yip et al., 2005), while rscS1 does not (data not shown). Second, unlike rscS1, multi-copy sypG fails to induce wrinkled colonies (Hussa and Visick, unpublished data). Although cognate response regulators and sensor kinases can induce distinct phenotypes (e.g. Yap et al., 2005), it is also possible that these two regulators control the expression of distinct subsets of genes. The syp cluster includes two additional two-component regulators, a putative sensor kinase (SypF) and a putative response regulator (SypE) that is not predicted to bind DNA, suggesting that syp regulation may be complex.
Why is a sensor kinase necessary for one of the earliest steps of colonization? Presumably RscS recognizes an environmental signal and subsequently induces syp transcription such that surface modification can occur prior to aggregation. Potentially, a component of the seawater itself could serve as the signal for RscS. However, our preliminary experiments have provided no evidence for seawater-mediated induction of syp transcription (Geszvain and Visick, unpublished data). Therefore, it is likely that the environmental signal is found in proximity to the squid but external to it.
The work presented here suggests that the syp cluster functions to modify the cell surface. Biotinylated lectin assays revealed that the surface polysaccharides produced by syp likely contain glucose or α-linked mannose residues. Intriguingly, the rscS1-induced matrix promotes the formation of a strong pellicle at the air-liquid interface similar to the cellulose-based pellicle produced by Gluconacetobacter xylinum (Ross et al., 1991). Cellulose biosynthetic genes are present in the V. fischeri genome [(Ruby et al., 2005); Visick, unpublished data]; however, our preliminary data suggest that the V. fischeri matrix is not composed of cellulose, as the addition of cellulase did not dissolve it and rscS1-containing cells failed to bind either Congo Red or calcofluor (Yip and Visick, unpublished data). It is worth noting that addition of a mannose analog to the seawater interfered with the ability of V. fischeri to initiate the symbiosis (McFall-Ngai et al., 1998), supporting a role for a mannose-based interaction between the bacteria and their host, or perhaps among the bacteria, during colonization.
The increased surface hydrophobicity we detected in cells expressing rscS1 may be relevant to the function of RscS and the syp cluster in symbiosis. In a number of systems, bacterial hydrophobicity has been correlated with adherence to plant and animal tissues (reviewed in (Doyle, 2000)). In the oral bacterium Streptococcus gordonii, fibrils composed of CshA render the cell surface hydrophobic and are required for colonization of the oral cavity in mice (McNab et al., 1999). In addition, the rugose phase variant of V. cholerae is more hydrophobic than the smooth variant due to the production of the VPSElTor exopolysaccharide (Yildiz et al., 2004). For V. fischeri, it is possible that hydrophobic interactions among the bacterial cells, between the bacterium and its host, or both, could provide a substantial advantage, since the initial interactions occur in the context of an aqueous seawater environment. Indeed, it has been estimated that seawater flows in and out of the mantle cavity of the juvenile squid every 0.3 seconds (Visick and McFall-Ngai, 2000), creating a difficult environment for specific, receptor-ligand-type interactions to occur between the two organisms.
After aggregation, V. fischeri cells must migrate through ducts into the light organ crypts in order to establish the symbiotic association. This migration was apparently not impaired in rscS1 cells, despite the increased cell-cell aggregation on the light organ surface that could potentially impair subsequent cell movement. However, in our aggregation experiments we detected rscS1-containing bacteria beginning to move into the ducts in continuous streams of cells (Fig. 7E). This streaming of the bacteria into the ducts has also been observed with wild-type cells (Nyholm et al., 2000), indicating that colonization by rscS1-containing cells proceeds normally after aggregate formation. Indeed, the presence of rscS1 does not impair the migration of V. fischeri through complex soft agar media (data not shown), supporting the conclusion that these cells can freely migrate away from the aggregate.
The ability of bacteria to form biofilm communities within their host organisms is thought to play a central role in both pathogenesis and symbiosis, by rendering the bacteria resistant to antimicrobial agents and enhancing adherence to host tissues, such as infection of lungs of cystic fibrosis patients by P. aeruginosa (Singh et al., 2000). Mutants which decrease the ability of pathogenic bacteria to form biofilms in culture show a concomitant decrease in virulence in animal models of infection (Kulasakara et al., 2006; Paranjpye and Strom, 2005). However, in many cases, these studies did not examine loss of biofilm within the host. Here, we report a mutualistic symbiosis where biofilm production in culture is directly linked to biofilm production in the host. Therefore, our work provides critical support for the importance of bacterial biofilm formation during the establishment of a natural bacteria-animal association.
Experimental Procedures
Strains, plasmids and media
Strains used in this study are listed in Table 1. V. fischeri strains constructed in this work were generated by conjugation as previously described (DeLoney et al., 2002; Visick and Skoufos, 2001). A bacterial isolate from E. scolopes, ES114 (Boettcher and Ruby, 1990), and its rifampicin-resistant derivative, ESR1 (Graf et al., 1994), were the symbiosis-competent parent strains used in this study. The E. coli strains DH5α, TOP10 (Invitrogen, Carlsbad, CA) and CC118λpir (Herrero et al., 1990) were used as hosts for cloning and conjugation. V. fischeri strains were grown in complex media [seawater tryptone, SWT (Yip et al., 2005) or LBS (Graf et al., 1994; Stabb et al., 2001)] or in Hepes minimal medium [HMM (Ruby and Nealson, 1977) containing 0.3% Casamino acids and 0.2% glucose (Yip et al., 2005)]. The following antibiotics were added, as needed, to the final concentrations indicated: chloramphenicol (Cm), 5 μg ml−1; erythromycin (Em) 5 μg ml−1; and tetracycline (Tet), 5 μg ml−1 in LBS, 30 μg ml−1 in SWT and HMM. Agar was added to a final concentration of 1.5% for solid media. Juvenile E. scolopes were maintained in artificial seawater (Instant Ocean; Aquarium Systems, Mentor, OH). In some experiments, natural seawater collected in E. scolopes-free waters off the coast of Oahu, Hawaii was used. This seawater does not contain sufficient V. fischeri cells to colonize the squid. For motility assays, bacteria were grown at 28°C to mid-exponential phase and inoculated onto SWT, TBS or TB-SW soft agar plates (DeLoney-Marino et al., 2003).
Molecular techniques
Plasmids were constructed using standard molecular biology techniques, with restriction and modifying enzymes obtained from New England Biolabs (Beverly, MA) or Promega (Madison, WI). Plasmids used or constructed in this study are shown in Table 1.
Random mutagenesis of rscS
A plasmid encoding wild-type rscS, pLMS33, was introduced into mutator strain CC130 [mutD5; (Manoil and Beckwith, 1985; Schaaper, 1988)] by transformation. Five independent transformations were performed to obtain a total of approximately 24,000 single colonies on selective media. Colonies were scraped off the plates into six separate pools; each pool of mutagenized plasmids was then conjugated into syp reporter strains KV1601 and KV1635 (Table 1). Transconjugants were selected for the presence of the plasmid and screened for induction of β-galactosidase expression on SWT Tet containing 80 μg ml−1 X-gal (Molecular Probes). After incubation at RT for two days, approximately 200 blue (i.e., syp-expressing) colonies were found among the ∼200,000 colonies plated. Plasmids were isolated from the six colonies that exhibited the greatest amount of syp expression (based on the intensity of blue color), the rscS gene from these plasmids was subcloned into unmutagenized vector (pKV69, Table 1) and the resulting constructs were examined again for their ability to induce syp transcription. Two subcloned rscS alleles (rscS1 and rscS2, encoded on plasmids pKG11 and pKG13, respectively) retained the ability to induce syp expression.
Sequencing
The mutations in rscS1 and rscS2 were identified by automated sequencing (MWG Biotech, Inc. Highpoint, NC) of the rscS gene present on pKG11 and pKG13, respectively, using primers specific to rscS. The rscS1 allele contains a C to T transition in the 6th position of the putative ribosome binding site (AGGAGC to AGGAGT) and a C to T transition at the wobble position within a Leu codon at amino acid 25 (CTC to CTT). The rscS2 allele contains a T to C transition immediately 3' of the ribosome binding site (AGGAGCT to AGGAGCC).
β-galactosidase assay
Strains were grown in HMM with Tet at 22°C overnight, then subcultured into fresh medium and grown for 24 h. Aliquots (1 ml) were removed and β-galactosidase activity in triplicate was assayed as described (Miller, 1972). Protein concentration was measured as described (Lowry et al., 1951).
Hydrophobicity assay
Strains were grown in HMM with Tet at 28°C overnight, subcultured into fresh medium and grown for 24 h. Next, an equal volume of hexadecane (Acros Organics, New Jersey) was added to the culture tubes and vortexed until the suspension was homogeneous (at least 1 min). Then, the hydrophobic (hexadecane) and hydrophilic fractions were allowed to separate. Partitioning of the cells to the hydrophobic layer was visualized as an increase in turbidity of the top layer (Araujo et al., 1994; Rosenberg et al., 1980).
Confocal microscopy
Cells expressing fluorescent proteins (RFP or GFP) were grown statically in HMM in 12 well microtiter plates with glass cover slips submerged partially into the culture medium. Cover slips were incubated with bacteria at RT for up to 30 h and removed at specific time points for biofilm examination. A Zeiss LSM 510 confocal microscope (65X objective) was used to collect xy plane and z sections (xz and yz plane) images of the biofilms. Images were prepared using the Zeiss LSM Image Browser software.
Scanning electron microscopy
Colonies grown on cellulose membranes overlaid on LBS tet plates, were prepared as described (Yildiz et al., 2001) with the following modifications: cells were fixed in 2% glutaraldehyde in 0.45 M cacodylate buffer (pH 7.3), washed with 0.45 M cacodylate buffer, postfixed with 1% osmium tetroxide for 1 h and washed again with cacodylate buffer. The samples were subjected to dehydration with ethanol, critical point drying and sputter-coating with gold-palladium. Finally, samples were examined with a scanning electron microscope (JEOL JSM-840A).
Transmission electron microscopy
Colonies were grown on LBS tet plates at RT for 3 day and fixed as described (Luft, 1971; Patterson et al., 1975) with the following modifications: colonies were fixed overnight in a solution containing 1.2% glutaraldehyde, 0.45 M cacodylate buffer (pH 7.3) and 0.1% (wt/vol) ruthenium red. After fixation, cells were washed, postfixed with 1% osmium tetroxide for 1 h, washed again and subjected to serial dehydration with ethanol. Samples were embedded in resin, thin-sectioned and stained with uranyl acetate and lead citrate. Finally, the samples were examined with a transmission electron microscope (Hitachi H-600) operating at an accelerating voltage of 75 kV.
Polysaccharide extraction
Polysaccharides were extracted as described by Enos-Berlage and McCarter (2000) with the following modifications: Bacterial strains were grown in a lawn on LBS tet until rscS1-containing WT cells became wrinkled. Cells scrapped off of the plates were resuspended in 5 ml of phosphate-buffered saline (20 mM sodium phosphate, 100 mM sodium chloride, pH 7.3). Cell suspensions were shaken at 28°C for 90 min, vortexed and shaken for another 90 min. Cultures were centrifuged at 10,000 × g for 15 min to remove cells and cellular debris. Supernatants were transferred to fresh tubes and incubated for 3h at 37°C with MgCl2, RNase (Sigma, St. Louis, MO) and RQ1 DNase (Promega, Madison, WI) at final concentrations of 10 mM, 50 μg/ml and 50 μg/ml, respectively. Proteinase K (Fisher, Fair Lawn, NJ) was subsequently added to a final concentration of 200 μg/ml and the samples were incubated at 37°C overnight. Polysaccharides were extracted using phenol/chloroform and precipitated with ethanol. Samples were boiled in loading buffer containing 10% β-mercaptoethanol prior to gel electrophoresis. However, only a portion of the samples were solubilized and thus loaded onto the gels for Stains-All (Kelley and Parker, 1981) staining and lectin binding assays.
Lectin binding assay
Polysaccharide extracts were evaluated using a panel of biotinylated lectins. Extracts were subjected to electrophoresis on a Tris glycine-polyacrylamide (14%) gel and then transferred onto a polyvinylidene fluoride (PVDF) membrane (Millipore Corp, Bedford, MA). The membrane was blocked with Tris-buffered saline containing 0.2% Tween (TBST) and 2% (vol/vol) bovine serum albumin (BSA) for 1 h. After blocking, the membrane was incubated in TBST for 45 min with 1 μg/ml (final concentration) of one of seven biotinylated lectins in the biotinylated lectin kit I, which contains ConA, DBA, PNA, RCA120, SBA, UEA I and WGA (Vector Lab., Burlingame, CA). Then, the membrane was washed with TBST three times for 10 min each and subsequently incubated with horseradish peroxidase-conjugated streptavidin (Jackson ImmunoResearch, West Grove, PA) for 30 min. Reactive carbohydrates were detected by incubating the membrane with an equal volume of solution 1 (2.5 mM luminal, 0.4 mM p-coumaric acid and 100 mM Tris pH 8.5) and solution 2 (0.02% H2O2, 100 mM Tris pH 8.5).
Colonization assays
Both single-strain and competition assays were performed as described (Lee and Ruby, 1994; Ruby, 1996; Visick and Skoufos, 2001). For the competition assays, vector and rscS1 were both introduced into V. fischeri strain KV1421, which is marked with an EmR cassette inserted at the Tn7 attachment site (DeLoney et al., 2002). Strains used in the colonization experiments were grown in SWT with Tet for about 4 h before inoculation; these conditions did not promote significant aggregation or growth defects in culture. These strains also exhibited no motility defect when tested for their ability to migrate through complex soft agar media (DeLoney-Marino et al., 2003). To test for a competitive defect, we inoculated 12 juveniles with a mixture of the EmR vector strain (KV2435, 1750 CFU/ml) and the EmS rscS1 strain (KV1956, 750 CFU/ml) (a ratio of 2.3:1). In a control experiment, 11 juveniles were inoculated with a mixture of cells with vector in the EmR strain background (KV2435, 2477 CFU/ml) and vector in the EmS strain (KV1844, 3283 CFU/ml) (a ratio of 0.75:1). Approximately 20 h after inoculation, the individual squid were monitored for luminescence, rinsed with bacteria-free artificial seawater and frozen at −80°C. The following day, the squid were homogenized and plated on SWT. In these experiments, all animals luminesced and were colonized to at least 104 CFU/squid. Between 100 and 200 colonies from each squid homogenate were patched onto LBS with Em to determine the ratio of EmS to EmR bacteria within the light organ.
Squid aggregation assays
Log-phase cells (OD600 0.3 to 0.6) were grown in 2 ml SWT medium at 28°C. Bacterial cells were then inoculated into unfiltered natural seawater or filtered artificial seawater at concentrations of between 106 and 108 cells per ml. Juvenile squid were then placed into inoculated seawater and the two organisms were allowed to associate at RT for between 2 and 6 h prior to dissection. At various times, the juvenile squid were removed to vials containing 2 ml filter-sterilized seawater and 5 μM of the counter-stain, Celltracker Orange (Molecular Probes, Eugene, Oregon). The animals were then anesthetized in seawater or filtered artificial seawater containing 2% ethanol. Each squid was placed ventral side up on a depression well slide and dissected to remove the mantle and funnel and expose the light organ. Fluorescently-labeled light organs (red) and GFP-labeled (green) bacteria were viewed using a Zeiss LSM 510 confocal microscope.
Acknowledgments
We thank Ned Ruby and Margaret McFall-Ngai for the use of E. scolopes juveniles and a confocal microscope and Eric Stabb for generous donation of plasmid vectors. We also thank Ned Ruby and Cheryl Whistler for helpful advice and Jon Visick, Joerg Graf and members of our lab for critical reading of our manuscript. This work was supported by NIH grant GM59690 awarded to K.L.V., by the Estate of William G. Potts in support of medical research at the Stritch School of Medicine at Loyola University Chicago, by the National Science Foundation under a Research Fellowship in Microbial Biology awarded in 2001 to C.R.D-M, and by the NIH under the Ruth L. Kirschstein National Research Service Award 1 F32 GM073523 from the NIGMS awarded to K.G.
References
- Araujo RS, Robleto EA, Handelsman J. A Hydrophobic mutant of Rhizobium etli altered in nodulation competitiveness and growth in the rhizosphere. Appl Environ Microbiol. 1994;60:1430–1436. doi: 10.1128/aem.60.5.1430-1436.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher KJ, Ruby EG. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol. 1990;172:3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Re B, Sgorbati B, Miglioli M, Palenzona D. Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum. Lett Appl Microbiol. 2000;31:438–442. doi: 10.1046/j.1365-2672.2000.00845.x. [DOI] [PubMed] [Google Scholar]
- DeLoney CR, Bartley TM, Visick KL. Role for phosphoglucomutase in Vibrio fischeri-Euprymna scolopes symbiosis. J Bacteriol. 2002;184:5121–5129. doi: 10.1128/JB.184.18.5121-5129.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLoney-Marino CR, Wolfe AJ, Visick KL. Chemoattraction of Vibrio fischeri to serine, nucleosides, and N-acetylneuraminic acid, a component of squid light-organ mucus. Appl Environ Microbiol. 2003;69:7527–7530. doi: 10.1128/AEM.69.12.7527-7530.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle RJ. Contribution of the hydrophobic effect to microbial infection. Microbes Infect. 2000;2:391–400. doi: 10.1016/s1286-4579(00)00328-2. [DOI] [PubMed] [Google Scholar]
- Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Microbiol. 2006;72:802–810. doi: 10.1128/AEM.72.1.802-810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enos-Berlage JL, McCarter LL. Relation of capsular polysaccharide production and colonial cell organization to colony morphology in Vibrio parahaemolyticus. J Bacteriol. 2000;182:5513–5520. doi: 10.1128/jb.182.19.5513-5520.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L, Kolter R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol. 2004a;186:4457–4465. doi: 10.1128/JB.186.14.4457-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol. 2004b;51:675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- Graf J, Dunlap PV, Ruby EG. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol. 1994;176:6986–6991. doi: 10.1128/jb.176.22.6986-6991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero M, DeLorenzo V, Timmis KN. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung DT, Zhu J, Sturtevant D, Mekalanos JJ. Bile acids stimulate biofilm formation in Vibrio cholerae. Mol Microbiol. 2006;59:193–201. doi: 10.1111/j.1365-2958.2005.04846.x. [DOI] [PubMed] [Google Scholar]
- Jackson KD, Starkey M, Kremer S, Parsek MR, Wozniak DJ. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J Bacteriol. 2004;186:4466–4475. doi: 10.1128/JB.186.14.4466-4475.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley JT, Parker CD. Identification and preliminary characterization of Vibrio cholerae outer membrane proteins. J Bacteriol. 1981;145:1018–1024. doi: 10.1128/jb.145.2.1018-1024.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirn TJ, Lafferty MJ, Sandoe CM, Taylor RK. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol Microbiol. 2000;35:896–910. doi: 10.1046/j.1365-2958.2000.01764.x. [DOI] [PubMed] [Google Scholar]
- Kulasakara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci U S A. 2006;103:2839–2844. doi: 10.1073/pnas.0511090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Ruby EG. Competition between Vibrio fischeri strains during initiation and maintenance of a light organ symbiosis. J Bacteriol. 1994;176:1985–1991. doi: 10.1128/jb.176.7.1985-1991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Luft JH. Ruthenium red and violet. I. Chemistry, purification, methods of use for electron microscopy and mechanism of action. Anat Rec. 1971;171:347–368. doi: 10.1002/ar.1091710302. [DOI] [PubMed] [Google Scholar]
- Manoil C, Beckwith J. Tn phoA: a transposon probe for protein export signals. Proc Natl Acad Sci U S A. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukawa M, Greenberg EP. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J Bacteriol. 2004;186:4449–4456. doi: 10.1128/JB.186.14.4449-4456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann J, Stabb EV, Millikan DS, Ruby EG. Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl Environ Microbiol. 2003;69:5928–5934. doi: 10.1128/AEM.69.10.5928-5934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M, Brennan C, Lamarcq L. Mannose adhesin-glycan interactions in the Euprymna scolopes-Vibrio fischeri symbiosis. Plenum Press; New York: 1998. [Google Scholar]
- McNab R, Forbes H, Handley PS, Loach DM, Tannock GW, Jenkinson HF. Cell wall-anchored CshA polypeptide (259 kilodaltons) in Streptococcus gordonii forms surface fibrils that confer hydrophobic and adhesive properties. J Bacteriol. 1999;181:3087–3095. doi: 10.1128/jb.181.10.3087-3095.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1972. [Google Scholar]
- Millikan DS, Ruby EG. Alterations in Vibrio fischeri motility correlate with a delay in symbiosis initiation and are associated with additional symbiotic colonization defects. Appl Environ Microbiol. 2002;68:2519–2528. doi: 10.1128/AEM.68.5.2519-2528.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millikan DS, Ruby EG. FlrA, a sigma54-dependent transcriptional activator in Vibrio fischeri, is required for motility and symbiotic light-organ colonization. J Bacteriol. 2003;185:3547–3557. doi: 10.1128/JB.185.12.3547-3557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. Establishment of an animal-bacterial association: recruiting symbiotic Vibrios from the environment. Proc Natl Acad Sci U S A. 2000;97:10231–10235. doi: 10.1073/pnas.97.18.10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm S, McFall-Ngai MJ. Dominance of Vibrio fischeri in secreted mucus outside the light organ of Euprymna scolopes: the first site of symbiont specificity. Appl Environ Microbiol. 2003 Jul;69(7):3932–7. doi: 10.1128/AEM.69.7.3932-3937.2003. 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai M. The winnowing: establishing the squid-Vibrio symbiosis. Nat Rev Microbiol. 2004;2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- Paranjpye RN, Strom MS. A Vibrio vulnificus type IV pilin contributes to biofilm formation, adherence to epithelial cells, and virulence. Infect Immun. 2005;73:1411–1422. doi: 10.1128/IAI.73.3.1411-1422.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson H, Irvin R, Costerton JW, Cheng KJ. Ultrastructure and adhesion properties of Ruminococcus albus. J Bacteriol. 1975;122:278–287. doi: 10.1128/jb.122.1.278-287.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U, Sierralta WD, Eriksson K, Normark S. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol Microbiol. 1998;28:249–264. doi: 10.1046/j.1365-2958.1998.00791.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg M, Gutnick D, Rosenberg E. Adherence of bacteria to hydrocarbons: A simple method for measuring cell-surface hydrophobicity. FEMS Micro Let. 1980;9:29–33. [Google Scholar]
- Ross P, Mayer R, Benziman M. Cellulose biosynthesis and function in bacteria. Microbiol Rev. 1991;55:35–58. doi: 10.1128/mr.55.1.35-58.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG, Nealson KH. Pyruvate production and excretion by the luminous marine bacteria. Appl Environ Microbiol. 1977;34:164–169. doi: 10.1128/aem.34.2.164-169.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby EG. Lessons from a cooperative, bacterial-animal association: the Vibrio fischeri-Euprymna scolopes light organ symbiosis. Annu Rev Microbiol. 1996;50:591–624. doi: 10.1146/annurev.micro.50.1.591. [DOI] [PubMed] [Google Scholar]
- Ruby EG, Urbanowski M, Campbell J, Dunn A, Faini M, Gunsalus R, Lostroh P, Lupp C, McCann J, Millikan D, Schaefer A, Stabb E, Stevens A, Visick K, Whistler C, Greenberg EP. Complete genome sequence of Vibrio fischeri: A symbiotic bacterium with pathogenic congeners. Proc Natl Acad Sci U S A. 2005;102:3004–3009. doi: 10.1073/pnas.0409900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper RM. Mechanisms of mutagenesis in the Escherichia coli mutator mutD5: role of DNA mismatch repair. Proc Natl Acad Sci U S A. 1988;85:8126–8130. doi: 10.1073/pnas.85.21.8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- Solano C, Garcia B, Valle J, Berasain C, Ghigo JM, Gamazo C, Lasa I. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol Microbiol. 2002;43:793–808. doi: 10.1046/j.1365-2958.2002.02802.x. [DOI] [PubMed] [Google Scholar]
- Sonnenburg JL, Angenent LT, Gordon JI. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat Immunol. 2004;5:569–573. doi: 10.1038/ni1079. [DOI] [PubMed] [Google Scholar]
- Stabb EV, Reich KA, Ruby EG. Vibrio fischeri genes hvnA and hvnB encode secreted NAD(+)-glycohydrolases. J Bacteriol. 2001;183:309–317. doi: 10.1128/JB.183.1.309-317.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabb EV, Ruby EG. New RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 2002;358:413–426. doi: 10.1016/s0076-6879(02)58106-4. [DOI] [PubMed] [Google Scholar]
- Visick KL, McFall-Ngai MJ. An exclusive contract: specificity in the Vibrio fischeri-Euprymna scolopes partnership. J Bacteriol. 2000;182:1779–1787. doi: 10.1128/jb.182.7.1779-1787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KL, Skoufos LM. Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J Bacteriol. 2001;183:835–842. doi: 10.1128/JB.183.3.835-842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadstrom T, Andersson K, Sydow M, Axelsson L, Lindgren S, Gullmar B. Surface properties of lactobacilli isolated from the small intestine of pigs. J Appl Bacteriol. 1987;62:513–520. doi: 10.1111/j.1365-2672.1987.tb02683.x. [DOI] [PubMed] [Google Scholar]
- Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- Woodcock DM, Crowther PJ, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith SS, Michael MZ, Graham MW. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 1989;17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap MN, Yang CH, Barak JD, Jahn CE, Charkowski AO. The Erwinia chrysanthemi type III secretion system is required for multicellular behavior. J Bacteriol. 2005;187:639–648. doi: 10.1128/JB.187.2.639-648.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Schoolnik GK. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci U S A. 1999;96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Dolganov NA, Schoolnik GK. VpsR, a Member of the Response Regulators of the Two-Component Regulatory Systems, Is Required for Expression of vps Biosynthesis Genes and EPS(ETr)-Associated Phenotypes in Vibrio cholerae O1 El Tor. J Bacteriol. 2001;183:1716–1726. doi: 10.1128/JB.183.5.1716-1726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz FH, Liu XS, Heydorn A, Schoolnik GK. Molecular analysis of rugosity in a Vibrio cholerae O1 El Tor phase variant. Mol Microbiol. 2004;53:497–515. doi: 10.1111/j.1365-2958.2004.04154.x. [DOI] [PubMed] [Google Scholar]
- Yip ES, Grublesky BT, Hussa EA, Visick KL. A novel, conserved cluster of genes promotes symbiotic colonization and sigma-dependent biofilm formation by Vibrio fischeri. Mol Microbiol. 2005;57:1485–1498. doi: 10.1111/j.1365-2958.2005.04784.x. [DOI] [PubMed] [Google Scholar]
- Zogaj X, Nimtz M, Rohde M, Bokranz W, Romling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol. 2001;39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]