Abstract
Background
Enterohemorrhagic E. coli (EHEC), a subgroup of Shiga toxin (Stx) producing E. coli (STEC), may cause severe enteritis and hemolytic uremic syndrome (HUS) and is transmitted orally via contaminated foods or from person to person. The infectious dose is known to be very low, which requires most of the bacteria to survive the gastric acid barrier. Acid resistance therefore is an important mechanism of EHEC virulence. It should also be a relevant characteristic of E. coli strains used for therapeutic purposes such as the probiotic E. coli Nissle 1917 (EcN). In E. coli and related enteric bacteria it has been extensively demonstrated, that the alternative sigma factor σS, encoded by the rpoS gene, acts as a master regulator mediating resistance to various environmental stress factors.
Methods
Using rpoS deletion mutants of a highly virulent EHEC O26:H11 patient isolate and the sequenced prototype EHEC EDL933 (ATCC 700927) of serotype O157:H7 we investigated the impact of a functional rpoS gene for orchestrating a satisfactory response to acid stress in these strains. We then functionally characterized rpoS of probiotic EcN and five rpoS genes selected from STEC isolates pre-investigated for acid resistance.
Results
First, we found out that ATCC isolate 700927 of EHEC EDL933 has a point mutation in rpoS, not present in the published sequence, leading to a premature stop codon. Moreover, to our surprise, one STEC strain as well as EcN was acid sensitive in our test environment, although their cloned rpoS genes could effectively complement acid sensitivity of an rpoS deletion mutant.
Conclusion
The attenuation of sequenced EHEC EDL933 might be of importance for anyone planning to do either in vitro or in vivo studies with this prototype strain. Furthermore our data supports recently published observations, that individual E. coli isolates are able to significantly modulate their acid resistance phenotype independent of their rpoS genotype.
Background
Enterohemorrhagic Escherichia coli (EHEC), a subgroup of Shiga toxin (Stx) producing E. coli (STEC), are enteric pathogens frequently causing severe illness in humans. EHEC infection may lead to non-bloody and bloody diarrhea and most dangerously, the extraintestinal complication hemolytic uremic syndrome (HUS) [1,2]. In order to cause gastrointestinal disease, bacteria must pass through the acidic gastric barrier. When taking into account the apparent low infectious dose of enterohemorrhagic E. coli, which may be as little as 100 viable organisms [3], it becomes obvious that acid resistance is an important virulence trait of EHEC. Investigation of the complex mechanisms conferring acid tolerance onto E. coli has revealed many new insights in the last years. Currently, four different acid stress protection systems are being discussed. At least three of these are controlled by σS, an alternative sigma subunit of RNA polymerase encoded by the rpoS gene [4]. σS, which is seen as a master regulator of general stress response, confers promoter specificity to the core RNA polymerase and is activated under a variety of stress conditions, as well as during stationary phase [5-7]. The glutamate-dependent acid resistance (GDAR) system is believed to provide best protection for bacterial cells below pH 3 [8]. Bhagwat et al. have recently reported, that in natural populations of pathogenic E. coli mutations in gadE exist which, in addition to mutant alleles of rpoS, may contribute to varying acid resistance phenotypes of EHEC [9]. gadE encodes the GadE protein, a regulatory molecule of the GDAR system [10].
In 1994 Small et al. already reported that the growth pH is important for expression of acid resistance in E. coli [11]. Waterman et al. who have investigated a large set of STEC for acid resistance [12], identified defective rpoS genes as cause for loss of acid resistance in individual E. coli isolates. Recently it was demonstrated by King et al., that modulation of genome usage enables regulatory diversity which contributes to strain variation in E. coli [13]. A similar observation was reported by Bhagwat et al., who investigated stress tolerance of EHEC and could observe a functional heterogeneity of RpoS [14].
Our study was initially focused on analyzing the role of σS in acid stress behavior of two different EHEC isolates used as model organisms in our laboratory: the sequenced prototype O157:H7 EHEC EDL933 (ATCC 700927) [15] and a very well characterized EHEC O26:H11 isolate from a HUS patient [16,17]. We expected these results to improve the interpretation of data obtained from in vitro and in vivo virulence experiments investigating pathogen-host interactions. Using homologous recombination and suicide vector technique we constructed unmarked isogenic rpoS deletion mutants of both strains. Resistance of wild type organisms and their mutants towards acidic conditions at pH 2.5 and 1.5 were tested as described by Lange et al. [18] with slight modifications. While the EHEC O26:H11 patient isolate was highly dependent on a functional rpoS gene for an adequate response to acid stress, surprisingly, deletion of rpoS in EHEC EDL933 had no measurable effect when compared to its wild type strain. Further investigation revealed a point mutation in the EHEC EDL933 rpoS gene which leads to a TAA stop codon being responsible for this phenotype.
We then evaluated another 39 isolates of human and porcine origin and functionally characterized a subgroup of five selected rpoS genes. Surprisingly, σS activity of STEC ED-68 [19,20] appeared to be dependent on a yet unknown regulatory mechanism that modulated its activity. In the following, we could also observe a similar phenomenon with the well characterized probiotic E. coli Nissle 1917 [21-23].
Results
Construction and confirmation of unmarked isogenic rpoS deletion mutants
After PCR screening of potential mutants one rpoS negative isolate of each EHEC wild type strain, EDL933a and 126814 (Table 1), was subjected to further testing by Southern blotting. They both showed DNA fragments of the expected size, after restriction digest of their genomic DNA with either XmnI or StuI [see Additional file 1] thereby indicating the correct insertion of the rpoS deletion mutation into the genome of the two EHEC strains. The mutants were termed E. coli MHH933-5 and MHH126-5 respectively. Additionally, the mutation was confirmed by DNA sequencing of a PCR product generated with primers RpoS 3 and RpoS 4. Antibiotic resistance patterns of the mutants did not differ from their parental strains. Biochemical reaction profiles of all strains, as determined with API 20 E strips, were quite similar [see Additional file 2]. EHEC EDL933a and its mutant, which both could not ferment sorbitol, were identified as 89.6 % E. coli according to their API profile index "5144172". Sorbitol positive EHEC 126814 and mutant strain E. coli MHH126-5 generated the profile index "5144562" which was 99.8 % specific for E. coli. Additionally, all E. coli O157:H7 isolates were able to ferment rhamnose, while the O26:H11 strains were not. After 1 h treatment with 30 mM hydrogen peroxide no viable bacteria could be recovered from cultures of E. coli MHH933-5, MHH126-5 and EHEC EDL933a, while EHEC 126814 and EDL933b (Table 1) still produced more than 60 % colony forming units (CFU) compared to the blank value (data not shown).
Table 1.
Bacterial strains investigated in this study.
| Laboratory code | stx genotype | Serotype | Reference |
| EDL933a (ATCC 700927) | stx1, stx2 | O157:H7 | [15] |
| EDL933b (ATCC 43895/LMG 15068) | stx1, stx2 | O157:H7 | [32] |
| 126814/97 | stx2 | O26:H11 | [16,17] |
| 86-24 | stx2 | O157:H7 | [37] |
| E-D53 | stx2e | O101: H- | [19,20] |
| E-D68 | stx2e | O101: H- | [19,20] |
| 288597/03 | stx2 | O157:H- | This study |
| Nissle 1917 | ∅ | O6:H1 | [21-23] |
All bacterial strains investigated in detail in this study are listed with their laboratory codes, stx genotype, serotype and references, if applicable.
Acid resistance assays
The strains became acid resistant from OD600 0.7 for EHEC 126814 and from OD600 1.2 for EHEC EDL933a, respectively. EHEC 126814 very effectively responded to acid stress (Fig. 1A) and showed resistance rates up to 115 % at pH 2.5, indicating bacterial growth at this low pH, and 75 % at pH 1.5. In contrast, EHEC EDL933a was only moderately acid resistant at pH 2.5 as was its isogenic ΔrpoS mutant (Fig. 1B). The survival of these strains was about 17 % at most, which appeared to be independent of the rpoS genotype. The behavior of a second clone of EHEC EDL933a, obtained from ATCC, was identical (data not shown). EHEC EDL933b however, was nearly as acid resistant as EHEC 126814 (Fig. 1A and 1B). Furthermore, E. coli MHH126-5 was completely unable to survive acidic growth conditions at pH 2.5 and pH 1.5 (Fig. 1A). By complementation of E. coli MHH126-5 with pSC1 bearing its own rpoS gene, a wild type like phenotype could be restored. At pH 2.5 it was even more resistant than the parental organism. At pH 1.5 its survival was still about 60 % (Fig. 1C). When complemented with pSC2, harboring rpoS*, the rpoS gene from EHEC EDL933a, E. coli MHH126-5 was completely sensitive to acidic growth conditions further on (data not shown). However, when E. coli MHH126-5 was transformed with plasmid pMH33, containing an rpoS* allele cured by site directed mutagenesis from the TAA stop codon at position 723, its acid resistance increased to more than 100 % (data not shown). Thus, we could unequivocally prove that the point mutation G721T in rpoS* was solely responsible for the acid sensitive phenotype of EHEC EDL933a.
Figure 1.
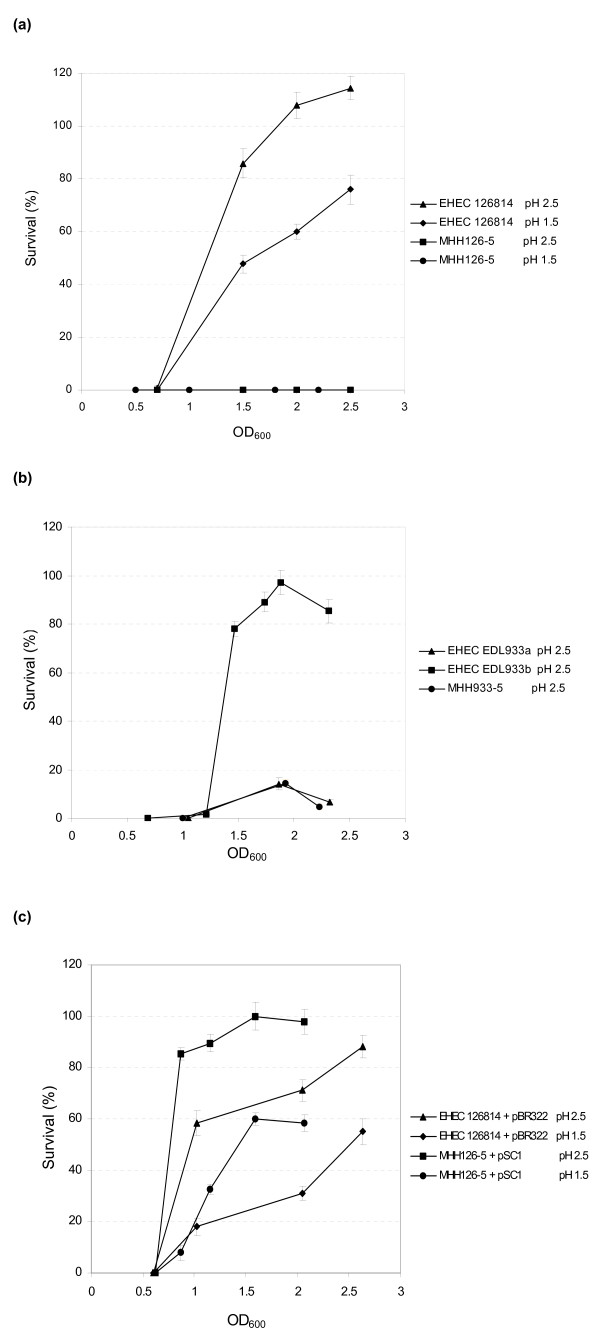
A: Acid resistance of EHEC 126814 and E. coli MHH126-5. Inducible acid resistance of EHEC wild type strain 126814 and its isogenic rpoS deletion mutant E. coli MHH126-5 was investigated after 2 h incubation in LB media at pH 2.5 or 1.5. The wild type strain showed a high level of acid resistance, which was induced from OD600 0.7 of the preparatory culture. It reached up to 115 % survival at OD600 2.5 of the starter culture, indicating that EHEC 126814 was able to grow under these conditions. In LB media with pH 1.5 up to 75 % of the inoculum survived. E. coli MHH126-5 was completely sensitive to acid treatment regardless of pH and OD600 of the preparatory culture. Percentage survival figures in relation to OD600 of one typical experiment are depicted. The means and standard deviations were calculated from three independent dilution series made at each individual measuring point. B: Acid resistance of EHEC EDL933a, EDL933b and E. coli MHH933-5. EHEC EDL933b was very acid resistant in all experiments. However, between EHEC wild type strain EDL933a and its mutant E. coli MHH933-5 no differences could be observed. Both isolates showed weak resistance under acidic growth conditions and showed a similar behavior in all other experiments. In contrast to EHEC 126814, acid resistance of the O157 isolates was induced at OD600 1.2 of the starter culture. One typical experiment has been shown as a representation of all independent tests carried out. The means and standard deviations were calculated from three independent dilution series prepared at each individual measuring point. C: Acid resistance of E. coli MHH126-5 complemented with pSC1. This figure depicts the inducible acid resistance in relation to OD600 of one typical experiment. Acid resistance was assayed at pH 2.5 and 1.5. By complementation of rpoS deletion mutant E. coli MHH126-5 with pSC1, containing its own rpoS gene cloned into plasmid pBR322, a phenotype could be restored that was even more resistant to acid stress than wild type strain EHEC 126814. OD600 of acid resistance induction was identical to values obtained with the wild type strain, shown in figure 1A. In order to compare growth conditions in LB media containing ampicillin, positive control EHEC 126814 had been transformed with plasmid pBR322. Compared to figure 1A, the antibiotic and/or pBR322 negatively influenced acid resistance of this strain. The percentage survival at pH 2.5 was below 90 %. One typical experiment is shown representative of independent tests. The means and standard deviations were calculated from three independent dilution series made at each individual measuring point.
Homology comparison of rpoS genes
Homology of all sequenced rpoS genes was compared with the software package BioEdit [see Additional file 3]. In the following all nucleotide positions are enumerated as described in materials and methods. We again sequenced the rpoS gene of EHEC EDL933b and used it as reference for all comparisons since the respective nucleotide data from the genome sequence NC_002655 had two sequencing errors, G57C and G61C. The main rpoS promoter rpoSp is located within the nlpD gene at position -568 to -566. The 35 and the -10 region are positioned at -601 to -596 and -578 to -573 respectively. Both, the highly acid resistant EHEC strain 126814 as well as the STEC isolate E-D53 had the point mutation G-570A, located in between the 10 region and rpoSp. Furthermore, these strains had point mutation A-521G leading to an amino acid exchange from threonine to alanine in NlpD. EHEC 126814 and 288597, STEC E-D53 and E-D68 and EcN also showed mutations T-306C in the nlpD gene, as well as A543C in rpoS. With the exception of EHEC 288597, these strains were mutated at position T387C as well. EHEC 126814 and the two STEC strains E-D53 and E-D68 also carried mutation A819G. Furthermore, EHEC 126814 and STEC E-D53 had the additional mutation G-465A in nlpD. STEC E-D68 showed four more point mutations: C-472T, which leads to an amino acid exchange from threonine to isoleucine in NlpD, C-293T, G-183T and A-162G. EcN carried the nucleotide exchange C-463T resulting in isoleucine instead of threonine in NlpD. Furthermore, the rpoS gene of EcN had point mutations T171C, C272T, T365G, T470C, T581C, and C995T. Except for the three amino acid exchanges in NlpD described above, all other point mutations observed in either rpoS or nlpD of all E. coli strains investigated were silent. In contrast, EHEC 86-24 had an 8 bp duplicate sequence (GAAGAGGA) in rpoS beginning at position 131 which caused a shift in the open reading frame and a stop codon 219 bp later.
Functional analysis of further rpoS genes
All experiments are illustrated in figure 2. Plasmids pUD2, pUD8 and pUD10 conferred a highly acid resistant phenotype on to E. coli MHH126-5, comparable to the respective wild type organisms EHEC EDL933b, EHEC 288597 and STEC E-D53. As expected, complementation with pUD4 did not mediate acid resistance to E. coli MHH126-5. This behavior was comparable to the parental strain EHEC 86-24. However, when the test strain E. coli MHH126-5 was transformed with pUD6, surprisingly it became strongly acid resistant. This was in sharp contrast to the manner of the corresponding wild type strain STEC E-D68, which only showed an acid resistance ≤ 0.1 %. This response becomes comprehensible considering the unaffected open reading frame in the rpoS sequence of pUD6. To confirm this effect we constructed pUD9, a second independent plasmid containing rpoS from STEC E-D68. With this plasmid the acid resistance of complemented E. coli MHH126-5 was identical to pUD6. Interestingly, we observed a similar phenomenon when we investigated acid tolerance of probiotic EcN. While survival of the wild type organism after pH 2.5 treatment was only around 5 %, test strain E. coli MHH126-5 became fully acid resistant, when complemented with plasmid pDS4 containing rpoS of EcN (Fig. 2).
Figure 2.
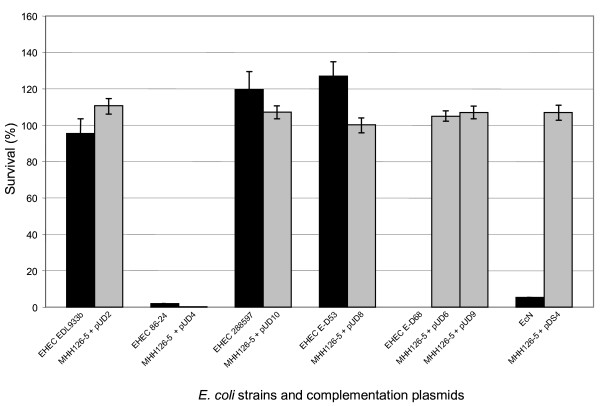
Functional analysis of further rpoS genes by complementation of E. coli MHH126-5. This figure shows resistance data of the rpoS deletion mutant E. coli MHH126-5 complemented with each of the plasmids pUD2 (pBR322 + rpoSEDL933b), pUD4 (pBR322 + rpoS86-24), pUD10 (pBR322 + rpoS288597), pUD8 (pBR322 + rpoSE-D53), pUD6 (pBR322 + rpoSE-D68), pUD9 (pBR322 + rpoSE-D68) or pDS4 (pBR322 + rpoSEcN) in comparison to the corresponding wild type EHEC strains EDL933b, 86-24 and 288597, STEC isolates E-D53 and E-D68 as well as EcN. All wild type strains are indicated by black bars, the complemented mutants by grey ones. Plasmids pUD2 and pUD4, pUD10 and pUD8 conferred an acid resistance phenotype to the mutant, which was comparable to the corresponding parental strain. Interestingly, upon complementation with pUD6 and pUD9, bearing the rpoS gene of STEC E-D68, E. coli MHH126-5 became strongly pH resistant. This was in sharp contrast to the behavior of the STEC E-D68 wild type strain. A similar phenomenon was observed when the acid resistance test strain E. coli MHH126-5 was complemented with pDS4 containing rpoS of EcN. The means and standard deviations were calculated from three independent dilution series in this exemplary experiment.
Discussion
Gastric acid is a natural barrier that all bacteria entering the lower intestine have to pass. With pH values ranging between 1.5 and 2.5, the stomach is one of the most inhospitable areas in the human body. A correlation between an infectious dose of enterobacteriaceae and their capacity to withstand acidic conditions is well known. With respect to the very low infectious dose of EHEC, of about 100 to 1000 organisms [3], it becomes obvious, that acid resistance is a key factor for virulence of these bacteria. E. coli have developed elegant regulatory systems, that enable their survival under such conditions [4,24]. The alternative sigma factor σS is instrumental in the regulation of acid protection mechanisms in this species of enterobacteriacae [4].
In this study we have investigated the role of the rpoS genotype on the acid stress response of a set of Shiga toxin producing E. coli as well as the widely used probiotic E. coli strain Nissle 1917 [21-23]. We first constructed unmarked isogenic rpoS deletion mutants of the highly virulent EHEC patient isolate 126814 [16,17] and of the completely sequenced prototype EHEC EDL933 [15], labeled EHEC EDL933a in this study. The patient isolate was highly dependent on a functional rpoS gene for an adequate response to acid stress. By complementation of its rpoS deletion mutant E. coli MHH126-5 with the rpoS gene from EHEC 126814 a wild type like phenotype could be restored. Surprisingly, EHEC EDL933a and its mutant did not exhibit major differences regarding their acid tolerance, but both appeared to be σS defective. Indeed, when we sequenced rpoS* from EHEC EDL933a we could identify a point mutation that caused a premature stop codon which was in conflict with the published database sequence [15]. In order to rule out that this mutation was an artifact generated in our laboratory, we purchased a second isolate of sequenced EHEC EDL933. It carried the identical point mutation, which may have occurred during passage prior to storage of the isolate at the strain collection. Allelic variations in the rpoS gene are not uncommon since it is localized in a highly mutable region of the E. coli genome [25,26]. In a large study Waterman et al. identified EHEC strains that were defective in their response to low pH [12]. They attributed this phenotype to a non functional σS as a consequence of rpoS mutations. Phenotypical characterization of EHEC EDL933b, a further clone of EDL933, and complementation experiments clearly showed that the stop codon in rpoS* causally determined the stress phenotype of ATCC strain 700927. When considering the significant impact of σS in the regulatory network of E. coli, which controls up to 10 % of the E. coli genes directly or indirectly [27], this observation is of importance for those working in the field of EHEC and planning to do both, in vitro or in vivo studies with this particular sequenced isolate.
Further investigation of six rpoS genes from Shiga toxin producing E. coli and a probiotic E. coli strain revealed that σS activity is not always dependent on the rpoS genotype. Despite having a functional rpoS gene, as shown by complementation experiments in a ΔrpoS background, STEC E-D68 as well as EcN behaved σS defective regarding their acid resistance. While STEC E-D68 was completely sensitive to acid stress, probiotic EcN exhibited about 5 % survival in the same test environment. With respect to EcN, this observation adds to an actual study of Bhagwat et al. where the authors observed functional heterogeneity of σS in food-borne and clinical EHEC isolates [14]. However, in STEC E-D68 as well as in EcN, mutations in the gene encoding the GDAR system regulator GadE [10] have to be ruled out. Such mutations have been reported by Bhagwat et al. as another reason for attenuated acid resistance in E. coli wild type isolates [9]. With regard to the varying σS activities in individual E. coli strains observed in our study, it should be mentioned that all isolates analyzed here carried glutamate at codon 33 (GAG) resulting in σS (33E). This seems to account for a higher variability in rpoS related phenotypes as recently described by Atlung et al. [25].
Price et al. have shown recently, that EHEC make use of their different acid resistance systems depending on the type of acid stress they are exposed to [28,29]. In either case σS was important for a sufficient acid stress response of EHEC EDL933 (ATCC 43895) in vitro as well as in vivo [29]. It should be investigated though, whether this impact of σS on the in vivo acid resistance of EHEC also allows assumptions about the virulence of a particular strain. In our laboratory environment for instance wild type EHEC 86-24 was highly virulent in an oral infection model with gnotobiotic piglets [16], although it has a non functional rpoS gene and is only weakly acid resistant as shown above. Krogfelt et al. have published an accomplished experiment which clearly demonstrates that rpoS gene function may be a disadvantage for E. coli colonizing the intestine [30]. The authors therefore conclude that the benefit of a functional rpoS regulon for E. coli depends on the actual growth phase of a particular strain. The apparent down regulation of σS activity in STEC E-D68 and EcN in our study seems to be another way of adapting the rpoS regulon to specific growth conditions. We have performed further experiments to investigate the impact of rpoS in EcN on regulation of potential host probiotic marker genes which we have recently identified [31]. Our long-term objective is to establish to what extent E. coli is able to modulate pathogenic but also beneficial properties using its rpoS regulatory network.
Conclusion
The results of our study clearly confirm the central role of σS as a key regulator for acid resistance in STEC and EcN. When interpreting in vitro or in vivo data generated with EHEC EDL933a it is important to realize, that this prototype EHEC has an attenuated σS phenotype. We could also show that rpoS gene function is modified in singular E. coli isolates by regulatory mechanisms that lead to an altered σS activity, as exemplified by STEC strain E-D68 and probiotic EcN. This is in line with the observations regarding functional heterogeneity of RpoS in stress tolerance of K-12 E. coli strains and EHEC isolates, King et al. and Bhagwat et al. have reported [13,14], and expands them to a commensal E. coli with beneficial traits.
Methods
Bacterial strains and media
All bacterial strains investigated in detail in this study are listed in table 1. Acid stress response assays were performed with EHEC 126814, a highly virulent patient isolate [16,17] and the prototype O157:H7 EHEC EDL933. Two independent clones of EHEC EDL933 were used, the sequenced strain ATCC 700927 [15], referred to as EHEC EDL933a and the original isolate ATCC 43895/BCCM LMG 15068 [32], termed EHEC EDL933b. Acid resistance was also evaluated using 39 human and porcine EHEC and STEC strains (data not shown) as well as the probiotic E. coli Nissle 1917 [21-23]. Unmarked isogenic rpoS negative mutants were produced from EHEC EDL933a and from EHEC 126814. E. coli SM 10 λpir is a λ lysogen of E. coli SM10 and contains the trans acting factors needed to replicate and mobilize all suicide plasmids [33] used in this study. E. coli DH5α (Invitrogen, Karlsruhe, Germany) was used as host strain for all other plasmids. Bacteria were grown in LB broth (Invitrogen) and on MacConkey agar (Oxoid, Wesel, Germany), MH agar (Becton Dickinson, Heidelberg, Germany) or LB agar plates (Invitrogen) at 37°C or on LB sucrose plates (10 % sucrose w/v, no NaCl) at room temperature. Ampicillin (Ratiopharm, Ulm, Germany) was added at a concentration of 200 μg/ml, where necessary. Concentrated HCl was used to adjust LB broth for acid resistance testing at pH 2.5 or 1.5. Chemicals were obtained from Sigma, Deisenhofen or Merck, Darmstadt, Germany.
Primers and sequencing
Table 2 lists all oligonucleotide primers used in this study. These were synthesized by MWG-Biotech (Ebersberg, Germany), who also completed the custom sequencing of PCR products and plasmids. Sequences of all nlpD-rpoS genes investigated in this study were generated by primer walking with two independent PCR products for which corresponding accession numbers are given in table 3. Nucleotide data was analyzed with the DNASTAR software package (Lasergene, DNASTAR, Madison, WI, USA), the BioEdit Sequence Alignment Editor 7.0.5.3. [34] and Clone Manager 6 (Scientific & Educational Software, Cary, NC, USA) and then submitted to GenBank. Nucleotide positions within rpoS, nlpD or the flanking regions are referenced to the open reading frame of rpoS. Positions 5' upstream of the rpoS start codon are indicated with a negative sign, positions 3' downstream of the stop codon with a positive one.
Table 2.
Oligonucleotide primers used in this study.
| Target gene | Name | Position | 5' Sequence 3' | ⇔ |
| rpoS | RpoS 3 | 4 – 24 | GCT CTA GAA GTC AGA ATA CGC TGA AAG TT XbaI |
s |
| rpoS | RpoS 4 | 989 – 969 | CCG AGC TCC TCG CGG AAC AGC GCT TCG AT SacI |
as |
| rpoS | RpoS 4c | 1129 – 1109 | CCC AAG CTT TTA CTC GCG GAA CAG CGC TTC HindIII |
as |
| rpoS | RpoS 5 | 572 – 589 | ATG AAC CAA GTG GG AAG | s |
| rpoS | RpoS 6 | 871 – 854 | ACA TCT TCC AGT GTT GCC | as |
| nlpD | RpoS 8 | (- 657) – (- 636) | CCC AAG CTT CGA CGC AGC AGA GCA AGG AGT T HindIII |
s |
| rpoS | RpoS 9 | 1141 – 1119 | CCC AAG CTT CAG CTG GTG CGC AAG ATG ATG AA HindIII |
as |
| rpoS | RpoS 17a | 706 – 736 | GAC ATC CTG GCC GAT GAA AAA GAG AAC GGT C | s |
| rpoS | RpoS 17b | 736 – 706 | GAC CGT TCT CTT TTT CAT CGG CCA GGA TGT C | as |
| rpoS | RpoS 22 | (+ 171) – (+ 151) | CCC AAG CTT GCC AAA TGT GAC GCT GAC GCG HindIII |
as |
Primers used in this study are listed with their 5' – 3' sequences. All nucleotide positions refer to the start codon of their target gene. Positions of RpoS primers within rpoS, nlpD or the flanking regions are referenced to the open reading frame of rpoS. Positions 5' upstream of the start codon are indicated with a negative sign, positions 3' downstream of the stop codon with a positive one. s = sense, as = antisense
Table 3.
Plasmids used in this study.
| Plasmid | Description | Host |
| pUC19 | • High copy plasmid [41]; accession # L09137 • AmpR |
• E. coli DH5α |
| pGP704 | • Suicide vector [33] • Backbone from pBR322, ori R6K instead of ori E1 from pBR322, mob region from pRP4 • Replicates in E. coli SM10λpir but not in EHEC strains used because ori R6K needs π-Protein coded by the pir gene for replication • AmpR |
• E. coli SM10λpir |
| pMHH1 | • Derivative of pGP704 • Has a positive selection system based on the sacB gene from Bacillus subtilis [38] |
• E. coli SM10λpir |
| pMHH6* | • Derivative of pUC19 • Harbors a 990 bp rpoS fragment from EHEC 86-24 amplified with primers RpoS 3/RpoS 4 and cloned via XbaI und SacI restriction sites |
• E. coli DH5α |
| pMHH7* | • Derivative of pMHH6 • Restriction digest with DraIII und BsaAI leads to a 390 bp deletion in rpoS |
• E. coli DH5α |
| pMHH8* | • Derivative of pMHH1 • Contains the mutagnized 600 bp rpoS fragment from pMHH7 cloned via XbaI and SacI restriction sites |
• E. coli SM10λpir |
| pBR322 | • Low copy plasmid [36]; accession # J01749 • AmpR, TetR |
• E. coli DH5α • EHEC 126814 |
| pSC1* | • Derivative of pBR322 • Harbors nlpD-rpoS from EHEC 126814 accession # DQ272417 amplified by PCR using primers RpoS 8/RpoS 9 and cloned via HindIII restriction sites |
• E. coli DH5α • E. coli MHH126-5 |
| pSC2* | • Derivative of pBR322 • Harbors nlpD-rpoS from EHEC EDL933a (rpoS*) accession # DQ272419 amplified by PCR with primers RpoS 8/RpoS 9 and cloned via HindIII restriction sites |
• E. coli DH5α • E. coli MHH126-5 |
| pMH33* | • Made from pSC2 by PCR mutagenesis using Primers RpoS 17a/RpoS 17b • Has point mutation rpoST721G which resolves the TAA stop codon at position 723 in |
• E. coli MHH126-5 |
| pUD2* | • Derivative of pBR322 • Harbors nlpD-rpoS from EHEC EDL933b accession # DQ272421 amplified by PCR with primers RpoS 8/RpoS 9 and cloned via HindIII restriction sites |
• E. coli MHH126-5 |
| pUD4* | • Derivative of pBR322 • Harbors nlpD-rpoS from EHEC 86-24 accession # DQ272418 amplified by PCR with primers RpoS 8/RpoS 9 and cloned via HindIII restriction sites |
• E. coli MHH126-5 |
| pUD8* | • Derivative of pBR322 • Harbors nlpD-rpoS from STEC E-D53 accession # DQ272423 amplified by PCR with primers RpoS 8/RpoS 9 and cloned via HindIII restriction sites |
• E. coli MHH126-5 |
| pUD6* | • Derivative of pBR322 • Harbors nlpD-rpoS from STEC E-D68 accession # DQ272424 amplified by PCR with primers RpoS 8/RpoS 4c and cloned via HindIII restriction sites |
• E. coli MHH126-5 |
| pUD9* | • Derivative of pBR322 • Harbors nlpD-rpoS from STEC E-D68 accession # DQ272424 amplified by PCR with primers RpoS 8/RpoS 4c and cloned via HindIII restriction sites |
• E. coli MHH126-5 |
| pUD10* | • Derivative of pBR322 • Harbors nlpD-rpoS from EHEC 288597 accession # DQ272422 amplified by PCR with primers RpoS 8/RpoS 9 and cloned via HindIII restriction sites |
• E. coli MHH126-5 |
| pDS4* | • Derivative of pBR322 • Harbors nlpD-rpoS from EcN amplified by PCR with primers RpoS 8/RpoS 22 and cloned via HindIII restriction sites |
• E. coli MHH126-5 |
All plasmids used in this study are listed with their names, descriptions and host strains. Sequenced rpoS genes were submitted to GenBank. Their accession numbers are given in the table. * = plasmid designed in this study
Plasmids and DNA preparation
All plasmids used in this study are listed in table 3. Allelic exchange experiments were conducted with the suicide vector pMHH8 derived from plasmid pGP704 [33]. A map of suicide vectors pMHH1, pMHH7 and pMHH8 is shown in figure 3. All other cloning was performed in pUC19 [35] or pBR322 [36]. Construction of all complementation plasmids is depicted in figure 4. Plasmid DNA was isolated using the Qiagen Plasmid Midi Kit (Qiagen, Hilden, Germany). Chromosomal DNA was purified with Qiagen-tip 100 columns and the Genomic DNA Buffer Set from Qiagen.
Figure 3.
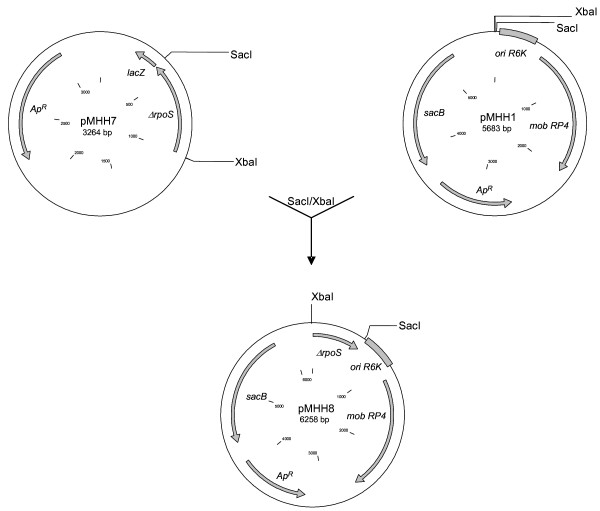
Construction of suicide vector pMHH8. The rpoS gene from EHEC 86-24 was amplified by PCR with primers RpoS 3/RpoS 4 and cloned into pUC19 after restriction digest with enzymes XbaI and SacI (pMHH6). A 390 bp sequence was deleted from the insert through restriction digest with enzymes DraIII and BsaAI leading to plasmid pMHH7. The mutated rpoS gene was then cloned into suicide vector pMHH1 after restriction digest with XbaI and SacI. The resulting plasmid was termed pMHH8. It was used in the construction of all rpoS deletion mutants described in this study.
Figure 4.
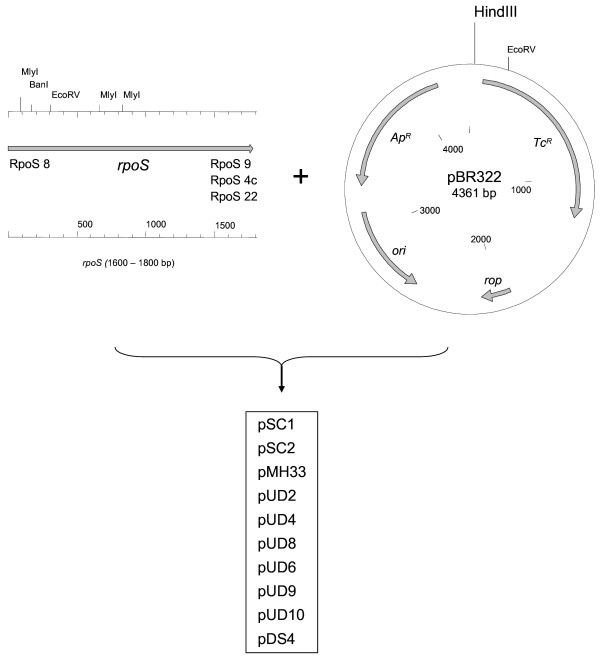
Construction of complementation plasmids. Complementation plasmids pSC1, pSC2, pUD2, pUD4, pUD8, pUD6, pUD9, pUD10 and pDS4 were constructed using low copy vector pBR322 as backbone. Each of them harbors one complete rpoS gene and the rpoSp promoter [39] from EHEC/STEC strains 126814 (pSC1), EDL933a (pSC2), EDL933b (pUD2), 86-24 (pUD4), E-D53 (pUD8) and 288597 (pUD10), amplified by PCR with primers RpoS 8/RpoS 9 containing 5' HindIII restriction sites. RpoS 8 and RpoS 4c were taken to synthesize the respective DNA fragment from STEC E-D68 in order to generate the two independent plasmids pUD6 and pUD9. pDS4 contained the cloned rpoS gene from EcN, amplified with primers RpoS 8 and RpoS 22. pMH33 was made from pSC2 by PCR mediated site specific mutagenesis, employing the mutation rpoST721G to resolve the TAA stop codon in rpoS*. RpoS negative mutant strain E. coli MHH126-5 was transformed with each of the complementation vectors by electroporation.
Suicide vector pMHH8
For construction of suicide vector pMHH8 the rpoS gene from EHEC 86-24 [37] was amplified by PCR using primers RpoS 3/RpoS 4 and cloned into pUC19. Upon restriction digest with enzymes DraIII and BsaI (New England Biolabs, Schwalbach, Germany) a 390 bp deletion in rpoS was created. The mutated gene was then subcloned into pMHH1, a derivative of suicide vector pGP704. The resulting plasmid was called pMHH8 (Fig. 3). It contained a positive selection system based on the sacB gene from Bacillus subtilis [38] to facilitate screening for potential mutants. Electroporation was used to transform E. coli SM 10 λpir with pMHH8.
Construction of complementation plasmids
For complementation experiments, an nlpD-rpoS DNA amplicon, containing the rpoS main promoter rpoSp [39] was generated by PCR using primers with 5' HindIII restriction sites (Table 2). RpoS 8 and RpoS 9 were taken to amplify nlpD-rpoS from EHEC strains 126814 (pSC1), EDL933a (pSC2), EDL933b (pUD2), 86-24 (pUD4), 288597 (pUD10) and from porcine STEC strain E-D53 (pUD8). RpoS 8 and RpoS 4c were needed to synthesize the respective DNA fragment from porcine STEC E-D68 (pUD6 and pUD9). Amplification of nlpD-rpoS from EcN (pDS4) was performed with primers RpoS 8 and RpoS 22. Following endonuclease digest, the amplicon was cloned into the low copy plasmid pBR322 linearized with HindIII (New England Biolabs). Plasmid pMH33 was produced from pSC2 by the PCR mediated nucleotide exchange T721G with primers RpoS 17a/RpoS 17b using the QuikChange Site Directed Mutagenesis Kit from Stratagene (Amsterdam, The Netherlands) according to manufacturer's instructions. rpoS deletion mutant E. coli MHH126-5 was then transformed with each of the complementation vectors by electroporation.
Construction of rpoS deletion mutants from EHEC EDL933a and EHEC 126814
Allelic exchange using suicide vector pMHH8 was performed in a two-step procedure as previously described [40]. A positive selection system based on the sacB gene from Bacillus subtilis [38] was used to facilitate screening for potential mutants.
Genotypical and phenotypical confirmation of mutant strains
The rpoS deletion mutation was confirmed by the sequencing of PCR products obtained with primers RpoS 3 and RpoS 4, as well as with Southern hybridizations of chromosomal DNA from EHEC EDL933a and EHEC 126814 wild type and mutant strains digested with either StuI or XmnI (New England Biolabs) [see Additional file 1]. Hybridization was performed under high stringency conditions with digoxigenin labelled probes using the Dig Labeling and Detection Kit from Roche Diagnostics (Mannheim, Germany). Specific binding was detected by autoradiography and chemoluminescence with CSPD (Applied Biosystems, Darmstadt, Germany) as substrate. API 20 E strips (bioMérieux Deutschland, Nürtingen, Germany) were used to analyze biochemical profiles of mutants and their parental strains. Antibiotic resistances of all isolates were determined with the MERLIN Microdilution Detection System (MICRONAUT-SB, MERLIN Diagnostika, Bornheim-Hersel, Germany). Resistance of EHEC EDL933a, EDL933b, 126814 and of the mutants E. coli MHH933-5 and MHH126-5 to 30 mM hydrogen peroxide was tested as described by Lange et al. [18]. Viability of bacteria was evaluated after 5, 10, 20, 30 and 60 minutes of H2O2 treatment.
Acid resistance assays
Inducible acid resistance of EHEC EDL933a, EHEC EDL933b, EHEC 126814, the mutants E. coli MHH933-5 and MHH126-5 as well as of E. coli MHH126-5 complemented with pSC1, pSC2 or pMH33, was assessed by inoculating 100 ml LB broth pH 7.0 1:1000 with an overnight culture from each bacterial strain. Bacteria were sampled at defined time points, as depicted in figure 1A to 1C, and subjected to incubation at pH 2.5 or 1.5 for two hours. Colony forming units (CFU) of untreated and acid treated cultures were determined. Acid resistance screening of further EHEC and STEC isolates, of EcN and of E. coli MHH126-5 complemented with pUD2, pUD4, pUD6, pUD8, pUD9, pUD10 or pDS4 was performed in a similar manner but as a one step test at pH 2.5, with cultures grown to an OD600 of 1.7 (Fig. 2). Acid resistance was calculated as percentage CFU recovered after acid exposure compared to untreated cultures. Resistance data has been based on three independent tests.
Complementation experiments with mutant E. coli MHH 126-5
E. coli strain MHH126-5 was transformed with each of the complementation plasmids by electroporation. Inducible resistance at pH 2.5 or 1.5 was assayed with E. coli MHH126-5 complemented with pSC1 (Fig. 1C), using the same experimental procedure as described above, except for adding 200 μg/ml ampicillin to all media. Functional activity of pSC2, pMH33 and all other cloned rpoS genes (Fig. 2) was evaluated with the one step test described above for acid resistance screening. EHEC 126814 transformed with pBR322 was used as reference strain for these experiments.
Abbreviations
ATCC, American Type Culture Collection; BCCM, Belgian Co-ordinated Collections of Microorganisms; CFU, colony forming units; EHEC, enterohemorrhagic E. coli; HUS, hemolytic uremic syndrome; EcN, E. coli Nissle 1917
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
SMC carried out all of the experiments described except for functional characterization of additional rpoS genes from STEC 288597, E-D53 and E-D68 and EcN, was involved in the interpretation of data, designed figures and tables and wrote the manuscript. MH constructed plasmid pMH33, used it for complementation experiments and participated in the analysis of sequenced rpoS genes. DSS sequenced and functionally characterized rpoS of EcN. UE screened a collection of STEC strains for acid resistance and has sequenced and functionally characterized rpoS genes from STEC strains 288597, E-D53 and E-D68. SNU has performed preparatory investigations to this study, which were instrumental in designing the subsequent experiments. FG is primary investigator, who conceived the study, helped to interpret the data and critically revised and finished the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
Genotypical confirmation of rpoS mutants by Southern blotting. In this figure genomic DNA is shown digested with either XmnI (A) or StuI (B). DNA from wild type strains EHEC EDL933a and 126814 is separated in lanes 1 and 3, DNA from the corresponding mutants E. coli MHH933-5 and E. coli MHH126-5 is running in lanes 2 and 4 respectively. The probes were hybridizing with fragments of a calculated size of 1042 bp (wild type) and 652 bp (mutants) after restriction digest with XmnI (A) and 841 bp (wild type) and 451 bp (mutant), when StuI (B) was used. No additional bands appeared. kb = DNA Molecular Weight Marker III, DIG-labelled (Roche Molecular Biochemicals, Mannheim, Germany).
Biochemical reaction profiles in the API 20 E test. This table displays the biochemical reactions of EHEC EDL933, EHEC 126814 and their isogenic rpoS negative mutants as determined with API 20 E strips.
Multiple alignment of rpoS alleles. A multiple nucleotide sequence alignment of all eight rpoS alleles functionally characterized and described in detail in this study is provided in BioEdit format. The file can be viewed with the sequence alignment editor BioEdit [34]. The program is available free of charge at [42].
Acknowledgments
Acknowledgements
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 621) to FG.
Contributor Information
Sina M Coldewey, Email: sina.coldewey@web.de.
Maike Hartmann, Email: hartmann.maike@mh-hannover.de.
Dorothea S Schmidt, Email: dorothea.s.schmidt@web.de.
Uta Engelking, Email: uta.engelking@gmx.de.
Sya N Ukena, Email: sya.ukena@helmholtz-hzi.de.
Florian Gunzer, Email: florian.gunzer@tu-dresden.de.
References
- Ray PE, Liu XH. Pathogenesis of Shiga toxin-induced hemolytic uremic syndrome. Pediatr Nephrol. 2001;16:823–839. doi: 10.1007/s004670100660. [DOI] [PubMed] [Google Scholar]
- Mead PS, Griffin PM. Escherichia coli O157:H7. Lancet. 1998;352:1207–1212. doi: 10.1016/S0140-6736(98)01267-7. [DOI] [PubMed] [Google Scholar]
- Lin J, Lee IS, Frey J, Slonczewski JL, Foster JW. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J Bacteriol. 1995;177:4097–4104. doi: 10.1128/jb.177.14.4097-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JW. Escherichia coli acid resistance: tales of an amateur acidophile. Nat Rev Microbiol. 2004;2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- Hengge-Aronis R. Back to log phase: sigma S as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol Microbiol. 1996;21:887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- Muffler A, Barth M, Marschall C, Hengge-Aronis R. Heat shock regulation of sigmaS turnover: a role for DnaK and relationship between stress responses mediated by sigmaS and sigma32 in Escherichia coli. J Bacteriol. 1997;179:445–452. doi: 10.1128/jb.179.2.445-452.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee IS, Lin J, Hall HK, Bearson B, Foster JW. The stationary-phase sigma factor sigma S (RpoS) is required for a sustained acid tolerance response in virulent Salmonella typhimurium. Mol Microbiol. 1995;17:155–167. doi: 10.1111/j.1365-2958.1995.mmi_17010155.x. [DOI] [PubMed] [Google Scholar]
- Hersh BM, Farooq FT, Barstad DN, Blankenhorn DL, Slonczewski JL. A glutamate-dependent acid resistance gene in Escherichia coli. J Bacteriol. 1996;178:3978–3981. doi: 10.1128/jb.178.13.3978-3981.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat AA, Chan L, Han R, Tan J, Kothary M, Jean-Gilles J, Tall BD. Characterization of enterohemorrhagic Escherichia coli strains based on acid resistance phenotypes. Infect Immun. 2005;73:4993–5003. doi: 10.1128/IAI.73.8.4993-5003.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Gong S, Richard H, Tucker DL, Conway T, Foster JW. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol Microbiol. 2003;49:1309–1320. doi: 10.1046/j.1365-2958.2003.03633.x. [DOI] [PubMed] [Google Scholar]
- Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski JL. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J Bacteriol. 1994;176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman SR, Small PL. Characterization of the acid resistance phenotype and rpoS alleles of shiga-like toxin-producing Escherichia coli. Infect Immun. 1996;64:2808–2811. doi: 10.1128/iai.64.7.2808-2811.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King T, Ishihama A, Kori A, Ferenci T. A regulatory trade-off as a source of strain variation in the species Escherichia coli. J Bacteriol. 2004;186:5614–5620. doi: 10.1128/JB.186.17.5614-5620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagwat AA, Tan J, Sharma M, Kothary M, Low S, Tall BD, Bhagwat M. Functional heterogeneity of RpoS in stress tolerance of enterohemorrhagic Escherichia coli strains. Appl Environ Microbiol. 2006;72:4978–4986. doi: 10.1128/AEM.02842-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna NT, Plunkett G, III, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- Gunzer F, Hennig-Pauka I, Waldmann KH, Sandhoff R, Gröne HJ, Kreipe HH, Matussek A, Mengel M. Gnotobiotic piglets develop thrombotic microangiopathy after oral infection with enterohemorrhagic Escherichia coli. Am J Clin Pathol. 2002;118:364–375. doi: 10.1309/UMW9-D06Q-M94Q-JGH2. [DOI] [PubMed] [Google Scholar]
- Bellin T, Pulz M, Matussek A, Hempen HG, Gunzer F. Rapid detection of enterohemorrhagic Escherichia coli by real-time PCR with fluorescent hybridization probes. J Clin Microbiol. 2001;39:370–374. doi: 10.1128/JCM.39.1.370-374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- Franke S, Harmsen D, Caprioli A, Pierard D, Wieler LH, Karch H. Clonal relatedness of Shiga-like toxin-producing Escherichia coli O101 strains of human and porcine origin. J Clin Microbiol. 1995;33:3174–3178. doi: 10.1128/jcm.33.12.3174-3178.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprioli A, Nigrelli A, Gatti R, Zavanella M, Blando AM, Minelli F, Donelli G. Characterisation of verocytotoxin-producing Escherichia coli isolated from pigs and cattle in northern Italy. Vet Rec. 1993;133:323–324. doi: 10.1136/vr.133.13.323. [DOI] [PubMed] [Google Scholar]
- Nissle A. Die antagonistische Behandlung chronischer Darmstörungen mit Colibakterien. Med Klin. 1918;2:29–30. [Google Scholar]
- Rembacken BJ, Snelling AM, Hawkey PM, Chalmers DM, Axon AT. Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: a randomised trial. Lancet. 1999;354:635–639. doi: 10.1016/S0140-6736(98)06343-0. [DOI] [PubMed] [Google Scholar]
- Kruis W, Fric P, Pokrotnieks J, Lukas M, Fixa B, Kascak M, Kamm MA, Weismueller J, Beglinger C, Stolte M, Wolff C, Schulze J. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut. 2004;53:1617–1623. doi: 10.1136/gut.2003.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge-Aronis R. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol Mol Biol Rev. 2002;66:373–395. doi: 10.1128/MMBR.66.3.373-395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlung T, Nielsen HV, Hansen FG. Characterisation of the allelic variation in the rpoS gene in thirteen K12 and six other non-pathogenic Escherichia coli strains. Mol Genet Genomics. 2002;266:873–881. doi: 10.1007/s00438-001-0610-0. [DOI] [PubMed] [Google Scholar]
- Ferenci T. What is driving the acquisition of mutS and rpoS polymorphisms in Escherichia coli? Trends Microbiol. 2003;11:457–461. doi: 10.1016/j.tim.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol. 2005;187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price SB, Cheng CM, Kaspar CW, Wright JC, DeGraves FJ, Penfound TA, Castanie-Cornet MP, Foster JW. Role of rpoS in acid resistance and fecal shedding of Escherichia coli O157:H7. Appl Environ Microbiol. 2000;66:632–637. doi: 10.1128/AEM.66.2.632-637.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price SB, Wright JC, DeGraves FJ, Castanie-Cornet MP, Foster JW. Acid resistance systems required for survival of Escherichia coli O157:H7 in the bovine gastrointestinal tract and in apple cider are different. Appl Environ Microbiol. 2004;70:4792–4799. doi: 10.1128/AEM.70.8.4792-4799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogfelt KA, Hjulgaard M, Sorensen K, Cohen PS, Givskov M. rpoS gene function is a disadvantage for Escherichia coli BJ4 during competitive colonization of the mouse large intestine. Infect Immun. 2000;68:2518–2524. doi: 10.1128/IAI.68.5.2518-2524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukena SN, Westendorf AM, Hansen W, Rohde M, Geffers R, Coldewey S, Suerbaum S, Buer J, Gunzer F. The host response to the probiotic Escherichia coli strain Nissle 1917: specific up-regulation of the proinflammatory chemokine MCP-1. BMC Med Genet. 2005;6:43. doi: 10.1186/1471-2350-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JG, Davis BR, Wachsmuth IK, Riley LW, Remis RS, Sokolow R, Morris GK. Laboratory investigation of hemorrhagic colitis outbreaks associated with a rare Escherichia coli serotype. J Clin Microbiol. 1983;18:512–520. doi: 10.1128/jcm.18.3.512-520.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Bolivar F, Rodriguez RL, Greene PJ, Betlach MC, Heyneker HL, Boyer HW. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. doi: 10.1016/0378-1119(77)90074-9. [DOI] [PubMed] [Google Scholar]
- Griffin PM, Ostroff SM, Tauxe RV, Greene KD, Wells JG, Lewis JH, Blake PA. Illnesses associated with Escherichia coli 0157:H7 infections. A broad clinical spectrum. Ann Intern Med. 1988;109:705–712. doi: 10.7326/0003-4819-109-9-705. [DOI] [PubMed] [Google Scholar]
- Gay P, Le Coq D, Steinmetz M, Ferrari E, Hoch JA. Cloning structural gene sacB, which codes for exoenzyme levansucrase of Bacillus subtilis: expression of the gene in Escherichia coli. J Bacteriol. 1983;153:1424–1431. doi: 10.1128/jb.153.3.1424-1431.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange R, Fischer D, Hengge-Aronis R. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the sigma S subunit of RNA polymerase in Escherichia coli. J Bacteriol. 1995;177:4676–4680. doi: 10.1128/jb.177.16.4676-4680.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunzer F, Bohn U, Fuchs S, Mühldorfer I, Hacker J, Tzipori S, Donohue-Rolfe A. Construction and characterization of an isogenic slt-II deletion mutant of enterohemorrhagic Escherichia coli. Infect Immun. 1998;66:2337–2341. doi: 10.1128/iai.66.5.2337-2341.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Hall TA. Biological sequence alignment editor BioEdit. 2005. http://www.mbio.ncsu.edu/BioEdit/bioedit.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genotypical confirmation of rpoS mutants by Southern blotting. In this figure genomic DNA is shown digested with either XmnI (A) or StuI (B). DNA from wild type strains EHEC EDL933a and 126814 is separated in lanes 1 and 3, DNA from the corresponding mutants E. coli MHH933-5 and E. coli MHH126-5 is running in lanes 2 and 4 respectively. The probes were hybridizing with fragments of a calculated size of 1042 bp (wild type) and 652 bp (mutants) after restriction digest with XmnI (A) and 841 bp (wild type) and 451 bp (mutant), when StuI (B) was used. No additional bands appeared. kb = DNA Molecular Weight Marker III, DIG-labelled (Roche Molecular Biochemicals, Mannheim, Germany).
Biochemical reaction profiles in the API 20 E test. This table displays the biochemical reactions of EHEC EDL933, EHEC 126814 and their isogenic rpoS negative mutants as determined with API 20 E strips.
Multiple alignment of rpoS alleles. A multiple nucleotide sequence alignment of all eight rpoS alleles functionally characterized and described in detail in this study is provided in BioEdit format. The file can be viewed with the sequence alignment editor BioEdit [34]. The program is available free of charge at [42].


