Myosin 5 is a two-headed motor protein that moves cargoes along actin filaments1, 2. Its tail ends in paired globular tail domains (GTDs) thought to bind cargo3. At nanomolar calcium, actin-activated ATPase is low and the molecule is folded. Micromolar calcium activates ATPase and the molecule unfolds3-6. We now describe the structure of folded myosin and the GTD's role in regulating activity. Electron microscopy shows the two heads lie either side of the tail, contacting the GTDs at a lobe of the motor domain (∼Pro117-Pro137) that contains conserved acidic side chains, suggesting ionic interactions between motor domain and GTD. Myosin 5 heavy meromyosin (HMM), a constitutively active fragment lacking the GTDs, is inhibited and folded by a dimeric GST-GTD fusion protein. Motility assays reveal that at nanomolar calcium HMM moves robustly on actin filaments whereas few myosins bind or move. These results combine to show that with no cargo, the GTDs bind in an intramolecular manner to the motor domains producing an inhibited and compact structure that binds weakly to actin and allows the molecule to recycle towards new cargoes.
Myosin 5 has two heavy chains, each contributing an N-terminal motor domain, an α-helical lever stabilised by six sequentially bound calmodulin light chains, a coiled coil region that dimerises with the other heavy chain to form the tail and finally the GTD3 (Fig. 1o). The unregulated high activity of myosin 5 HMM implicates the GTD in down-regulation of activity at nanomolar calcium. Our initial images of the inhibited state showed a compact conformation, but identification of substructure was ambiguous4. The structures seen by other groups were less compact5, 6. To understand the mechanism of regulation of cargo transport by myosins requires understanding the structure and properties of the inhibited state. Therefore we have studied the structure of folded myosin 5, and the GTD's mechanism of inhibition.
Figure 1.
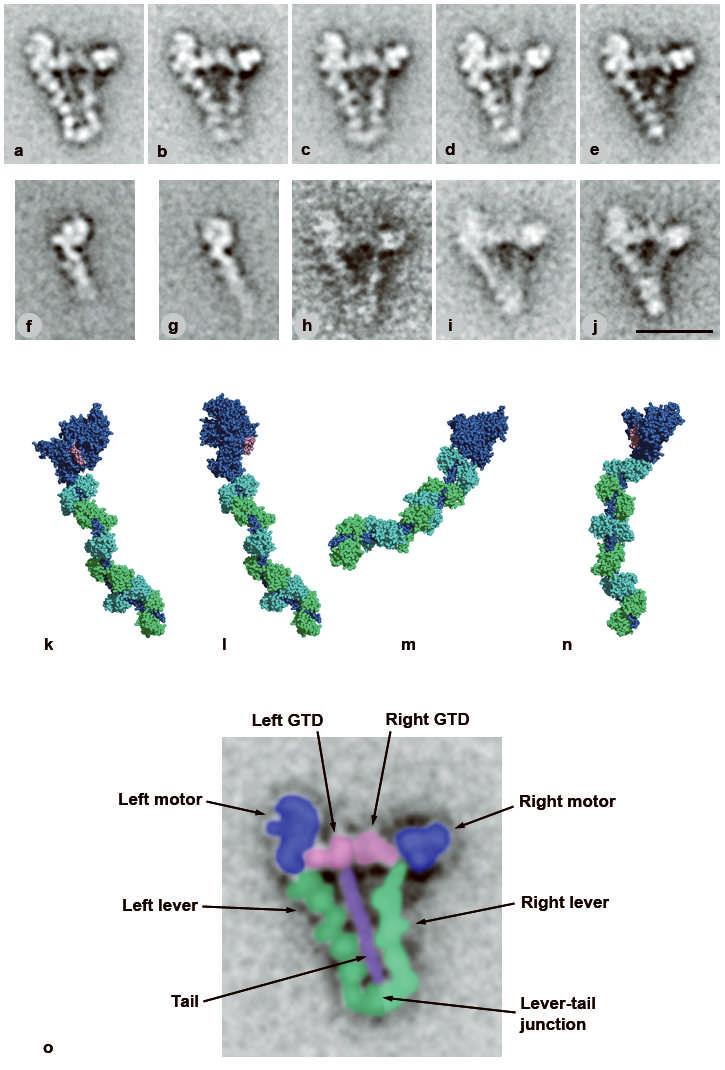
Structure of switched-off myosin 5 and HMM-GTD complex. a-e, averaged images of negative-stained, folded whole myosin molecules; 46-53 molecules per class. f, g, averaged images of myosin 5 S1 stained in the presence of ATP; f shows the pre-powerstroke conformation (63 molecules), g the post-rigor conformation (58 molecules). h, i, averaged images of myosin in presence of ADP or no nucleotide, respectively; 21 and 55 molecules. j, averaged images of the complex of myosin 5 HMM and GST-GTD dimer; 28 molecules. Scale bar in j is 20 nm and applies to panels a-j. k-n, atomic models of myosin 5 head; heavy chain shaded dark blue, the six calmodulins shaded alternately cyan and green, the putative GTD-binding region of the motor domain (Pro117-Pro137) shaded pink. k, model using scallop motor domain containing ADP.vanadate11 oriented to try to match left head of folded myosin in image averages. l, model using myosin 5 motor domain10 containing ADP.BeFx oriented to match left head. m, same model as l, oriented to match right head. Pro117-Pro137 lies behind the converter subdomain in this view. n, same model as k, oriented to try to match right head motor domain appearance. k-n created using PyMOL (DeLano Scientific). o, enlargement of a, coloured and labelled to show domains within folded myosin 5.
Single particle image processing7 of folded myosin 5 molecules negative-stained in ATP at nanomolar calcium reveals detailed substructure. Briefly, molecules abstracted from raw images (Supplementary Fig. 1) are aligned together, grouped into classes based on image features, and an average image calculated for each class. Most features are consistent between the class averages (Fig. 1a-e), and identification of substructure is now straightforward (Fig. 1o). The two heads lie either side of an uninterrupted proximal 17nm segment of the tail, with the head on the left of the tail more clearly delineated and closer to the tail. Within each head, calmodulin subunits of the lever domain are seen, and the levers overlap at their junction with the tail. Overlap is more extensive in some classes (Fig. 1e) suggesting levers are flexible. The tail length was highly variable in extended, rotary-shadowed molecules at high ionic strength3. In both raw and averaged negatively stained images of the folded state, we see only ∼30% of the total predicted coiled coil3. Thus, portions of the coiled coil may be unstable and function as elastic linkers between motor and cargo8. Preliminary studies of molecules prepared in ADP or nucleotide-free conditions indicate that their structures are similar to those in ATP (Fig. 1h,i).
The motor domains have structural detail that can be reconciled with image averages of single heads and atomic structures. Visual inspection suffices for these comparisons since we have a single 2D view of the molecule, and the features are distinct. The left head closely resembles a proportion of heads of myosin 5 HMM9 and single heads (S1) (Fig. 1g) in ATP. It also matches well an atomic model built using the crystal structure of the myosin 5 motor domain in the ‘post-rigor’ conformation with an ATP analogue in the active site10 (see Supplementary Methods; Fig. 1l). It does not match the pre-powerstroke conformation of myosin 5 HMM9 or S1 (Fig. 1f) or a pre-powerstroke11 atomic model oriented to show the same motor domain features (Fig. 1k), primarily because the motor is differently oriented on the lever. Differences in lever shape between folded molecules and models of heads are expected since folding may exert intramolecular strain and the lever is flexible9 (see following paragraph and Supplementary Information).
The right head motor domain looks different. Its smaller size, strong stain exclusion and pronounced surrounding stain deposit indicate it is approximately end-on. Its triangular shape is well matched by atomic models with the motor domain almost end-on (Fig. 1m,n). Although the appearance of the right lever resembles the pre-powerstroke model (Fig. 1n), the emergence point of the lever from the motor more accurately matches the post-rigor model (Fig. 1m). Moreover right heads of molecules in ADP or nucleotide-free resemble those in ATP (Fig. 1h,i), yet are unlikely to occupy the pre-powerstroke state. This head is therefore probably in the same post-rigor conformation as the left head. Differences between images and the post-rigor model arise from lever flexibility9 (see also Supplementary Information). Thus neither head appears to be in the pre-powerstroke conformation, despite this being the commonest conformation of heads of unregulated HMM in ATP9.
The GTDs appear as two globules lying between the motor domains. A smaller lobe of the GTD, which touches the motor domain of the left head close to the lever, can be identified because it is absent from images of the head alone (Fig. 1a-e cf Fig. 1g; see also Fig. 1o). The length of each GTD (∼7nm) is similar to that of isolated, crystallised GTD from yeast Myo2p (8.8nm; ref 12). The consistent location of the smaller lobe indicates specific binding. Comparison with the atomic model (see Supplementary Information) shows the GTD-binding site on the motor domain appears restricted to a specific segment, chiefly residues Pro117-Pro137 (pink in Fig. 1k-n and Fig. 2a,b), and possibly extending to include His138-Glu154 (i.e. helix E; ref13). The small lobe of GTD associated with the right head is less prominent, but extends into the junction between motor domain and lever, which is the location of Pro117-Pro137 in right head models. Within the motor domain, Pro117-Pro137 abuts the entrance to the ATP-binding pocket, loop 1 and the β-bulge of the transducer10, but is far from the actin-binding surface. GTD-induced conformational changes could readily modify nucleotide binding or hydrolysis and, through modifying the properties of the central 7-strand β-sheet, inhibit actin binding and stimulation of ATPase activity (see Supplementary Information). Thus each GTD binds at an equivalent site and acts allosterically, not by physically blocking binding to actin.
Figure 2.
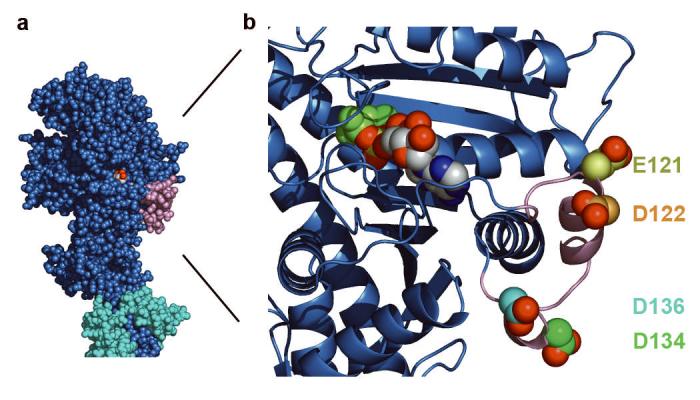
GTD-binding region of myosin 5 motor domain. a, motor domain region of myosin 5.ADP.BeFx complex, with putative GTD-binding region pink and proximal calmodulin cyan. b, enlargement of GTD-binding region of the motor domain of a to show spatial relationship to bound nucleotide; polypeptide chain shown in cartoon form, except the four acidic residues (E121, D122, D134, D136) shown in spacefill with carbons yellow, orange, green and cyan respectively and carboxylate oxygens red; bound MgADP.BeFx as spacefilling model, ADP in CPK colours, BeFx olive, Mg2+ green. Created using PyMOL (DeLano Scientific).
We aligned amino acid sequences at the putative GTD-binding region of the motor domain for myosins 5a, b and c and compared this with other myosin classes (Supplementary Fig. 2). There are 3-4 acidic residues and no basic residues in any myosin 5a or 5b. The C-terminal GDMDP sequence is conserved among vertebrate myosins 5, with conservative substitutions in invertebrates. Other myosin classes have non-conservative substitutions, indicating this may be a site of specificity for myosin 5 regulation. The acidic residues face outwards (Fig. 2b), suggesting ionic interactions with basic residues of the GTD. The densest cluster of basic residues in the GTD is 5 within Arg1800-Lys1810. This is homologous sequence to helix 13 of the GTD of yeast Myo2p, located at the distal end of the GTD12, and therefore well-suited to match the morphology we see. Ionic interactions are consistent with the salt-sensitivity of folding4-6.
The role of the GTD in regulation was studied using a glutathione S-transferase (GST)-47 kDa GTD fusion protein. GST is dimeric so the GTD is paired, as in myosin. (GST-GTD)2 inhibits actin-activated MgATPase of myosin 5 HMM at nanomolar calcium with a Ki of 0.53±0.14μM (n=5), but not at micromolar calcium (Fig. 3a). GST2 alone has no effect. Variable residual activity at GTD)2 partly derives from contaminant single-headed HMM4. saturating (GST. This is supported by finding that (GST-GTD)2 inhibits S1 more weakly (Ki 7.5μM; Fig. 3b). The GTD is therefore sufficient to inhibit activity: the intervening sequence between the HMM coiled coil and the GTD is not required. S1 inhibition shows that paired heads are not required for inhibition, but do enhance the effectiveness. Electron microscopy shows that under inhibitory conditions, the HMM-(GST-GTD)2 complex is a compact, triangular shape very similar to folded myosin 5 (Fig. 1j). Similarities include the difference between the shape of the two heads, and the shape and location of the GTDs, consistent with them binding the same sites as in myosin. (GST)2, of mass (51kDa) similar to a single GTD, is not visible, possibly being obscured by the central pool of stain.
Figure 3.
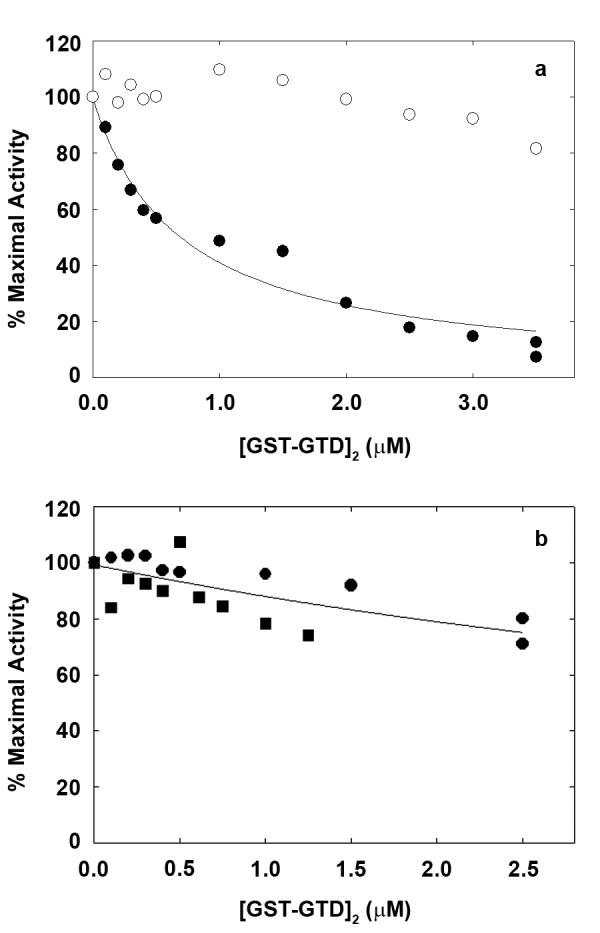
Regulation of actin-activated myosin 5 ATPase activity by GST-GTD dimers. a, myosin 5 HMM; solid circles at nanomolar calcium, open circles with 0.1 mM free calcium. Fitted line gives Ki 0.75μM. b, myosin 5 S1 at nanomolar calcium, squares and circles represent independent experiments. Ca2+ did not greatly affect the Km values for actin activation of HMM MgATPase which were generally in the range of 0.3-0.7 μM in the presence of 50 mM KCl.
It is a long-standing paradox that at nanomolar calcium, where ATPase is inhibited, myosin 5 bound to a coverslip moves actin better than at higher calcium3, and single molecule motility assays show individual myosin 5 molecules moving along actin14-16. Discovery of the folded conformation at nanomolar calcium did not solve this puzzle, since folding echoes the ‘off’-state of myosin 2 and kinesin17, 18. We addressed this paradox using two-colour TIRF (total internal reflection fluorescence) microscopy to measure simultaneously the single molecule motility of GFP-HMM and Cy3-myosin in mixtures interacting with the same actin filaments. Observing both species interacting with the same actin filaments overcomes difficulties of comparison between separate experiments. Without ATP, the filaments became heavily decorated with both proteins (Supplementary Fig. 3). With ATP, labelling was lighter with rapid movements of both proteins (see Supplementary Movies). The frequency of movements of each protein showed HMM movements (560 events/100pM HMM/min) were much more frequent than myosin movements (14 events/100pM myosin/min). This indicates that the great majority of myosin molecules do not move on actin, due to being switched off. Including 2.5μM (GST-GTD)2 in the mixtures reduced HMM movements from 660±50 to 317±26 events±SD/100pM/min, but myosin movements were unaffected (16±6 to 15±6 events/100pM/min), showing the assay senses inhibition of activity. Myosins that move are probably either a small proportion of molecules that cannot fold or molecules that are unfolded when they collide with actin, due to the folded-unfolded equilibrium. Movement of a small proportion of myosin molecules explains the residual actin activation of myosin 5 ATPase (ref 3, 4, 6) and the movements seen in earlier studies, and thus largely explains the paradox. In vitro, calcium disrupts the folded state and stimulates ATPase activity. Since neither motor nor GTD binds calcium, structural changes linked to calcium binding by calmodulins in the lever are the likely cause. In particular, dissociation of calcium-calmodulin from the lever, weakening it, is implicated in lowering motility6, 19. This explains why the few active molecules at nanomolar calcium move actin better. Calcium-induced calmodulin dissociation may also disrupt the geometry required for inhibitory motor domain-GTD interactions, explaining how calcium stimulates ATPase activation by actin.
TIRF shows few, if any, long-lived (>1sec) stationary transient attachments of myosin to actin in ATP. Moreover, electron microscopy of myosin-actin mixtures under these conditions shows detached, folded myosin and undecorated actin (Supplementary Fig. 4a). In contrast, with no nucleotide or with ADP or ADP+phosphate, TIRF shows heavy decoration, and electron microscopy shows that bundles of myosin-decorated actin filaments are immediately formed in solution (Supplementary Fig. 4b-d). Thus the folded molecules in ATP bind only weakly to actin with a fast off rate, but under other conditions they bind more tightly.
The mechanism of myosin 5 regulation we have described differs from myosin 2, which uses head-head and head-proximal tail interactions17, 20, 21 and therefore has regulated HMM20. Myosin 5 has more similarities with kinesin, in which a GTD binds the motor domain, slows ADP release and reduces microtubule affinity18.
The GTD binds cargo receptor proteins as well as the motor domain. Activation of myosin 5 in vivo may therefore entail receptor proteins binding to the GTD, weakening the GTD-motor interactions we have described and allowing the heads to interact with actin22. Our data indicate that the folded molecule binds weakly to actin, so would be able to diffuse freely in cells. After myosin 5 dissociates from cargo at the cell periphery, folding would allow recycling to bind new cargo elsewhere in the cell.
METHODS
Proteins
GST-GTD fusion protein comprised the C-terminal 414 residues of myosin 5a, encompassing the entire GTD, that was amplified by PCR with Ultra High Fidelity Pfu (Stratagene) with Bam H1/Eco R1 ends, cloned into pGex-4T1 (Amersham) and expressed and purified by standard techniques. Other protein preparations are detailed in Supplementary Methods.
MgATPase assay
Actin-activated MgATPase assays were performed as previously described23 using 20-40nM HMM or S1, 10μM actin, 50mM KCl, 2mM MgCl2, 1mM ATP, 0.1mM EGTA, 10mM MOPS (pH 7.0), 40 units/ml lactate dehydrogenase, 1mM phosphoenolpyruvate, 200 units/ml pyruvate kinase, 200μM NADH, 2μM calmodulin at 25°C. CaCl2 if added was 0.2mM.
Single molecule motility assay
Single molecule TIRF assays were performed as described previously23, except that a two colour observation system (Dual view system, Optical Insights, LLC, AL) was used to simultaneously view both GFP-HMM and Cy3-labeled myosin. 6-75pM GFP-HMM and 60-4,000pM Cy3-myosin were added into a flow chamber in the presence of ATP and recorded at a frame rate of 2s−1. The number of moving spots on all actin filaments was counted in a 22×44μm2 area on both channels. Further detail is given in Supplementary Methods.
Electron microscopy and image processing
Typically, 40nM myosin, 480nM calmodulin, 100mM KCl, 1.5mM MgCl2, 20μM ATP, 0.1mM EGTA, 2mM Kphosphate, 10mM MOPS, pH 7.0 at 20°C was applied to a carbon-coated electron microscope grid and immediately stained with uranyl acetate24. In some experiments ATP was replaced by hexokinase+glucose treated ADP (1.25mM) or the same plus Kphosphate (25mM), or myosin was treated with apyrase. Actin when present was 0.75μM, pre-treated with hexokinase+glucose or apyrase as needed. HMM-GTD complex comprised 40nM HMM, 60nM GST-GTD dimer, 55μM ATP in the above buffer. Images were processed using SPIDER as described24. Further detail is given in Supplementary Methods.
Supplementary Material
Acknowledgements
We thank S. A. Burgess for expert advice and assistance with image processing and The Wellcome Trust for support. TS was supported by a fellowship from the Japanese Society for the Promotion of Science.
Footnotes
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
References
- 1.Vale RD. Myosin V motor proteins: marching stepwise towards a mechanism. J. Cell Biol. 2003;163:445–450. doi: 10.1083/jcb.200308093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sellers JR, Veigel C. Walking with myosin V. Curr. Opin. Cell Biol. 2006;18:68–73. doi: 10.1016/j.ceb.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 3.Cheney RE, et al. Brain myosin-V is a 2-headed unconventional myosin with motor-activity. Cell. 1993;75:13–23. doi: 10.1016/S0092-8674(05)80080-7. [DOI] [PubMed] [Google Scholar]
- 4.Wang F, et al. Regulated conformation of myosin V. J. Biol. Chem. 2004;279:2333–2336. doi: 10.1074/jbc.C300488200. [DOI] [PubMed] [Google Scholar]
- 5.Li XD, Mabuchi K, Ikebe R, Ikebe M. Ca2+-induced activation of ATPase activity of myosin Va is accompanied with a large conformational change. Biochem. Biophys. Res. Comm. 2004;315:538–545. doi: 10.1016/j.bbrc.2004.01.084. [DOI] [PubMed] [Google Scholar]
- 6.Krementsov DN, Krementsova EB, Trybus KM. Myosin V: regulation by calcium, calmodulin, and the tail domain. J. Cell Biol. 2004;164:877–886. doi: 10.1083/jcb.200310065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 8.Schilstra MJ, Martin SR. An elastically tethered viscous load imposes a regular gait on the motion of myosin-V. Simulation of the effect of transient force relaxation on a stochastic process. J. R. Soc. Interface. 2006;3:153–165. doi: 10.1098/rsif.2005.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burgess S, et al. The prepower stroke conformation of myosin V. J. Cell Biol. 2002;159:983–991. doi: 10.1083/jcb.200208172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coureux PD, Sweeney HL, Houdusse A. Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J. 2004;23:4527–4537. doi: 10.1038/sj.emboj.7600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houdusse A, Szent-Györgyi AG, Cohen C. Three conformational states of scallop myosin S1. Proc. Natl. Acad. Sci. USA. 2000;97:11238–11243. doi: 10.1073/pnas.200376897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pashkova N, Jin Y, Ramaswamy S, Weisman LS. Structural basis for myosin V discrimination between distinct cargoes. EMBO J. 2006;25:693–700. doi: 10.1038/sj.emboj.7600965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cope M, Whisstock J, Rayment I, Kendrick-Jones J. Conservation within the myosin motor domain: Implications for structure and function. Structure. 1996;4:969–987. doi: 10.1016/s0969-2126(96)00103-7. [DOI] [PubMed] [Google Scholar]
- 14.Sakamoto T, Amitani I, Yokota E, Ando T. Direct observation of processive movement by individual myosin V molecules. Biochem. Biophys. Res. Comm. 2000;272:586–590. doi: 10.1006/bbrc.2000.2819. [DOI] [PubMed] [Google Scholar]
- 15.Yildiz A, et al. Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 16.Forkey JN, Quinlan ME, Shaw MA, Corrie JET, Goldman YE. Three-dimensional structural dynamics of myosin V by single-molecule fluorescence polarization. Nature. 2003;422:399–404. doi: 10.1038/nature01529. [DOI] [PubMed] [Google Scholar]
- 17.Wendt T, Taylor D, Trybus KM, Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc. Natl. Acad. Sci. USA. 2001;98:4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hackney DD, Stock MF. Kinesin's IAK tail domain inhibits initial microtubule-stimulated ADP release. Nature Cell Biol. 2000;2:257–260. doi: 10.1038/35010525. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen H, Higuchi H. Motility of myosin V regulated by the dissociation of single calmodulin. Nat. Struct. Mol. Biol. 2005;12:127–132. doi: 10.1038/nsmb894. [DOI] [PubMed] [Google Scholar]
- 20.Sellers JR. In: Myosins. Sheterline P, editor. Oxford: Oxford University Press; 1999. [Google Scholar]
- 21.Ankrett RJ, Rowe AJ, Cross RA, Kendrick-Jones J, Bagshaw CR. A folded (10 S) conformer of myosin from a striated muscle and its implications for regulation of ATPase activity. J. Mol. Biol. 1991;217:323–335. doi: 10.1016/0022-2836(91)90546-i. [DOI] [PubMed] [Google Scholar]
- 22.Li XD, Ikebe R, Ikebe M. Activation of myosin Va function by melanophilin, a specific docking partner of myosin Va. J. Biol. Chem. 2005;280:17815–17822. doi: 10.1074/jbc.M413295200. [DOI] [PubMed] [Google Scholar]
- 23.Sakamoto T, et al. Neck length and processivity of myosin V. J. Biol. Chem. 2003;278:29201–29207. doi: 10.1074/jbc.M303662200. [DOI] [PubMed] [Google Scholar]
- 24.Burgess SA, Walker ML, Thirumurugan K, Trinick J, Knight PJ. Use of negative stain and single-particle image processing to explore dynamic properties of flexible macromolecules. J. Struct. Biol. 2004;147:247–258. doi: 10.1016/j.jsb.2004.04.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


