Abstract
To date, only the H1 MAPT haplotype has been consistently associated with risk of developing the neurodegenerative disease progressive supranuclear palsy (PSP). We hypothesized that additional genetic loci may be involved in conferring risk of PSP that could be identified through a pooling-based genomewide association study of >500,000 SNPs. Candidate SNPs with large differences in allelic frequency were identified by ranking all SNPs by their probe-intensity difference between cohorts. The MAPT H1 haplotype was strongly detected by this methodology, as was a second major locus on chromosome 11p12-p11 that showed evidence of association at allelic (P<.001), genotypic (P<.001), and haplotypic (P<.001) levels and was narrowed to a single haplotype block containing the DNA damage-binding protein 2 (DDB2) and lysosomal acid phosphatase 2 (ACP2) genes. Since DNA damage and lysosomal dysfunction have been implicated in aging and neurodegenerative processes, both genes are viable candidates for conferring risk of disease.
Progressive supranuclear palsy (PSP [MIM 601104]) is the second-most-common form of parkinsonism, with a population prevalence rate of 6–6.4 per 100,000.1,2 Clinical features include vertical-gaze palsy and postural instability.3,4 PSP is characterized neuropathologically by neuronal and glial inclusions composed of aggregated microtubule associated protein tau (MAPT) in the basal ganglia and brain stem.5,6 Mutations in the MAPT (MIM 157140) gene have been identified in patients with a clinical presentation of PSP.7–14 A recent report also described linkage to chromosome 1q31.1 in a family with autosomal dominant PSP.15 However, only the MAPT locus has been consistently associated with increased risk for idiopathic PSP.16–20 The MAPT locus exists as two major haplotype groups, termed “H1” and “H2”16 in European populations, with the H2 haplotype defined by >100 SNPs that are inherited in strong linkage disequilibrium (LD) with each other, reflecting the total absence of H1-H2 recombination.21 Inheritance of two copies of the H1 haplotype (H1/H1) is a major genetic risk factor for PSP.16 A large collection of pathologically confirmed PSP samples was used recently to fine map PSP risk on H1 chromosomes in PSP cases and controls. 22,23 PSP risk was associated with an extended subhaplotype, and narrowing the region for PSP risk to a 22-kb region in intron 0 of MAPT was accomplished by examining younger patients with, presumably, a larger genetic component to their disease.22,23 The most likely explanation of the association with the MAPT H1 haplotype and PSP is that variants in the H1 (and H2) haplotypes confer risk of (protect against) disease by altering expression at the locus, with the risky H1 haplotypes expressing higher levels of MAPT.22–26
Calculations of population-attributable risk suggest that only ∼68% of the risk of PSP can be accounted for by the MAPT H1 haplotype, suggesting there may be additional risk genes involved in PSP. We hypothesized that additional genetic loci involved in conferring risk of PSP could be identified through genomewide association (GWA) methods. The cost of performing an association study that involved individual genotyping of thousands of SNPs for a series this size was prohibitive, so, instead, we used a pooled-DNA approach to identify additional risk factors. Whereas a pooling-based genomewide scan of thousands of SNPs has been proposed in principle, in large part, these studies have not been used for the discovery of genes predisposing to complex diseases,27,28 likely because of technical concerns or lack of technology and analysis tools.
The patients used in the initial pooling study, the “original” series, were largely derived from pathologically confirmed subjects collected by the PSP Society and sent to D.W.D. for brain autopsy. As described elsewhere, the patient samples in this brain bank were donated from the United States and Canada.29 The patient series is similar to the one that we employed in previous studies to fine map the H1 genetic risk,22 with 288 subjects with a primary pathological diagnosis of PSP used to create the pool of PSP-affected patients. A total of 344 age- and sex-matched cognitively normal control individuals were obtained through the Normal and Pathological Aging protocol at the Mayo Clinic (Scottsdale),30,31 to create the pool of control individuals. All patient and control individuals were white from the United States and Canada, and institutional review board (IRB)–approved protocols were used in the collection of all samples.
Replicate pools of patients with PSP and control individuals were created as described elsewhere.32 Samples were genotyped on 20 replicate Affymetrix 500K arrays and 20 Affymetrix 100K, in accordance with the Affymetrix protocols, whereby each of the five replicate pools was genotyped on two replicate arrays. This design therefore yielded probe-intensity data for both platforms on 10 replicate arrays per cohort. Data were analyzed using GenePool software (TGen Bioinformatics Research Unit).32 In brief, probe-intensity data were directly read from cell-intensity (CEL) files, and relative allele signal (RAS) values were calculated for each quartet. These values yield independent measures of different hybridization events and are consequently treated as individual data points. We used a silhouette statistic to rank all genotyped SNPs,33 because it avoids introducing unnecessary variance by averaging probe-intensity data from probes with different hybridization properties. Silhouette scores range from 1, where significant separation between data points has been achieved and cluster assignment can be made with confidence, to −1, where differences in allelic frequencies are less reliable. Poorly performing SNPs were identified by Affymetrix as unreliable in the transition to Mendel3 libraries or exhibited high variance between replicate arrays and were removed from the analysis; 428,867 SNPs remained. SNPs were ranked on the basis of silhouette score, whereby the SNP with the highest score was ranked 1 and the SNP with the lowest score was ranked 428,867, with use of Affymetrix’s Mendel3 libraries for the Affymetrix 500K arrays and HindIII and XbaI libraries for the Affymetrix 100K arrays, then were sorted by chromosome and physical position. With this ranking, it is assumed that SNPs approaching a rank of 1 will have larger differences in allelic frequency. With each sample ranked by silhouette score, we calculated a sliding-window statistic of the mean rank for consecutively neighboring SNPs across a fixed window size. Window sizes from 2 to 31 were used.
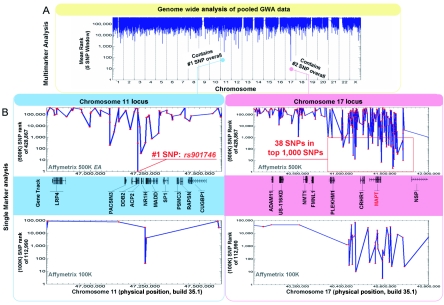
Since the MAPT H1 haplotype is associated with disease with a haplotypic odds ratio (OR) of ∼3–4,16,22,23,34 it served as an internal positive control for the study. For analysis, we used the 500K data to identify chromosomal regions of interest (i.e., those with small mean-rank scores). The 100K data were then used to confirm that a region identified in the 500K analysis contained SNPs with large allelic frequency differences. The SNP with the single best statistical rank on the 500K chip was rs901746 on chromosome 11p12, and the second-best SNP was rs17662235, near MAPT. The top 1,000 SNPs, based on individual statistical rank, are given in table 1. Multimarker statistics also identified both chromosome 11p12 and chromosome 17q21 (MAPT) regions with sliding windows of multiple sizes. Although we recognize that this type of statistic is biased because of genomewide LD, it allowed us to identify clusters of high-ranking SNPs that neighbor one another, which reduced the possibility of technical errors influencing the results. Shown in figure 1A, the MAPT locus, labeled as having the #2 SNP overall, showed the greatest evidence of differences between case and control pools with use of the sliding-window analysis, largely because of 38 SNPs within the top 1,000 SNPs overall and a total of 75 SNPs in the region with a rank score of <10,000 (fig. 1B and table 1). Examination of the individual SNPs with high rank scores over this locus showed SNPs that were derived from a region covering the full extent of the MAPT H1 haplotype, spanning nearly 1 Mb (fig. 1B).36 All of the 75 SNPs with genotype-frequency data in the database resembled MAPT H2 variants (which differentiate between H1 and H2 MAPT haplotypes) rather than H1 variants (which differentiate between H1 subhaplotypes); this is because, in white populations, the SNP minor-allele frequency was ∼0.2, whereas the minor allele of the SNP was absent or rare in Asian populations and African populations.36,37 In addition, two of the SNPs with low rank scores (rs12150111 and rs807072) were identified definitively as MAPT H2 variants from prior MAPT genomic sequencing efforts.22
Table 1. .
Predicted Allelic Frequencies for the Top 1,000 SNPs
| Frequencyc (SD) of Predicted Allele | |||||
| SNP Ranka | dbSNP Number | Chromosome | Positionb | Case | Control |
| 1 | rs901746 | 11 | 47216895 | 61.3 (.042) | 81.1 (.027) |
| 2 | rs17662235 | 17 | 41589553 | 2.7 (.019) | 15.3 (.028) |
| 3 | rs2699808 | 7 | 26175747 | 60.5 (.039) | 45.2 (.043) |
| 4 | rs17662853 | 17 | 41604625 | 26.7 (.068) | 14.0 (.039) |
| 5 | rs11898241 | 2 | 115847267 | 75.6 (.057) | 61.0 (.042) |
| 6 | rs17563501 | 17 | 41157478 | 96.4 (.050) | 79.9 (.021) |
| 7 | rs2141299 | 17 | 41596763 | 94.7 (.035) | 77.7 (.023) |
| 8 | rs17574228 | 17 | 41460355 | 12.4 (.016) | 24.8 (.035) |
| 9 | rs2732706 | 17 | 41707463 | 8.5 (.036) | 22.1 (.044) |
| 10 | rs7139545 | 13 | 24677429 | 74.2 (.022) | 85.7 (.039) |
| 11 | rs6450530 | 5 | 59095265 | 30.6 (.047) | 51.3 (.041) |
| 12 | rs2668695 | 17 | 41647903 | 13.3 (.034) | 27.3 (.038) |
| 13 | rs3759297 | 12 | 32029119 | 63.6 (.033) | 80.0 (.030) |
| 14 | rs10798036 | 1 | 182784619 | 45.0 (.045) | 61.1 (.046) |
| 15 | rs10213502 | 4 | 11624923 | 93.7 (.023) | 79.4 (.039) |
| 16 | rs7700506 | 5 | 56032355 | 65.5 (.029) | 85.5 (.031) |
| 17 | rs7198242 | 16 | 68205996 | 24.0 (.083) | 5.9 (.048) |
| 18 | rs2532286 | 17 | 41597441 | 96.6 (.045) | 89.6 (.048) |
| 19 | rs7534271 | 1 | 47406001 | 55.7 (.035) | 68.6 (.051) |
| 20 | rs9649052 | 7 | 134311099 | 49.4 (.012) | 58.6 (.034) |
| 21 | rs17604700 | 15 | 29922129 | 51.6 (.053) | 34.6 (.078) |
| 22 | rs11623728 | 14 | 96882409 | 27.6 (.019) | 18.6 (.018) |
| 23 | rs1557996 | 7 | 7430544 | 93.4 (.039) | 77.9 (.054) |
| 24 | rs2202445 | 5 | 7381176 | 57.7 (.047) | 74.5 (.056) |
| 25 | rs1527036 | 23 | 3215185 | 50.3 (.035) | 63.7 (.039) |
| 26 | rs2035515 | 8 | 107825837 | 92.1 (.025) | 82.4 (.019) |
| 27 | rs11745421 | 5 | 168123644 | 11.8 (.044) | 0.7 (.017) |
| 28 | rs429988 | 3 | 41081069 | 63.2 (.033) | 74.0 (.028) |
| 29 | rs10838681 | 11 | 47231640 | 67.6 (.030) | 80.8 (.041) |
| 30 | rs10517938 | 4 | 168729149 | 77.6 (.044) | 84.9 (.028) |
| 31 | rs12918956 | 16 | 70781836 | 42.0 (.070) | 57.0 (.059) |
| 32 | rs9567416 | 13 | 43806741 | 36.3 (.100) | 15.2 (.044) |
| 33 | rs2107699 | 7 | 49367808 | 19.6 (.082) | 45.4 (.182) |
| 34 | rs4794984 | 17 | 29333048 | 66.1 (.037) | 78.8 (.037) |
| 35 | rs8088596 | 18 | 72620941 | 65.2 (.032) | 50.1 (.049) |
| 36 | rs2116457 | 8 | 129876983 | 53.9 (.039) | 67.3 (.029) |
| 37 | rs762038 | 14 | 20304608 | 71.0 (.028) | 83.3 (.035) |
| 38 | rs1336433 | 10 | 113348065 | 32.8 (.038) | 46.2 (.039) |
| 39 | rs7620394 | 3 | 55206368 | 36.1 (.035) | 23.8 (.039) |
| 40 | rs6590039 | 11 | 123446989 | 72.7 (.029) | 57.0 (.040) |
| 41 | rs10824896 | 10 | 51463093 | 81.7 (.046) | 89.2 (.018) |
| 42 | rs354636 | 1 | 91184514 | 86.2 (.044) | 95.7 (.051) |
| 43 | rs13020820 | 2 | 33892179 | 48.8 (.042) | 59.9 (.039) |
| 44 | rs520605 | 1 | 181527436 | 68.8 (.021) | 84.7 (.058) |
| 45 | rs11926691 | 3 | 83064778 | 64.6 (.027) | 55.1 (.011) |
| 46 | rs16936441 | 9 | 18201655 | 70.9 (.068) | 88.7 (.045) |
| 47 | rs8036777 | 15 | 48208710 | 46.8 (.060) | 66.3 (.030) |
| 48 | rs1299704 | 3 | 108688432 | 32.0 (.022) | 42.7 (.047) |
| 49 | rs1550532 | 2 | 234046848 | 58.4 (.042) | 69.4 (.038) |
| 50 | rs9394550 | 6 | 38960939 | 56.3 (.059) | 41.7 (.035) |
| 51 | rs12717171 | 6 | 112800652 | 53.2 (.027) | 62.0 (.032) |
| 52 | rs2548879 | 16 | 77200307 | 54.7 (.081) | 27.9 (.077) |
| 53 | rs10256927 | 7 | 125167859 | 71.8 (.021) | 79.4 (.022) |
| 54 | rs7407281 | 18 | 55756601 | 67.4 (.045) | 82.4 (.032) |
| 55 | rs1122452 | 3 | 144910038 | 77.7 (.039) | 63.4 (.029) |
| 56 | rs12962651 | 18 | 56476045 | 47.9 (.052) | 61.0 (.040) |
| 57 | rs2070929 | 1 | 47395627 | 30.0 (.045) | 17.0 (.028) |
| 58 | rs2036535 | 17 | 28775126 | 47.9 (.046) | 38.8 (.022) |
| 59 | rs11211670 | 2 | 1346042 | 42.7 (.041) | 29.4 (.024) |
| 60 | rs4372225 | 1 | 211830838 | 75.3 (.070) | 92.3 (.048) |
| 61 | rs17660065 | 17 | 41518102 | 97.2 (.045) | 82.1 (.081) |
| 62 | rs1725604 | 7 | 101507570 | 51.8 (.035) | 41.7 (.017) |
| 63 | rs2194711 | 5 | 154994268 | 17.8 (.043) | 7.2 (.044) |
| 64 | rs6753459 | 2 | 190890259 | 25.0 (.041) | 14.2 (.039) |
| 65 | rs1041834 | 21 | 41841892 | 40.9 (.034) | 31.7 (.012) |
| 66 | rs6847349 | 4 | 13717192 | 27.6 (.040) | 45.2 (.061) |
| 67 | rs6101206 | 20 | 59050033 | 59.8 (.040) | 41.0 (.032) |
| 68 | rs2732675 | 17 | 41635965 | 1.6 (.048) | 16.9 (.046) |
| 69 | rs7396713 | 11 | 88020266 | 39.0 (.073) | 27.5 (.030) |
| 70 | rs11835314 | 12 | 106412032 | 75.3 (.034) | 57.2 (.062) |
| 71 | rs1520961 | 15 | 35523947 | 85.5 (.056) | 71.8 (.067) |
| 72 | rs4740919 | 9 | 7844892 | 38.0 (.042) | 20.2 (.039) |
| 73 | rs8088963 | 18 | 57198294 | 48.5 (.057) | 37.6 (.017) |
| 74 | rs695271 | 22 | 32327285 | 59.2 (.050) | 47.9 (.057) |
| 75 | rs9377831 | 6 | 58761209 | 14.3 (.037) | 6.3 (.013) |
| 76 | rs9955849 | 18 | 3052581 | 84.2 (.042) | 70.0 (.027) |
| 77 | rs10791979 | 11 | 55916546 | 77.2 (.068) | 55.7 (.079) |
| 78 | rs2805386 | 9 | 110761555 | 69.4 (.018) | 56.6 (.072) |
| 79 | rs4845237 | 1 | 187114452 | 82.1 (.062) | 61.3 (.061) |
| 80 | rs4440119 | 3 | 61936314 | 40.0 (.092) | 60.2 (.080) |
| 81 | rs1289951 | 1 | 117625061 | 87.2 (.021) | 76.5 (.061) |
| 82 | rs4669550 | 2 | 10273437 | 15.3 (.055) | 7.2 (.016) |
| 83 | rs6712370 | 2 | 69766745 | 35.7 (.072) | 20.3 (.057) |
| 84 | rs6469264 | 8 | 110594779 | 20.0 (.089) | 6.4 (.041) |
| 85 | rs323376 | 8 | 30851071 | 15.5 (.071) | 39.5 (.084) |
| 86 | rs916793 | 17 | 41310477 | 95.3 (.068) | 78.9 (.027) |
| 87 | rs210069 | 20 | 52162352 | 62.5 (.022) | 71.8 (.016) |
| 88 | rs1826763 | 4 | 177179201 | 27.1 (.059) | 46.4 (.062) |
| 89 | rs12427267 | 12 | 90826440 | 49.4 (.079) | 70.7 (.035) |
| 90 | rs2442497 | 8 | 6314156 | 11.5 (.052) | 22.7 (.046) |
| 91 | rs2583513 | 4 | 116275788 | 71.1 (.023) | 64.0 (.029) |
| 92 | rs2191265 | 7 | 12678307 | 42.2 (.039) | 56.7 (.040) |
| 93 | rs2163385 | 18 | 38452575 | 8.5 (.021) | 0.5 (.022) |
| 94 | rs11071075 | 15 | 29550901 | 68.0 (.042) | 80.2 (.037) |
| 95 | rs12819116 | 12 | 12537788 | 59.2 (.036) | 70.2 (.028) |
| 96 | rs7940239 | 11 | 55880498 | 19.4 (.056) | 35.1 (.067) |
| 97 | rs1400174 | 3 | 52174 | 36.6 (.050) | 23.4 (.048) |
| 98 | rs17563787 | 17 | 41169023 | 6.9 (.064) | 22.4 (.022) |
| 99 | rs10829618 | 10 | 131380410 | 72.8 (.067) | 89.9 (.064) |
| 100 | rs9710693 | 2 | 239504258 | 34.1 (.033) | 20.7 (.040) |
| 101 | rs11157865 | 14 | 51436825 | 52.1 (.063) | 67.9 (.041) |
| 102 | rs1678719 | 6 | 38901529 | 77.5 (.043) | 95.5 (.033) |
| 103 | rs1692506 | 18 | 24511864 | 61.7 (.030) | 73.5 (.037) |
| 104 | rs10928098 | 2 | 141715827 | 42.1 (.046) | 23.8 (.046) |
| 105 | rs10975882 | 9 | 689065 | 43.1 (.034) | 50.4 (.024) |
| 106 | rs578987 | 1 | 17948686 | 38.3 (.041) | 25.2 (.042) |
| 107 | rs2274403 | 13 | 94645021 | 61.8 (.047) | 48.4 (.024) |
| 108 | rs3807404 | 7 | 38325866 | 61.0 (.062) | 77.7 (.036) |
| 109 | rs2532275 | 17 | 41602774 | 95.7 (.033) | 84.0 (.048) |
| 110 | rs6920684 | 6 | 8306390 | 66.6 (.044) | 56.3 (.054) |
| 111 | rs3911781 | 10 | 67516022 | 77.9 (.046) | 89.6 (.036) |
| 112 | rs1304527 | 21 | 35459060 | 94.4 (.048) | 77.0 (.067) |
| 113 | rs10021851 | 4 | 40209776 | 85.4 (.058) | 58.0 (.109) |
| 114 | rs1445517 | 4 | 138159604 | 40.1 (.034) | 48.7 (.056) |
| 115 | rs11119752 | 1 | 208064160 | 65.9 (.041) | 54.1 (.015) |
| 116 | rs17577094 | 17 | 41543275 | 2.7 (.098) | 28.7 (.093) |
| 117 | rs2532316 | 17 | 41569489 | 90.6 (.043) | 80.2 (.021) |
| 118 | rs7177316 | 15 | 48209122 | 43.6 (.049) | 58.8 (.059) |
| 119 | rs2696600 | 17 | 41572003 | 89.8 (.036) | 79.7 (.031) |
| 120 | rs5751592 | 22 | 21822228 | 75.7 (.047) | 91.0 (.019) |
| 121 | rs9354193 | 6 | 65733294 | 59.6 (.047) | 47.7 (.041) |
| 122 | rs6581596 | 12 | 63670237 | 31.8 (.059) | 46.1 (.037) |
| 123 | rs722979 | 10 | 83952201 | 66.8 (.051) | 54.6 (.038) |
| 124 | rs4943389 | 13 | 36008899 | 36.4 (.049) | 51.8 (.048) |
| 125 | rs10931378 | 2 | 189420130 | 29.9 (.049) | 40.1 (.047) |
| 126 | rs919262 | 5 | 151246277 | 38.9 (.045) | 26.1 (.044) |
| 127 | rs1399564 | 8 | 76646411 | 37.1 (.034) | 49.7 (.052) |
| 128 | rs12043736 | 1 | 215866548 | 57.0 (.047) | 72.5 (.057) |
| 129 | rs17353336 | 3 | 134155679 | 54.1 (.032) | 39.8 (.024) |
| 130 | rs9371835 | 6 | 155327752 | 39.1 (.038) | 48.7 (.030) |
| 131 | rs1635216 | 23 | 3214080 | 57.9 (.075) | 80.9 (.053) |
| 132 | rs1159788 | 4 | 103664727 | 35.5 (.049) | 25.0 (.030) |
| 133 | rs153518 | 5 | 142937957 | 46.4 (.021) | 40.3 (.037) |
| 134 | rs17074118 | 13 | 29420217 | 17.8 (.026) | 4.3 (.043) |
| 135 | rs206034 | 23 | 39291012 | 56.2 (.032) | 74.3 (.066) |
| 136 | rs9287986 | 2 | 176312475 | 51.9 (.027) | 36.9 (.046) |
| 137 | rs1562495 | 3 | 144910247 | 31.1 (.025) | 43.8 (.051) |
| 138 | rs1993329 | 6 | 68992781 | 43.9 (.062) | 28.3 (.068) |
| 139 | rs6060965 | 20 | 29869759 | 29.5 (.032) | 18.1 (.030) |
| 140 | rs1957779 | 14 | 62739400 | 62.5 (.027) | 43.1 (.059) |
| 141 | rs10745813 | 12 | 96767932 | 11.6 (.033) | 24.0 (.055) |
| 142 | rs2064855 | 20 | 59058742 | 35.4 (.048) | 16.9 (.045) |
| 143 | rs7321718 | 13 | 72257693 | 52.6 (.036) | 66.2 (.025) |
| 144 | rs1220627 | 5 | 72906819 | 29.9 (.033) | 20.0 (.029) |
| 145 | rs1952251 | 1 | 180714144 | 36.5 (.079) | 23.8 (.040) |
| 146 | rs4420176 | 10 | 127666188 | 51.2 (.032) | 40.7 (.061) |
| 147 | rs338565 | 1 | 181508758 | 22.5 (.112) | 7.8 (.034) |
| 148 | rs776965 | 8 | 107768142 | 47.9 (.066) | 30.1 (.052) |
| 149 | rs4132608 | 13 | 109898267 | 68.4 (.030) | 80.8 (.021) |
| 150 | rs1841043 | 4 | 69889478 | 10.9 (.034) | 30.4 (.058) |
| 151 | rs11259272 | 10 | 6586077 | 34.3 (.058) | 61.5 (.106) |
| 152 | rs7901651 | 10 | 85617227 | 49.3 (.048) | 63.1 (.057) |
| 153 | rs7999230 | 13 | 106749278 | 27.0 (.033) | 39.2 (.039) |
| 154 | rs12144146 | 1 | 151338046 | 40.1 (.081) | 22.3 (.038) |
| 155 | rs10842885 | 12 | 27291048 | 63.8 (.054) | 47.7 (.056) |
| 156 | rs17173853 | 7 | 149243724 | 81.3 (.037) | 93.4 (.026) |
| 157 | rs1547597 | 7 | 147404398 | 39.5 (.096) | 2.3 (.064) |
| 158 | rs1549411 | 12 | 29684407 | 75.5 (.070) | 92.6 (.070) |
| 159 | rs586457 | 10 | 6537428 | 41.6 (.042) | 25.6 (.042) |
| 160 | rs10894846 | 11 | 133954025 | 23.6 (.074) | 10.7 (.048) |
| 161 | rs10459826 | 16 | 5680786 | 87.5 (.070) | 70.6 (.039) |
| 162 | rs256447 | 5 | 79375707 | 1.6 (.029) | 9.7 (.064) |
| 163 | rs476906 | 3 | 177685396 | 68.6 (.040) | 78.8 (.017) |
| 164 | rs4633144 | 9 | 32494294 | 52.3 (.058) | 71.6 (.065) |
| 165 | rs694444 | 13 | 63372180 | 73.3 (.066) | 83.3 (.018) |
| 166 | rs332735 | 2 | 119167473 | 79.3 (.056) | 90.6 (.028) |
| 167 | rs12424367 | 12 | 90826168 | 55.4 (.079) | 73.1 (.048) |
| 168 | rs12281742 | 11 | 68060205 | 30.8 (.050) | 16.1 (.060) |
| 169 | rs2809658 | 1 | 42719489 | 35.8 (.047) | 53.7 (.049) |
| 170 | rs1249426 | 2 | 141775551 | 67.9 (.039) | 88.3 (.060) |
| 171 | rs2709733 | 7 | 20728610 | 56.7 (.052) | 37.6 (.096) |
| 172 | rs1878248 | 10 | 113311751 | 32.1 (.060) | 21.1 (.028) |
| 173 | rs4730720 | 7 | 115489186 | 34.1 (.048) | 16.2 (.053) |
| 174 | rs9321395 | 6 | 133642955 | 8.3 (.057) | 24.2 (.042) |
| 175 | rs2167287 | 12 | 61068734 | 85.2 (.040) | 92.7 (.014) |
| 176 | rs973406 | 2 | 176935238 | 76.4 (.047) | 63.9 (.041) |
| 177 | rs10795321 | 10 | 15901281 | 14.4 (.061) | 30.4 (.044) |
| 178 | rs10832515 | 11 | 15725228 | 33.6 (.038) | 24.8 (.034) |
| 179 | rs6538018 | 9 | 135465832 | 50.3 (.044) | 75.4 (.095) |
| 180 | rs1692507 | 18 | 24511941 | 63.6 (.042) | 75.2 (.027) |
| 181 | rs798544 | 7 | 2536343 | 32.9 (.048) | 19.3 (.025) |
| 182 | rs11708068 | 3 | 63429650 | 68.9 (.055) | 86.0 (.038) |
| 183 | rs2399547 | 10 | 10719044 | 62.2 (.055) | 78.2 (.020) |
| 184 | rs2059741 | 2 | 218719965 | 26.2 (.067) | 11.9 (.053) |
| 185 | rs7610580 | 3 | 179446096 | 59.9 (.017) | 50.9 (.045) |
| 186 | rs4705890 | 5 | 130882899 | 28.3 (.071) | 14.4 (.050) |
| 187 | rs6949651 | 7 | 68983791 | 82.2 (.045) | 72.1 (.070) |
| 188 | rs10944721 | 6 | 94937693 | 91.5 (.051) | 80.5 (.062) |
| 189 | rs4865206 | 4 | 53047922 | 13.1 (.073) | 26.6 (.026) |
| 190 | rs2532292 | 17 | 41592845 | 93.2 (.029) | 81.3 (.078) |
| 191 | rs807339 | 14 | 98483425 | 21.4 (.057) | 5.0 (.051) |
| 192 | rs2830191 | 21 | 26664758 | 1.8 (.049) | 12.6 (.044) |
| 193 | rs980214 | 6 | 116996269 | 57.8 (.073) | 77.1 (.057) |
| 194 | rs1868629 | 14 | 80117727 | 37.5 (.043) | 16.1 (.045) |
| 195 | rs2589559 | 10 | 9101376 | 94.9 (.084) | 76.3 (.081) |
| 196 | rs402642 | 1 | 18332121 | 56.8 (.038) | 70.1 (.021) |
| 197 | rs10823706 | 10 | 72678863 | 40.6 (.028) | 54.3 (.032) |
| 198 | rs2227072 | 22 | 33662312 | 47.3 (.054) | 33.1 (.030) |
| 199 | rs1460167 | 8 | 80402202 | 33.9 (.054) | 24.7 (.026) |
| 200 | rs6470355 | 8 | 126529717 | 74.1 (.032) | 89.1 (.034) |
| 201 | rs4877253 | 9 | 83194004 | 58.2 (.050) | 73.5 (.049) |
| 202 | rs17641413 | 5 | 177706306 | 65.7 (.025) | 52.6 (.048) |
| 203 | rs11869264 | 17 | 28135096 | 45.7 (.035) | 63.8 (.051) |
| 204 | rs17617724 | 8 | 40283622 | 74.0 (.029) | 89.7 (.057) |
| 205 | rs4795653 | 17 | 27163735 | 6.8 (.043) | 18.0 (.050) |
| 206 | rs2053844 | 4 | 64740967 | 61.3 (.056) | 76.3 (.050) |
| 207 | rs7649638 | 3 | 61936012 | 40.5 (.094) | 66.5 (.104) |
| 208 | rs2916755 | 8 | 6356758 | 80.3 (.069) | 61.6 (.066) |
| 209 | rs1257669 | 14 | 98561945 | 52.6 (.026) | 42.8 (.027) |
| 210 | rs1635217 | 23 | 3214104 | 32.7 (.077) | 16.5 (.077) |
| 211 | rs11241644 | 5 | 121831417 | 73.1 (.034) | 80.3 (.009) |
| 212 | rs594731 | 1 | 178226185 | 21.3 (.048) | 13.5 (.028) |
| 213 | rs7750190 | 6 | 67325355 | 45.5 (.058) | 33.6 (.028) |
| 214 | rs10826304 | 10 | 60940671 | 43.3 (.035) | 55.1 (.038) |
| 215 | rs13419861 | 2 | 139837545 | 23.0 (.031) | 14.0 (.018) |
| 216 | rs12936006 | 17 | 2628917 | 71.8 (.026) | 61.9 (.035) |
| 217 | rs12434898 | 14 | 100831794 | 15.1 (.018) | 22.0 (.031) |
| 218 | rs17100010 | 14 | 62022876 | 18.6 (.068) | 37.0 (.031) |
| 219 | rs307022 | 4 | 124375261 | 38.0 (.105) | 15.6 (.080) |
| 220 | rs4945261 | 11 | 77667908 | 21.9 (.042) | 8.0 (.042) |
| 221 | rs11609955 | 12 | 641938 | 73.5 (.024) | 79.4 (.047) |
| 222 | rs7270791 | 20 | 42558543 | 86.3 (.028) | 94.3 (.025) |
| 223 | rs228832 | 20 | 49493555 | 69.5 (.047) | 78.2 (.044) |
| 224 | rs5770111 | 22 | 47957002 | 74.2 (.036) | 84.2 (.030) |
| 225 | rs11596456 | 10 | 120562236 | 20.4 (.041) | 5.1 (.040) |
| 226 | rs796459 | 3 | 119980825 | 42.9 (.073) | 28.7 (.036) |
| 227 | rs4537652 | 1 | 213626581 | 37.4 (.047) | 46.2 (.055) |
| 228 | rs5755439 | 22 | 33662286 | 51.1 (.031) | 38.7 (.029) |
| 229 | rs741245 | 12 | 4097509 | 44.2 (.082) | 28.3 (.028) |
| 230 | rs2128404 | 3 | 65075198 | 65.9 (.058) | 48.0 (.042) |
| 231 | rs4276705 | 8 | 131631298 | 6.7 (.030) | 19.9 (.048) |
| 232 | rs2532276 | 17 | 41602401 | 4.9 (.059) | 19.9 (.029) |
| 233 | rs17098942 | 1 | 76590177 | 24.5 (.034) | 16.6 (.048) |
| 234 | rs9320784 | 6 | 121334748 | 95.9 (.021) | 89.2 (.028) |
| 235 | rs17650063 | 17 | 41358383 | 95.8 (.032) | 82.3 (.051) |
| 236 | rs17659881 | 17 | 41513416 | 89.8 (.038) | 77.4 (.035) |
| 237 | rs9296268 | 6 | 39007463 | 76.1 (.029) | 64.8 (.025) |
| 238 | rs12708283 | 9 | 107956429 | 35.2 (.057) | 20.8 (.054) |
| 239 | rs9372465 | 6 | 116968576 | 56.5 (.034) | 69.4 (.036) |
| 240 | rs11043827 | 12 | 18109374 | 87.7 (.103) | 68.4 (.072) |
| 241 | rs1760931 | 14 | 20017108 | 71.5 (.046) | 54.8 (.068) |
| 242 | rs2037276 | 15 | 60349273 | 70.2 (.047) | 80.5 (.033) |
| 243 | rs1178355 | 7 | 17978051 | 67.7 (.055) | 82.2 (.044) |
| 244 | rs10892044 | 11 | 116272109 | 15.6 (.069) | 5.5 (.031) |
| 245 | rs1356266 | 12 | 37689848 | 56.0 (.051) | 40.0 (.040) |
| 246 | rs4358170 | 2 | 42027555 | 62.3 (.061) | 77.4 (.060) |
| 247 | rs241212 | 1 | 4541639 | 39.2 (.053) | 26.7 (.026) |
| 248 | rs1902408 | 10 | 33913367 | 20.5 (.059) | 8.1 (.047) |
| 249 | rs17040424 | 3 | 14882981 | 16.7 (.046) | 3.3 (.055) |
| 250 | rs7491764 | 13 | 113618608 | 49.0 (.041) | 38.0 (.034) |
| 251 | rs7849137 | 9 | 18132898 | 36.9 (.126) | 9.2 (.067) |
| 252 | rs5934262 | 23 | 15451678 | 50.1 (.037) | 36.3 (.064) |
| 253 | rs11650055 | 17 | 11643618 | 45.2 (.055) | 56.9 (.023) |
| 254 | rs13414618 | 2 | 31488285 | 23.4 (.052) | 41.7 (.028) |
| 255 | rs9825224 | 3 | 181158256 | 78.8 (.039) | 69.6 (.045) |
| 256 | rs9861677 | 3 | 29472993 | 45.5 (.051) | 57.5 (.031) |
| 257 | rs6058847 | 20 | 30787796 | 17.9 (.039) | 28.4 (.037) |
| 258 | rs386871 | 4 | 127938037 | 5.4 (.033) | 15.0 (.033) |
| 259 | rs11835989 | 12 | 90827221 | 40.2 (.049) | 27.5 (.037) |
| 260 | rs3865177 | 16 | 81127053 | 52.7 (.064) | 69.4 (.046) |
| 261 | rs11598814 | 10 | 120562175 | 71.0 (.089) | 93.6 (.068) |
| 262 | rs6746542 | 2 | 141692156 | 34.5 (.085) | 18.6 (.042) |
| 263 | rs3924849 | 18 | 5422330 | 18.3 (.043) | 31.5 (.057) |
| 264 | rs2501856 | 1 | 209245380 | 49.2 (.053) | 57.1 (.022) |
| 265 | rs7748820 | 6 | 112048849 | 61.3 (.025) | 66.3 (.020) |
| 266 | rs1347638 | 15 | 32729249 | 50.7 (.058) | 63.2 (.053) |
| 267 | rs6548038 | 2 | 30949136 | 59.5 (.036) | 71.8 (.037) |
| 268 | rs5993875 | 22 | 18289880 | 54.6 (.046) | 41.8 (.056) |
| 269 | rs2483535 | 10 | 116303825 | 27.3 (.025) | 18.7 (.033) |
| 270 | rs2292006 | 3 | 172341108 | 68.8 (.062) | 81.5 (.039) |
| 271 | rs2696425 | 17 | 41022689 | 10.5 (.054) | 22.6 (.027) |
| 272 | rs10242225 | 7 | 67089936 | 38.0 (.081) | 14.8 (.061) |
| 273 | rs11895224 | 2 | 226748363 | 70.9 (.027) | 81.0 (.026) |
| 274 | rs11080702 | 18 | 13933768 | 91.2 (.045) | 78.4 (.106) |
| 275 | rs258619 | 5 | 75429046 | 72.0 (.050) | 84.0 (.040) |
| 276 | rs721010 | 6 | 16680699 | 46.4 (.044) | 60.8 (.065) |
| 277 | rs2321743 | 3 | 158491298 | 61.8 (.054) | 46.5 (.053) |
| 278 | rs1458246 | 4 | 69998842 | 0.6 (.037) | 11.8 (.037) |
| 279 | rs9929519 | 16 | 77365884 | 45.0 (.041) | 28.6 (.074) |
| 280 | rs968027 | 17 | 41137033 | 96.1 (.025) | 85.3 (.034) |
| 281 | rs4732082 | 7 | 134175660 | 58.5 (.063) | 43.2 (.040) |
| 282 | rs12454115 | 18 | 55731532 | 69.2 (.025) | 57.3 (.055) |
| 283 | rs475444 | 7 | 54325257 | 31.6 (.045) | 17.0 (.048) |
| 284 | rs17330110 | 2 | 36021585 | 24.1 (.047) | 16.5 (.031) |
| 285 | rs11761356 | 7 | 21016758 | 39.3 (.036) | 23.8 (.046) |
| 286 | rs2241392 | 19 | 6636983 | 28.8 (.025) | 41.7 (.046) |
| 287 | rs10175397 | 2 | 154779198 | 74.9 (.050) | 85.6 (.051) |
| 288 | rs3777994 | 6 | 116846716 | 63.3 (.059) | 80.4 (.034) |
| 289 | rs1229718 | 5 | 136771691 | 37.7 (.049) | 23.1 (.028) |
| 290 | rs7690944 | 4 | 44765392 | 84.4 (.043) | 99.1 (.032) |
| 291 | rs2765315 | 13 | 100470046 | 82.0 (.035) | 89.0 (.021) |
| 292 | rs2511775 | 11 | 130709025 | 47.7 (.124) | 70.4 (.059) |
| 293 | rs11846959 | 14 | 93916069 | 64.2 (.038) | 77.1 (.046) |
| 294 | rs7116710 | 11 | 15218897 | 44.6 (.038) | 36.8 (.039) |
| 295 | rs6468953 | 8 | 88748007 | 24.5 (.115) | 41.2 (.049) |
| 296 | rs1558226 | 17 | 9747805 | 61.9 (.022) | 55.1 (.026) |
| 297 | rs1965805 | 8 | 73818016 | 89.8 (.024) | 74.9 (.036) |
| 298 | rs9836282 | 3 | 168225293 | 37.7 (.037) | 52.8 (.056) |
| 299 | rs4965458 | 15 | 96496047 | 76.3 (.039) | 82.4 (.026) |
| 300 | rs6740906 | 2 | 213658228 | 76.4 (.031) | 67.7 (.062) |
| 301 | rs206942 | 6 | 34363454 | 12.6 (.045) | 23.5 (.063) |
| 302 | rs166812 | 3 | 194908758 | 65.3 (.070) | 43.5 (.089) |
| 303 | rs199436 | 17 | 42144468 | 9.8 (.052) | 23.5 (.046) |
| 304 | rs6078524 | 20 | 12268155 | 4.0 (.067) | 26.1 (.071) |
| 305 | rs12620374 | 2 | 162024500 | 63.4 (.065) | 76.3 (.049) |
| 306 | rs6918297 | 6 | 19507476 | 30.3 (.053) | 48.1 (.067) |
| 307 | rs1232487 | 20 | 11827248 | 66.4 (.058) | 52.9 (.052) |
| 308 | rs1583758 | 2 | 209886318 | 21.7 (.052) | 38.7 (.053) |
| 309 | rs282802 | 15 | 77859875 | 63.7 (.055) | 78.2 (.015) |
| 310 | rs241301 | 1 | 225269162 | 26.4 (.040) | 45.4 (.048) |
| 311 | rs892535 | 13 | 104007916 | 62.8 (.044) | 73.8 (.038) |
| 312 | rs10817882 | 9 | 116225295 | 71.2 (.040) | 81.7 (.034) |
| 313 | rs2766542 | 6 | 35815668 | 58.6 (.049) | 42.4 (.035) |
| 314 | rs10038760 | 5 | 134924732 | 18.5 (.060) | 10.2 (.025) |
| 315 | rs10043237 | 5 | 24890906 | 81.7 (.018) | 87.3 (.021) |
| 316 | rs223185 | 1 | 18330326 | 51.3 (.095) | 67.6 (.034) |
| 317 | rs11863694 | 16 | 12561363 | 38.0 (.161) | 13.5 (.034) |
| 318 | rs305039 | 15 | 67873370 | 69.5 (.076) | 87.6 (.041) |
| 319 | rs9525291 | 13 | 113962630 | 83.6 (.064) | 93.8 (.027) |
| 320 | No rs numberd | 17 | 41472085 | 9.2 (.076) | 25.5 (.029) |
| 321 | rs241031 | 17 | 41090087 | 4.1 (.033) | 14.9 (.040) |
| 322 | rs3127572 | 6 | 160652629 | 14.6 (.067) | 4.1 (.016) |
| 323 | rs10420904 | 19 | 34394929 | 30.7 (.050) | 42.1 (.041) |
| 324 | rs10777224 | 12 | 89010777 | 89.4 (.030) | 75.1 (.061) |
| 325 | rs6418604 | 23 | 28557998 | 49.4 (.045) | 60.7 (.032) |
| 326 | rs4245562 | 7 | 54178194 | 65.2 (.035) | 76.4 (.053) |
| 327 | rs6670619 | 1 | 204831917 | 57.9 (.067) | 78.8 (.080) |
| 328 | rs9806222 | 15 | 66671258 | 53.6 (.027) | 47.0 (.024) |
| 329 | rs6797081 | 3 | 24973727 | 50.9 (.064) | 38.0 (.043) |
| 330 | rs7181055 | 15 | 97852959 | 21.7 (.041) | 9.0 (.033) |
| 331 | rs12485961 | 3 | 163179791 | 30.4 (.032) | 22.6 (.024) |
| 332 | rs237744 | 20 | 47336183 | 36.7 (.036) | 27.2 (.035) |
| 333 | rs10956287 | 8 | 127256564 | 34.4 (.066) | 23.3 (.049) |
| 334 | rs1635135 | 12 | 111917616 | 74.8 (.023) | 81.7 (.028) |
| 335 | rs13313479 | 15 | 55959003 | 58.7 (.086) | 30.8 (.077) |
| 336 | rs6803992 | 3 | 10597941 | 64.8 (.067) | 52.8 (.051) |
| 337 | rs1289745 | 3 | 109408948 | 22.2 (.042) | 11.5 (.019) |
| 338 | rs1343657 | 6 | 127305227 | 69.3 (.062) | 52.8 (.067) |
| 339 | rs7136648 | 12 | 48911089 | 60.5 (.058) | 48.7 (.033) |
| 340 | rs4426313 | 15 | 77590382 | 37.7 (.057) | 53.1 (.053) |
| 341 | rs2013867 | 11 | 47216848 | 74.1 (.066) | 96.6 (.053) |
| 342 | rs1158811 | 6 | 69090289 | 67.2 (.032) | 75.3 (.032) |
| 343 | rs10853854 | 19 | 57923940 | 17.1 (.079) | 33.0 (.034) |
| 344 | rs12273933 | 11 | 78628220 | 53.8 (.067) | 68.8 (.031) |
| 345 | rs5920468 | 23 | 144018371 | 47.9 (.061) | 61.0 (.023) |
| 346 | rs1247487 | 10 | 77676143 | 49.8 (.045) | 63.6 (.047) |
| 347 | rs9831276 | 3 | 142995721 | 82.1 (.051) | 70.5 (.026) |
| 348 | rs10932486 | 2 | 214210418 | 69.1 (.037) | 53.2 (.076) |
| 349 | rs10203659 | 2 | 152777095 | 91.1 (.029) | 80.0 (.044) |
| 350 | rs1482108 | 4 | 52968985 | 17.0 (.037) | 30.5 (.031) |
| 351 | rs6998348 | 8 | 122706022 | 33.9 (.046) | 22.2 (.055) |
| 352 | rs12612116 | 2 | 227618234 | 37.1 (.063) | 47.7 (.036) |
| 353 | rs10507428 | 13 | 35274755 | 96.5 (.108) | 65.0 (.111) |
| 354 | rs2248161 | 10 | 13056534 | 51.7 (.043) | 62.2 (.054) |
| 355 | rs12607476 | 18 | 26841275 | 73.5 (.040) | 87.6 (.025) |
| 356 | rs10922273 | 1 | 194457030 | 88.3 (.055) | 77.8 (.045) |
| 357 | rs2024266 | 7 | 150825612 | 55.1 (.053) | 40.9 (.064) |
| 358 | rs10276128 | 7 | 8632452 | 53.9 (.051) | 66.0 (.064) |
| 359 | rs6000790 | 22 | 20740851 | 33.9 (.062) | 21.7 (.016) |
| 360 | rs4701563 | 5 | 22072008 | 15.2 (.028) | 28.1 (.021) |
| 361 | rs11695415 | 2 | 217963990 | 75.0 (.053) | 87.5 (.030) |
| 362 | rs3743761 | 16 | 75785385 | 69.3 (.062) | 58.6 (.033) |
| 363 | rs12546080 | 8 | 137129495 | 60.6 (.054) | 48.6 (.045) |
| 364 | rs7776346 | 6 | 111968058 | 71.9 (.038) | 83.6 (.049) |
| 365 | rs11005115 | 10 | 57357276 | 42.5 (.054) | 30.1 (.033) |
| 366 | rs6560011 | 9 | 87486191 | 68.5 (.035) | 58.2 (.055) |
| 367 | rs9859834 | 3 | 143589049 | 57.4 (.037) | 74.4 (.041) |
| 368 | rs1564117 | 8 | 107688729 | 99.5 (.035) | 87.7 (.047) |
| 369 | rs9398069 | 6 | 106707858 | 69.5 (.052) | 50.7 (.078) |
| 370 | rs10461198 | 4 | 116786149 | 71.6 (.066) | 55.3 (.059) |
| 371 | rs9309981 | 3 | 85588634 | 88.5 (.049) | 75.7 (.076) |
| 372 | rs1656400 | 2 | 233253288 | 27.1 (.040) | 7.5 (.066) |
| 373 | rs16845213 | 2 | 141641328 | 29.4 (.054) | 15.1 (.053) |
| 374 | rs3946526 | 17 | 40897439 | 86.3 (.075) | 76.5 (.082) |
| 375 | rs11799420 | 1 | 168384635 | 23.4 (.039) | 15.5 (.030) |
| 376 | rs12570751 | 10 | 5480932 | 26.0 (.058) | 10.1 (.053) |
| 377 | rs660465 | 10 | 78855117 | 64.8 (.060) | 52.5 (.074) |
| 378 | rs659670 | 18 | 30903493 | 12.9 (.031) | 19.0 (.025) |
| 379 | rs318931 | 10 | 131967734 | 61.2 (.031) | 48.7 (.042) |
| 380 | rs6134031 | 20 | 10700610 | 15.9 (.044) | 29.4 (.035) |
| 381 | No rs numberd | 2 | 36099185 | 83.0 (.024) | 91.4 (.023) |
| 382 | rs1506929 | 12 | 59784657 | 74.9 (.046) | 64.1 (.036) |
| 383 | rs9803850 | 1 | 85782465 | 70.6 (.041) | 79.3 (.047) |
| 384 | rs12730116 | 1 | 117845272 | 20.8 (.100) | 12.4 (.022) |
| 385 | rs11078927 | 17 | 35317931 | 48.4 (.030) | 58.0 (.039) |
| 386 | rs11199686 | 10 | 122728541 | 52.2 (.042) | 43.3 (.060) |
| 387 | rs5958390 | 23 | 122976032 | 60.4 (.081) | 77.0 (.046) |
| 388 | rs10215677 | 7 | 140538698 | 87.7 (.056) | 72.1 (.063) |
| 389 | rs17135662 | 7 | 100838409 | 74.1 (.072) | 85.6 (.033) |
| 390 | rs7037570 | 9 | 132325895 | 29.1 (.035) | 19.7 (.024) |
| 391 | rs10815701 | 9 | 7861163 | 33.8 (.044) | 18.3 (.029) |
| 392 | rs4796017 | 17 | 23099118 | 33.2 (.038) | 46.8 (.069) |
| 393 | rs601962 | 8 | 29306465 | 66.1 (.059) | 79.1 (.038) |
| 394 | rs2069278 | 1 | 66347392 | 65.3 (.045) | 74.4 (.028) |
| 395 | rs2566970 | 3 | 114944813 | 36.8 (.028) | 24.7 (.054) |
| 396 | rs840600 | 2 | 187985256 | 75.1 (.040) | 62.9 (.053) |
| 397 | rs8034444 | 15 | 56323687 | 46.7 (.047) | 29.0 (.063) |
| 398 | rs17167930 | 7 | 13961171 | 93.7 (.028) | 87.5 (.039) |
| 399 | rs9564057 | 13 | 62927711 | 19.7 (.034) | 31.3 (.049) |
| 400 | rs6895344 | 5 | 55607343 | 73.7 (.060) | 56.7 (.085) |
| 401 | rs7219245 | 17 | 60547161 | 7.5 (.074) | 24.0 (.753) |
| 402 | rs17379636 | 4 | 154764283 | 67.1 (.040) | 56.8 (.039) |
| 403 | rs4931243 | 12 | 30072654 | 61.1 (.057) | 48.5 (.030) |
| 404 | rs2018971 | 17 | 26896751 | 55.6 (.044) | 67.8 (.036) |
| 405 | rs1002007 | 15 | 69369104 | 67.7 (.082) | 85.2 (.076) |
| 406 | rs2673559 | 8 | 133548347 | 48.2 (.152) | 15.7 (.055) |
| 407 | rs12230555 | 12 | 4289045 | 46.7 (.050) | 34.4 (.058) |
| 408 | rs2878802 | 7 | 158027122 | 15.9 (.056) | 5.8 (.025) |
| 409 | rs11639949 | 16 | 12561909 | 69.6 (.093) | 85.2 (.032) |
| 410 | rs2075425 | 1 | 210514249 | 32.0 (.042) | 43.2 (.046) |
| 411 | rs2469564 | 15 | 74277010 | 86.4 (.060) | 98.3 (.044) |
| 412 | rs6999868 | 8 | 5502341 | 18.9 (.057) | 12.1 (.018) |
| 413 | rs9388860 | 6 | 131215145 | 71.8 (.124) | 83.3 (.025) |
| 414 | rs2977070 | 8 | 75275324 | 35.4 (.118) | 24.5 (.027) |
| 415 | No rs numberd | 7 | 90549644 | 79.5 (.120) | 96.3 (.045) |
| 416 | rs12711664 | 2 | 124517464 | 59.7 (.034) | 70.0 (.025) |
| 417 | rs1331574 | 6 | 115747405 | 47.4 (.039) | 34.5 (.032) |
| 418 | rs2838571 | 21 | 44722797 | 43.2 (.069) | 57.9 (.034) |
| 419 | rs4382714 | 1 | 234742585 | 48.8 (.058) | 34.6 (.038) |
| 420 | No rs numberd | 4 | 4652651 | 73.3 (.060) | 83.7 (.042) |
| 421 | rs1886543 | 13 | 51599783 | 37.2 (.044) | 19.1 (.056) |
| 422 | rs879278 | 10 | 72671959 | 57.4 (.039) | 51.9 (.027) |
| 423 | rs1038990 | 2 | 238329392 | 39.6 (.068) | 30.0 (.055) |
| 424 | rs1496483 | 18 | 24511520 | 32.3 (.048) | 20.8 (.041) |
| 425 | rs696851 | 3 | 155586013 | 83.0 (.028) | 92.9 (.012) |
| 426 | rs17053864 | 9 | 88033026 | 88.2 (.075) | 99.0 (.026) |
| 427 | rs4585704 | 8 | 27766421 | 38.2 (.054) | 48.8 (.052) |
| 428 | rs17306695 | 10 | 87333513 | 13.2 (.037) | 20.1 (.022) |
| 429 | rs884329 | 4 | 106281075 | 29.4 (.079) | 49.4 (.047) |
| 430 | rs2071967 | 1 | 4606465 | 8.1 (.052) | 22.8 (.118) |
| 431 | rs13423714 | 2 | 227722157 | 35.4 (.045) | 48.4 (.016) |
| 432 | rs10977527 | 9 | 9134115 | 65.1 (.029) | 80.6 (.088) |
| 433 | rs10875641 | 5 | 148273115 | 30.6 (.062) | 42.4 (.054) |
| 434 | rs285753 | 15 | 91019081 | 26.6 (.022) | 16.7 (.026) |
| 435 | rs190034 | 5 | 95043287 | 51.2 (.040) | 36.6 (.039) |
| 436 | rs17085569 | 6 | 155030335 | 12.1 (.110) | 34.9 (.056) |
| 437 | rs2676419 | 8 | 33475428 | 14.2 (.098) | 45.0 (.092) |
| 438 | no rs# | 17 | 41492888 | 91.2 (.061) | 82.3 (.063) |
| 439 | rs6659522 | 1 | 204587495 | 56.7 (.033) | 66.4 (.014) |
| 440 | rs17192114 | 12 | 88673906 | 86.3 (.042) | 74.1 (.035) |
| 441 | rs7442929 | 5 | 24262649 | 27.6 (.047) | 17.0 (.041) |
| 442 | rs10769846 | 11 | 7860814 | 91.9 (.054) | 75.8 (.050) |
| 443 | rs780331 | 8 | 127129712 | 75.5 (.042) | 63.7 (.055) |
| 444 | rs872411 | 5 | 116089281 | 57.1 (.027) | 63.8 (.036) |
| 445 | rs5909876 | 23 | 120947783 | 59.4 (.074) | 77.4 (.041) |
| 446 | rs11764997 | 7 | 115242171 | 37.2 (.022) | 51.1 (.034) |
| 447 | rs2191057 | 17 | 29915167 | 26.9 (.063) | 13.5 (.042) |
| 448 | rs6050164 | 20 | 24863295 | 26.7 (.109) | 6.4 (.050) |
| 449 | rs12265750 | 10 | 130742160 | 42.1 (.094) | 27.5 (.058) |
| 450 | rs10950297 | 7 | 71018779 | 73.2 (.023) | 82.0 (.034) |
| 451 | rs205737 | 7 | 130118083 | 53.2 (.085) | 65.4 (.064) |
| 452 | rs10501321 | 11 | 47251202 | 60.7 (.041) | 71.7 (.045) |
| 453 | rs16910869 | 8 | 140616426 | 53.0 (.055) | 36.0 (.036) |
| 454 | No rs numberd | 7 | 19368364 | 55.5 (.061) | 71.6 (.047) |
| 455 | rs2426936 | 20 | 58909276 | 36.1 (.027) | 22.7 (.062) |
| 456 | rs9261489 | 6 | 30217461 | 73.0 (.056) | 85.5 (.047) |
| 457 | rs17784515 | 11 | 44831086 | 62.6 (.063) | 71.1 (.027) |
| 458 | rs4943770 | 13 | 39808388 | 39.9 (.034) | 50.3 (.024) |
| 459 | rs17637161 | 8 | 138940595 | 26.6 (.049) | 16.1 (.056) |
| 460 | rs11170782 | 12 | 37796650 | 10.5 (.043) | 29.2 (.046) |
| 461 | rs11006702 | 10 | 19066613 | 83.7 (.043) | 73.2 (.064) |
| 462 | rs419097 | 9 | 111791298 | 71.6 (.095) | 45.7 (.044) |
| 463 | rs4880568 | 10 | 303504 | 29.4 (.041) | 41.5 (.060) |
| 464 | rs10894157 | 11 | 99289243 | 80.1 (.042) | 69.8 (.056) |
| 465 | rs6495126 | 15 | 72962079 | 92.3 (.107) | 73.0 (.075) |
| 466 | rs2495408 | 1 | 184528663 | 61.0 (.084) | 43.1 (.066) |
| 467 | rs2402057 | 7 | 115491047 | 28.2 (.019) | 12.5 (.049) |
| 468 | rs17649553 | 17 | 41350476 | 87.8 (.074) | 75.7 (.091) |
| 469 | rs9403813 | 6 | 147112198 | 72.6 (.071) | 89.0 (.034) |
| 470 | rs2244284 | 20 | 24696432 | 66.0 (.053) | 47.9 (.030) |
| 471 | rs477233 | 23 | 23464832 | 49.8 (.029) | 60.0 (.056) |
| 472 | rs11695811 | 2 | 234997133 | 46.7 (.028) | 59.9 (.037) |
| 473 | rs1266734 | 23 | 107257424 | 25.7 (.042) | 11.4 (.054) |
| 474 | rs6450023 | 5 | 68561819 | 74.3 (.027) | 84.4 (.030) |
| 475 | rs10492331 | 12 | 113026864 | 81.9 (.062) | 89.9 (.024) |
| 476 | rs12487 | 15 | 72923747 | 33.5 (.031) | 43.9 (.066) |
| 477 | rs41295 | 7 | 28439876 | 65.2 (.044) | 53.4 (.026) |
| 478 | rs234440 | 14 | 96892646 | 30.4 (.062) | 12.4 (.036) |
| 479 | rs5916245 | 23 | 5521272 | 81.2 (.056) | 67.2 (.050) |
| 480 | rs743444 | 21 | 38978129 | 37.8 (.078) | 24.6 (.051) |
| 481 | rs2040920 | 7 | 147228429 | 29.4 (.045) | 21.8 (.021) |
| 482 | rs1862175 | 5 | 121560760 | 69.8 (.026) | 63.2 (.022) |
| 483 | rs7701573 | 5 | 59053652 | 52.2 (.049) | 39.0 (.042) |
| 484 | rs2986425 | 11 | 34872332 | 75.4 (.068) | 61.1 (.080) |
| 485 | rs4264335 | 15 | 59000266 | 55.1 (.043) | 67.3 (.031) |
| 486 | rs1911573 | 3 | 102336872 | 59.5 (.022) | 70.8 (.053) |
| 487 | rs273981 | 7 | 137076530 | 59.5 (.061) | 44.8 (.057) |
| 488 | rs2240737 | 17 | 9731884 | 19.4 (.062) | 10.4 (.039) |
| 489 | rs492125 | 11 | 125949932 | 51.0 (.044) | 59.9 (.059) |
| 490 | rs4420962 | 4 | 120378074 | 73.4 (.048) | 61.1 (.071) |
| 491 | rs12605509 | 18 | 52998303 | 36.9 (.039) | 26.6 (.031) |
| 492 | rs764180 | 1 | 168531993 | 59.9 (.098) | 43.4 (.087) |
| 493 | rs11640077 | 16 | 19432372 | 34.6 (.043) | 21.4 (.046) |
| 494 | rs1152472 | 14 | 55840960 | 48.2 (.031) | 34.6 (.063) |
| 495 | No rs numberd | 2 | 203333169 | 73.9 (.081) | 53.3 (.133) |
| 496 | rs1975634 | 16 | 54756806 | 63.5 (.082) | 45.9 (.085) |
| 497 | rs6817763 | 4 | 139567306 | 35.6 (.119) | 66.3 (.072) |
| 498 | rs10893347 | 11 | 124568317 | 61.6 (.086) | 86.4 (.112) |
| 499 | rs4973285 | 2 | 230871448 | 79.6 (.033) | 69.0 (.033) |
| 500 | rs5971587 | 23 | 31450636 | 24.9 (.047) | 39.0 (.074) |
| 501 | rs9484261 | 6 | 139686988 | 78.3 (.048) | 65.4 (.039) |
| 502 | rs1934534 | 6 | 150125146 | 71.4 (.041) | 58.6 (.070) |
| 503 | rs9504093 | 6 | 4371770 | 43.6 (.024) | 31.2 (.047) |
| 504 | rs2394807 | 10 | 72971347 | 50.6 (.065) | 32.6 (.037) |
| 505 | rs4122017 | 6 | 148347645 | 62.1 (.054) | 47.8 (.035) |
| 506 | rs11587621 | 1 | 208056795 | 11.6 (.035) | 36.0 (.116) |
| 507 | rs17363368 | 1 | 160090491 | 9.0 (.064) | 0.7 (.023) |
| 508 | rs4119152 | 2 | 18599315 | 83.7 (.078) | 63.4 (.039) |
| 509 | rs6907995 | 6 | 127304781 | 41.5 (.047) | 52.1 (.042) |
| 510 | rs1448871 | 2 | 6893288 | 68.6 (.058) | 53.1 (.048) |
| 511 | rs1356951 | 12 | 117486021 | 52.9 (.056) | 61.8 (.021) |
| 512 | rs1049296 | 3 | 134977052 | 73.4 (.217) | 83.7 (.117) |
| 513 | rs2405783 | 1 | 221333402 | 53.3 (.032) | 61.5 (.035) |
| 514 | rs10917825 | 1 | 160431297 | 50.2 (.057) | 37.7 (.022) |
| 515 | rs1884014 | 14 | 53934713 | 27.0 (.041) | 12.5 (.051) |
| 516 | rs4685846 | 3 | 538399 | 69.5 (.091) | 43.1 (.044) |
| 517 | rs7141809 | 14 | 49630695 | 72.8 (.034) | 58.5 (.057) |
| 518 | rs12925573 | 16 | 76662867 | 56.4 (.086) | 73.3 (.070) |
| 519 | rs2836531 | 21 | 38870860 | 57.1 (.038) | 64.9 (.050) |
| 520 | rs7763880 | 6 | 152894411 | 47.8 (.074) | 65.5 (.049) |
| 521 | rs1414491 | 10 | 43632364 | 79.9 (.030) | 90.7 (.062) |
| 522 | rs4675490 | 2 | 205768066 | 56.8 (.045) | 70.6 (.025) |
| 523 | rs1437933 | 2 | 211862181 | 29.9 (.046) | 43.7 (.075) |
| 524 | rs1699001 | 6 | 38714589 | 30.6 (.038) | 37.2 (.077) |
| 525 | rs12432184 | 14 | 55753214 | 37.8 (.060) | 49.3 (.057) |
| 526 | rs4740904 | 9 | 7672860 | 44.7 (.054) | 29.5 (.052) |
| 527 | rs469326 | 5 | 102192017 | 41.9 (.068) | 28.1 (.043) |
| 528 | rs17783630 | 14 | 92025138 | 53.8 (.049) | 43.5 (.044) |
| 529 | rs6463270 | 7 | 45040529 | 24.1 (.046) | 10.3 (.046) |
| 530 | rs4759061 | 12 | 52024481 | 66.6 (.055) | 77.5 (.053) |
| 531 | rs5017082 | 12 | 37600831 | 49.2 (.058) | 40.2 (.035) |
| 532 | rs2694874 | 12 | 124527344 | 67.1 (.033) | 73.8 (.048) |
| 533 | rs17684894 | 8 | 138604979 | 78.5 (.083) | 90.7 (.038) |
| 534 | rs1588706 | 4 | 69795828 | 3.3 (.057) | 19.0 (.033) |
| 535 | rs7706831 | 5 | 116087496 | 60.8 (.078) | 75.1 (.050) |
| 536 | rs379643 | 14 | 87719507 | 69.3 (.020) | 59.7 (.029) |
| 537 | rs17005517 | 1 | 215992848 | 26.2 (.040) | 16.7 (.048) |
| 538 | rs17040460 | 3 | 14889969 | 71.0 (.103) | 86.0 (.043) |
| 539 | rs2504458 | 1 | 107939403 | 54.8 (.042) | 70.4 (.055) |
| 540 | rs10503818 | 8 | 27731666 | 48.0 (.100) | 31.0 (.087) |
| 541 | rs2016459 | 8 | 82067869 | 76.3 (.050) | 59.4 (.064) |
| 542 | rs7068699 | 10 | 129132012 | 49.5 (.044) | 58.0 (.046) |
| 543 | rs8019096 | 14 | 47033221 | 5.1 (.065) | 26.5 (.056) |
| 544 | rs12865636 | 13 | 109097925 | 8.8 (.070) | 27.3 (.116) |
| 545 | rs13022222 | 2 | 226573182 | 18.7 (.035) | 12.8 (.028) |
| 546 | rs6035539 | 20 | 19987506 | 63.9 (.060) | 80.1 (.061) |
| 547 | rs2878722 | 18 | 38437616 | 78.9 (.020) | 86.6 (.028) |
| 548 | rs12701769 | 7 | 39626870 | 9.9 (.054) | 21.3 (.059) |
| 549 | rs6701341 | 1 | 150787694 | 56.4 (.037) | 68.8 (.043) |
| 550 | rs6462825 | 7 | 38067354 | 26.3 (.045) | 36.9 (.095) |
| 551 | rs2280884 | 8 | 27670057 | 50.1 (.033) | 59.6 (.028) |
| 552 | rs7357405 | 8 | 79183255 | 49.8 (.065) | 30.0 (.093) |
| 553 | rs9826951 | 3 | 132921144 | 82.5 (.057) | 94.2 (.024) |
| 554 | rs1408575 | 6 | 147264593 | 75.0 (.055) | 62.5 (.051) |
| 555 | rs2283792 | 22 | 20455679 | 57.8 (.082) | 44.6 (.061) |
| 556 | rs2316640 | 13 | 54542925 | 84.8 (.043) | 71.0 (.062) |
| 557 | rs16858997 | 1 | 229811908 | 54.6 (.073) | 69.4 (.040) |
| 558 | rs8070993 | 17 | 39109982 | 49.5 (.031) | 34.9 (.013) |
| 559 | rs7836489 | 8 | 21460484 | 42.0 (.043) | 51.3 (.040) |
| 560 | rs9381307 | 6 | 44361369 | 27.8 (.048) | 17.9 (.028) |
| 561 | rs12942155 | 17 | 46608140 | 41.2 (.049) | 25.2 (.042) |
| 562 | rs588933 | 9 | 2527815 | 82.3 (.029) | 73.0 (.028) |
| 563 | rs10785334 | 12 | 40906644 | 38.3 (.045) | 27.0 (.043) |
| 564 | rs7084059 | 10 | 93828325 | 42.3 (.022) | 32.8 (.044) |
| 565 | rs6473008 | 8 | 78185065 | 39.3 (.047) | 57.6 (.057) |
| 566 | rs7424879 | 2 | 181884034 | 55.7 (.058) | 43.3 (.058) |
| 567 | rs459162 | 6 | 1161376 | 41.2 (.045) | 26.0 (.046) |
| 568 | rs4629541 | 4 | 142381927 | 70.2 (.061) | 80.7 (.031) |
| 569 | rs2056411 | 4 | 83859712 | 58.8 (.046) | 72.7 (.056) |
| 570 | rs13427835 | 2 | 23142654 | 21.8 (.089) | 12.1 (.019) |
| 571 | No rs numberd | 4 | 25764488 | 39.2 (.048) | 54.5 (.051) |
| 572 | rs503897 | 9 | 37372099 | 59.9 (.049) | 74.5 (.037) |
| 573 | rs7795573 | 7 | 12693015 | 33.1 (.112) | 4.7 (.122) |
| 574 | rs1564274 | 1 | 34351855 | 71.3 (.065) | 81.1 (.043) |
| 575 | rs10822753 | 10 | 67648865 | 33.3 (.036) | 48.2 (.055) |
| 576 | rs2254535 | 10 | 72207879 | 88.7 (.035) | 96.8 (.016) |
| 577 | rs7926553 | 11 | 76470079 | 47.6 (.040) | 33.6 (.022) |
| 578 | rs17805522 | 20 | 59174920 | 29.5 (.088) | 17.3 (.016) |
| 579 | rs17361077 | 7 | 75083414 | 64.5 (.045) | 73.7 (.064) |
| 580 | rs308901 | 2 | 86758147 | 62.8 (.037) | 70.1 (.030) |
| 581 | rs260454 | 19 | 63458980 | 49.4 (.058) | 66.7 (.054) |
| 582 | rs13184396 | 5 | 149669366 | 63.3 (.037) | 77.5 (.022) |
| 583 | rs11558511 | 1 | 164745508 | 79.4 (.026) | 74.2 (.037) |
| 584 | rs6563655 | 13 | 38433775 | 74.8 (.031) | 93.0 (.060) |
| 585 | rs2179593 | 20 | 42093700 | 64.6 (.086) | 81.6 (.067) |
| 586 | rs9385717 | 6 | 135500116 | 75.2 (.033) | 79.0 (.035) |
| 587 | rs10000203 | 4 | 78633738 | 80.2 (.086) | 93.1 (.024) |
| 588 | rs11034770 | 11 | 38459187 | 48.9 (.063) | 78.9 (.111) |
| 589 | rs265619 | 12 | 128636559 | 61.0 (.030) | 69.5 (.030) |
| 590 | rs6470418 | 8 | 127311456 | 70.5 (.065) | 82.9 (.048) |
| 591 | rs7327426 | 13 | 60510741 | 63.3 (.040) | 73.1 (.035) |
| 592 | rs10800485 | 1 | 166578804 | 59.8 (.064) | 74.6 (.058) |
| 593 | rs10167944 | 2 | 115857630 | 15.0 (.081) | 28.7 (.077) |
| 594 | rs11685401 | 2 | 100435203 | 42.4 (.046) | 52.7 (.048) |
| 595 | rs1389709 | 3 | 86572955 | 23.6 (.038) | 39.8 (.069) |
| 596 | rs4730470 | 7 | 110202088 | 59.5 (.064) | 74.6 (.055) |
| 597 | rs1899384 | 18 | 49357887 | 91.5 (.040) | 80.3 (.046) |
| 598 | rs10467913 | 15 | 45505944 | 25.5 (.037) | 8.6 (.057) |
| 599 | rs1703975 | 8 | 8614749 | 86.7 (.036) | 96.4 (.024) |
| 600 | rs9378540 | 6 | 8280374 | 14.3 (.045) | 30.2 (.067) |
| 601 | No rs numberd | 1 | 176703994 | 86.8 (.047) | 79.5 (.050) |
| 602 | rs11803410 | 1 | 107853520 | 33.6 (.063) | 46.4 (.033) |
| 603 | rs10149991 | 14 | 46638818 | 51.6 (.036) | 63.3 (.046) |
| 604 | rs981579 | 4 | 28221496 | 79.4 (.056) | 67.6 (.040) |
| 605 | rs573301 | 6 | 24030685 | 40.7 (.045) | 56.5 (.055) |
| 606 | rs1918788 | 17 | 41623394 | 6.4 (.023) | 16.0 (.027) |
| 607 | rs2081748 | 12 | 113780260 | 99.7% (1.982) | 89.4 (.066) |
| 608 | rs16978462 | 18 | 41551134 | 26.0 (.058) | 9.6 (.044) |
| 609 | rs10518973 | 15 | 56438872 | 72.7 (.079) | 80.5 (.035) |
| 610 | rs12288391 | 11 | 124629292 | 32.1 (.064) | 21.9 (.040) |
| 611 | rs7774349 | 6 | 137517551 | 58.1 (.058) | 49.0 (.043) |
| 612 | rs2972284 | 16 | 5008153 | 60.8 (.110) | 79.6 (.032) |
| 613 | rs10885211 | 10 | 113314292 | 58.4 (.031) | 50.4 (.028) |
| 614 | rs11124178 | 2 | 239700789 | 35.9 (.070) | 26.6 (.029) |
| 615 | rs10491524 | 9 | 119336808 | 65.3 (.156) | 87.2 (.050) |
| 616 | rs890067 | 2 | 60089993 | 47.6 (.050) | 37.0 (.058) |
| 617 | rs12980971 | 19 | 56872738 | 51.6 (.072) | 63.2 (.071) |
| 618 | rs17283761 | 3 | 152070880 | 36.7 (.047) | 45.4 (.054) |
| 619 | rs1220620 | 5 | 72813638 | 33.1 (.036) | 22.8 (.058) |
| 620 | rs1944761 | 9 | 18178242 | 70.9 (.049) | 85.7 (.039) |
| 621 | rs1397148 | 16 | 54655880 | 46.3 (.043) | 60.2 (.053) |
| 622 | rs5958281 | 23 | 122515806 | 44.3 (.099) | 25.3 (.033) |
| 623 | rs17363814 | 19 | 4795633 | 35.3 (.072) | 29.2 (.024) |
| 624 | rs9372118 | 6 | 106708048 | 65.8 (.058) | 49.1 (.063) |
| 625 | rs10935960 | 3 | 155136125 | 78.8 (.050) | 71.2 (.050) |
| 626 | rs13120622 | 4 | 139911721 | 44.0 (.052) | 31.1 (.063) |
| 627 | rs7769478 | 6 | 103393153 | 73.2 (.097) | 49.6 (.054) |
| 628 | rs600702 | 11 | 124569824 | 14.2 (.056) | 29.3 (.064) |
| 629 | rs2136531 | 11 | 108406015 | 83.2 (.069) | 97.1 (.036) |
| 630 | rs11832772 | 12 | 41145855 | 60.0 (.046) | 50.3 (.038) |
| 631 | rs16990692 | 4 | 34868232 | 79.2 (.032) | 69.4 (.042) |
| 632 | rs2434245 | 5 | 175139805 | 24.2 (.066) | 9.1 (.056) |
| 633 | rs8007728 | 14 | 81242457 | 15.7 (.042) | 31.4 (.059) |
| 634 | rs404801 | 3 | 83534051 | 62.6 (.063) | 50.4 (.032) |
| 635 | rs2350811 | 6 | 115984204 | 88.5 (.044) | 71.9 (.063) |
| 636 | rs7655209 | 4 | 158408990 | 91.2 (.036) | 78.1 (.054) |
| 637 | rs9852802 | 3 | 8116722 | 46.3 (.033) | 31.7 (.029) |
| 638 | rs3784678 | 15 | 34733595 | 47.0 (.060) | 60.7 (.086) |
| 639 | rs1711004 | 3 | 189348121 | 53.0 (.034) | 37.3 (.043) |
| 640 | rs1363530 | 5 | 147324564 | 15.4 (.047) | 29.4 (.059) |
| 641 | rs7799704 | 7 | 92874191 | 38.3 (.065) | 52.3 (.047) |
| 642 | rs1922185 | 2 | 52353433 | 38.9 (.057) | 23.3 (.074) |
| 643 | rs6473164 | 8 | 80567257 | 58.2 (.040) | 41.1 (.032) |
| 644 | rs734489 | 15 | 55435304 | 71.9 (.040) | 57.0 (.040) |
| 645 | rs2607924 | 12 | 570936 | 71.1 (.043) | 81.9 (.058) |
| 646 | rs925111 | 15 | 78872538 | 21.8 (.028) | 34.5 (.018) |
| 647 | rs1483198 | 1 | 4627764 | 29.2 (.070) | 11.8 (.060) |
| 648 | rs13424077 | 2 | 35455013 | 76.4 (.056) | 68.9 (.033) |
| 649 | rs6069892 | 20 | 54820158 | 53.3 (.038) | 43.3 (.060) |
| 650 | rs10992674 | 9 | 93042669 | 59.1 (.047) | 72.4 (.058) |
| 651 | rs1790318 | 11 | 70777862 | 36.1 (.032) | 48.4 (.041) |
| 652 | rs4316515 | 11 | 68065416 | 31.9 (.070) | 17.8 (.038) |
| 653 | rs4747385 | 10 | 7096022 | 53.3 (.038) | 65.1 (.073) |
| 654 | rs207078 | 6 | 66826775 | 39.1 (.056) | 52.3 (.042) |
| 655 | rs7221595 | 17 | 3954343 | 26.1 (.042) | 14.4 (.045) |
| 656 | rs7241635 | 18 | 31568422 | 69.1 (.071) | 54.6 (.042) |
| 657 | rs917500 | 17 | 29884677 | 69.3 (.035) | 77.5 (.035) |
| 658 | rs2274201 | 14 | 30716992 | 53.7 (.055) | 71.5 (.044) |
| 659 | rs10113968 | 9 | 2445202 | 51.2 (.059) | 39.4 (.038) |
| 660 | rs4714890 | 6 | 46003658 | 58.7 (.059) | 73.7 (.035) |
| 661 | rs6469756 | 8 | 119625611 | 26.7 (.050) | 18.6 (.053) |
| 662 | rs10979211 | 9 | 107950576 | 59.8 (.036) | 75.7 (.052) |
| 663 | rs12716267 | 5 | 170356139 | 60.1 (.069) | 69.6 (.034) |
| 664 | rs3913972 | 6 | 87217610 | 50.3 (.057) | 40.1 (.025) |
| 665 | rs252811 | 5 | 106764256 | 78.7 (.156) | 95.0 (.088) |
| 666 | rs11136949 | 8 | 5792133 | 44.1 (.043) | 56.3 (.034) |
| 667 | rs9380791 | 6 | 38938388 | 51.1 (.052) | 28.9 (.036) |
| 668 | rs6486303 | 11 | 16593899 | 31.8 (.097) | 13.0 (.051) |
| 669 | rs11103117 | 9 | 135753858 | 16.3 (.089) | 32.4 (.098) |
| 670 | rs4706205 | 6 | 85241292 | 47.0 (.030) | 56.8 (.036) |
| 671 | rs7048135 | 9 | 1776681 | 78.3 (.064) | 97.4 (.049) |
| 672 | rs12551465 | 9 | 4565673 | 65.5 (.023) | 55.3 (.023) |
| 673 | rs3910449 | 16 | 5750350 | 28.4 (.037) | 41.7 (.067) |
| 674 | rs433395 | 7 | 23175195 | 55.0 (.054) | 65.4 (.049) |
| 675 | rs4446347 | 4 | 62889346 | 53.2 (.060) | 42.1 (.057) |
| 676 | rs4641528 | 12 | 70673609 | 44.9 (.077) | 56.5 (.034) |
| 677 | rs17651822 | 3 | 28695130 | 20.5 (.035) | 11.2 (.017) |
| 678 | rs4939008 | 11 | 55446947 | 43.8 (.056) | 34.1 (.021) |
| 679 | rs3741755 | 12 | 68358746 | 30.1 (.098) | 43.6 (.060) |
| 680 | rs169173 | 21 | 30926217 | 62.3 (.036) | 69.4 (.016) |
| 681 | rs1893854 | 11 | 105574558 | 91.7 (.059) | 80.4 (.040) |
| 682 | rs1996537 | 11 | 36707703 | 8.4 (.135) | 3.6 (.042) |
| 683 | rs1159929 | 5 | 77804576 | 84.0 (.051) | 75.5 (.035) |
| 684 | rs2256143 | 4 | 166903785 | 72.8 (.067) | 56.8 (.042) |
| 685 | rs1550099 | 2 | 233253652 | 70.5 (.045) | 87.0 (.029) |
| 686 | rs1454535 | 2 | 221055427 | 50.6 (.020) | 41.9 (.041) |
| 687 | rs17753780 | 7 | 4275526 | 62.7 (.034) | 73.7 (.052) |
| 688 | rs697550 | 5 | 14125453 | 7.9 (.070) | 21.0 (.047) |
| 689 | rs3777995 | 6 | 116846696 | 61.9 (.065) | 78.4 (.042) |
| 690 | rs11613448 | 12 | 37811416 | 48.7 (.048) | 33.2 (.060) |
| 691 | rs3737880 | 1 | 199778332 | 81.6 (.054) | 91.4 (.033) |
| 692 | rs11787001 | 8 | 25654515 | 67.2 (.024) | 76.4 (.035) |
| 693 | rs7656178 | 4 | 173829976 | 33.0 (.108) | 21.2 (.067) |
| 694 | rs9990848 | 4 | 32885412 | 8.9 (.045) | 17.9 (.029) |
| 695 | rs685478 | 11 | 112182092 | 49.1 (.060) | 32.9 (.031) |
| 696 | rs7213273 | 17 | 40511440 | 59.2 (.067) | 71.9 (.030) |
| 697 | rs201480 | 7 | 101341010 | 56.8 (.025) | 70.3 (.058) |
| 698 | rs7649496 | 3 | 61936045 | 56.0 (.070) | 40.9 (.080) |
| 699 | rs17673165 | 2 | 9385739 | 32.9 (.021) | 25.5 (.024) |
| 700 | rs17284017 | 7 | 26356960 | 26.8 (.065) | 46.8 (.105) |
| 701 | rs6089071 | 20 | 29842045 | 79.9 (.038) | 70.0 (.059) |
| 702 | rs7944870 | 11 | 68065684 | 64.1 (.098) | 82.3 (.051) |
| 703 | rs10751296 | 11 | 77954925 | 73.8 (.041) | 81.8 (.059) |
| 704 | rs7791862 | 7 | 47516496 | 88.2 (.034) | 75.7 (.036) |
| 705 | rs4373552 | 8 | 139055105 | 43.6 (.046) | 64.0 (.060) |
| 706 | rs570549 | 3 | 173541823 | 17.5 (.024) | 24.8 (.024) |
| 707 | rs3784131 | 14 | 68032257 | 17.8 (.028) | 10.4 (.024) |
| 708 | rs142167 | 17 | 42150418 | 22.8 (.057) | 36.0 (.070) |
| 709 | rs10236767 | 7 | 7385089 | 89.9 (.069) | 71.0 (.118) |
| 710 | rs7636734 | 3 | 37721855 | 61.5 (.067) | 50.2 (.073) |
| 711 | rs1457623 | 12 | 13432691 | 15.8 (.036) | 6.9 (.048) |
| 712 | rs2848519 | 11 | 97618389 | 33.0 (.037) | 22.6 (.048) |
| 713 | rs4677106 | 3 | 72296777 | 71.9 (.054) | 82.5 (.040) |
| 714 | rs10095813 | 8 | 78201617 | 69.4 (.041) | 56.4 (.039) |
| 715 | rs3106653 | 2 | 155401068 | 84.6 (.044) | 73.0 (.038) |
| 716 | rs17544750 | 4 | 170548087 | 27.0 (.021) | 18.6 (.031) |
| 717 | rs1250388 | 18 | 36833774 | 36.4 (.051) | 25.8 (.061) |
| 718 | rs7860079 | 9 | 2096237 | 24.3 (.039) | 17.1 (.025) |
| 719 | rs10895266 | 11 | 101533105 | 45.7 (.090) | 24.7 (.071) |
| 720 | rs10899285 | 11 | 76191470 | 38.9 (.081) | 4.6 (.047) |
| 721 | rs17762565 | 14 | 87726574 | 26.2 (.050) | 15.9 (.056) |
| 722 | rs4629211 | 2 | 139112236 | 95.0 (.045) | 85.7 (.065) |
| 723 | rs10445581 | 19 | 56275540 | 45.3 (.039) | 30.6 (.033) |
| 724 | rs1437539 | 15 | 59173372 | 55.3 (.032) | 66.3 (.034) |
| 725 | rs11844114 | 14 | 93530282 | 48.5 (.027) | 36.1 (.032) |
| 726 | rs7143971 | 14 | 89650542 | 35.8 (.060) | 20.8 (.073) |
| 727 | rs7211982 | 17 | 15335097 | 54.4 (.084) | 35.0 (.069) |
| 728 | rs11639621 | 16 | 81545373 | 49.0 (.049) | 42.2 (.027) |
| 729 | rs7938161 | 11 | 5169852 | 51.9 (.093) | 23.5 (.091) |
| 730 | rs10754052 | 1 | 189871903 | 66.0 (.035) | 76.5 (.032) |
| 731 | rs6929790 | 6 | 99152470 | 73.7 (.080) | 89.9 (.033) |
| 732 | rs7727239 | 5 | 124084840 | 23.2 (.074) | 16.1 (.023) |
| 733 | rs11961994 | 6 | 127691043 | 61.9 (.025) | 74.4 (.039) |
| 734 | rs10979500 | 9 | 108641021 | 18.4 (.081) | 6.5 (.023) |
| 735 | rs7113069 | 11 | 55939582 | 51.4 (.039) | 36.0 (.024) |
| 736 | rs798601 | 3 | 119995613 | 38.9 (.055) | 29.6 (.031) |
| 737 | rs2836365 | 21 | 38690144 | 39.8 (.029) | 29.2 (.022) |
| 738 | rs7006032 | 8 | 140328379 | 55.8 (.028) | 66.4 (.046) |
| 739 | rs831791 | 5 | 56385520 | 67.5 (.029) | 57.7 (.048) |
| 740 | rs6425430 | 1 | 174229105 | 38.5 (.050) | 50.5 (.059) |
| 741 | rs1078671 | 8 | 142140160 | 35.7 (.053) | 24.1 (.053) |
| 742 | rs4644762 | 13 | 85511389 | 50.2 (.028) | 65.8 (.066) |
| 743 | rs4981220 | 14 | 33669097 | 38.5 (.045) | 26.0 (.035) |
| 744 | rs6898765 | 5 | 50910013 | 44.6 (.065) | 29.4 (.043) |
| 745 | rs4734037 | 8 | 102740588 | 7.3 (.061) | 21.0 (.016) |
| 746 | rs870992 | 5 | 52228994 | 7.0 (.055) | 1.3 (.018) |
| 747 | rs12189287 | 5 | 148286759 | 27.4 (.040) | 36.0 (.037) |
| 748 | rs1588705 | 4 | 69795733 | 16.0 (.050) | 28.4 (.035) |
| 749 | rs3824070 | 7 | 1348618 | 58.9 (.046) | 69.5 (.025) |
| 750 | rs10806221 | 6 | 82133833 | 81.8 (.079) | 64.6 (.107) |
| 751 | rs4628676 | 11 | 68238806 | 61.1 (.023) | 71.3 (.040) |
| 752 | rs17450420 | 13 | 103837147 | 95.5 (.021) | 88.3 (.026) |
| 753 | rs777574 | 6 | 166426507 | 11.1 (.027) | 19.4 (.041) |
| 754 | rs4130337 | 3 | 107848773 | 2.8 (.037) | 10.8 (.035) |
| 755 | rs2491649 | 9 | 126108064 | 56.9 (.079) | 70.5 (.077) |
| 756 | rs6995156 | 8 | 82494218 | 68.8 (.059) | 54.6 (.055) |
| 757 | rs6472348 | 8 | 68945433 | 31.6 (.043) | 43.9 (.053) |
| 758 | rs4448276 | 8 | 10092338 | 47.4 (.058) | 36.0 (.053) |
| 759 | rs7582215 | 2 | 33139818 | 67.0 (.088) | 49.0 (.061) |
| 760 | rs7201414 | 16 | 76762567 | 11.7 (.066) | 21.9 (.076) |
| 761 | rs12947718 | 17 | 40848884 | 1.7 (.086) | 17.3 (.067) |
| 762 | rs1468995 | 2 | 154842933 | 86.4 (.049) | 78.7 (.038) |
| 763 | rs2532259 | 17 | 41609141 | 14.2 (.030) | 24.7 (.027) |
| 764 | rs17439526 | 2 | 154656842 | 23.1 (.026) | 35.9 (.067) |
| 765 | rs16841426 | 2 | 157942808 | 95.4 (.034) | 84.3 (.053) |
| 766 | rs2272789 | 22 | 34004505 | 64.9 (.039) | 74.9 (.047) |
| 767 | rs878815 | 14 | 65561860 | 58.7 (.024) | 71.9 (.062) |
| 768 | rs3885957 | 5 | 75884149 | 67.2 (.046) | 75.8 (.029) |
| 769 | rs11599418 | 10 | 113408949 | 19.2 (.054) | 39.5 (.082) |
| 770 | rs7441878 | 4 | 118680094 | 11.9 (.074) | 5.3 (.031) |
| 771 | rs360234 | 2 | 126757785 | 25.7 (.026) | 15.9 (.026) |
| 772 | rs2696640 | 17 | 41007016 | 44.5 (.048) | 54.3 (.053) |
| 773 | rs963666 | 1 | 230165486 | 26.9 (.021) | 31.9 (.025) |
| 774 | rs16869658 | 8 | 103702938 | 65.7 (.099) | 81.0 (.053) |
| 775 | rs17836505 | 16 | 86279192 | 59.3 (.065) | 48.8 (.039) |
| 776 | rs4873584 | 8 | 52802297 | 89.6 (.061) | 78.5 (.064) |
| 777 | rs13095558 | 3 | 77827384 | 36.2 (.037) | 54.9 (.052) |
| 778 | rs11742145 | 5 | 166764657 | 37.6 (.064) | 58.7 (.077) |
| 779 | rs8181728 | 12 | 115462617 | 23.6 (.066) | 7.2 (.051) |
| 780 | rs4806891 | 19 | 2898152 | 77.0 (.061) | 90.1 (.049) |
| 781 | rs6543297 | 2 | 105529677 | 68.1 (.030) | 57.9 (.032) |
| 782 | rs16936517 | 10 | 37773387 | 15.0 (.032) | 7.8 (.023) |
| 783 | rs2245201 | 16 | 77092871 | 68.4 (.105) | 83.7 (.020) |
| 784 | rs9524017 | 13 | 92803806 | 23.1 (.022) | 31.2 (.035) |
| 785 | rs7803877 | 7 | 146543556 | 33.9 (.091) | 21.6 (.044) |
| 786 | rs4815812 | 20 | 5352526 | 34.6 (.113) | 13.4 (.073) |
| 787 | rs10926796 | 1 | 239122055 | 23.6 (.117) | 7.0 (.036) |
| 788 | rs430306 | 1 | 112364061 | 17.5 (.028) | 27.0 (.028) |
| 789 | rs3749237 | 3 | 49745036 | 60.9 (.073) | 75.6 (.048) |
| 790 | rs16927557 | 9 | 1315179 | 84.3 (.058) | 95.7 (.034) |
| 791 | rs10808523 | 8 | 123579918 | 60.4 (.051) | 52.6 (.040) |
| 792 | rs10933144 | 2 | 227088913 | 21.7 (.061) | 7.4 (.042) |
| 793 | rs17182039 | 13 | 105595620 | 87.0 (.069) | 95.5 (.036) |
| 794 | rs6051846 | 20 | 3351824 | 34.8 (.049) | 21.5 (.044) |
| 795 | rs9923322 | 16 | 77401370 | 75.3 (.034) | 80.2 (.040) |
| 796 | rs8007849 | 14 | 84494886 | 10.2 (.049) | 18.7 (.035) |
| 797 | rs10787472 | 10 | 114771287 | 54.8 (.038) | 46.7 (.028) |
| 798 | rs13016130 | 2 | 217799783 | 52.8 (.031) | 42.5 (.038) |
| 799 | rs12408987 | 1 | 42721564 | 99.9 (.033) | 88.7 (.038) |
| 800 | rs4837264 | 9 | 128281449 | 33.6 (.038) | 15.1 (.046) |
| 801 | rs10898503 | 11 | 85970170 | 58.9 (.060) | 40.6 (.088) |
| 802 | rs16867195 | 2 | 9940040 | 64.6 (.148) | 96.1 (.103) |
| 803 | rs7811301 | 7 | 66876971 | 31.0 (.053) | 19.0 (.026) |
| 804 | rs9366215 | 6 | 170591998 | 34.1 (.027) | 25.5 (.025) |
| 805 | rs330609 | 2 | 150811273 | 44.0 (.056) | 57.3 (.047) |
| 806 | rs4749580 | 10 | 31014180 | 70.1 (.049) | 51.9 (.096) |
| 807 | rs2639974 | 18 | 71057723 | 55.5 (.035) | 68.8 (.041) |
| 808 | rs265632 | 12 | 128600215 | 73.3 (.035) | 59.8 (.027) |
| 809 | rs4236335 | 7 | 32840370 | 82.9 (.044) | 71.3 (.057) |
| 810 | rs7367380 | 1 | 243492784 | 35.8 (.043) | 24.0 (.063) |
| 811 | rs10858925 | 12 | 88649902 | 77.4 (.052) | 67.5 (.055) |
| 812 | rs4302660 | 6 | 38953337 | 41.2 (.048) | 55.0 (.044) |
| 813 | rs852102 | 20 | 17102385 | 15.2 (.055) | 4.2 (.036) |
| 814 | rs792106 | 2 | 5482130 | 40.9 (.067) | 52.4 (.069) |
| 815 | rs12885270 | 14 | 86242583 | 62.7 (.049) | 76.4 (.060) |
| 816 | rs711635 | 3 | 4465513 | 66.5 (.047) | 60.8 (.069) |
| 817 | rs2554830 | 18 | 72256690 | 47.9 (.035) | 34.8 (.040) |
| 818 | rs7703173 | 5 | 51796987 | 92.6 (.056) | 76.1 (.029) |
| 819 | rs12614495 | 2 | 172902347 | 17.7 (.074) | 34.0 (.089) |
| 820 | rs6945478 | 7 | 149862999 | 98.3 (.034) | 89.5 (.060) |
| 821 | rs34872 | 3 | 10483495 | 53.8 (.058) | 44.9 (.025) |
| 822 | rs4796420 | 17 | 7320341 | 79.8 (.063) | 93.1 (.035) |
| 823 | rs9646322 | 16 | 77722352 | 27.4 (.043) | 17.1 (.027) |
| 824 | rs11924195 | 3 | 109409208 | 18.7 (.064) | 11.1 (.023) |
| 825 | rs2814064 | 1 | 223996368 | 39.4 (.075) | 26.9 (.035) |
| 826 | rs7810044 | 7 | 25718910 | 29.1 (.029) | 38.3 (.037) |
| 827 | rs12418302 | 11 | 102844087 | 93.7 (.015) | 88.4 (.027) |
| 828 | rs9573193 | 13 | 72902025 | 29.8 (.030) | 21.5 (.057) |
| 829 | rs1188445 | 1 | 30832969 | 70.3 (.099) | 94.0 (.055) |
| 830 | rs6089195 | 20 | 30194984 | 77.3 (.144) | 99.9 (.071) |
| 831 | rs7406828 | 17 | 77229631 | 54.9 (.060) | 38.0 (.027) |
| 832 | rs2065914 | 1 | 236496837 | 48.8 (.048) | 62.1 (.041) |
| 833 | rs597566 | 15 | 31671915 | 26.3 (.031) | 33.9 (.048) |
| 834 | rs1346173 | 12 | 107137927 | 44.9 (.040) | 32.0 (.026) |
| 835 | rs7598978 | 2 | 66074063 | 84.4 (.038) | 73.8 (.040) |
| 836 | rs748005 | 16 | 53882623 | 38.8 (.050) | 33.3 (.043) |
| 837 | rs11173732 | 12 | 59750049 | 73.7 (.041) | 54.1 (.039) |
| 838 | rs927297 | 6 | 91081015 | 58.3 (.026) | 69.9 (.049) |
| 839 | rs36059 | 3 | 136215430 | 37.1 (.057) | 25.8 (.035) |
| 840 | rs1793220 | 11 | 130414497 | 27.7 (.041) | 16.9 (.027) |
| 841 | rs4397372 | 8 | 95061743 | 58.7 (.045) | 45.2 (.050) |
| 842 | rs6703991 | 1 | 231011001 | 63.7 (.078) | 72.9 (.045) |
| 843 | rs1005478 | 22 | 35802754 | 45.1 (.040) | 36.1 (.037) |
| 844 | rs11846868 | 14 | 48857807 | 58.7 (.042) | 65.3 (.047) |
| 845 | rs9905258 | 17 | 5847985 | 53.0 (.064) | 40.5 (.044) |
| 846 | rs1156154 | 8 | 72646537 | 37.4 (.055) | 28.0 (.027) |
| 847 | rs508521 | 6 | 116853934 | 62.5 (.042) | 75.3 (.054) |
| 848 | rs2075520 | 16 | 21130173 | 40.1 (.077) | 22.3 (.056) |
| 849 | rs2033050 | 7 | 134391023 | 39.2 (.065) | 29.4 (.040) |
| 850 | rs10893032 | 11 | 122751364 | 73.9 (.073) | 60.4 (.062) |
| 851 | rs6852460 | 4 | 183245512 | 52.2 (.059) | 65.4 (.075) |
| 852 | rs16954586 | 13 | 97306879 | 79.8 (.048) | 91.2 (.065) |
| 853 | rs11199884 | 10 | 123053164 | 81.8 (.120) | 63.6 (.145) |
| 854 | rs7073611 | 10 | 126225492 | 41.2 (.117) | 20.7 (.049) |
| 855 | rs545735 | 3 | 29093778 | 94.0 (.027) | 89.5 (.056) |
| 856 | rs13419175 | 2 | 168813446 | 22.0 (.078) | 9.2 (.050) |
| 857 | rs340989 | 11 | 22452794 | 70.6 (.027) | 59.5 (.039) |
| 858 | rs10033498 | 4 | 31038208 | 50.9 (.084) | 69.6 (.046) |
| 859 | rs348901 | 5 | 83457623 | 79.7 (.054) | 67.4 (.058) |
| 860 | rs2693371 | 17 | 41011471 | 42.8 (.101) | 59.2 (.066) |
| 861 | rs6791314 | 3 | 179505092 | 54.6 (.049) | 38.7 (.078) |
| 862 | rs11579848 | 1 | 204517250 | 65.8 (.073) | 86.8 (.084) |
| 863 | rs274946 | 9 | 23575350 | 68.3 (.121) | 42.7 (.097) |
| 864 | rs4888846 | 16 | 77363264 | 68.2 (.069) | 91.4 (.058) |
| 865 | rs10763586 | 10 | 60181765 | 66.2 (.028) | 78.3 (.084) |
| 866 | rs1440355 | 8 | 73798241 | 74.1 (.033) | 64.4 (.027) |
| 867 | rs10075995 | 5 | 148273622 | 33.1 (.063) | 49.5 (.016) |
| 868 | rs12100904 | 14 | 35554731 | 42.9 (.032) | 32.3 (.020) |
| 869 | rs1023497 | 22 | 40665008 | 95.2 (.037) | 81.3 (.078) |
| 870 | rs10975412 | 9 | 6039547 | 99.3 (.062) | 75.6 (.061) |
| 871 | rs184556 | 15 | 62100208 | 19.2 (.062) | 0.4 (.034) |
| 872 | rs2973869 | 5 | 91133551 | 60.3 (.040) | 73.0 (.049) |
| 873 | rs17814201 | 11 | 36225647 | 18.1 (.098) | 5.2 (.043) |
| 874 | rs13204990 | 6 | 122731099 | 51.6 (.054) | 65.9 (.052) |
| 875 | rs6065533 | 20 | 40828800 | 72.4 (.144) | 97.6 (.033) |
| 876 | rs1549622 | 5 | 151246242 | 68.3 (.051) | 74.9 (.028) |
| 877 | rs7096546 | 10 | 115234052 | 16.1 (.050) | 4.5 (.027) |
| 878 | rs357758 | 2 | 71276935 | 34.5 (.060) | 27.9 (.043) |
| 879 | rs1501605 | 3 | 172449118 | 22.8 (.044) | 16.6 (.040) |
| 880 | rs4810113 | 20 | 56010505 | 78.1 (.063) | 87.5 (.029) |
| 881 | rs1870419 | 12 | 68653898 | 66.8 (.095) | 74.9 (.031) |
| 882 | rs344540 | 19 | 6638768 | 45.8 (.083) | 27.7 (.055) |
| 883 | rs6864641 | 5 | 14803251 | 32.5 (.124) | 8.9 (.076) |
| 884 | rs2804386 | 23 | 68759939 | 68.3 (.041) | 58.0 (.055) |
| 885 | rs11938968 | 4 | 112100356 | 51.3 (.069) | 77.2 (.111) |
| 886 | rs7869905 | 9 | 132680938 | 39.1 (.030) | 30.9 (.022) |
| 887 | rs864643 | 3 | 39530584 | 74.4 (.043) | 81.4 (.019) |
| 888 | rs2074948 | 22 | 28232404 | 71.6 (.057) | 78.8 (.045) |
| 889 | rs2172712 | 1 | 161819150 | 34.6 (.046) | 45.2 (.041) |
| 890 | rs8030411 | 15 | 74486910 | 72.2 (.062) | 52.9 (.070) |
| 891 | rs1707956 | 3 | 39498520 | 25.5 (.059) | 18.0 (.021) |
| 892 | rs1461349 | 11 | 37157502 | 45.2 (.044) | 30.3 (.033) |
| 893 | rs4364968 | 10 | 6543737 | 55.8 (.075) | 71.7 (.097) |
| 894 | rs16830611 | 2 | 135019123 | 27.5 (.093) | 15.9 (.034) |
| 895 | rs557373 | 6 | 44366302 | 70.1 (.037) | 81.9 (.032) |
| 896 | rs13440446 | 9 | 28241541 | 20.9 (.065) | 30.7 (.050) |
| 897 | rs11059856 | 12 | 127735009 | 38.6 (.049) | 46.2 (.026) |
| 898 | rs17727982 | 18 | 45689548 | 81.7 (.026) | 89.8 (.038) |
| 899 | rs2722683 | 19 | 49610498 | 10.2 (.054) | 23.7 (.103) |
| 900 | rs9309783 | 3 | 78033636 | 82.2 (.052) | 73.1 (.039) |
| 901 | rs788216 | 10 | 27550340 | 84.2 (.074) | 62.1 (.068) |
| 902 | rs10911886 | 1 | 183269248 | 95.3 (.018) | 87.4 (.050) |
| 903 | rs3748538 | 1 | 237020043 | 25.5 (.055) | 35.8 (.029) |
| 904 | rs6529924 | 23 | 5978160 | 23.6 (.023) | 39.3 (.032) |
| 905 | rs13263328 | 8 | 21396041 | 33.6 (.046) | 45.6 (.046) |
| 906 | rs7560229 | 2 | 214274149 | 73.7 (.036) | 55.6 (.084) |
| 907 | rs10814831 | 9 | 4073863 | 60.8 (.087) | 77.8 (.059) |
| 908 | rs1949807 | 2 | 227717366 | 95.5 (.084) | 64.3 (.044) |
| 909 | rs17118133 | 8 | 13848795 | 3.1 (.051) | 20.6 (.057) |
| 910 | rs10845852 | 12 | 13918404 | 88.0 (.082) | 75.2 (.060) |
| 911 | rs6428237 | 1 | 191155604 | 35.3 (.096) | 46.7 (.065) |
| 912 | rs4689558 | 4 | 7038538 | 54.5 (.052) | 41.2 (.040) |
| 913 | rs10513456 | 9 | 125799403 | 67.8 (.040) | 80.8 (.024) |
| 914 | rs1035695 | 7 | 134301230 | 48.0 (.055) | 28.5 (.115) |
| 915 | rs2099821 | 5 | 3276744 | 83.3 (.046) | 98.2 (.052) |
| 916 | rs9590631 | 13 | 41197653 | 80.5 (.047) | 67.7 (.078) |
| 917 | rs9321570 | 6 | 137137372 | 60.5 (.040) | 69.0 (.028) |
| 918 | rs925848 | 2 | 115802862 | 85.4 (.075) | 66.7 (.064) |
| 919 | rs6954522 | 7 | 141300966 | 62.9 (.053) | 46.1 (.112) |
| 920 | rs3936080 | 6 | 120900452 | 39.8 (.031) | 48.7 (.031) |
| 921 | rs11969564 | 6 | 52889519 | 72.5 (.055) | 86.8 (.021) |
| 922 | rs12989577 | 2 | 107656407 | 27.8 (.031) | 43.3 (.061) |
| 923 | rs7808122 | 7 | 22571320 | 34.2 (.048) | 46.1 (.041) |
| 924 | rs7757213 | 6 | 67053332 | 80.3 (.032) | 88.8 (.038) |
| 925 | rs3766396 | 1 | 27905696 | 42.9 (.046) | 27.2 (.051) |
| 926 | rs10053917 | 5 | 56858271 | 34.7 (.137) | 8.2 (.059) |
| 927 | rs17790217 | 18 | 63944299 | 38.3 (.056) | 50.0 (.047) |
| 928 | rs10052199 | 5 | 130544267 | 17.4 (.054) | 7.0 (.032) |
| 929 | rs4072865 | 16 | 10931606 | 45.2 (.052) | 55.3 (.058) |
| 930 | rs12477582 | 2 | 125980163 | 66.1 (.078) | 76.3 (.036) |
| 931 | rs4681577 | 3 | 151316687 | 87.2 (.030) | 92.0 (.030) |
| 932 | rs942703 | 1 | 197824636 | 36.9 (.079) | 21.6 (.037) |
| 933 | rs12037277 | 1 | 79983359 | 56.4 (.022) | 69.1 (.031) |
| 934 | rs8104146 | 19 | 34220766 | 47.2 (.047) | 58.1 (.030) |
| 935 | rs4837082 | 9 | 126267377 | 26.1 (.060) | 14.6 (.048) |
| 936 | rs2280732 | 2 | 28730018 | 78.8 (.042) | 88.1 (.037) |
| 937 | rs7233739 | 18 | 6718488 | 37.7 (.046) | 50.9 (.033) |
| 938 | rs11131719 | 4 | 68357286 | 41.1 (.043) | 17.8 (.061) |
| 939 | rs7994224 | 13 | 49450185 | 18.2 (.076) | 4.5 (.042) |
| 940 | rs1446893 | 2 | 72731680 | 88.0 (.040) | 99.1 (.024) |
| 941 | rs7401404 | 14 | 62684209 | 29.0 (.028) | 35.6 (.036) |
| 942 | rs4851854 | 2 | 105842256 | 51.5 (.027) | 44.8 (.041) |
| 943 | rs1467616 | 10 | 102631681 | 38.6 (.050) | 26.6 (.045) |
| 944 | rs9822959 | 3 | 185681145 | 37.8 (.051) | 26.9 (.022) |
| 945 | rs8021814 | 14 | 46735752 | 50.6 (.032) | 61.6 (.039) |
| 946 | rs12921623 | 16 | 16003634 | 87.8 (.070) | 63.2 (.073) |
| 947 | rs4961688 | 9 | 16252191 | 46.7 (.062) | 33.8 (.041) |
| 948 | rs2837297 | 21 | 40225051 | 23.3 (.038) | 13.0 (.037) |
| 949 | rs4309752 | 3 | 136416314 | 49.3 (.055) | 38.2 (.046) |
| 950 | rs500586 | 1 | 40744302 | 59.7 (.030) | 70.6 (.047) |
| 951 | rs2577001 | 15 | 94555158 | 75.9 (.099) | 88.3 (.026) |
| 952 | rs2662389 | 5 | 58162103 | 50.4 (.021) | 40.5 (.021) |
| 953 | rs264705 | 3 | 65839896 | 34.2 (.047) | 27.6 (.053) |
| 954 | rs275838 | 19 | 53558026 | 28.6 (.033) | 21.0 (.033) |
| 955 | rs17345528 | 3 | 102777520 | 35.3 (.071) | 14.2 (.072) |
| 956 | rs7541193 | 1 | 150485320 | 34.6 (.079) | 53.0 (.064) |
| 957 | rs960537 | 2 | 124629747 | 14.6 (.077) | 5.6 (.023) |
| 958 | rs1332011 | 6 | 39016289 | 25.1 (.052) | 15.6 (.033) |
| 959 | rs875964 | 14 | 95802744 | 77.8 (.135) | 95.1 (.087) |
| 960 | rs10016578 | 4 | 17742615 | 22.7 (.048) | 3.3 (.041) |
| 961 | rs6056754 | 20 | 9540598 | 45.5 (.035) | 36.9 (.015) |
| 962 | rs2480490 | 9 | 89387779 | 44.2 (.052) | 32.3 (.034) |
| 963 | rs3857501 | 6 | 92348639 | 63.8 (.057) | 74.3 (.033) |
| 964 | rs2084414 | 8 | 481735 | 72.6 (.034) | 85.2 (.039) |
| 965 | rs2909824 | 5 | 116091320 | 69.2 (.087) | 86.7 (.063) |
| 966 | rs1954916 | 6 | 162418023 | 91.1 (.055) | 78.6 (.061) |
| 967 | rs17755177 | 2 | 187794592 | 71.0 (.054) | 79.9 (.043) |
| 968 | rs7965234 | 12 | 126595604 | 62.1 (.086) | 78.7 (.034) |
| 969 | rs6131684 | 20 | 15572156 | 56.2 (.062) | 46.6 (.057) |
| 970 | rs17712923 | 11 | 39149865 | 16.6 (.172) | 52.6 (.158) |
| 971 | rs2252550 | 6 | 38649462 | 29.2 (.040) | 39.6 (.046) |
| 972 | rs6908475 | 6 | 22680777 | 67.1 (.032) | 58.8 (.041) |
| 973 | rs11176031 | 12 | 64763239 | 71.5 (.132) | 53.3 (.146) |
| 974 | rs3770536 | 2 | 216664326 | 69.7 (.029) | 57.4 (.052) |
| 975 | rs10027488 | 4 | 187482998 | 29.2 (.059) | 44.7 (.048) |
| 976 | rs2218248 | 3 | 179591920 | 64.5 (.091) | 79.3 (.090) |
| 977 | rs17077348 | 5 | 173699036 | 71.0 (.044) | 87.0 (.061) |
| 978 | rs974491 | 14 | 24835423 | 76.1 (.040) | 62.1 (.070) |
| 979 | rs7751500 | 6 | 37697775 | 32.3 (.078) | 14.7 (.060) |
| 980 | rs11127608 | 3 | 78033812 | 82.2 (.044) | 68.6 (.042) |
| 981 | rs975974 | 2 | 43851917 | 15.4 (.048) | 26.6 (.040) |
| 982 | rs3845948 | 3 | 37798704 | 75.6 (.043) | 85.5 (.034) |
| 983 | rs611052 | 7 | 68601918 | 70.2 (.031) | 62.0 (.039) |
| 984 | rs7930841 | 11 | 38466276 | 56.0 (.053) | 68.9 (.039) |
| 985 | rs12433343 | 14 | 46617494 | 7.7 (.036) | 17.8 (.023) |
| 986 | rs1281978 | 6 | 153565346 | 38.5 (.051) | 51.1 (.047) |
| 987 | rs7077548 | 10 | 113350071 | 76.6 (.083) | 58.8 (.036) |
| 988 | rs7127507 | 11 | 27671460 | 25.2 (.059) | 38.3 (.076) |
| 989 | rs4971342 | 2 | 926687 | 88.8 (.076) | 67.8 (.056) |
| 990 | rs1683119 | 14 | 96572498 | 33.5 (.040) | 22.5 (.041) |
| 991 | rs2482527 | 9 | 25554648 | 81.6 (.057) | 72.5 (.054) |
| 992 | rs6894312 | 5 | 18712040 | 13.7 (.044) | 24.3 (.030) |
| 993 | rs4716908 | 7 | 155060486 | 77.2 (.054) | 96.5 (.039) |
| 994 | rs1900455 | 10 | 55266508 | 50.5 (.043) | 62.9 (.060) |
| 995 | rs2391870 | 7 | 30314272 | 77.4 (.102) | 58.3 (.079) |
| 996 | rs771443 | 6 | 83452859 | 72.3 (.055) | 57.4 (.090) |
| 997 | rs3091260 | 7 | 76475046 | 22.9 (.032) | 34.4 (.053) |
| 998 | rs10878366 | 12 | 64708641 | 79.6 (.055) | 68.7 (.112) |
| 999 | rs255026 | 5 | 178717241 | 48.6 (.035) | 56.2 (.039) |
| 1,000 | rs5750373 | 22 | 35749531 | 73.5 (.064) | 59.6 (.036) |
The rank order of the SNPs is based on a silhouette-test statistic calculated on probe intensity measures from replicate arrays, implemented in GenePool 0.81 (TGen Bioinformatics Research Unit). Only SNPs for which allelic frequencies could be calculated using the k-correction method35 from a training database of 1,000 individually genotyped samples are included.
Chromosomal positions for each SNP are based on National Center for Biotechnology Information build 35.1.
Frequencies are expressed as percentages. Predicted allelic frequencies are the median frequency of all replicate arrays and refer to the A allele as defined by Affymetrix 500K. The median value is used, since an extreme outlier is occasionally observed.
SNP has no rs number, only an Affymetrix identification number.
Figure 1. .
Two loci showing strong support for association by pooled analysis. A, Genomewide plot of the mean rank of five consecutive SNPs, calculated to identify clusters of high-ranking SNPs. The single best region was on chromosome 17, neighboring MAPT, and the second best region was on chromosome 11p12. Chromosome 11p12 also harbored the SNP that ranked #1 overall by single-marker statistics. B, Single-marker rank statistics for SNPs over the MAPT (left) and DDB2/ACP2 (right) loci. SNPs deemed less reliable or showing high variability among replicates were removed, and the remaining SNPs were ranked in order from 1 (showing the greatest difference between cases and controls) to 428,867 (showing the least difference between cases and controls) with use of a silhouette-test statistic in GenePool software (TGen Bioinformatics Research Unit). Rank scores are plotted versus chromosomal position. Genes within the plotted chromosomal region are shown below. SNPs on the Affymetrix 500K platform are shown above, and SNPs on the Affymetrix 100K platform are shown below. EA = Early Access.
The chromosome 11p12 region that showed the highest rank SNP by single-marker statistics and multimarker sliding-window analysis was a novel locus and therefore was examined in greater detail (fig. 1B). The top overall ranked SNP, rs901746, a SNP in intron 9 of the DNA damage-binding protein 2 (DDB2 [MIM 600811]) gene, was chosen for follow-up in the individual samples comprising the pooled DNA. A significant increase of 10% in the G allele frequency was seen in cases versus controls (P=.0002) (table 2). The SNP was then genotyped in a second U.S. series to confirm the association. This “replication” sample (n=161) was made up of both pathologically confirmed (n=97) and clinically defined PSP case individuals (n=64), as described in Rademakers et al.22 A total of 165 age- and sex-matched cognitively normal control individuals were obtained from the Normal and Pathological Aging Protocol at the Mayo Clinic (Scottsdale).30,31 In addition, for the rs901746 and rs2167079 analysis, additional pathologically confirmed cases (n=41) and clinically defined PSP case indivuals (n=22) were genotyped, and 252 age- and sex-matched cognitively normal control individuals collected at Mayo Clinic Jacksonville were used as a second source of controls.22 All case and control individuals in this set were white from the United States and Canada, and IRB-approved protocols were used in the collection of all samples.
Table 2. .
Association Analysis of rs901746 in Original and Replication Series
| No. (%) of |
|||||||||
| Alleles |
Genotypes |
GG versus AG and AA |
|||||||
| Population | n | A | G | AA | AG | GG | OR | 95% CI | P |
| Control combined | 735 | 1,011 (75) | 335 (25) | 377 (56) | 257 (38) | 39 (6) | … | … | … |
| Control original | 344 | 438 (78) | 126 (22) | 166 (58) | 106 (37) | 10 (4) | … | … | … |
| Control replication | 391 | 573 (73) | 209 (27) | 211 (54) | 151 (39) | 29 (7) | … | … | … |
| PSP combined | 501 | 661 (68) | 317 (32) | 231 (47) | 199(41) | 59 (12) | 2.2 | 1.5–3.4 | .0001 |
| PSP original | 288 | 374 (68) | 178 (32) | 131 (47) | 112 (41) | 33 (12) | 4.0 | 1.9–8.3 | .0001 |
| PSP replication | 213 | 287 (67) | 139 (33) | 100 (47) | 87 (41) | 26 (12) | 1.7 | 1.0–3.0 | .05 |
When allele frequencies at rs901746 were examined in the replication sample set, a 6% increase in the frequency of the G allele in subjects with PSP was observed; however, because of the smaller sample size, this allele frequency difference is borderline significant (P=.05). When genotype distributions were examined in both PSP case-control series, the frequencies were very similar, with an increase from 4% to 12% in the GG genotype in the original population and an increase from 7% to 12% in the replication set. The allelic frequency difference in both series is explained by an apparent doubling of the GG frequency in subjects with PSP compared with controls, suggesting that risk at this locus acts in a recessive manner. We explicitly tested dominant, recessive, and additive models at this locus, and the model that best fit the data was a recessive one (P<.0001). The OR for harboring an rs901746 GG genotype versus all other genotypes in the original series was 3.7 (95% CI 1.2–3.9) and was 1.7 (95% CI 1.0–3.0) for the replication series. When these individuals in both of these series were combined and analyzed, the combined OR for the GG genotype compared with all other genotypes in the series was 2.2 (95% CI 1.4–3.4). To confirm that the rs901746 association observed is not a control frequency artifact, we examined allele frequencies for rs901746 in 250 cognitively normal controls recently published in a Parkinson disease (PD [MIM 168600]) GWA study.38 We found that the frequency of the rs901746 G allele in this independent control series was 0.27, consistent with our observed control frequencies (0.22 and 0.27).
The genomic context near rs901746 was examined by downloading the CEPH-from-Utah SNP genotypes for 100 kb around rs901746 from the HapMap genome browser and by examining the LD patterns and haplotype-block structure of the region with use of the Haploview software.39 rs901746 lies in the middle of a haplotype block encompassing at least two genes—the DDB2 gene and the lysosomal acid phosphatase 2 (ACP2 [MIM 171650]) gene—and can extend into the 3′ of another gene, nuclear receptor subfamily 1, group H, member 3 (NR1H3 [MIM 602423]), depending on the type of haplotype-block definition used.39 Variation in this 100-kb region could be fully described by 16 additional tag SNPs. These tag SNPs were genotyped in all PSP series, and both single and multimarker analysis was performed on the combined series (table 3). Single-marker analysis showed that nine tag SNPs showed significant allelic association. Of these, five tag SNP associations were highly significant (P values ⩽.003), with rs10501320 showing the greatest association after rs901746 (P<.0001).
Table 3. .
Single-Marker Analysis of Tag SNPs in the Combined Series and in Both Young and Old Patient Populations[Note]
| All Cases(n=448) |
Youngb Cases(n=162) |
Oldb Cases(n=182) |
|||||
| tagIDa (SNP), and Allele |
No. (%) of Alleles in Controls (n=532) |
No. (%) of Alleles |
P | No. (%) of Alleles |
P | No. (%) of Alleles |
P |
| 1 (rs11039130): | .003 | .02 | .25 | ||||
| C | 600 (69) | 614 (75) | 224 (76) | 245 (72) | |||
| T | 274 (31) | 202 (25) | 72 (24) | 95 (28) | |||
| 2 (rs4647709): | .5 | .57 | .88 | ||||
| C | 806 (91) | 787 (90) | 292 (90) | 331 (91) | |||
| T | 78 (9) | 85 (10) | 32 (10) | 31 (9) | |||
| 3 (rs2291120): | .0004 | .003 | .006 | ||||
| T | 781 (92) | 859 (87) | 280 (86) | 317 (87) | |||
| C | 67 (8) | 115 (13) | 44 (14) | 47 (13) | |||
| 4 (rs10742797): | .81 | .72 | .98 | ||||
| A | 591 (81) | 572 (81) | 212 (82) | 243 (80) | |||
| T | 143 (19) | 134 (19) | 48 (18) | 59 (20) | |||
| 5 (rs1685404): | .72 | .97 | .96 | ||||
| G | 598 (68) | 560 (67) | 213 (68) | 237 (68) | |||
| C | 282 (32) | 274 (33) | 101 (32) | 111 (32) | |||
| 6 (rs7395581): | .03 | .02 | .07 | ||||
| A | 378 (71) | 437 (65) | 157 (63) | 180 (65) | |||
| G | 152 (29) | 233 (35) | 93 (37) | 96 (35) | |||
| 7 (rs11039138): | .01 | .02 | .37 | ||||
| G | 470 (56) | 442 (62) | 168 (64) | 173 (59) | |||
| A | 372 (44) | 268 (38) | 94 (36) | 121 (41) | |||
| 8 (rs2957873): | .22 | .52 | .28 | ||||
| A | 728 (83) | 679 (81) | 257 (81) | 281 (80) | |||
| G | 150 (17) | 163 (19) | 59 (19) | 69 (20) | |||
| 9 (rs4647736): | .03 | .04 | .12 | ||||
| C | 807 (91) | 736 (88) | 273 (88) | 305 (89) | |||
| T | 75 (9) | 98 (12) | 39 (13) | 39 (11) | |||
| 10 (rs2013867): | .004 | .006 | .02 | ||||
| T | 657 (74) | 549 (66) | 206 (66) | 236 (67) | |||
| C | 229 (26) | 279 (34) | 106 (34) | 114 (33) | |||
| 11 (rs901746): | <.0001 | .003 | .004 | ||||
| A | 659 (76) | 570 (67) | 204 (65) | 242 (68) | |||
| G | 213 (24) | 282 (33) | 110 (35) | 116 (32) | |||
| 12 (rs1050244): | .53 | .89 | .44 | ||||
| C | 851 (97) | 823 (96) | 307 (97) | 343 (96) | |||
| T | 29 (3) | 33 (4) | 11 (3) | 15 (4) | |||
| 13 (rs11039143): | .87 | .64 | .52 | ||||
| T | 830 (98) | 782 (98) | 293 (98) | 319 (98) | |||
| G | 18 (2) | 16 (2) | 5 (2) | 5 (2) | |||
| 14 (rs7118396): | .16 | .27 | .13 | ||||
| C | 741 (86) | 688 (84) | 253 (84) | 287 (83) | |||
| T | 117 (14) | 132 (16) | 49 (16) | 59 (17) | |||
| 15 (rs12577530): | .0009 | .005 | .04 | ||||
| G | 784 (88) | 701 (82) | 260 (82) | 296 (84) | |||
| C | 106 (12) | 149 (18) | 58 (18) | 58 (16) | |||
| 16 (rs7114704): | .01 | .3 | .0006 | ||||
| C | 813 (93) | 803 (96) | 288 (95) | 343 (98) | |||
| T | 61 (7) | 35 (4) | 16 (5) | 7 (2) | |||
| 17 (rs10501320): | <.0001 | .003 | .16 | ||||
| G | 609 (70) | 641 (78) | 230 (79) | 255 (74) | |||
| C | 265 (30) | 179 (22) | 62 (21) | 91 (26) | |||
Note.— Significant P values are shown in bold.
Tag SNPs were chosen on the basis of the tagging algorithm in Haploview v3.32 software,39 with the “Pairwise Tagging Only” option selected and the r2 threshold set at .8.
Subjects aged <76 years were classified as “young”; subjects aged ⩾76 were classified as “old.”
We examined the DDB2/ACP2 tag SNP data set in different age groups in our combined series of cases and controls to see whether looking at younger cases might help further refine the associated region, as it had for the MAPT locus, where younger cases show a stronger association with the H1/H1 genotype.22 Pathologically confirmed cases were divided into “young” and “old” groups on the basis of median age at death (75 years), and single-marker allelic association statistics were calculated using 2×2 contingency tables and were examined using χ2 tests. On the whole, all of the SNPs that show significant association in the combined PSP case set also show significant association (P<.05) with the younger case subset, whereas there is less-significant association observed for the older cases.
To refine the disease-associated region, we performed haplotype-inference analysis in the young cases versus all controls, using a sliding-window approach.40 As described elsewhere, this type of approach was key in refining the associated region on the MAPT H1 haplotype.22 However, as figure 2 displays, when data from all the tag SNPs were included in the analysis, there was no obvious resolution of the associated region when the young cases were considered separately. This may reflect the fact that the contribution to the overall signal of the association at this locus was not as great with the younger cases as had been seen with the MAPT locus; therefore, the sample size and/or the number of informative SNPs was inadequate to detect a smaller associated region.
Figure 2. .
Haplotype sliding window–analysis results. The haplotype score–based method of Schaid et al.40 was used to investigate evidence of association of haplotypes with case-control status. Only haplotypes with an estimated overall frequency of ⩾5% were considered for these analyses. Reported P values are based on asymptotic assumptions but were verified by simulating P values derived from 1,000 permutations of case and control labels and were found to be consistent. Global P values for each 4-marker haplotype are denoted as lines at the −log10P. Only young pathologically confirmed PSP cases (death at age <76 years) were used for the analysis. All individuals in the control group were used in all analyses, since no single SNP showed significantly different allelic frequency distribution in controls when stratified by age. Global P=.01 is denoted by a dashed line. SNP numbers are as noted in table 3.
Since a haplotype-inference approach was unsuccessful in narrowing the associated region, we decided to identify additional novel SNPs that may represent functional variant(s) accounting for increased risk of disease by sequencing a series of 18 subjects with PSP who had the various genotypes at rs901746, the majority of whom carried the risky GG genotype (n=11 GG, n=5 AG, and n=2 AA) (table 4). Primers were designed to fully sequence coding exons of both DDB2 and ACP2. Only one SNP, found 63 bp downstream of exon 3 in ACP2, was not already in the dbSNP database; however, this SNP appeared to be in near-complete LD with nearby rs10838677 in exon 5 of ACP2, encoding a silent change (L165L). Interestingly, a number of SNPs identified through sequencing appeared to be in near-complete LD with rs901746, including rs2167079, a coding SNP in ACP2 in which the minor allele changes the amino acid at position 29 from an arginine to a glutamine (R29Q). This converts the protein sequence to the mouse amino acid residue at the equivalent position. Interestingly, this position in ACP2 is predicted to encode the signal peptidase cleavage site,41 suggesting that carriers of the minor allele encoding glutamine at position 29 may have altered cleavage of the signal peptide compared with those encoding arginine at that position. Since this SNP could affect function of the protein, we genotyped it through the combined series. Results from this analysis are shown in table 5. Overall, the LD between rs901746 and rs2167079 was high (cases r2=0.97; controls r2=0.94). As expected, significant allelic association was observed with rs2167079 (P=.002); however, this was not any greater than the association observed with rs901746, suggesting that rs2167079 is unlikely to fully explain the association at the DDB2/ACP2 locus. We tested the R29Q variant for dominant, recessive, and additive models, and the additive model best fit the data (P<.0001).
Table 4. .
SNP Discovery Results from Sequencing DDB2 and ACP2 in 18 Subjects with PSP
| Genotype at DDB25′→3′ |
Genotype at ACP23′→5′ |
||||||||
| Intron 8 |
Intron 9 |
3′ UTR |
Intron 6 |
Exon 5 |
Intron 3 |
Exon 1 |
|||
| Sample | rs326222 | rs901746 | rs1050244 | rs11039146 | rs2242261 | rs10838677a | ss68362654b | rs4752973 | rs2167079c |
| 1 | GG | GG | CT | CT | AA | AG | AG | AG | AA |
| 2 | GG | GG | CC | CC | AA | GG | GG | AA | AA |
| 3 | GG | GG | CC | CC | CC | GG | GG | GG | AA |
| 4 | GG | GG | CC | CC | AC | GG | GG | AG | AA |
| 5 | GG | GG | CC | CC | AC | GG | GG | AG | AA |
| 6 | GG | GG | CC | CC | CC | GG | GG | GG | AA |
| 7 | GG | GG | CC | CC | AA | GG | GG | AA | AA |
| 8 | GG | GG | CT | CT | AA | AG | AG | AG | AA |
| 9 | AG | AG | CC | CC | AA | GG | GG | AA | AG |
| 10 | GG | GG | CC | CC | AC | GG | GG | AG | AA |
| 11 | GG | GG | CC | CC | AC | GG | GG | AG | AA |
| 12 | GG | GG | CC | CC | AC | GG | GG | AG | AA |
| 13 | AG | AG | CC | CC | AA | GG | GG | AA | AG |
| 14 | AG | AG | CC | CC | AA | GG | GG | AA | GG |
| 15 | AG | AG | CC | CC | AA | GG | GG | AA | AG |
| 16 | AG | AG | CC | CC | AA | GG | GG | AA | AG |
| 17 | AA | AA | CC | CC | AA | GG | GG | AA | GG |
| 18 | AA | AA | CC | CC | AA | GG | GG | AA | GG |
Encodes synonymous change L165L.
No rs number; submitted to dbSNP.
Encodes nonsynonymous change R29Q.
Table 5. .
Association Analysis of rs2167079 in the Combined Series[Note]
| No. (%) of Alleles |
|||
| SNP and Allele |
All Controls (n=735) |
All Patients (n=501) |
P |
| rs901746: | <.0001 | ||
| A | 1,011 (75) | 661 (68) | |
| G | 335 (25) | 317 (32) | |
| rs2167079: | .002 | ||
| G | 918 (73) | 598 (67) | |
| A | 332 (27) | 292 (33) | |
Note.— Results include the additional cases and controls used in the replication series.
In an alternative method for determining the gene responsible for disease risk at this locus, expression analysis was performed on the DDB2 and ACP2 genes. Analysis was performed, using real-time Taqman expression assays (Applied Biosystems), on mRNA extracted from the cerebella of 20 rs901746 AA and 20 rs901746 GG genotype carriers, to determine whether risk variants at the DDB2/ACP2 locus have a direct effect on gene expression. Unfortunately, although DDB2 transcript levels are slightly increased in cases with a GG genotype, no significant differences were observed between the cases with AA and GG genotypes for either DDB2 or ACP2 mRNA levels (for DDB2, P=.29; for ACP2, P=.48) (fig. 3).
Figure 3. .
Relative mRNA expression with TATA-binding protein as an endogenous control. Plotted are relative levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH); tyrosine 3-monooxygenase/tryptophan 5-mono-oxygenase activation protein, zeta polypeptide (YWHAZ); ACP2 (assay Hs00155636_m1 [Applied Biosystems]); and DDB2 (assay Hs00172068_m1 [Applied Biosystems]) for 20 carriers of the rs901746 AA (neutral) genotype and 20 carriers of the rs901746 GG (risky) genotype. SE is denoted by the error bars. None of the comparisons between AA and GG carriers reach the level of statistical significance (P values noted below each graph). Similar results are seen when GAPDH or YWHAZ was used as the endogenous control (data not shown). RQ = relative quantity.
GWA studies are appealing because of their lack of bias, in that they represent a model-free approach for identification of new and novel genes that are involved in a disease process that may never be identified using other methodologies. However, even now, individually genotyping hundreds of individuals to perform a “traditional” GWA is not feasible for many rarer diseases, including PSP, because of the lack of available funding. Therefore, this type of pooled genomewide approach potentially represents a fast and economical initial solution to this problem. Pooling methods lack the analytical flexibility inherent in a traditional genomewide study because it is not possible to reanalyze the data with use of subgroups of cases or controls or to perform true haplotype-scanning analyses. However, there is still some uncertainty about how best to analyze the large amounts of individual genotype data used in GWA studies. An early GWA study of the PD showed problems in replication of results, potentially because of problems in study design.42–47
Although pooling methods clearly have limitations, the analysis procedures we used in the GenePool software (TGen Bioinformatics Research Unit) were developed using individual genotype data from samples that were also pooled, thereby allowing the algorithms to be adjusted until they predicted SNP ranks on the basis of what was known from the individual genotype data.32 In the present analysis, we had prior knowledge that the MAPT H1 haplotype is associated with disease, so it could serve as a positive control for the genomewide analysis.
The identification of a new risk locus for PSP on chromosome 11 from the pooled genomewide approach was confirmed in a second U.S. PSP case-control series, with similar allele and genotype frequencies. Closer examination of this locus by dense SNP genotyping suggests that the association spans the entire haplotype block containing the DDB2 and ACP2 genes. Examination of potential functional variants yielded no definitive explanation for the observed association.
Both ACP2 and DDB2 are reasonable candidate genes that highlight previously implicated pathways for neurodegenerative disease. There are many lines of evidence suggesting a role for lysosomes and autophagic processes in neurodegeneration. Autophagy has been implicated in the clearance of protein aggregates, a common feature of many neurodegenerative disorders.48,49 Interestingly, patients with lysosomal-storage disorders often exhibit neurological phenotypes with pathology similar to that seen in PSP.50–52 Two lines of evidence implicate ACP2 in neurodegeneration. First, it has been reported that, in brains of subjects with Alzheimer disease (AD [MIM 104300]), microglia surrounding the amyloid plaques stain strongly for ACP2.53 In addition, cerebrospinal fluid from half of the examined subjects with AD showed evidence of ACP2 activity, whereas patients not affected with AD showed no activity.53 These results leave open the question of whether ACP2 in AD is just a secondary marker of neurodegeneration or perhaps plays a more active role in the neurodegenerative process. Second, knockouts and mutations of Acp2 in mice have neurological phenotypes.54,55 Neuropathology of Acp2−/− tissue showed increased lysosomal staining (as detected by lamp-1 and cathepsin D immunoreactivity), primarily in glial cells. Interestingly, ∼7% of these Acp2−/− mice presented with generalized seizures after age 8 wk, and it has been suggested that this phenotype may be correlated with the defective lysosomal storage observed in glial cells.54 The observation that loss of Acp2 causes deficits in glial lysosomal storage in the Acp2−/− mice may also be significant, given that, in PSP, there is abundant MAPT-inclusion pathology within glia (astrocytes and olgodendroglia), as well as in neurons.56
Mutations in the DDB2 gene are responsible for xeroderma pigmentosum (XP) complementation group E (XPE [MIM 278740]). Interestingly, some mutations in the nucleotide excision–repair pathway that cause the diseases XP and Cockayne syndrome (MIM 216400) present with neurological phenotypes; however, XPE does not seem to be one of them.57,58 DDB2 forms a ubiquitin E3-ligase complex, with DNA damage-binding protein 1 (DDB1 [MIM 600045]) and Cullin 4a (CUL4A [MIM 603137]), that binds damaged DNA. Both histone H2A (H2AA [MIM 603137]) and XP complementation group C (XPC [MIM 278720]) proteins have been implicated as substrates for the DDB1/DDB2/CUL4A complex upon activation.59,60 Ubiquitination of histone H2A may change local chromatin configuration at the damage site, thereby allowing access to other DNA-repair proteins farther down the pathway.60 The accumulation of damaged DNA in aging brain suggests that DNA-repair capacity is reduced as we age and appears to be selective to genes important in learning and memory. Interestingly, there is evidence of brain-specific alternatively spliced forms of DDB2 that splice out either exons 4–7 or exons 4 and 6 alone.61 The proteins encoded by these alternatively spliced transcripts act as dominant negative inhibitors of DNA repair, when tested in an in vitro system.61 It will be interesting to tease apart which gene or genes at this locus are involved in conferring risk of PSP, but functional studies, rather than genetic ones, will probably be required to address these issues.
Given the size of the association seen at the DDB2/ACP2 locus, the fact that the described PSP series represents the largest collection of PSP-affected subjects worldwide, and the fact that our U.S. replication series is underpowered to detect changes with an OR <2.0, we may be at the limit of what can be consistently detected and confirmed using the case-control populations available. Six additional weaker loci were identified in the genomewide screen that still need to be analyzed in detail, and it will be interesting to examine this potential power issue in closer detail. This genomewide analysis has identified a novel second locus implicated in PSP risk, accelerating research and the hope of identifying effective therapeutics for this devastating disease.
Acknowledgments
We are grateful to the patients and their families for their cooperation in this project. We also thank the Society for Progressive Supranuclear Palsy brain bank for providing samples. We thank John Gonzalez and Monica Castanedes-Casey in the Neuropathology Laboratory at Mayo Clinic Jacksonville for technical assistance. This research was funded by National Institutes of Aging grant PO1 AG 17216 (to M.H., D.W.D., and M.J.F.); the Mayo Foundation, USA; The Society for Progressive Supranuclear Palsy; and a Robert and Clarice Smith Fellowship. S.M. was funded by the National Research Service Award postdoctoral fellowship AG24030. Funding was also provided by a foundation grant from the Stardust Foundation (to D.W.C.) and the National Institutes of Health Neuroscience Blueprint grant 1U24NS043571 (to D.A.S.).
Web Resources
The URLs for data presented herein are as follows:
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PSP, MAPT, DDB2, PD, ACP2, NR1H3, AD, XPE, Cockayne syndrome, DDB1, CUL4A, H2AA, and XPC)
- TGen Bioinformatics Research Unit, http://bioinformatics.tgen.org/ (for the GenePool source code)
References
- 1.Steele JC, Richardson JC, Olszewski J (1964) Progressive supranuclear palsy: a heterogeneous degeneration involving the brain stem, basal ganglia and cerebellum with vertical gaze and pseudobulbar palsy, nuchal dystonia and dementia. Arch Neurol 10:333–359 [DOI] [PubMed] [Google Scholar]
- 2.Litvan I (2004) Update on progressive supranuclear palsy. Curr Neurol Neurosci Rep 4:296–302 [DOI] [PubMed] [Google Scholar]
- 3.Litvan I, Agid Y, Calne D, Campbell G, Dubois B, Duvoisin RC, Goetz CG, Golbe LI, Grafman J, Growdon JH, et al (1996) Clinical research criteria for the diagnosis of progressive supranuclear palsy (Steele-Richardson-Olszewski syndrome): report of the NINDS-SPSP International Workshop. Neurology 47:1–9 [DOI] [PubMed] [Google Scholar]
- 4.Litvan I, Agid Y, Jankovic J, Goetz C, Brandel JP, Lai EC, Wenning G, D’Olhaberriague L, Verny M, Chaudhuri KR, et al (1996) Accuracy of clinical criteria for the diagnosis of progressive supranuclear palsy (Steele-Richardson-Olszewski syndrome). Neurology 46:922–930 [DOI] [PubMed] [Google Scholar]
- 5.Hauw JJ, Daniel SE, Dickson D, Horoupian DS, Jellinger K, Lantos PL, McKee A, Tabaton M, Litvan I (1994) Preliminary NINDS neuropathologic criteria for Steele-Richardson-Olszewski syndrome (progressive supranuclear palsy). Neurology 44:2015–2019 [DOI] [PubMed] [Google Scholar]
- 6.Litvan I, Hauw JJ, Bartko JJ, Lantos PL, Daniel SE, Horoupian DS, McKee A, Dickson D, Bancher C, Tabaton M, et al (1996) Validity and reliability of the preliminary NINDS neuropathologic criteria for progressive supranuclear palsy and related disorders. J Neuropathol Exp Neurol 55:97–105 [DOI] [PubMed] [Google Scholar]
- 7.Rojo A, Pernaute RS, Fontan A, Ruiz PG, Honnorat J, Lynch T, Chin S, Gonzalo I, Rabano A, Martinez A, et al (1999) Clinical genetics of familial progressive supranuclear palsy. Brain 122:1233–1245 (erratum 123:419) 10.1093/brain/122.7.1233 [DOI] [PubMed] [Google Scholar]
- 8.Ros R, Thobois S, Streichenberger N, Kopp N, Sanchez MP, Perez M, Hoenicka J, Avila J, Honnorat J, de Yebenes JG (2005) A new mutation of the τ gene, G303V, in early-onset familial progressive supranuclear palsy. Arch Neurol 62:1444–1450 10.1001/archneur.62.9.1444 [DOI] [PubMed] [Google Scholar]
- 9.Spillantini MG, Goedert M (2000) Tau mutations in familial frontotemporal dementia. Brain 123:857–859 10.1093/brain/123.5.857 [DOI] [PubMed] [Google Scholar]
- 10.Stanford PM, Halliday GM, Brooks WS, Kwok JB, Storey CE, Creasey H, Morris JG, Fulham MJ, Schofield PR (2000) Progressive supranuclear palsy pathology caused by a novel silent mutation in exon 10 of the tau gene: expansion of the disease phenotype caused by tau gene mutations. Brain 123:880–893 10.1093/brain/123.5.880 [DOI] [PubMed] [Google Scholar]
- 11.Wszolek ZK, Tsuboi Y, Uitti RJ, Reed L, Hutton ML, Dickson DW (2001) Progressive supranuclear palsy as a disease phenotype caused by the S305S tau gene mutation. Brain 124:1666–1670 10.1093/brain/124.8.1666 [DOI] [PubMed] [Google Scholar]
- 12.Pastor P, Pastor E, Carnero C, Vela R, Garcia T, Amer G, Tolosa E, Oliva R (2001) Familial atypical progressive supranuclear palsy associated with homozigosity for the delN296 mutation in the tau gene. Ann Neurol 49:263–267 [DOI] [PubMed] [Google Scholar]
- 13.Oliva R, Pastor P (2004) Tau gene delN296 mutation, Parkinson’s disease, and atypical supranuclear palsy. Ann Neurol 55:448–449 10.1002/ana.20025 [DOI] [PubMed] [Google Scholar]
- 14.Rossi G, Gasparoli E, Pasquali C, Di Fede G, Testa D, Albanese A, Bracco F, Tagliavini F (2004) Progressive supranuclear palsy and Parkinson’s disease in a family with a new mutation in the tau gene. Ann Neurol 55:448 10.1002/ana.20006 [DOI] [PubMed] [Google Scholar]
- 15.Ros R, Gomez Garre P, Hirano M, Tai YF, Ampuero I, Vidal L, Rojo A, Fontan A, Vazquez A, Fanjul S, et al (2005) Genetic linkage of autosomal dominant progressive supranuclear palsy to 1q31.1. Ann Neurol 57:634–641 10.1002/ana.20449 [DOI] [PubMed] [Google Scholar]
- 16.Baker M, Litvan I, Houlden H, Adamson J, Dickson D, Perez-Tur J, Hardy J, Lynch T, Bigio E, Hutton M (1999) Association of an extended haplotype in the tau gene with progressive supranuclear palsy. Hum Mol Genet 8:711–715 10.1093/hmg/8.4.711 [DOI] [PubMed] [Google Scholar]
- 17.Conrad C, Andreadis A, Trojanowski JQ, Dickson DW, Kang D, Chen X, Wiederholt W, Hansen L, Masliah E, Thal LJ, et al (1997) Genetic evidence for the involvement of τ in progressive supranuclear palsy. Ann Neurol 41:277–281 10.1002/ana.410410222 [DOI] [PubMed] [Google Scholar]
- 18.Higgins JJ, Golbe LI, De Biase A, Jankovic J, Factor SA, Adler RL (2000) An extended 5′-tau susceptibility haplotype in progressive supranuclear palsy. Neurology 55:1364–1367 [DOI] [PubMed] [Google Scholar]
- 19.Morris HR, Janssen JC, Bandmann O, Daniel SE, Rossor MN, Lees AJ, Wood NW (1999) The tau gene A0 polymorphism in progressive supranuclear palsy and related neurodegenerative diseases. J Neurol Neurosurg Psychiatry 66:665–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliva R, Tolosa E, Ezquerra M, Molinuevo JL, Valldeoriola F, Burguera J, Calopa M, Villa M, Ballesta F (1998) Significant changes in the tau A0 and A3 alleles in progressive supranuclear palsy and improved genotyping by silver detection. Arch Neurol 55:1122–1124 10.1001/archneur.55.8.1122 [DOI] [PubMed] [Google Scholar]
- 21.Cruts M, Rademakers R, Gijselinck I, van der Zee J, Dermaut B, de Pooter T, de Rijk P, Del-Favero J, van Broeckhoven C (2005) Genomic architecture of human 17q21 linked to frontotemporal dementia uncovers a highly homologous family of low-copy repeats in the tau region. Hum Mol Genet 14:1753–1762 10.1093/hmg/ddi182 [DOI] [PubMed] [Google Scholar]
- 22.Rademakers R, Melquist S, Cruts M, Theuns J, Del-Favero J, Poorkaj P, Baker M, Sleegers K, Crook R, De Pooter T, et al (2005) High-density SNP haplotyping suggests altered regulation of tau gene expression in progressive supranuclear palsy. Hum Mol Genet 14:3281–3292 10.1093/hmg/ddi361 [DOI] [PubMed] [Google Scholar]
- 23.Pittman AM, Myers AJ, Abou-Sleiman P, Fung HC, Kaleem M, Marlowe L, Duckworth J, Leung D, Williams D, Kilford L, et al (2005) Linkage disequilibrium fine mapping and haplotype association analysis of the tau gene in progressive supranuclear palsy and corticobasal degeneration. J Med Genet 42:837–846 10.1136/jmg.2005.031377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwok JB, Teber ET, Loy C, Hallupp M, Nicholson G, Mellick GD, Buchanan DD, Silburn PA, Schofield PR (2004) Tau haplotypes regulate transcription and are associated with Parkinson’s disease. Ann Neurol 55:329–334 10.1002/ana.10826 [DOI] [PubMed] [Google Scholar]
- 25.Caffrey TM, Joachim C, Paracchini S, Esiri MM, Wade-Martins R (2006) Haplotype-specific expression of exon 10 at the human MAPT locus. Hum Mol Genet 15:3529–3537 10.1093/hmg/ddl429 [DOI] [PubMed] [Google Scholar]
- 26.Myers AJ, Pittman AM, Zhao AS, Rohrer K, Kaleem M, Marlowe L, Lees A, Leung D, McKeith IG, Perry RH, et al (2006) The MAPT H1c risk haplotype is associated with increased expression of tau and especially of 4 repeat containing transcripts. Neurobiol Dis (electronically published ahead of print December 14, 2006; accessed February 7, 2007) [DOI] [PubMed] [Google Scholar]
- 27.Bansal A, van den Boom D, Kammerer S, Honisch C, Adam G, Cantor CR, Kleyn P, Braun A (2002) Association testing by DNA pooling: an effective initial screen. Proc Natl Acad Sci USA 99:16871–16874 10.1073/pnas.262671399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meaburn E, Butcher LM, Schalkwyk LC, Plomin R (2006) Genotyping pooled DNA using 100K SNP microarrays: a step towards genomewide association scans. Nucleic Acids Res 34:e28 10.1093/nar/gnj027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Josephs KA, Dickson DW (2003) Diagnostic accuracy of progressive supranuclear palsy in the Society for Progressive Supranuclear Palsy brain bank. Mov Disord 18:1018–1026 10.1002/mds.10488 [DOI] [PubMed] [Google Scholar]
- 30.Caselli RJ, Osborne D, Reiman EM, Hentz JG, Barbieri CJ, Saunders AM, Hardy J, Graff-Radford NR, Hall GR, Alexander GE (2001) Preclinical cognitive decline in late middle-aged asymptomatic apolipoprotein E-e4/4 homozygotes: a replication study. J Neurol Sci 189:93–98 10.1016/S0022-510X(01)00577-9 [DOI] [PubMed] [Google Scholar]
- 31.Caselli RJ, Hentz JG, Osborne D, Graff-Radford NR, Barbieri CJ, Alexander GE, Hall GR, Reiman EM, Hardy J, Saunders AM (2002) Apolipoprotein E and intellectual achievement. J Am Geriatr Soc 50:49–54 10.1046/j.1532-5415.2002.50007.x [DOI] [PubMed] [Google Scholar]
- 32.Pearson JV, Huentelman MJ, Halperin RF, Tembe WD, Melquist S, Homer N, Brun M, Szelinger S, Coon KD, Zismann VL, et al (2007) Identification of the genetic basis for complex disorders by use of pooling-based genomewide single-nucleotide–polymorphism association studies. Am J Hum Genet 80:126–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lovmar L, Ahlford A, Jonsson M, Syvanen AC (2005) Silhouette scores for assessment of SNP genotype clusters. BMC Genomics 6:35 10.1186/1471-2164-6-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pittman AM, Myers AJ, Duckworth J, Bryden L, Hanson M, Abou-Sleiman P, Wood NW, Hardy J, Lees A, de Silva R (2004) The structure of the tau haplotype in controls and in progressive supranuclear palsy. Hum Mol Genet 13:1267–1274 10.1093/hmg/ddh138 [DOI] [PubMed] [Google Scholar]
- 35.Craig DW, Huentelman MJ, Hu-Lince D, Zismann VL, Kruer MC, Lee AM, Puffenberger EG, Pearson JM, Stephan DA (2005) Identification of disease causing loci using an array-based genotyping approach on pooled DNA. BMC Genomics 6:138 10.1186/1471-2164-6-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, Barnard J, Baker A, Jonasdottir A, Ingason A, Gudnadottir VG, et al (2005) A common inversion under selection in Europeans. Nat Genet 37:129–137 10.1038/ng1508 [DOI] [PubMed] [Google Scholar]
- 37.Evans W, Fung HC, Steele J, Eerola J, Tienari P, Pittman A, Silva R, Myers A, Vrieze FW, Singleton A, et al (2004) The tau H2 haplotype is almost exclusively Caucasian in origin. Neurosci Lett 369:183–185 10.1016/j.neulet.2004.05.119 [DOI] [PubMed] [Google Scholar]
- 38.Fung HC, Scholz S, Matarin M, Simon-Sanchez J, Hernandez D, Britton A, Gibbs JR, Langefeld C, Stiegert ML, Schymick J, et al (2006) Genome-wide genotyping in Parkinson’s disease and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol 5:911–916 10.1016/S1474-4422(06)70578-6 [DOI] [PubMed] [Google Scholar]
- 39.Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 40.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA (2002) Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet 70:425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pohlmann R, Krentler C, Schmidt B, Schroder W, Lorkowski G, Culley J, Mersmann G, Geier C, Waheed A, Gottschalk S, et al (1988) Human lysosomal acid phosphatase: cloning, expression and chromosomal assignment. EMBO J 7:2343–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maraganore DM, de Andrade M, Lesnick TG, Strain KJ, Farrer MJ, Rocca WA, Pant PV, Frazer KA, Cox DR, Ballinger DG (2005) High-resolution whole-genome association study of Parkinson disease. Am J Hum Genet 77:685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clarimon J, Scholz S, Fung H-C, Hardy J, Eerola J, Hellstrom O, Chen C-M, Wu Y-R, Tienari PJ, Singleton A (2006) Conflicting results regarding the semaphorin gene (SEMA5A) and the risk for Parkinson disease. Am J Hum Genet 78:1082–1084 (author reply 78:1092–1094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farrer MJ, Haugarvoll K, Ross OA, Stone JT, Milkovic NM, Cobb SA, Whittle AJ, Lincoln SJ, Hulihan MM, Heckman MG, et al (2006) Genomewide association, Parkinson disease, and PARK10. Am J Hum Genet 78:1084–1088 (author reply 78:1092–1094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goris A, Williams-Gray CH, Foltynie T, Compston DAS, Barker RA, Sawcer SJ (2006) No evidence for association with Parkinson disease for 13 single-nucleotide polymorphisms identified by whole-genome association screening. Am J Hum Genet 78:1088–1090 (author reply 78:1092–1094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Rowland C, Schrodi S, Laird W, Tacey K, Ross D, Leong D, Catanese J, Sninsky J, Grupe A (2006) A case-control association study of the 12 single-nucleotide polymorphisms implicated in Parkinson disease by a recent genome scan. Am J Hum Genet 78:1090–1092 (author reply 1092–1094) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Myers DR (2006) Considerations for genomewide association studies in Parkinson disease. Am J Hum Genet 78:1081–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC (2003) α-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem 278:25009–25013 10.1074/jbc.M300227200 [DOI] [PubMed] [Google Scholar]
- 49.Qin ZH, Wang Y, Kegel KB, Kazantsev A, Apostol BL, Thompson LM, Yoder J, Aronin N, DiFiglia M (2003) Autophagy regulates the processing of amino terminal huntingtin fragments. Hum Mol Genet 12:3231–3244 10.1093/hmg/ddg346 [DOI] [PubMed] [Google Scholar]
- 50.Auer IA, Schmidt ML, Lee VM, Curry B, Suzuki K, Shin RW, Pentchev PG, Carstea ED, Trojanowski JQ (1995) Paired helical filament tau (PHFtau) in Niemann-Pick type C disease is similar to PHFtau in Alzheimer’s disease. Acta Neuropathol (Berl) 90:547–551 [DOI] [PubMed] [Google Scholar]
- 51.Love S, Bridges LR, Case CP (1995) Neurofibrillary tangles in Niemann-Pick disease type C. Brain 118:119–129 10.1093/brain/118.1.119 [DOI] [PubMed] [Google Scholar]
- 52.Suzuki K, Parker CC, Pentchev PG, Katz D, Ghetti B, D’Agostino AN, Carstea ED (1995) Neurofibrillary tangles in Niemann-Pick disease type C. Acta Neuropathol (Berl) 89:227–238 [DOI] [PubMed] [Google Scholar]
- 53.Omar R, Pappolla M, Argani I, Davis K (1993) Acid phosphatase activity in senile plaques and cerebrospinal fluid of patients with Alzheimer’s disease. Arch Pathol Lab Med 117:166–169 [PubMed] [Google Scholar]
- 54.Saftig P, Hartmann D, Lullmann-Rauch R, Wolff J, Evers M, Koster A, Hetman M, von Figura K, Peters C (1997) Mice deficient in lysosomal acid phosphatase develop lysosomal storage in the kidney and central nervous system. J Biol Chem 272:18628–18635 10.1074/jbc.272.30.18628 [DOI] [PubMed] [Google Scholar]
- 55.Mannan AU, Roussa E, Kraus C, Rickmann M, Maenner J, Nayernia K, Krieglstein K, Reis A, Engel W (2004) Mutation in the gene encoding lysosomal acid phosphatase (Acp2) causes cerebellum and skin malformation in mouse. Neurogenetics 5:229–238 10.1007/s10048-004-0197-9 [DOI] [PubMed] [Google Scholar]
- 56.Ishizawa K, Dickson DW (2001) Microglial activation parallels system degeneration in progressive supranuclear palsy and corticobasal degeneration. J Neuropathol Exp Neurol 60:647–657 [DOI] [PubMed] [Google Scholar]
- 57.Cleaver JE (2005) Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat Rev Cancer 5:564–573 10.1038/nrc1652 [DOI] [PubMed] [Google Scholar]
- 58.Robbins JH, Brumback RA, Moshell AN (1993) Clinically asymptomatic xeroderma pigmentosum neurological disease in an adult: evidence for a neurodegeneration in later life caused by defective DNA repair. Eur Neurol 33:188–190 [DOI] [PubMed] [Google Scholar]
- 59.Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, Tanaka K, et al (2005) UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 121:387–400 10.1016/j.cell.2005.02.035 [DOI] [PubMed] [Google Scholar]
- 60.Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapic-Otrin V, Levine AS (2006) The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc Natl Acad Sci USA 103:2588–2593 10.1073/pnas.0511160103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Inoki T, Yamagami S, Inoki Y, Tsuru T, Hamamoto T, Kagawa Y, Mori T, Endo H (2004) Human DDB2 splicing variants are dominant negative inhibitors of UV-damaged DNA repair. Biochem Biophys Res Commun 314:1036–1043 10.1016/j.bbrc.2004.01.003 [DOI] [PubMed] [Google Scholar]