Abstract
Previous linkage analyses of families with multiple cases of schizophrenia by us and others have confirmed the involvement of the chromosome 11q22-24 region in the etiology of schizophrenia, with LOD scores of 3.4 and 3.1. We now report fine mapping of a susceptibility gene in the 11q22-24 region, determined on the basis of a University College London (UCL) sample of 496 cases and 488 supernormal controls. Confirmation was then performed by the study of an Aberdeen sample consisting of 858 cases and 591 controls (for a total of 2,433 individuals: 1,354 with schizophrenia and 1,079 controls). Seven microsatellite or single-nucleotide polymorphism (SNP) markers localized within or near the FXYD6 gene showed empirically significant allelic associations with schizophrenia in the UCL sample (for D11S1998, P=.021; for rs3168238, P=.009; for TTTC20.2, P=.048; for rs1815774, P=.049; for rs4938445, P=.010; for rs4938446, P=.025; for rs497768, P=.023). Several haplotypes were also found to be associated with schizophrenia; for example, haplotype Hap-F21 comprising markers rs10790212-rs4938445-rs497768 was found to be associated with schizophrenia, by a global permutation test (P=.002). Positive markers in the UCL sample were then genotyped in the Aberdeen sample. Two of these SNPs were found to be associated with schizophrenia in the Scottish sample (for rs4938445, P=.044; for rs497768, P=.037). The Hap-F21 haplotype also showed significant association with schizophrenia in the Aberdeen sample, with the same alleles being associated (P=.013). The FXYD6 gene encodes a protein called “phosphohippolin” that is highly expressed in regions of the brain thought to be involved in schizophrenia. The protein functions by modulating the kinetic properties of Na,K-ATPase to the specific physiological requirements of the tissue. Etiological base-pair changes in FXYD6 or in associated promoter/control regions are likely to cause abnormal function or expression of phosphohippolin and to increase genetic susceptibility to schizophrenia.
Schizophrenia (SCZD [MIM #181500]) is a disease characterized by self-neglect, thought disorder, auditory hallucinations, delusions, and bizarre behavior.1 It has a lifetime prevalence of 0.85% in the United Kingdom. Family, twin, and adoption studies have confirmed that the disorder is highly heritable. More than 20 whole-genome linkage scans of SCZD have demonstrated that there is heterogeneity of linkage, involving multiple loci on different chromosomal regions.2 It is not yet known whether these loci interact. The linkage studies are interpretable only if it is accepted that at least some of these loci act independently in different families.
Chromosome 11q22 was first implicated in SCZD in a linkage study of two Japanese pedigrees in which positive LOD scores between 1.0 and 1.5 were reported with marker D11S35.3 This was confirmed in a separate study of a large Canadian pedigree, in which a maximum LOD score of 3.4 was obtained using the same genetic marker, D11S35.4 Later, we published further evidence of linkage to SCZD at 11q23.3 in a genomewide scan of very large multiply affected families.5 Overall, we found a 3-point LOD score of 3.1 with markers D11S925 and D11S934. A LOD score of 3.2 was found in a single large Icelandic pedigree, again supporting the existence of locus heterogeneity. The chromosome 11q22-24 region has been shown to be one of the most well-established linkages to SCZD by a meta-analysis of 20 genome scans that employed nonparametric rank-order statistics to show that evidence of linkage at this region was nonrandom.2 Chromosome 11q22.3-q24.1 was ranked third of all regions, and, when the different sample sizes of the 20 SCZD genome scans were taken into account, it was ranked fourth. This provided considerable confidence that an SCZD susceptibility locus was likely to exist in the 11q22-24 region.
Material and Methods
Allelic association studies were performed to fine map an 11q22-24 SCZD susceptibility gene by detecting linkage disequilibrium (LD). The University College London (UCL) case-control sample consisted of 496 unrelated cases and 488 ancestrally matched controls. Research subjects were selected only if both parents were English, Scottish, or Welsh, with at least three grandparents having the same origins. Subjects were also included if the fourth grandparent was of another white European origin but were excluded if one grandparent was of Jewish or non–European Union (EU) (before the enlargement of the EU in 2004) ancestry. These data were recorded in an ancestry questionnaire, with confirmation from family histories noted on medical records. U.K. National Health Service (NHS) multicenter and local research ethics–committee approval was obtained, and all participating subjects signed an approved consent form after reading an information sheet. Each schizophrenic research subject had received a diagnosis and assessment by NHS psychiatrists as part of routine clinical diagnosis and treatment. Those with short-term drug-induced psychoses, psychoses with either learning disability or head injury, and other symptomatic psychoses were excluded. Schizophrenic subjects were recruited on the basis of having an International Classification of Diseases version 10 (ICD10) diagnosis of SCZD recorded in medical case-history notes after clinical interview by NHS psychiatrists. The diagnoses were confirmed by a senior psychiatrist, usually within 1 wk. Schedule for Affective Disorders and Schizophrenia–Lifetime Version (SADS-L) interview was completed for all cases and controls by a research psychiatrist.6 Schizophrenic subjects were then chosen on the basis of having received a diagnosis at the “probable level” of the Research Diagnostic Criteria (RDC).7 Patients with schizoaffective bipolar disorder or schizomania were not included. All screened control individuals were also interviewed by a psychiatrist, specifically for the study, and were selected for having no family history of SCZD, alcoholism, or bipolar disorder, according to self-report by the research subject. They were then interviewed with the SADS-L schedule and were included only if they were found to have no present or lifetime history of any RDC-defined mental disorder. The UCL case-control sample has been used in studies of other chromosomal regions and has implicated the chromosome 5q33 epsin 4 (CLINT1) gene encoding enthoprotin, the chromosome 8p22 PCM1 gene, and the chromosome 1q23 UHMK1 gene as the cause of SCZD.8–10 The sample has also been used to exclude association between SCZD and markers at the chromosome 1q23 RGS4 and CAPON (NOS1AP) genes.11,12
Genomic DNA for the UCL case-control sample was extracted from whole-blood samples by use of standard cell lysis, proteinase K digestion, and phenol/chloroform ethanol precipitation method, as described elsewhere.8 All DNA samples were quantified with Picogreen (Molecular Probes) by use of fluorimetry. Fifteen reference microsatellite markers at chromosomal loci not thought to be involved in SCZD were genotyped in a subset of samples (200 cases and 300 controls). This was done to detect genetic heterogeneity between the cases and controls and to confirm that the samples were genetically well matched. In addition, a statistical test with use of CHECKHET was employed to detect subjects with an atypical genetic background.13 No evidence of genetic heterogeneity between cases and controls was found with use of the reference markers. The CHECKHET test detected two schizophrenic subjects with abnormal genotypes. These were xcluded from the sample before any chromosome 11 markers were genotyped.
The Aberdeen sample consisted of 858 subjects with SCZD and 591 controls. The cases were recruited through Scottish psychiatric hospitals and met DSM-III-R or DSM-IV criteria of SCZD, with use of an operational criteria checklist (OPCRIT). Diagnosis of SCZD was confirmed through agreement by two independent senior psychiatrists on the basis of structured clinical interviews for DSM-III-R/DSM-IV (SCID) and inspection of psychiatric case notes. All controls were volunteers recruited through general practices from the same region of Scotland and were ethnically matched. The control samples were screened for absence of psychiatric illness. Informed consent was obtained from all patients and control individuals. A proportion of the Aberdeen sample was employed elsewhere in tests for genetic association between NRG1 and SCZD.14
Random markers on chromosome 11q22-24, close to the maximum LOD score position in the UCL SCZD family-linkage study, were initially selected for fine mapping.5 Five of these random microsatellite markers were genotyped before positive association with D11S1998 was found. Further nonrandom microsatellite and SNP markers close to D11S1998 were then chosen for genotyping. Microsatellite marker TTTC20.2 was identified from a simple repeat found in the UCSC Genome Browser database May 2004 assembly. The forward (5′-AGGACCACACTCAGCCTCAC-3′) and reverse (5′-GGGAAGGAAGGAGAGAGAGAG-3′) primers were designed using Primer3.15 The primer sequences for D11S29, D11S4127, D11S1998, D11S939, D11S976, D11S4195, D11S925, D11S964, D11S836, and D11S934 were obtained from the GDB Human Genome Database and Ensembl. Microsatellite markers were genotyped in an initial sample of 443 controls and 480 schizophrenic patients in the UCL sample. PCR amplification of these markers was performed using an M13-tailed primer and a second nontailed primer. A third universal M13 sequence primer labeled with infrared dye IRD700 or IRD800 was used to hybridize against the M13-tailed locus-specific primer. Microsatellite-marker fragment sizes were separated and visualized with either of the two infrared dyes on LiCor 4200L sequencers. Genotyping was accomplished using SAGA-GT genotyping software (LiCor). The genotypes were then checked by eye, with all allele calls verified by a second independent person. Any discrepant genotypes were repeated. SNPs rs11999, rs869789, rs529623, rs479991, rs2019655, rs10790212, rs3087563, rs1815774, rs876798, rs876797, r1s4938445, rs4938446, rs11216598, and rs631898 were chosen from Ensembl. Further SNPs—rs4579962, rs678776, rs11216567, rs10892181, rs3168238, rs564989, rs12363888, rs11608153, rs512481, rs476130, rs7121573, rs873713, rs10790218, rs11605223, rs3809043, rs3809042, and rs497768—were then selected from the International HapMap Project with use of the Haploview tagger function.16,17 All SNP markers were determined using KASPar (KBioscience), a modified Amplifluor SNP-genotyping method. Of the samples, 17% from each microtitre plate were reduplicated to detect possible errors and to confirm the reproducibility of genotypes. The data were then analyzed to confirm Hardy-Weinberg equilibrium (HWE). Any markers lacking HWE were repeated using an alternative Taqman method. Before any association analysis, the genotype data were assessed using the SCANGROUP program in GENECOUNTING.18–20 This program highlights any potential genotyping errors by identifying significant differences in haplotypic frequencies between any single 96-well microtitre plate and haplotypic frequencies in all other plates combined. It is argued that genotyping errors can show up as rare haplotypes occurring on just a single plate, whereas true haplotypic associations with disease will appear as differences in haplotype frequencies between cases and controls spread across a number of plates.
Allelic association analysis with SCZD was performed using a simple χ2 test for biallelic SNPs and by using CLUMP for microsatellite markers. CLUMP employs an empirical Monte Carlo test of significance that does not require further correction for multiple alleles.21 Subtests of the CLUMP program include T1, Pearson’s χ2 statistic of the “raw” contingency table; T2, the χ2 statistic of a table with rare alleles grouped together to prevent small expected cell counts; T3, the largest of the χ2 statistics of 2×2 tables, each of which compares one allele against the rest grouped together; and T4, the largest of the χ2 statistics of all possible 2×2 tables, which compares any combination of alleles against the rest. To select regions of interest for further investigation, we took account of the output of the most significant of the four statistics by use of CLUMP. Likewise, we considered the pointwise P values for markers individually rather than attempting to derive an overall P value for the region on the basis of combining results from multiple markers. The multilocus genotypes were then analyzed for haplotypic association with SCZD by use of GENECOUNTING, which computes maximum-likelihood estimates of haplotype frequencies from phase-unknown case-control data. The significance of any overall haplotypic association with SCZD was then obtained with a permutation test.18–20 GENECOUNTING was also used to calculate pairwise LD between all markers with use of the LDPAIRS program; this was visualized using LocusView 2.0.
Results
One the five initial randomly selected microsatellites in the 11q22-24 region, D11S1998, showed allelic association with SCZD (P=.021; CLUMP T2) (tables 1 and 2). After this, other markers to be genotyped were selected nonrandomly by virtue of being very likely to be in LD with marker D11S1998. From these nonrandom microsatellite markers, TTTC20.2 was also found to be associated with SCZD (P=.048; CLUMP T4). None of the other genotyped microsatellite markers localized to chromosome 11q22-24 showed association with SCZD (P values are for CLUMP T1 statistic: for D11S29, P=.899; for D11S4127, P=.521; for D11S939, P=.460; for D11S976, P=.393; for D11S4195, P=.287; for D11S925, P=.379; for D11S964, P=.132; for D11S836, P=.423; for D11S934, P=.561). After microsatellite analysis, SNP markers localized near positive markers D11S1998 and TTTC20.2 were selected for genotyping. SNPs rs3168238 (P=.009; odds ratio [OR] 1.64), rs1815774 (P=.049; OR 1.21), rs4938445 (P=.010; OR 1.31), rs4938446 (P=.025; OR 1.26), and rs497768 (P=.023; OR 1.24) were all found to be associated with SCZD, with six further SNPs (for rs869789, P=.097; for rs11216567, P=.072; for rs7121573, P=.076; for rs876797, P=.086; for rs873713, P=.091; and for rs10790218, P=.099) showing a trend toward association.
Table 1. .
Allelic Association Tests with SCZD at the FXYD6 Locus in the UCL Sample
| No. of Observed Alleles for Allelic Base |
||||||||
| Marker and Sample |
Marker Location (bp) |
Previous Marker Distance (bp) | χ2 | Pa | A | C | G | T |
| rs11999: | 117196128 | .91 | .340 | |||||
| Control | 542 | 300 | ||||||
| Case | 521 | 261 | ||||||
| rs869789: | 117196596 | 468 | 2.75 | .097 | ||||
| Control | 107 | 753 | ||||||
| Case | 138 | 772 | ||||||
| rs4579962: | 117197544 | 948 | .03 | .860 | ||||
| Control | 274 | 688 | ||||||
| Case | 266 | 680 | ||||||
| rs529623: | 117198465 | 921 | .60 | .440 | ||||
| Control | 473 | 395 | ||||||
| Case | 475 | 427 | ||||||
| rs678776: | 117201954 | 3,489 | .00 | .975 | ||||
| Control | 285 | 671 | ||||||
| Case | 282 | 666 | ||||||
| rs479991: | 117202168 | 214 | 1.17 | .280 | ||||
| Control | 665 | 197 | ||||||
| Case | 715 | 187 | ||||||
| D11S1998 | 117202941 | 773 | 12.60 | .021b | ||||
| rs11216567: | 117203693 | 752 | 3.25 | .072 | ||||
| Control | 880 | 70 | ||||||
| Case | 829 | 89 | ||||||
| rs10892181: | 117204796 | 1,103 | .49 | .484 | ||||
| Control | 351 | 609 | ||||||
| Case | 320 | 594 | ||||||
| rs2019655: | 117205088 | 292 | .60 | .437 | ||||
| Control | 208 | 664 | ||||||
| Case | 230 | 674 | ||||||
| rs10790212: | 117207900 | 2,812 | .47 | .493 | ||||
| Control | 640 | 240 | ||||||
| Case | 666 | 232 | ||||||
| rs3087563: | 117213147 | 5,247 | .09 | .763 | ||||
| Control | 415 | 451 | ||||||
| Case | 422 | 472 | ||||||
| rs3168238: | 117214855 | 1,708 | 6.80 | .009c | ||||
| Control | 76 | 874 | ||||||
| Case | 47 | 887 | ||||||
| rs564989: | 117214964 | 109 | .30 | .584 | ||||
| Control | 222 | 742 | ||||||
| Case | 226 | 712 | ||||||
| rs12363888: | 117218397 | 3,433 | .04 | .843 | ||||
| Control | 907 | 55 | ||||||
| Case | 909 | 53 | ||||||
| rs11608153: | 117221873 | 3,476 | .48 | .489 | ||||
| Control | 821 | 129 | ||||||
| Case | 805 | 115 | ||||||
| rs512481: | 117224090 | 2,217 | .90 | .344 | ||||
| Control | 452 | 494 | ||||||
| Case | 425 | 507 | ||||||
| rs476130: | 117233093 | 9,003 | .64 | .423 | ||||
| Control | 890 | 70 | ||||||
| Case | 853 | 77 | ||||||
| TTTC20.2 | 117234550 | 1,457 | 17.54 | .048d | ||||
| rs7121573: | 117235040 | 490 | 3.15 | .076 | ||||
| Control | 382 | 564 | ||||||
| Case | 331 | 579 | ||||||
| rs1815774: | 117236649 | 1,609 | 3.87 | .049e | ||||
| Control | 342 | 528 | ||||||
| Case | 311 | 583 | ||||||
| rs876798: | 117242650 | 6,001 | .45 | .503 | ||||
| Control | 125 | 739 | ||||||
| Case | 123 | 797 | ||||||
| rs876797: | 117242957 | 307 | 2.94 | .086 | ||||
| Control | 207 | 651 | ||||||
| Case | 186 | 712 | ||||||
| rs873713: | 117245126 | 2,169 | 2.86 | .091 | ||||
| Control | 230 | 710 | ||||||
| Case | 197 | 733 | ||||||
| rs10790218: | 117247205 | 2,079 | 2.73 | .099 | ||||
| Control | 374 | 576 | ||||||
| Case | 334 | 602 | ||||||
| rs4938445: | 117250213 | 3,008 | 6.68 | .010f | ||||
| Control | 299 | 579 | ||||||
| Case | 250 | 632 | ||||||
| rs4938446: | 117250259 | 46 | 5.00 | .025g | ||||
| Control | 296 | 570 | ||||||
| Case | 271 | 655 | ||||||
| rs11216598: | 117253662 | 3,403 | .07 | .798 | ||||
| Control | 727 | 149 | ||||||
| Case | 771 | 153 | ||||||
| rs631898: | 117253688 | 26 | 2.31 | .128 | ||||
| Control | 419 | 451 | ||||||
| Case | 466 | 434 | ||||||
| rs11605223: | 117254054 | 366 | 1.41 | .235 | ||||
| Control | 490 | 470 | ||||||
| Case | 462 | 494 | ||||||
| rs3809043: | 117254325 | 271 | .57 | .452 | ||||
| Control | 929 | 29 | ||||||
| Case | 898 | 34 | ||||||
| rs3809042: | 117254330 | 5 | .21 | .646 | ||||
| Control | 27 | 925 | ||||||
| Case | 23 | 899 | ||||||
| rs497768: | 117255950 | 1,620 | 5.21 | .023h | ||||
| Control | 416 | 500 | ||||||
| Case | 371 | 553 | ||||||
Two-tailed significance from 2×2 χ2, with 1 df.
P from CLUMP Monte Carlo subtest T2.
OR 1.64.
P from CLUMP Monte Carlo subtest T4.
OR 1.21.
OR 1.31.
OR 1.26.
OR 1.24.
Table 2. .
Fragment Sizes of Microsatellite Markers at the FXYD6 Locus in the UCL Sample
| No. of Observed Alleles by Population |
||
| Marker and Fragment Size (bp) |
Control | Case |
| D11S1998: | ||
| 150 | 60 | 86 |
| 158 | 1 | 0 |
| 162 | 2 | 0 |
| 166 | 38 | 27 |
| 170 | 405 | 428 |
| 174 | 275 | 239 |
| 178 | 75 | 60 |
| 182 | 7 | 6 |
| 186 | 1 | 0 |
| TTTC20.2: | ||
| 232 | 8 | 4 |
| 236 | 3 | 14 |
| 240 | 22 | 16 |
| 242 | 3 | 5 |
| 244 | 130 | 130 |
| 246 | 11 | 10 |
| 248 | 97 | 137 |
| 250 | 54 | 47 |
| 252 | 113 | 104 |
| 254 | 65 | 77 |
| 256 | 75 | 69 |
| 258 | 60 | 62 |
| 260 | 31 | 51 |
| 262 | 60 | 57 |
| 264 | 12 | 14 |
| 266 | 46 | 32 |
| 268 | 3 | 4 |
| 270 | 17 | 23 |
| 274 | 9 | 5 |
| 278 | 2 | 1 |
| 282 | 1 | 0 |
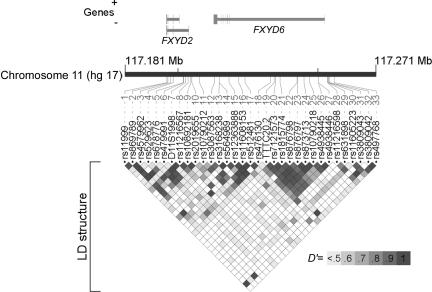
Tests of three-marker haplotypic association with use of rs11216598, rs11605223, and rs497768 (Hap-F7) provided significant evidence of association with SCZD, with a global permutation significance of P=.0013. Several other two-, three-, and four-marker tests of haplotypic association were also significantly positive, as shown in table 3. Nearly all the genotyped markers that show significant association with SCZD or a positive trend are localized within the FXYD domain–containing ion transport regulator 6 (FXYD6 [MIM *606683]) gene. The exceptions to these are markers rs869789, D11S1998, and rs11216567, situated on FXYD2. These three markers show significant LD with several of the markers in FXYD6. Results of pairwise LD statistics between all markers genotyped and their respective locations are shown in figure 1.
Table 3. .
Haplotypic Association Tests with SCZD at the FXYD6 Locus in the UCL Sample
| EstimatedHaplotypeFrequency(%) |
|||||
| No. of Markers and Haplotype Reference |
Haplotype | Global Empirical Pa | Alleles Increasing in Cases | Controls | Cases |
| 2: | |||||
| HAP-F1 | rs3168238-rs497768 | .0028 | T-G | 47.9 | 56.0 |
| 3: | |||||
| HAP-F2 | rs564989-rs476130-rs876797 | .0043 | C-C-C | 11.4 | 14.9 |
| HAP-F3 | rs564989-rs476130-rs497768 | .0048 | T-C-G | 41.6 | 48.7 |
| HAP-F4 | rs564989-rs876797-rs873713 | .0010 | C-C-T | 16.9 | 23.8 |
| HAP-F5 | rs476130-rs11605223-rs497768 | .0027 | C-G-G | 11.3 | 17.1 |
| HAP-F6 | rs1815774-rs4938445-rs4938446 | .0066 | G-G-T | 58.1 | 64.0 |
| HAP-F7 | rs11216598-rs11605223-rs497768 | .0013 | A-G-G | 11.4 | 16.8 |
| HAP-F8 | rs11605223-rs3809043-rs497768 | .0021 | G-C-G | 11.5 | 17.3 |
| 4: | |||||
| HAP-F9 | rs3168239-rs564989-rs876797-rs873713 | .0013 | T-C-C-T | 17.9 | 23.1 |
| HAP-F10 | rs3168238-rs12363888-rs11605223-rs497768 | .0057 | T-C-G-G | 10.8 | 17.2 |
| HAP-F11 | rs3168238-rs476130-rs11605223-rs497768 | .0026 | T-C-G-G | 10.7 | 16.9 |
| HAP-F12 | rs3168238-rs11605223-rs3809043-rs497768 | .0016 | T-G-C-G | 10.8 | 17.3 |
| HAP-F13 | rs3168238-rs11605223-rs3809042-rs497768 | .0035 | T-G-G-G | 10.9 | 17.2 |
| HAP-F14 | rs1815774-rs10790218-rs4938445-rs11605223 | .0055 | G-G-G-G | 40.5 | 46.7 |
| HAP-F15 | rs1815774-rs4938445-rs4938446-rs11605223 | .0159 | G-G-T-G | 40.6 | 47.2 |
| HAP-F16 | rs10790218-rs4938445-rs4938446-rs11605223 | .0066 | G-G-T-G | 40.8 | 46.3 |
| HAP-F17 | rs11216598-rs11605223-rs3809043-rs497768 | .0019 | A-G-C-G | 11.4 | 16.8 |
Haplotype-permutation test empirical P, based on 9,999 permutations.
Figure 1. .
LD between markers genotyped in the FXYD6 gene locus in the UCL sample. LD structure (D′) between marker pairs is indicated by the shaded matrices. Marker positions are given in table 1, and their locations are shown relative to both FXYD genes displayed above the chromosome bar. The gene structures contain vertical lines representing exons. Genomic positions are in accordance with the National Center for Biotechnology Information build 35 hg 17 of the human genome assembly. The figure was generated using LocusView 2.0.
To verify the significance of the positive allelic and haplotypic association found in the UCL sample, replication was attempted in an Aberdeen case-control sample with use of SNPs rs869789, rs11216567, rs10790212, rs3168238, rs876797, rs4938445, and rs497768. With a one-tailed test of significance, two SNPs were found to show association with SCZD in the Aberdeen sample (for rs4938445, P=.044; for rs497768, P=.037) (table 4). The allele-frequency increase for SNP rs3168238 (P=.074), although not significantly associated with SCZD, displayed a trend in the direction opposite to that found in the UCL sample. When the data from the UCL and the Aberdeen samples were combined, three SNP markers showed significant allelic association (for rs869789, P=.023; for rs4938445, P=.002; and for rs497768, P=.005), with two further SNPs displaying a trend toward association (for rs11216567, P=.058, and for rs876797, P=.063) (table 4).
Table 4. .
Allelic Association Tests with SCZD at the FXYD6 Locus in the UCL, Aberdeen, and Combined Samples
| UCLa |
Aberdeen |
Combined (UCL and Aberdeen) |
|||||||||||||||||
| No. (%) of Observed Alleles for Allelic Base |
No. (%) of Observed Alleles for Allelic Base |
No. (%) of Observed Alleles for Allelic Base |
|||||||||||||||||
| Marker and Sample | Marker Location (bp) |
A | C | G | T | χ2 | Pb | A | C | G | T | χ2 | Pc | A | C | G | T | χ2 | Pb |
| rs869789: | 117196596 | 2.75 | .097 | 1.996 | .079 | 5.170 | .023 | ||||||||||||
| Control | 107 (.12) | 753 (.88) | 173 (.15) | 985 (.85) | 280 (.14) | 1,738 (.86) | |||||||||||||
| Case | 138 (.15) | 772 (.85) | 282 (.17) | 1384 (.83) | 420 (.16) | 2156 (.84) | |||||||||||||
| rs11216567: | 117203693 | 3.25 | .072 | 2.477 | .058 | 3.585 | .058 | ||||||||||||
| Control | 880 (.93) | 70 (.07) | 1040 (.92) | 108 (.08) | 1920 (.92) | 178 (.08) | |||||||||||||
| Case | 829 (.90) | 89 (.10) | 1528 (.92) | 128 (.08) | 2357 (.92) | 217 (.08) | |||||||||||||
| rs10790212: | 117207900 | .47 | .493 | .040 | .420 | .435 | .510 | ||||||||||||
| Control | 640 (.73) | 240 (.27) | 860 (.74) | 296 (.26) | 1500 (.74) | 536 (.26) | |||||||||||||
| Case | 666 (.74) | 232 (.26) | 1242 (.75) | 420 (.25) | 1908 (.75) | 652 (.25) | |||||||||||||
| rs3168238: | 117214855 | 6.80 | .009 | 3.184 | .074b | .501 | .479 | ||||||||||||
| Control | 76 (.08) | 874 (.92) | 47 (.04) | 1103 (.96) | 123 (.06) | 1977 (.94) | |||||||||||||
| Case | 47 (.05) | 887 (.95) | 92 (.06) | 1558 (.94) | 139 (.05) | 2445 (.95) | |||||||||||||
| rs876797: | 117242957 | 2.94 | .086 | .823 | .182 | 3.449 | .063 | ||||||||||||
| Control | 207 (.24) | 651 (.76) | 250 (.22) | 882 (.78) | 457 (.23) | 1533 (.77) | |||||||||||||
| Case | 186 (.21) | 712 (.79) | 337 (.21) | 1295 (.79) | 523 (.21) | 2007 (.79) | |||||||||||||
| rs4938445: | 117250213 | 6.68 | .010 | 2.936 | .043 | 9.610 | .002 | ||||||||||||
| Control | 299 (.34) | 579 (.66) | 376 (.33) | 770 (.67) | 675 (.33) | 1,349 (.67) | |||||||||||||
| Case | 250 (.28) | 632 (.72) | 485 (.30) | 1145 (.70) | 735 (.29) | 1,777 (.71) | |||||||||||||
| rs497768: | 117255950 | 5.21 | .023 | 3.211 | .037 | 7.866 | .005 | ||||||||||||
| Control | 416 (.45) | 500 (.55) | 521 (.45) | 633 (.55) | 937 (.45) | 1,133 (.55) | |||||||||||||
| Case | 371 (.40) | 553 (.60) | 676 (.42) | 944 (.58) | 1,047 (.41) | 1,497 (.59) | |||||||||||||
Data as shown in tables 1.
Two-tailed significance (P) from 2×2 χ2, with 1 df.
One-tailed significance (P) from 2×2 χ2, with 1 df.
The three-marker haplotype rs10790212-rs4938445-rs497768 (Hap-F21), which produced a global empirical association in the UCL sample (empirical P=.002), was also found to be replicated in the Aberdeen sample (empirical P=.013). When data from both samples were combined, haplotype Hap-F21 exhibited strengthened permutation significance for haplotypic association with SCZD (empirical P=.0005) (table 5). Other significantly associated haplotypes in the UCL sample were also found to be replicated in the Aberdeen sample and when both samples were combined.
Table 5. .
Tests of Haplotypic Association with SCZD at the FXYD6 Locus for Comparison of UCL, Aberdeen, and Combined Samples
| EstimatedHaplotypeFrequency(%) |
||||
| No. of Markers, Haplotype Reference (Haplotype), and Sample |
Global Empirical Pa | Alleles Increasing in Cases | Controls | Cases |
| 2: | ||||
| HAP-F18 (rs10790212-rs4938445): | ||||
| UCL | .0183 | C-G | 44.7 | 48.9 |
| Aberdeen | .0266 | C-G | 44.6 | 49.7 |
| Combined | .0093 | C-G | 44.7 | 49.4 |
| HAP-F1 (rs3168238-rs497768): | ||||
| UCL | .0028 | T-G | 47.9 | 56.0 |
| Aberdeen | .1173 | T-G | 52.0 | 54.2 |
| Combined | .0157 | T-G | 50.1 | 54.9 |
| 3: | ||||
| HAP-F19 (rs11216567-rs10790212-rs4938445): | ||||
| UCL | .0038 | A-C-G | 40.2 | 44.4 |
| Aberdeen | .0453 | A-C-G | 38.6 | 44.2 |
| Combined | .0687 | A-C-G | 39.1 | 44.1 |
| HAP-F20 (rs11216567-rs10790212-rs497768): | ||||
| UCL | .0442 | A-C-G | 35.2 | 40.3 |
| Aberdeen | .0104 | A-C-G | 39.3 | 40.0 |
| Combined | .0348 | A-C-G | 37.5 | 40.0 |
| HAP-F21 (rs10790212-rs4938445-rs497768): | ||||
| UCL | .0024 | C-G-G | 23.4 | 27.9 |
| Aberdeen | .0127 | C-G-G | 25.5 | 27.6 |
| Combined | .0005 | C-G-G | 24.7 | 27.7 |
Haplotype permutation test empirical P, based on 9,999 permutations.
Discussion
The observation of allelic and haplotypic association in the UCL sample with use of nominal empirically derived P values and the fact that the same alleles and haplotypes were associated in the second Aberdeen sample suggest that it is unlikely that the association between FXYD6 and SCZD is a false-positive result. The strongest allelic and haplotypic associations were found with markers within FXYD6 and not in neighboring genes, which leads us to believe that this gene was the most likely to be involved in the etiology of SCZD. One microsatellite marker in FXYD2 was found to be associated with SCZD, but none of the SNPs within FXYD2 were significantly associated in either the UCL or the Aberdeen samples. We found consistency as to which alleles and haplotypes were associated with SCZD in the UCL and Aberdeen samples. Even though an allele at marker rs3168238 showed a trend to be associated with SCZD with an allele different from the one that showed significant association in the UCL sample, it was nevertheless found that the same allele was in the haplotype showing significant association with SCZD in the Aberdeen sample.
FXYD6 encodes the protein phosphohippolin and is part of a family of seven FXYD genes.22 The FXYD proteins share homology for a single common transmembrane domain.23 Each FXYD protein is expressed in a tissue-specific manner and functions by altering the kinetic activity of Na,K-ATPase.24 This involves changes in the active Na+ and K+ transports that are based on the specific requirements of different cells. The effects of FXYD proteins on kinetic parameters such as K1/2 Na+, K1/2 K+, and Vmax are usually approximately twofold.25–27 However, these moderate effects are likely to have long-term physiological importance in maintaining cation homeostasis.28 Regulation of Na,K-ATPase activity in a tissue- and isoform-specific manner is essential for tissue functions such as muscle contraction and neuronal excitability, because of the modification of its transport properties under changing physiological conditions.29
Phosphohippolin in rats has been found to be expressed in the neuronal fibers of the lateral habenula nucleus, hypothalamus, amygdaloid body, hippocampus, thalamus, stria terminalis, cingulum, olfactory bulb, cerebral cortex, and cerebellum.22,30 The distribution in the cerebellum is unique, with a predominant expression pattern in the granule layer of the posterior lobe. Expression studies of the brain during development show the greatest amount of phosphohippolin in postnatal 3-wk-old rat brain, with substantial capacity of phosphohippolin still existing in the adult brain. This suggests that phosphohippolin may play an important role in neuronal excitability of the CNS during postnatal development and in the adult brain.22 The Novartis gene-expression-atlas database (GNF SymAtlas) shows that the expression of FXYD6 in humans is primarily in the brain, with the highest level of expression found in the prefrontal cortex, amygdala, hypothalamus, and occipital lobe. The prominent levels of expression in regions of the brain thought to be involved in SCZD, as identified by brain-imaging abnormalities, provide strong support that the FXYD6 gene increases genetic susceptibility to SCZD.
Confirmation of genetic association with SCZD requires attention to the statistical power of future case-control replication studies. The critical issue is the proven presence of locus heterogeneity for SCZD. The linkage studies of SCZD have shown strong evidence of heterogeneity, with different subgroups of families containing different susceptibility loci. Therefore, any one susceptibility gene will increase susceptibility to SCZD in only a minority of cases. The UCL SCZD sample has the power to detect allelic association of 0.99 at P<.05 and of 0.91 at P<.001, with the assumption of complete LD between markers and disease alleles when the minor marker-allele frequency is <10% and with a difference of 5% in allele frequencies between cases and controls. If the allele frequency is as high as 50%, the power is 0.90 at P=.05, with a 10% allele-frequency difference. The power of the sample is only 0.41 at P=.05 for a 5% allele-frequency difference. Further attempts at replication should aim to have 0.8 power for a significance of .05; this may require 1,300 cases and 1,300 controls, even with the assumption of complete LD between markers and disease mutations.
The FXYD6 gene should now be sequenced in schizophrenic subjects from our sample who have inherited the alleles and haplotypes contributing to the positive association that has been found. This could identify etiological base-pair changes or mutations affecting the expression or function of FXYD6 that are involved in the molecular pathology of SCZD. These may be “private” mutations not yet identified or genetic variants that are already in databases.
Acknowledgments
This research was funded by the Neuroscience Research Charitable Trust. The sample collection in Scotland was funded by the University of Aberdeen and GlaxoSmithKline. We have no commercial conflict of interest. We thank the numerous NHS doctors who helped collect the samples.
Web Resources
The URLs for data presented herein are as follows:
- Ensembl, http://www.ensembl.org/
- GDB Human Genome Database, http://www.gdb.org/
- GNF SymAtlas, http://symatlas.gnf.org/SymAtlas/
- International Haplotype Project, http://www.hapmap.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for SCZD and FXYD6)
- UCSC Genome Browser, http://genome.ucsc.edu/
References
- 1.Andreasen NC (1995) Symptoms, signs, and diagnosis of schizophrenia. Lancet 346:477–481 10.1016/S0140-6736(95)91325-4 [DOI] [PubMed] [Google Scholar]
- 2.Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, Williams NM, Schwab SG, Pulver AE, Faraone SV, et al (2003) Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: schizophrenia. Am J Hum Genet 73:34–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nanko S, Gill M, Owen M, Takazawa N, Moridaira J, Kazamatsuri H (1992) Linkage study of schizophrenia with markers on chromosome 11 in two Japanese pedigrees. Jpn J Psychiatry Neurol 46:155–159 [DOI] [PubMed] [Google Scholar]
- 4.Maziade M, Raymond V, Cliche D, Fournier JP, Caron C, Garneau Y, Nicole L, Marcotte P, Couture C, Simard C, et al (1995) Linkage results on 11Q21-22 in eastern Quebec pedigrees densely affected by schizophrenia. Am J Med Genet 60:522–528 10.1002/ajmg.1320600607 [DOI] [PubMed] [Google Scholar]
- 5.Gurling HMD, Kalsi G, Brynjolfson J, Sigmundsson T, Sherrington R, Mankoo BS, Read T, Murphy P, Blaveri E, McQuillin A, et al (2001) Genomewide genetic linkage analysis confirms the presence of susceptibility loci for schizophrenia, on chromosomes 1q32.2, 5q33.2, and 8p21-22 and provides support for linkage to schizophrenia, on chromosomes 11q23.3-24 and 20q12.1-11.23. Am J Hum Genet 68:661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spitzer RL, Endicott J (1977) The Schedule for Affective Disorders and Schizophrenia, Lifetime Version, 3rd ed. New York State Psychiatric Institute, New York [Google Scholar]
- 7.Spitzer RL, Endicott J, Robins E (1978) Research Diagnostic Criteria for a selected group of functional disorders, 3rd ed. New York State Psychiatric Institute, New York [Google Scholar]
- 8.Pimm J, McQuillin A, Thirumalai S, Lawrence J, Quested D, Bass N, Lamb G, Moorey H, Datta SR, Kalsi G, et al (2005) The Epsin 4 gene on chromosome 5q, which encodes the clathrin-associated protein enthoprotin, is involved in the genetic susceptibility to schizophrenia. Am J Hum Genet 76:902–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gurling HM, Critchley H, Datta SR, McQuillin A, Blaveri E, Thirumalai S, Pimm J, Krasucki R, Kalsi G, Quested D, et al (2006) Genetic association and brain morphology studies and the chromosome 8p22 pericentriolar material 1 (PCM1) gene in susceptibility to schizophrenia. Arch Gen Psychiatry 63:844–854 10.1001/archpsyc.63.8.844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puri V, McQuillin A, Choudhury K, Datta S, Pimm J, Thirumalai S, Krasucki R, Lawrence J, Quested D, Bass N, et al (2006) Fine mapping by genetic association implicates the chromosome 1q23.3 gene UHMK1, encoding a serine/threonine protein kinase, as a novel schizophrenia susceptibility gene. Biol Psychiatry (electronically published September 13, 2006; accessed February 19, 2007) (http://www.journals.elsevierhealth.com/periodicals/bps/article/PIIS0006322306008067/abstract) [DOI] [PubMed] [Google Scholar]
- 11.Puri V, McQuillin A, Thirumalai S, Lawrence J, Krasucki R, Choudhury K, Datta S, Kerwin S, Quested D, Bass N, et al (2006) Failure to confirm allelic association between markers at the CAPON gene locus and schizophrenia in a British sample. Biol Psychiatry 59:195–197 10.1016/j.biopsych.2005.08.015 [DOI] [PubMed] [Google Scholar]
- 12.Rizig MA, McQuillin A, Puri V, Choudhury K, Datta S, Thirumalai S, Lawrence J, Quested D, Pimm J, Bass N, et al (2006) Failure to confirm genetic association between schizophrenia and markers on chromosome 1q23.3 in the region of the gene encoding the regulator of G-protein signaling 4 protein (RGS4). Am J Med Genet B Neuropsychiatr Genet 141:296–300 [DOI] [PubMed] [Google Scholar]
- 13.Curtis D, North BV, Gurling HM, Blaveri E, Sham PC (2002) A quick and simple method for detecting subjects with abnormal genetic background in case-control samples. Ann Hum Genet 66:235–244 10.1046/j.1469-1809.2002.00109.x [DOI] [PubMed] [Google Scholar]
- 14.Stefansson H, Sarginson J, Kong A, Yates P, Steinthorsdottir V, Gudfinnsson E, Gunnarsdottir S, Walker N, Petursson H, Crombie C, et al (2003) Association of neuregulin 1 with schizophrenia confirmed in a Scottish population. Am J Hum Genet 72:83–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386 [DOI] [PubMed] [Google Scholar]
- 16.Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 17.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D (2005) Efficiency and power in genetic association studies. Nat Genet 37:1217–1223 10.1038/ng1669 [DOI] [PubMed] [Google Scholar]
- 18.Zhao JH, Curtis D, Sham PC (2000) Model-free analysis and permutation tests for allelic associations. Hum Hered 50:133–139 10.1159/000022901 [DOI] [PubMed] [Google Scholar]
- 19.Zhao JH, Lissarrague S, Essioux L, Sham PC (2002) GENECOUNTING: haplotype analysis with missing genotypes. Bioinformatics 18:1694–1695 10.1093/bioinformatics/18.12.1694 [DOI] [PubMed] [Google Scholar]
- 20.Curtis D, Knight J, Sham PC (2006) Program report: GENECOUNTING support programs. Ann Hum Genet 70:277–279 10.1111/j.1529-8817.2005.00225.x [DOI] [PubMed] [Google Scholar]
- 21.Sham PC, Curtis D (1995) Monte Carlo tests for associations between disease and alleles at highly polymorphic loci. Ann Hum Genet 59:97–105 [DOI] [PubMed] [Google Scholar]
- 22.Kadowaki K, Sugimoto K, Yamaguchi F, Song T, Watanabe Y, Singh K, Tokuda M (2004) Phosphohippolin expression in the rat central nervous system. Brain Res Mol Brain Res 125:105–112 10.1016/j.molbrainres.2004.03.021 [DOI] [PubMed] [Google Scholar]
- 23.Sweadner KJ, Rael E (2000) The FXYD gene family of small ion transport regulators or channels: cDNA sequence, protein signature sequence, and expression. Genomics 68:41–56 10.1006/geno.2000.6274 [DOI] [PubMed] [Google Scholar]
- 24.Crambert G, Geering K (2003) FXYD proteins: new tissue-specific regulators of the ubiquitous Na,K-ATPase. Sci STKE 2003:RE1 [DOI] [PubMed] [Google Scholar]
- 25.Beguin P, Crambert G, Monnet-Tschudi F, Uldry M, Horisberger JD, Garty H, Geering K (2002) FXYD7 is a brain-specific regulator of Na,K-ATPase α1-β isozymes. EMBO J 21:3264–3273 10.1093/emboj/cdf330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crambert G, Fuzesi M, Garty H, Karlish S, Geering K (2002) Phospholemman (FXYD1) associates with Na,K-ATPase and regulates its transport properties. Proc Natl Acad Sci USA 99:11476–11481 10.1073/pnas.182267299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crambert G, Li C, Claeys D, Geering K (2005) FXYD3 (Mat-8), a new regulator of Na,K-ATPase. Mol Biol Cell 16:2363–2371 10.1091/mbc.E04-10-0878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garty H, Karlish SJ (2006) Role of FXYD proteins in ion transport. Annu Rev Physiol 68:431–459 10.1146/annurev.physiol.68.040104.131852 [DOI] [PubMed] [Google Scholar]
- 29.Geering K (2006) FXYD proteins: new regulators of Na-K-ATPase. Am J Physiol Renal Physiol 290:F241–F250 10.1152/ajprenal.00126.2005 [DOI] [PubMed] [Google Scholar]
- 30.Yamaguchi F, Yamaguchi K, Tai Y, Sugimoto K, Tokuda M (2001) Molecular cloning and characterization of a novel phospholemman-like protein from rat hippocampus. Brain Res Mol Brain Res 86:189–192 10.1016/S0169-328X(00)00213-8 [DOI] [PubMed] [Google Scholar]