Abstract
A susceptibility locus for coronary artery disease (CAD) has been mapped to chromosome 3q13-21 in a linkage study of early-onset CAD. We completed an association-mapping study across the 1-LOD–unit-down supporting interval, using two independent white case-control data sets (CATHGEN, initial and validation) to evaluate association under the peak. Single-nucleotide polymorphisms (SNPs) evenly spaced at 100-kb intervals were screened in the initial data set (N=468). Promising SNPs (P<.1) were then examined in the validation data set (N=514). Significant findings (P<.05) in the combined initial and validation data sets were further evaluated in multiple independent data sets, including a family-based data set (N=2,954), an African American case-control data set (N=190), and an additional white control data set (N=255). The association between genotype and aortic atherosclerosis was examined in 145 human aortas. The peakwide survey found evidence of association in SNPs from multiple genes. The strongest associations were found in three SNPs from the kalirin (KALRN) gene, especially in patients with early-onset CAD (P=.00001–00028 in the combined CATHGEN data sets). In-depth investigation of the gene found that an intronic SNP, rs9289231, was associated with early-onset CAD in all white data sets examined (P<.05). In the joint analysis of all white early-onset CAD cases (N=332) and controls (N=546), rs9289231 was highly significant (P=.00008), with an odds-ratio estimate of 2.1. Furthermore, the risk allele of this SNP was associated with atherosclerosis burden (P=.03) in 145 human aortas. KALRN is a protein with many functions, including the inhibition of inducible nitric oxide synthase and guanine-exchange-factor activity. KALRN and two other associated genes identified in this study (CDGAP and MYLK) belong to the Rho GTPase–signaling pathway. Our data suggest the importance of the KALRN gene and the Rho GTPase–signaling pathway in the pathogenesis of CAD.
Cardiovascular disease—in particular, coronary artery disease (CAD)—is the leading cause of death and disability worldwide.1 Like other complex diseases, the etiology of CAD is multifactorial. Classic epidemiologic studies have revealed many risk factors for CAD, including age, sex, hypertension, dyslipidemia, diabetes mellitus, smoking, and physical inactivity. However, numerous studies have repeatedly shown that a positive family history is a robust predictor of CAD, even after adjustment for all known risk factors,2–6 which suggests that there is a strong genetic component underlying this complex disease.
Nine genomewide linkage scans for CAD and myocardial infarction (MI) have been conducted,7–15 including the GENECARD study.12 However, within the linkage regions identified in these studies, only the TNFSF4 gene on chromosome 1,16 the VAMP8 gene on chromosome 2,17 and the ALOX5AP gene on chromosome 1313 have been reported as putative CAD-susceptibility genes.
To increase the genetic effect in the families and to attempt to produce a more homogeneous sample, the GENECARD study focused on siblings with early-onset CAD (MIM %608901) (males aged <51 years and females aged <56 years). The age cutoff for GENECARD was chosen on the basis of the estimates of the genetic effect on CAD by several studies.6 The most significant evidence of linkage in the GENECARD study was found on chromosome 3q13-21 (multipoint LOD score 3.5), with a peak near the microsatellite marker D3S2460. The 1-LOD–unit-down supporting interval is large, extending from 115 Mb to 135 Mb (National Center for Biotechnology Information [NCBI] build 35). Using ordered subset analysis methodology, we found that the linkage evidence for this peak was significantly increased in families with high high-density lipoprotein cholesterol levels.18 Recently, in support of this region’s importance for CAD, Bowden et al.19 also reported linkage to the same GENECARD peak marker—that is, D3S2460—for CAD and stroke in the Diabetes Heart Study, with a similar broad linkage region (multipoint LOD score 2.5).
To fine map the chromosome 3 linkage peak, we conducted a peakwide association mapping study, using SNPs, in two independent white case-control data sets. Significant results were then subsequently tested in additional data sets.
Material and Methods
Subjects
Several independent data sets were used at different stages of this study. The Duke Institutional Review Board approved all studies, and all subjects provided signed informed consent.
CATHGEN data sets (initial, validation, and African American)
CATHGEN subjects were recruited through the cardiac catheterization laboratories at Duke University Hospital. All subjects undergoing catheterization were offered the opportunity to participate in the study. To reduce confounding by population substructure, the initial and validation data sets (each with independent cases and controls ascertained sequentially) were composed of whites only, with an additional case-control data set constructed using all CATHGEN African American subjects.
Alzheimer disease controls
Subjects for a third white control data set were collected through an ongoing study of susceptibility genes for Alzheimer disease (M.A.P.-V., principal investigator) at the Duke Center for Human Genetics. Individuals in this group were recruited from community meetings and unrelated family members (e.g., spouses) of Alzheimer disease–affected families. All individuals were aged >60 years and were self-reported whites.
GENECARD data set
The sample collection and study design of the GENECARD study have been reported.12 In brief, GENECARD samples were collected through a collaborative effort from investigators affiliated with Duke and five other international sites. The family-based GENECARD data set was composed of families with at least two affected siblings who met the criteria for early-onset CAD. The majority (>90%) of the GENECARD subjects were white. Unlike the CATHGEN samples, angiographic data were not available.
Aorta data set
Human aortas were collected from heart-transplant donors, and each aorta was graded for atherosclerosis burden as described elsewhere.20 All donors of the aortas used in this study were white.
Classification Criteria
CATHGEN controls and cases were chosen on the basis of their extent of CAD, as measured by the CAD index (CADi). CADi is a numerical summary of coronary angiographic data that incorporates the extent and anatomical distribution of coronary disease.21 CATHGEN cases had a CADi ⩾32, and CATHGEN controls had an age at catheterization >60 years and no diseased vessels, documented MI, or interventional cardiac procedures. The CADi threshold used to determine affection status was chosen a priori on the basis of a previous study.22 A CADi ⩾32 was equivalent to having at least one major epicardial vessel with ⩾95% stenosis, ensuring a highly consistent CAD phenotype. Cases with an age at onset <56 years for females and <51 years for males were classified as “young affected,” whereas individuals with an age at onset ⩾56 years for females and ⩾51 years for males were designated as “old affected,” following the initial GENECARD criteria. In the old affected group, we wanted to adjust for the higher baseline extent of CAD. Since there is no study suggesting an appropriate adjustment for this purpose, we chose the cutoff of CADi ⩾74, since that represents the highest quartile of CADi in the patients ascertained at the Duke University Health System cardiac catheterization laboratories. The age at onset of CAD was defined as the age of first documented surgical or percutaneous coronary revascularization procedure, MI, or cardiac catheterization meeting the above CADi thresholds. The detailed classification criteria for the GENECARD samples were published elsewhere.23 In brief, the affected individuals had a CADi event at age <51 for males and <56 for females. Selected control individuals with Alzheimer disease had no history of MI, diabetes, stroke, or peripheral vascular disease, as determined by a detailed questionnaire for medical history. Results of their Modified Mini-Mental Status exam24 were normal. The aorta samples were assigned case or control status by evaluation of raised lesions by use of the Pathobiological Determinants of Atherosclerosis in Youth Study methodology.25 Samples with a raised lesion score >0 were classified as cases, and all others were controls.
SNP Genotyping
DNA was extracted from whole blood or tissue (aorta), as described elsewhere,26 by the Duke Center for Human Genetics DNABANK. SNP genotyping was performed using the TaqMan Allelic Discrimination assay. For the purpose of quality control, 1 blank, 2 CEPH-pedigree individuals,27 and 12 quality-control samples were included for every quadrant of the 384-well plate. The quality-control samples were used to provide duplicated samples within one quadrant, across quadrants within one plate, and across plates. Results of the CEPH and quality-control samples were compared, to identify possible sample-plating errors and genotype-calling inconsistencies. Hardy-Weinberg equilibrium (HWE) testing was performed for all markers. SNPs that showed mismatches on quality-control samples or that failed the HWE test (P<.05) in white controls were reviewed by an independent genotyping supervisor for potential genotyping errors. All examined SNPs were successfully genotyped for ⩾95% of the individuals in the study. On the basis of >26,000 duplicate genotypes, genotyping error–rate estimates for SNPs meeting the quality-control benchmarks were <0.2%.
Study Design
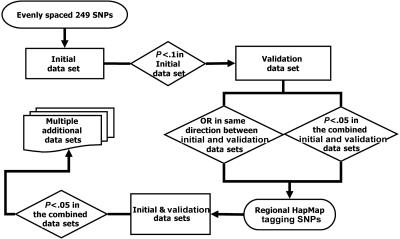
The overall study design is illustrated in figure 1. At first, to screen the large linkage peak region for association and to minimize genotyping labor and cost, the iterated association methodology (IAM) of Oliveira et al.26 was used. Briefly, SNPs at an average of 100-kb intervals across the 1-LOD–unit-down region of 3q13-21 were chosen for screening. SNPs were chosen using the software program SNPSelector.28 If none of these initial SNPs demonstrated significant association, then the SNP density was increased to 50 kb, then to 25 kb, and so on.
Figure 1. .
Flow chart of staged study design
Then, to evaluate the significance of the multiple SNPs used in the peakwide screening, we applied a staged design. The initial goal was to identify the highest priority SNPs for follow-up genotyping with all surrounding HapMapII tagging SNPs (International HapMap Project). In the first stage, we screened SNPs at the chosen IAM density in the CATHGEN initial data set. Any significant SNPs were then tested in the second CATHGEN validation data set. Since we wished to be inclusive rather than exclusive at this stage, all the SNPs that had nominal P values <.1 in the CATHGEN initial data set were analyzed in the CATHGEN validation data set. The odds-ratio (OR) estimates for each SNP between the two data sets were then compared, to identify those SNPs with the most-consistent association results. Since recent studies have suggested that analyzing one larger data set has more power and is more reliable in detecting the true genetic association for complex disease than is examining two smaller data sets separately as replication sets,29,30 joint analysis of the CATHGEN initial and validation data sets was then performed. SNPs that had a P value <.05 in this combined CATHGEN data set and that displayed the same direction of association (i.e., ORs) in both the initial and validation data sets were considered priority SNPs. Regions flanking the priority SNPs were evaluated with all surrounding HapMap tagging SNPs (minor-allele frequency >2%; r2<0.7) in the CATHGEN combined data set.
Finally, the most significant SNPs from these tagging-SNP studies in the CATHGEN combined data set were further evaluated in the GENECARD family-based data set, the CATHGEN African American case-control data set, the Alzheimer white control data set, and the human aorta data set.
Statistical Analysis
Case-control association was examined using logistic regression analysis. A simple logistic model was fitted, with adjustment for sex and age at exam in the overall analysis. Since age at onset is not applicable for the control individuals, we used age at exam as a covariable in the statistical model, to adjust for age-related differences that may impact risk factors measured at the time of catheterization, as well as disease risk between the case and control groups. In the subset analysis of early-onset and late-onset CAD, only sex was included as a covariable. This was necessary because of the different age distributions in cases and controls that is inherent in the sample selection. A multivariable logistic-regression model was also used, which included sex, hypertension (clinically significant hypertension by history), diabetes mellitus (previous physician diagnosis), BMI, dyslipidemia (previous diagnosis and/or treatment of hypercholesterolemia by a physician), and smoking history (more than half a pack per day) as covariables. For the human aorta samples, age at harvest was included in the logistic regression analysis, because we did not preselect cases and controls on the basis of age. The association in the presence of linkage (APL)31 test was implemented to evaluate family-based association in the GENECARD samples. The Graphical Overview of Linkage Disequilibrium (GOLD) program32 and Haploview33 were used to assess and display linkage disequilibrium (LD) between SNPs. To evaluate the interaction between genes that work in the same biological pathway, we analyzed the most-significant SNPs from each gene in both the additive and multiplicative models, using logistic regression. To evaluate the independence of the associated markers, backward stepwise logistic regression was performed. SAS 9.0 was used for statistical analyses. Raw P values from each association test were reported in this study. In place of correcting P values for multiple testing, we relied on the staged design and validation of association in multiple independent data sets to minimize the possibility of a false-positive finding (see the “Study Design” section). The Linkage and Association Modeling in Pedigrees test34 was used to evaluate whether the linkage signal on chromosome 3q13-21 could be explained by any of the significant SNPs identified in our study.
Results
Clinical Characteristics of Study Subjects
The CATHGEN initial data set consisted of 117 white controls and 351 cases (184 young affected individuals and 167 old affected individuals). The CATHGEN validation data set included an additional 174 white controls and 340 cases (148 young affected individuals and 192 old affected individuals). The combined CATHGEN data set was composed of 691 cases (332 young affected individuals and 359 old affected individuals) and 291 controls. The CATHGEN African American case-control data set comprised 77 controls and 173 cases (114 young affected individuals and 59 old affected individuals). Because of the small number of African American cases in the old affected subgroup, that group was not analyzed for association. The indications for cardiac catheterization in the CATHGEN control groups was possible ischemic heart disease (66%), followed by valvular heart disease (8%), congenital heart disease (<1%), and other indications (including evaluation for fatigue, preoperative clearance, and asymptomatic decreased ejection fraction). Baseline clinical characteristics are reported in table 1. As expected, the case groups had a higher prevalence of traditional CAD risk factors than did the controls. The Alzheimer disease control data set was composed of 255 whites, with an average age at examination of 74 years. The clinical characteristics of the GENECARD samples were reported elsewhere.12,35 This data set consisted of 1,101 families, with 2,260 affected individuals, 603 unaffected individuals, and 91 individuals of unknown status. The human aorta data set was composed of 35 cases and 110 controls.
Table 1. .
Clinical Characteristics of CATHGEN Data Sets[Note]
| White Initial Data Set |
White Validation Data Set |
African American Data Set |
|||||||
| Characteristic | Young Affected (N=184) |
Old Affected (N=167) |
Control (N=117) |
Young Affected (N=148) |
Old Affected (N=192) |
Control (N=174) |
Young Affected (N=114) |
Old Affected (N=59) |
Control (N=77) |
| Mean age (in years): | |||||||||
| At exam | 50.2 (7.6)a | 66.1 (10.5)a | 69.6 (6.7) | 53.9 (9.1)a | 68.9 (9.1) | 69.8 (6.7) | 48.7 (6.5)a | 63.8 (10.5)a | 69.4 (6.2) |
| At onset | 43.4 (5.9) | 60.5 (8.9) | NA | 44.5 (5.4) | 61.6 (8.8) | NA | 45.2 (5.7) | 60 (8.9) | NA |
| Mean CADi | 56.3 (21.6)a | 72.1 (19.2)a | 7.3 (9.9) | 61.1 (21.4)a | 79.1 (16.1)a | 5.4 (9) | 50.6 (18.9)a | 69.5 (20.3)a | 5.7 (9.3) |
| Mean BMI | 31 (6.3)a | 29.2 (6.6)a | 27.6 (6.3) | 30.3 (6.4)a | 28.7 (5.6) | 28.5 (6) | 31.6 (7.3) | 29.2 (7.3) | 31.8 (7.9) |
| Ever smoked (%) | 73.9a | 59.3a | 41.0 | 71.0a | 57.8a | 39.1 | 57.9 | 57.6 | 46.1 |
| Diabetic (%) | 29.4a | 32.9a | 7.7 | 35.8a | 25.0 | 19.0 | 41.2 | 42.4 | 46.1 |
| Hypertensive (%) | 60.9 | 73.7 | 63.3 | 68.2 | 70.3 | 67.2 | 79.8 | 88.1a | 73.7 |
| Male (%) | 79.9a | 83.8a | 46.2 | 77.0a | 85.4a | 40.8 | 54.4 | 72.9a | 44.7 |
| Hyperlipidemic (%) | 73.9a | 73.1a | 39.3 | 79.7a | 70.3a | 51.7 | 68.4a | 69.5a | 43.4 |
Note.— Numbers within parentheses are SD. NA = not applicable.
There were significant differences between cases and controls (P<.05) within each CATHGEN data set. Analysis of variance was performed by χ2 test for categorical variables and by t test for numeric variables.
IAM SNP Screening in the Initial Data Set
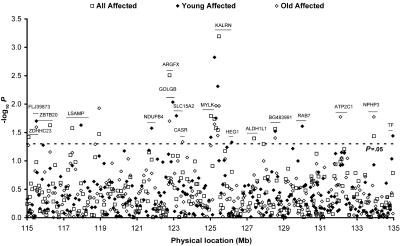
To survey the 1-LOD–unit-down region of the chromosome 3q13-21 peak, 249 evenly spaced SNPs were genotyped, with an average density of one SNP every 84 kb in the initial CATHGEN data set. Pairwise LD was estimated, and most SNPs (>90%) were not in high LD (r2<0.7) with each other. Evidence of association (P<.05) was observed for 15 SNPs, with the most significant association at SNP rs2272486, which resides in the kalirin (KALRN) gene (P=.0005) (fig. 2). Since the GENECARD study focused on early-onset CAD, we divided the analysis into separate groups of young affected and old affected subjects. In the analysis of young affected subset, evidence of association (P<.05) was found for 13 SNPs; 5 of them were also significant in the overall initial–data set analysis. In the analysis of the old affected subset, evidence of association (P<.05) was found for 12 SNPs; 8 of them were also significant in the overall initial–data set analysis (fig. 2).
Figure 2. .
Association tests of 249 SNPs in the CATHGEN initial data set. Each point represents an association test in the initial data set on one SNP in all affected (square), young affected (blackened diamond), and old affected (unblackened diamond) subgroups. Genes containing SNPs with evidence of association (P<.05) are labeled. The line segment illustrates the approximate location of those genes on chromosome 3. Forty-four SNPs with P values <.1 (−log10 P value >1.0) were chosen for validation genotyping (figs. 3 and 4).
Evaluation of Association in the Validation Data Set
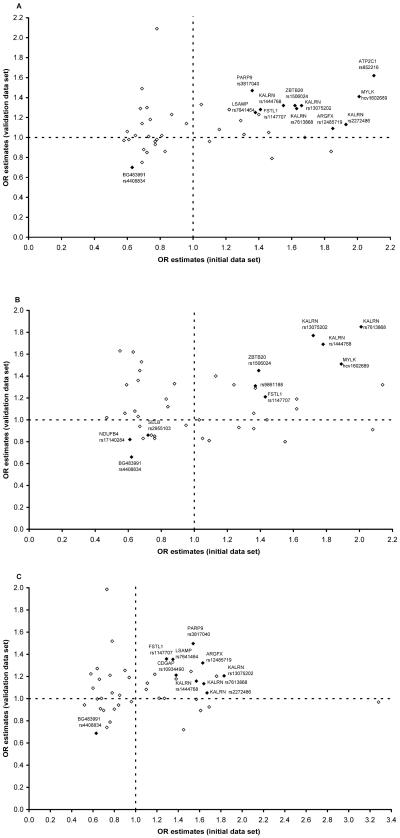
To evaluate the significance of the multiple SNPs tested in the initial data set, SNPs that had a nominal P value <.1 were examined in the CATHGEN validation case-control data set. Among the 44 SNPs tested at this stage, 28 (64%), 23 (52%), and 27 (61%) of the SNPs displayed the same direction of OR between the initial and the validation data sets in the overall, young affected, and old affected analyses, respectively (fig. 3). Among these consistent SNPs, SNPs rs1444768 (P=.0376) in the KALRN gene and rs4408834 (P=.0186) in the EST BG483991 were significant in the validation data set in the overall analysis. In the analysis of the young affected cases, three SNPs (rs7613868, rs13075202, and rs1444768) (P=.0007–.0039) in the KALRN gene and rs4408834 (P=.0115) met the significant level of .05 in the validation data set. None of the consistent SNPs were significant in the analysis of the old affected cases in the validation data set alone (data not shown).
Figure 3. .
OR estimates of the 44 selected SNPs in the CATHGEN initial and validation data sets. Each diamond represents one of the selected 44 SNPs (P<.1) from the initial peakwide screening on 3q13-21. OR estimates were calculated separately for each SNP in the CATHGEN initial and validation data sets. SNPs and genes that had the same genetic effect direction in the initial and the validation data sets and had a P value <.05 in the joint analysis of the combined CATHGEN data set are displayed as blackened diamonds and are labeled. A, All affected subjects. B, Young affected subjects. C, Old affected subjects.
Analysis of the Combined Data Sets
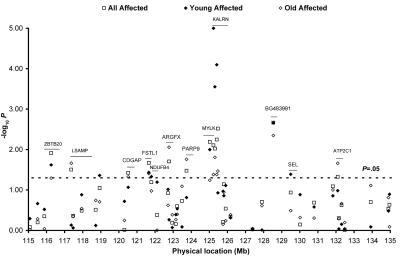
Those SNPs that provided consistent OR estimates between the initial and validation data sets were tested for association in the combined CATHGEN initial and validation data sets. In the overall analysis, SNPs rs12485719 in the arginine-fifty homeobox (ARGFX); rs10934490 in the Cdc42 GTPase–activating protein (CDGAP); rs1147707 in the follistatinlike 1 (FSTL1); rs7613868, rs13075202, rs1444768, and rs2272486 in the KALRN gene; rs7641464 in the limbic system–associated membrane protein (LSAMP); hcv1602689 in the myosin light chain kinase (MYLK); rs3817040 in the Poly (adenosine diphosphate–ribose) polymerase family member 9 (PARP9); rs1506024 in the Zinc finger and BTB domain containing 20 (ZBTB20) gene; and rs4408834 in the EST gene BG483991 were associated with CAD risk (P=.002–.038) (fig. 4).
Figure 4. .
Joint analysis of the 44 selected SNPs in the combined CATHGEN data set (initial and validation data sets). Each point represents the association test on one of the selected 44 SNPs (P<.1) from the initial peakwide screening on 3q13-21. Joint analyses were performed separately for the CATHGEN combined data set in all affected (square), young affected (blackened diamond), and old affected (unblackened diamond). Genes containing SNPs with evidence of association (P<.05) in the CATHGEN combined data set are labeled. The line segment illustrates the approximate location of those genes on chromosome 3.
We then analyzed the two subsets (young affected and old affected) in this combined data set. The strongest associations in this young affected data set were found at rs7613868, rs13075202, and rs1444768 (P=.00001–.00028) (fig. 4). All three SNPs reside in the KALRN gene, with limited LD between them (r2=0.29–0.81) (fig. 5). In the analysis of the old affected cases, the strongest associations were found with rs4408834 in BG483991 (P=.0105) (fig. 4).
Figure 5. .
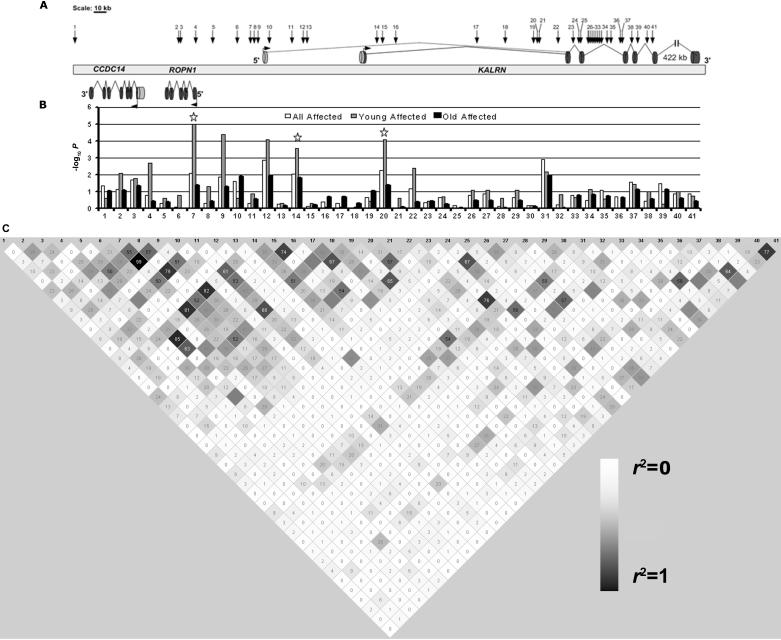
HapMap tagging SNPs in the KALRN gene region. Thirty-eight HapMap tagging SNPs in the KALRN gene region plus the three significant SNPs from the peakwide survey are displayed (A). The locations of the SNPs are indicated by the arrows with numbers (1–41). The gene structure is shown in genomic context, with the CCDC14 and ROPN1 genes on the reverse strand and the KALRN gene on the forward strand. Exons and introns are depicted as dark cylinders and solid lines, respectively. Alternative exons and introns are depicted as light cylinders and dotted lines, respectively. −log10 P values for association test are displayed on a bar graph (B). Association tests were performed separately for the combined CATHGEN data sets in all affected (white bar), young affected (gray bar), and old affected (black bar) cases. The three significant SNPs identified from peakwide screening are SNPs 7, 14, and 20 (labeled with a star). Pairwise r2 was estimated in the controls and is displayed by gray-shaded square with the hundredths of r2 value inside (HaploView)33 (C). The shades of gray are proportional to the r2 values, with the darkest gray being the highest r2 value. Similar LD relationships were seen in the affected individuals and are not shown.
Additional HapMap Tagging SNP Genotyping
Given the fact that the strongest association was found with SNPs rs7613868, rs13075202, and rs1444768, we examined additional HapMap tagging SNPs around those three SNPs. The three highly significant SNPs were located within a 200-kb stretch of DNA. We extended the region by 100 kb on each side and identified 38 tagging SNPs in that 400-kb block. The average density for the 38 SNPs was 1 SNP every 10 kb. Figure 5 illustrates the association tests in the combined CATHGEN data set on those tagging SNPs and the three highly significant SNPs from peakwide association mapping. The corresponding SNP rs numbers for the SNP identification numbers used in figure 5 are listed in table 2. In the overall analysis, eight SNPs displayed evidence of association (P=.001–.043). In the subset analysis, nine SNPs were associated with the young affected subset (P=.00004–.047), and four SNPs were associated with the old affected data set (P=.011–.045). The strongest associations were found in the subset of young affected cases—that is, SNP 9 and SNP 12 (raw P=.00004 and .00008, respectively), which remained significant even after correcting for multiple testing of the 38 new SNPs at this stage by Bonferroni correction (corrected P<.001). No SNPs in the overall analysis or the subset analysis of the old affected cases survived this stringent correction. SNPs 9 and 12 also remained significant after adjusting for known CAD risk factors in the young affected subset (P=.0002 and .00002, respectively) (table 2).
Table 2. .
Association of the HapMap Tagging SNPs in the KALRN Gene Region in the Young Affected Subset of the Combined CATHGEN Data Sets[Note]
| Allele Frequency(%) |
Association Test P (OR) |
|||||||
| SNP | Identification | Base-Pair Location | Locus | Allele | Young Affected n=332 |
Control n=291 |
Basic Model | Full Model |
| rs6766344 | 1 | 125110476 | CCDC14 | A | 5 | 7 | .2442 (.7) | .3929 (.8) |
| rs6810298 | 2 | 125177665 | ROPN1 | G | 34 | 27 | .0082 (1.4) | .0064 (1.5) |
| rs17376453 | 3 | 125179139 | ROPN1 | C | 18 | 12 | .017 (1.5) | .0094 (1.7) |
| rs7434266 | 4 | 125188532 | ROPN1 | A | 23 | 16 | .002 (1.6) | .0027 (1.7) |
| rs7429448 | 5 | 125198542 | ROPN1 | A | 12 | 9 | .2459 (1.3) | .512 (1.2) |
| rs9813249 | 6 | 125215401 | KALRN | C | 9 | 7 | .1691 (1.4) | .3344 (1.3) |
| rs7613868 | 7 | 125224719 | KALRN | A | 35 | 24 | 1×10−5 (1.9) | 7×10−5 (1.9) |
| rs12634530 | 8 | 125227160 | KALRN | T | 21 | 16 | .0474 (1.4) | .3313 (1.2) |
| rs12637456 | 9 | 125227353 | KALRN | A | 35 | 24 | 4×10−5- (1.8) | .0002 (1.8) |
| rs869585 | 10 | 125236654 | KALRN | A | 3 | 1 | .2468 (1.8) | .2546 (1.9) |
| rs2332690 | 11 | 125250612 | KALRN | A | 18 | 15 | .1396 (1.3) | .4009 (1.2) |
| rs9289231 | 12 | 125256768 | KALRN | G | 13 | 7 | 8×10−5 (2.4) | 2×10−5 (3.0) |
| rs16835038 | 13 | 125259296 | KALRN | G | 5 | 5 | .5086 (.8) | .428 (.8) |
| rs13075202 | 14 | 125304977 | KALRN | G | 30 | 21 | .0003 (1.7) | .0004 (1.8) |
| rs11714061 | 15 | 125308212 | KALRN | A | 11 | 9 | .4785 (1.2) | .4537 (1.2) |
| rs1316375 | 16 | 125316304 | KALRN | A | 10 | 8 | .7958 (1.1) | .5739 (1.2) |
| rs16835137 | 17 | 125368709 | KALRN | A | 8 | 7 | .7951 (1.1) | .8193 (1.1) |
| rs16835162 | 18 | 125386881 | KALRN | G | 16 | 15 | .7869 (1.0) | .738 (1.1) |
| rs7627053 | 19 | 125404954 | KALRN | A | 7 | 6 | .3573 (1.3) | .3955 (1.3) |
| rs1444768 | 20 | 125406612 | KALRN | G | 41 | 29 | 8×10−5 (1.7) | .0002 (1.8) |
| rs1444767 | 21 | 125407493 | KALRN | T | 10 | 8 | .2524 (1.3) | .4137 (1.2) |
| rs1444754 | 22 | 125420907 | KALRN | C | 43 | 34 | .0042 (1.4) | .0217 (1.4) |
| rs9833095 | 23 | 125430323 | KALRN | A | 5 | 4 | .369 (1.3) | .6735 (1.1) |
| rs13066449 | 24 | 125433827 | KALRN | T | 40 | 36 | .2095 (1.2) | .1442 (1.2) |
| rs1920629 | 25 | 125435293 | KALRN | T | 4 | 4 | .9869 (1.0) | .5881 (.8) |
| rs4678095 | 26 | 125439286 | KALRN | C | 49 | 55 | .0818 (.8) | .1155 (.8) |
| rs1158012 | 27 | 125440085 | KALRN | T | 45 | 51 | .0824 (.8) | .3365 (.9) |
| rs11720960 | 28 | 125440632 | KALRN | C | 7 | 8 | .2536 (.8) | .3849 (.8) |
| rs7616435 | 29 | 125442275 | KALRN | G | 7 | 5 | .0835 (1.6) | .1392 (1.5) |
| rs4678097 | 30 | 125443420 | KALRN | G | 17 | 16 | .6955 (1.1) | .9644 (1.0) |
| rs4234218 | 31 | 125443900 | KALRN | G | 41 | 33 | .0067 (1.4) | .044 (1.3) |
| rs6794049 | 32 | 125444174 | KALRN | T | 2 | 3 | .1567 (.5) | .2525 (.6) |
| rs4608635 | 33 | 125444186 | KALRN | C | 34 | 38 | .2281 (.9) | .5178 (.9) |
| rs4678100 | 34 | 125452115 | KALRN | C | 8 | 5 | .0771 (1.5) | .3252 (1.3) |
| rs6438837 | 35 | 125453375 | KALRN | G | 10 | 12 | .2864 (.8) | .5601 (.9) |
| rs17221479 | 36 | 125460168 | KALRN | T | 10 | 9 | .9454 (1.0) | .6881 (.9) |
| rs11929003 | 37 | 125462011 | KALRN | G | 45 | 50 | .0364 (.8) | .1839 (.8) |
| rs9813731 | 38 | 125468728 | KALRN | G | 5 | 3 | .098 (1.7) | .3906 (1.4) |
| rs9868324 | 39 | 125472134 | KALRN | A | 20 | 21 | .5441 (.9) | .6839 (.9) |
| rs2332769 | 40 | 125485918 | KALRN | A | 35 | 40 | .0982 (.8) | .3415 (.9) |
| rs1444746 | 41 | 125494623 | KALRN | C | 42 | 47 | .181 (.8) | .3987 (.9) |
Note.— The physical location of each SNP is based on NCBI build 35. Genotype-based association tests were performed in the young affected cases versus controls in the combined CATHGEN data sets, with adjustment for sex (basic model) or adjustment for sex, hypertension, diabetes mellitus, BMI, dyslipidemia, and smoking history (full model). P values <.05 are shown in bold.
The majority of the significant SNPs and the most significant SNPs reside in the 5′ end of the KALRN gene. Three significant SNPs—2, 3, and 4—also mapped to a small gene—Ropporin (ROPN1)—that resides next to the KALRN gene (fig. 5). Figure 5 also illustrates the LD structure among the 38 HapMap tagging SNPs and the three highly significant SNPs, from peakwide association mapping. There was no strong LD between them (r2=0–0.85) except for the LD between SNPs 7 and 9 (r2=0.99). In an attempt to evaluate whether the multiple significant signals in this region are independent of each other, backward stepwise logistic regression analysis was performed. This analysis found that SNP 12 remained highly significant (P=.0001), and two other SNPs—22 and 31—were marginally significant (P=.07 and .09, respectively), even after taking into account other SNPs and variants in the model, which suggests that the multiple strong associations in the KALRN gene region are driven mainly by the SNPs in the KALRN gene, especially SNP 12. In the combined CATHGEN data set, for SNP 12, we counted 0, 39, and 245 individuals with the GG, GT, and TT genotypes, respectively, in controls, and 5, 76, and 246 individuals with the GG, GT, and TT genotypes, respectively, in young affected individuals.
Validation in Multiple Additional Data Sets
Since the linkage peak was originally identified in families with early-onset CAD in GENECARD samples and the strongest evidence of association in the CATHGEN samples was found with the young affected cases who have the same age-at-onset criteria as those included in the GENECARD study, we further evaluated the association of the nine significant HapMap tagging SNPs and the three highly significant SNPs identified from peakwide association mapping in multiple additional data sets focusing on early-onset CAD (table 3). First, we constructed an independent set of white controls, using control subjects from an ongoing Alzheimer disease study. The combined young affected cases from the CATHGEN initial and validation data sets were compared with this independent control set. SNPs 4 (ROPN1), 7, 9, 12, and 14 (KALRN) were significant in this analysis (P=.0030–.0261). Second, we investigated the association of these SNPs in the CATHGEN African American young affected case-control data set. SNPs 12 (P=.0323) and 22 (P=.0298) were found to be associated with young affected cases in the African American data set. Third, we evaluated the SNP associations in the family-based GENECARD data set and found that SNP 12 was significant in this analysis as well (P=.0223). Finally, we examined whether any SNP genotypes were associated with atherosclerosis burden in human aortas. In this analysis, SNPs 7 (P=.0197) and 12 (P=.0339) were significant. Therefore, SNP 12, the SNP that seemed to account for most of the association signal in the KALRN gene region, was significant in all the data sets we examined. In addition, the direction of the genetic effect and allele frequency were highly consistent throughout the multiple data sets except for African Americans (table 3).
Table 3. .
Validation of the Significant Tagging SNPs in the KALRN Gene Region, with Early-Onset CAD or Atherosclerosis in Multiple Data Sets[Note]
| CATHGEN Initial Data Set(N=301) |
CATHGEN ValidationData Set(N=322) |
ADCa (N=255) versus Combined CATHGEN Cases (N=332) |
African American Data Set(N=190) |
Aorta Data Set(N=145) |
||||||||||||||
| Frequency(%) |
Frequency(%) |
Frequency(%) |
Frequency(%) |
Frequency(%) |
||||||||||||||
| SNP | Identification | Allele | Case | Control | Association P (OR) | Case | Control | Association P (OR) | Case | Control | Association P (OR) | Case | Control | Association P (OR) | Case | Control | Association P (OR) | GENECARD Data Set P (N=2,954) |
| rs6810298 | 2 | G | 32 | 28 | .1866 (1.3) | 33 | 26 | .0194 (1.6) | 32 | 30 | .5336 (1.1) | 59 | 66 | .1193 (.7) | 48 | 34 | .1930 (1.7) | .5648 |
| rs17376453 | 3 | C | 18 | 13 | .1054 (1.5) | 16 | 11 | .1223 (1.5) | 17 | 15 | .4288 (1.2) | 4 | 7 | .1244 (.5) | 26 | 12 | .1034 (2.1) | .1122 |
| rs7434266 | 4 | A | 24 | 17 | .0160 (1.8) | 22 | 16 | .0654 (1.5) | 23 | 17 | .0141 (1.5) | 15 | 20 | .2301 (.7) | 26 | 17 | .1653 (1.8) | .2708 |
| rs7613868 | 7 | A | 34 | 22 | .0015 (2.0) | 37 | 26 | .0010 (1.8) | 36 | 27 | .0030 (1.6) | 76 | 81 | .4410 (.8) | 44 | 28 | .0197 (2.6) | .1534 |
| rs12634530 | 8 | T | 18 | 15 | .6224 (1.1) | 23 | 16 | .0160 (1.7) | 21 | 18 | .5005 (1.1) | 41 | 34 | .1470 (1.4) | 24 | 19 | .5090 (1.3) | .8172 |
| rs12637456 | 9 | A | 33 | 22 | .0039 (1.9) | 37 | 26 | .0020 (1.8) | 35 | 27 | .0101 (1.5) | 67 | 69 | .7277 (.9) | 41 | 27 | .0622 (2.2) | .1300 |
| rs9289231 | 12 | G | 14 | 6 | .0011 (3.1) | 13 | 7 | .0269 (2.0) | 13 | 9 | .0184 (1.7) | 21 | 32 | .0323 (.6) | 16 | 6 | .0339 (3.1) | .0223 |
| rs13075202 | 14 | G | 27 | 19 | .0177 (1.7) | 33 | 23 | .0024 (1.8) | 30 | 24 | .0261 (1.4) | 68 | 72 | .6050 (.9) | 41 | 25 | .0519 (2.1) | .1830 |
| rs1444768 | 20 | G | 39 | 28 | .0049 (1.8) | 42 | 29 | .0045 (1.7) | 41 | 35 | .0640 (1.3) | 74 | 77 | .3868 (.8) | 42 | 34 | .1185 (1.8) | .4532 |
| rs1444754 | 22 | C | 42 | 34 | .0500 (1.4) | 44 | 35 | .0304 (1.4) | 43 | 39 | .1304 (1.2) | 62 | 50 | .0298 (1.6) | 45 | 37 | .1349 (1.7) | .3445 |
| rs4234218 | 31 | G | 43 | 36 | .0311 (1.5) | 40 | 32 | .1634 (1.3) | 42 | 42 | .6664 (.9) | 21 | 16 | .4781 (1.2) | 40 | 38 | .3766 (1.4) | .7422 |
| rs11929003 | 37 | G | 41 | 52 | .0193 (.7) | 49 | 48 | .6819 (.9) | 45 | 41 | .3826 (1.1) | 49 | 45 | .3005 (1.3) | 44 | 43 | .8322 (0.9) | .2010 |
Note.— Logistic regression analysis was performed for case-control data sets composed of young affected cases and controls. The APL test was performed for the family transmission data set (GENECARD). P values <.05 are shown in bold.
ADC = Alzheimer disease control.
Since the association of SNP 12 (rs9289231) was primarily with patients with early-onset CAD in the multiple data sets we tested, we evaluated the genetic effect of this SNP on early-onset CAD, using all white young affected (N=332) and control (N=546) subjects, to maximize the precision of the OR estimate. In this analysis, rs9289231 was highly significant (P=.00008), with an OR estimate of 2.1. This SNP is estimated to explain 12.1% of early-onset CAD in the sample of case-control subjects used in this study. However, we did not observe any statistical evidence suggesting that any association individually accounted for the linkage signal in the GENECARD samples.
Discussion
We found evidence of association with CAD in multiple genes lying under the 3q13-21 peak, with the KALRN SNP rs9289231 being the most highly significant in our CATHGEN data sets, as well as in multiple independent case-control data sets and a family-based association data set. Further, the risk allele of rs9289231 was significantly correlated with atherosclerosis burden in human aortas (table 3).
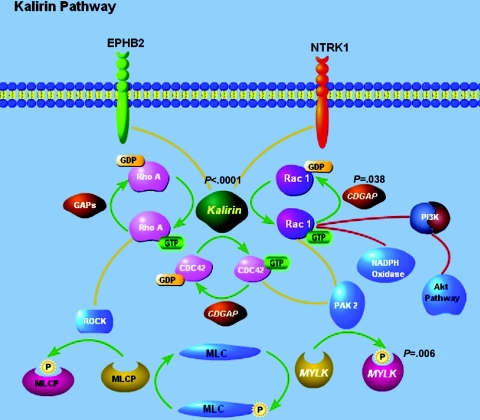
Remarkably, two other genes that display evidence of association with CAD in the combined CATHGEN initial and validation data set—that is, CDGAP (rs10934490) (P=.038) and MYLK (hcv1602689) (P=.006) (fig. 4)—are closely tied to KALRN in the same biological pathway, the Rho GTPase signal–transduction pathway (fig. 6). We did not observe evidence of statistical interaction between SNPs residing in these genes, but our sample size is likely too small for detecting such an interaction. The Rho GTPase is a family of small G proteins that regulate a wide range of cellular activity—including cell proliferation/migration/adhesion, cytoskeletal dynamics, oncogenic transformation, and inflammatory responses—by stimulating downstream signaling pathway.36–39 Some of the Rho GTPases, such as RhoA and its downstream effector Rho-kinase (ROCK), have been indicated in many cardiovascular activities.37,40–43 It has been reported that the guanine exchange factor (GEF) domain of KALRN—or unc-73 (KALRN’s homolog in Caenorhabditis elegans)—activates Rac1 by catalyzing the exchange of guanosine diphosphate (GDP) with guanosine triphosphate (GTP).44,45 There is mounting evidence that Rac1 plays an important role in the vascular cell signaling through regulation on the NADPH oxidases and the Rac1-dependent production of reactive oxygen species.46–48
Figure 6. .
Illustration of key proteins in the Rho GTPase signal–transduction pathway, focusing on the KALRN, CDGAP, and MYLK genes. The Rho GTPase signal–transduction pathway focusing on the three associated genes on chromosome 3q13-21—KALRN, CDGAP, and MYLK—is illustrated. Displayed P values are the smallest P value for each gene from the joint analysis of the combined CATHGEN initial and validation data sets.
The highly associated SNP rs9289231 resides in the first intron of an alternative transcript of the KALRN gene (fig. 5). The KALRN gene was originally identified through its interaction with peptidylglyine α-amidating monooxygenase (PAM) with use of a rat hippocampal cDNA library.49 PAM catalyzes amidation of peptide, a key step in synthesizing bioactive peptides such as neuropeptides and hormones. The KALRN gene encodes many transcript isoforms through alternative promoters, different internal transcription start sites, and alternative splicing at the 3′ end.50,51 The largest isoform is 11 kb in size and consists of 60 exons.51 Full-length KALRN possesses multiple functional protein domains, including an Sec14p domain, nine spectrinlike repeats, two GEF domains, two src homology 3 (SH3) domains, two pleckstrin-homology (PH) domains, and a putative Ser/Thr protein kinase domain.52 It has been documented that KALRN interacts with PAM, Huntington-associated protein 1 (HAP1), and inducible nitric oxide synthase (iNOS) through the spectrinlike repeats.49,53,54 On binding to iNOS, KALRN inhibits iNOS activity via prevention of homodimerization of the enzyme.54 Given the observed cardiovascular activity of nitric oxide,55,56 one possible mechanism for CAD susceptibility involving KALRN could be through its interaction with iNOS.
It is worth pointing out that our association studies provide appealing but not definitive evidence that KALRN is the CAD susceptibility gene, since other genes lie in the association region. The KALRN gene was identified through an unbiased association mapping based on SNPs. It is possible that rs9289231 is a proxy for the causative variant that is in a high degree of LD with it. According to HapMap phase II data (International HapMap Project), 10 SNPs are tagged by rs9289231. Each one of them has the potential to be the real variant that confers genetic risk for CAD. Of the 10 SNPs, 8 are located within the KALRN exon boundaries and upstream promoter region, with 2 mapped to the adjacent CCDC14 and ROPN1 genes. Without further analysis including functional evaluation of these SNPs, we cannot rule out the possibility that the CCDC14 and ROPN1 genes are the genes driving the observed association. However, several points argue against it: (1) the KALRN gene has known functions that provide a plausible biological mechanism for its role in atherosclerosis pathogenesis, (2) KALRN and the other significant genes identified in this study—MYLK and CDGAP—share a common Rho GTPase–signaling pathway, suggesting a potential interacting pathway for CAD pathophysiology, and (3) the majority of the associated SNPs reside in the KALRN gene. Interestingly, the ROPN1 gene was initially identified as an interacting protein with the protein Rhophilin, which binds specifically to the active form of Rho GTPase.57,58 Therefore, it is also possible that both ROPN1 and KALRN contribute to the CAD susceptibility through the Rho GTPase–signaling pathway.
One of the most discussed questions in association studies is how to evaluate the significance of multiple testing of SNPs. To this end, different methods have been proposed, including Bonferroni correction, correction by false-discovery rate, and replication in a second data set. Recent data generated from the Genetic Analysis Workshop 14 found that a multiple-testing correction is usually too conservative, whereas replication of a nominal P value in a second data set is less stringent.29 On the other hand, analysis of markers in one large data set seems to be the most effective approach in identification of the true-positive signals.29 Therefore, we applied a staged design enhanced by joint analysis in the combined data sets in our peakwide association mapping. In addition, we sought to validate the most promising associations in multiple independent data sets, to filter out the false-positive association. However, it is worth noting that the strong association in the KALRN gene region would survive all of these methods for multiple-testing corrections.
Another major concern in a case-control association study is the fact that population stratification could lead to false-positive findings. The strength of our study relies on the validation of association in multiple independent data sets, including a family-based data set, which is more robust against population stratification than the case-control design. Therefore, although we cannot exclude the possibility, it is unlikely that the consistent association reported here was caused by population stratification. Constrained by the available clinical data, the cases were defined by somewhat different but well-accepted criteria for CAD between data sets. The variation in case definition could potentially diminish the evidence of association and lead to false-negative results because of heterogeneity. The fact that we observed associations in all early-onset CAD groups suggests that the genetic effect is indeed robust. Our data merit studies of additional data sets, to further validate this novel finding. Given that the strongest genetic effect was seen in the subset of patients with early-onset CAD, future association studies should focus on early-onset CAD. Since we do not have definitive evidence suggesting that rs9289231 is the causative SNP, it is important that future studies test all the significant SNPs in the KALRN gene region that are not in high LD (r2<0.7), as well as the significant SNPs in the CDGAP and MYLK genes.
The finding of multiple associated genes under one large linkage peak for a complex trait, as seen in the present study, is interesting. The genetic basis for a common complex disease like CAD is believed to be polygenic and highly heterogeneous. The observed variability of disease phenotype is likely the contribution from many genes. It is quite possible that the presence of multiple susceptibility genes in the same region makes the region more likely to be detected in the linkage study. Interestingly, despite the multiple associations identified for 3q13-21, we did not find statistical evidence suggesting that any single SNP accounts for a significant amount of linkage signal in 3q13-21. One possible explanation for this discrepancy is that the observed linkage comes from the contribution of multiple loci and, therefore, no single locus can reach statistical significance. Alternatively, it is still possible that we missed the genetic variants that are responsible for the linkage signal, because of incomplete coverage of genetic contents in the region. Future studies, including deep resequencing studies, are needed to catalog all the genetic variations in this important region that have accumulated several lines of evidence of CAD risk.
In summary, we conducted a peakwide association mapping on 3q13-21 and identified multiple candidate genes for CAD susceptibility. The association with CAD in the KALRN gene was the strongest in this analysis and was validated in multiple data sets with different study designs and disease models. The association with the KALRN gene was significant even after adjustment for most known CAD risk factors, suggesting that the genetic effect is robust and confers susceptibility beyond that confirmed by known risk factors. In addition, two genes with positive association (CDGAP and MYLK) identified in this study belong to the same biological pathway; this suggests the importance of the Rho GTPase signal–transduction pathway in the pathogenesis of CAD and merits further study.
Acknowledgments
We are grateful to all of the patients, cardiology fellows, and staff who participated in this study. We thank the Genomics Research Cores of the Center for Human Genetics at Duke University Medical Center for their tremendous technical support. This research was supported by National Institutes of Health grants P01 HL73042 (to P.J.G.-C.), R01 HL073389-01 (to E.R.H.), R01 AG021547 (to M.A.P.-V.), and R01 AG019757 (to M.A.P.-V.) and by the Duke University Department of Medicine.
Web Resources
The URLs for data presented herein are as follows:
- International HapMap Project, http://www.hapmap.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for early-onset CAD)
Reference
- 1.Murray CJ, Lopez AD (1997) Mortality by cause for eight regions of the world: Global Burden of Disease Study. Lancet 349:1269–1276 10.1016/S0140-6736(96)07493-4 [DOI] [PubMed] [Google Scholar]
- 2.Shea S, Ottman R, Gabrieli C, Stein Z, Nichols A (1984) Family history as an independent risk factor for coronary artery disease. J Am Coll Cardiol 4:793–801 [DOI] [PubMed] [Google Scholar]
- 3.Ten Kate LP, Boman H, Daiger SP, Motulsky AG (1982) Familial aggregation of coronary heart disease and its relation to known genetic risk factors. Am J Cardiol 50:945–953 10.1016/0002-9149(82)90400-3 [DOI] [PubMed] [Google Scholar]
- 4.Murabito JM, Pencina MJ, Nam BH, D’Agostino RB Sr, Wang TJ, Lloyd-Jones D, Wilson PW, O’Donnell CJ (2005) Sibling cardiovascular disease as a risk factor for cardiovascular disease in middle-aged adults. JAMA 294:3117–3123 10.1001/jama.294.24.3117 [DOI] [PubMed] [Google Scholar]
- 5.Thomas CB, Cohen BH (1955) The familial occurrence of hypertension and coronary artery disease, with observations concerning obesity and diabetes. Ann Intern Med 42:90–127 [DOI] [PubMed] [Google Scholar]
- 6.Rissanen AM (1979) Familial occurrence of coronary artery diease: effect of age at diagnosis. Am J Cardiol 44:60–66 10.1016/0002-9149(79)90251-0 [DOI] [PubMed] [Google Scholar]
- 7.Harrap SB, Zammit KS, Wong ZY, Williams FM, Bahlo M, Tonkin AM, Anderson ST (2002) Genome-wide linkage analysis of the acute coronary syndrome suggests a locus on chromosome 2. Arterioscler Thromb Vasc Biol 22:874–878 10.1161/01.ATV.0000016258.40568.F1 [DOI] [PubMed] [Google Scholar]
- 8.Broeckel U, Hengstenberg C, Mayer B, Holmer S, Martin LJ, Comuzzie AG, Blangero J, Nurnberg P, Reis A, Riegger GA, et al (2002) A comprehensive linkage analysis for myocardial infarction and its related risk factors. Nat Genet 30:210–214 10.1038/ng827 [DOI] [PubMed] [Google Scholar]
- 9.Francke S, Manraj M, Lacquemant C, Lecoeur C, Lepretre F, Passa P, Hebe A, Corset L, Yan SL, Lahmidi S, et al (2001) A genome-wide scan for coronary heart disease suggests in Indo-Mauritians a susceptibility locus on chromosome 16p13 and replicates linkage with the metabolic syndrome on 3q27. Hum Mol Genet 10:2751–2765 10.1093/hmg/10.24.2751 [DOI] [PubMed] [Google Scholar]
- 10.Pajukanta P, Cargill M, Viitanen L, Nuotio I, Kareinen A, Perola M, Terwilliger JD, Kempas E, Daly M, Lilja H, et al (2000) Two loci on chromosomes 2 and X for premature coronary heart disease identified in early- and late-settlement populations of Finland. Am J Hum Genet 67:1481–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q, Rao S, Shen G-Q, Li L, Moliterno DJ, Newby LK, Rogers WJ, Cannata R, Zirzow E, Elston RC, et al (2004) Premature myocardial infarction novel susceptibility locus on chromosome 1P34-36 identified by genomewide linkage analysis. Am J Hum Genet 74:262–271 (erratum 74:1080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hauser ER, Crossman DC, Granger CB, Haines JL, Jones CJ, Mooser V, McAdam B, Winkelmann BR, Wiseman AH, Muhlestein JB, et al (2004) A genomewide scan for early-onset coronary artery disease in 438 families: the GENECARD Study. Am J Hum Genet 75:436–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helgadottir A, Manolescu A, Thorleifsson G, Gretarsdottir S, Jonsdottir H, Thorsteinsdottir U, Samani NJ, Gudmundsson G, Grant SF, Thorgeirsson G, et al (2004) The gene encoding 5-lipoxygenase activating protein confers risk of myocardial infarction and stroke. Nat Genet 36:233–239 10.1038/ng1311 [DOI] [PubMed] [Google Scholar]
- 14.BHF Family Heart Study Research Group (2005) A genomewide linkage study of 1,933 families affected by premature coronary artery disease: the British Heart Foundation (BHF) Family Heart Study. Am J Hum Genet 77 :1011–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farrall M, Green FR, Peden JF, Olsson PG, Clarke R, Hellenius ML, Rust S, Lagercrantz J, Franzosi MG, Schulte H, et al (2006) Genome-wide mapping of susceptibility to coronary artery disease identifies a novel replicated locus on chromosome 17. PLoS Genet 2:e72 10.1371/journal.pgen.0020072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Ria M, Kelmenson PM, Eriksson P, Higgins DC, Samnegard A, Petros C, Rollins J, Bennet AM, Wiman B, et al (2005) Positional identification of TNFSF4, encoding OX40 ligand, as a gene that influences atherosclerosis susceptibility. Nat Genet 37:365–372 10.1038/ng1524 [DOI] [PubMed] [Google Scholar]
- 17.Shiffman D, Rowland CM, Louie JZ, Luke MM, Bare LA, Bolonick JI, Young BA, Catanese JJ, Stiggins CF, Pullinger CR, et al (2006) Gene variants of VAMP8 and HNRPUL1 are associated with early-onset myocardial infarction. Arterioscler Thromb Vasc Biol 26:1613–1618 10.1161/01.ATV.0000226543.77214.e4 [DOI] [PubMed] [Google Scholar]
- 18.Shah S, Kraus W, Crossman D, Granger C, Haines J, Jones C, Mooser V, Huang L, Haynes C, Dowdy E, et al (2006) Serum lipids in the GENECARD study of coronary artery disease identify quantitative trait loci and phenotypic subsets on chromosomes 3q and 5q. Ann Hum Genet 70:738–748 10.1111/j.1469-1809.2006.00288.x [DOI] [PubMed] [Google Scholar]
- 19.Bowden DW, Rudock M, Ziegler J, Lehtinen AB, Xu J, Wagenknecht LE, Herrington D, Rich SS, Freedman BI, Carr JJ, et al (2006) Coincident linkage of type 2 diabetes, metabolic syndrome, and measures of cardiovascular disease in a genome scan of the diabetes heart study. Diabetes 55:1985–1994 10.2337/db06-0003 [DOI] [PubMed] [Google Scholar]
- 20.Seo D, Wang T, Dressman H, Herderick EE, Iversen ES, Dong C, Vata K, Milano CA, Rigat F, Pittman J, et al (2004) Gene expression phenotypes of atherosclerosis. Arterioscler Thromb Vasc Biol 24:1922–1927 10.1161/01.ATV.0000141358.65242.1f [DOI] [PubMed] [Google Scholar]
- 21.Smith LR, Harrell FE Jr, Rankin JS, Califf RM, Pryor DB, Muhlbaier LH, Lee KL, Mark DB, Jones RH, Oldham HN (1991) Determinants of early versus late cardiac death in patients undergoing coronary artery bypass graft surgery. Circulation Suppl 5 84:III245–III253 [PubMed] [Google Scholar]
- 22.Felker GM, Shaw LK, O’Connor CM (2002) A standardized definition of ischemic cardiomyopathy for use in clinical research. J Am Coll Cardiol 39:210–218 10.1016/S0735-1097(01)01738-7 [DOI] [PubMed] [Google Scholar]
- 23.Hauser ER, Mooser V, Crossman DC, Haines JL, Jones CH, Winkelmann BR, Schmidt S, Scott WK, Roses AD, Pericak-Vance MA, et al (2003) Design of the genetics of early onset cardiovascular disease (GENECARD) study. Am Heart J 145:602–613 10.1067/mhj.2003.13 [DOI] [PubMed] [Google Scholar]
- 24.Teng EL, Chui HC (1987) The modified Mini-Mental State (3MS) examination. J Clin Psychiatry 48:314–318 [PubMed] [Google Scholar]
- 25.Pathobiological Determinants of Atherosclerosis in Youth (PDAY) Research Group (1993) Natural history of aortic and coronary atherosclerotic lesions in youth: findings from the PDAY Study. Arterioscler Thromb 13:1291–1298 [DOI] [PubMed] [Google Scholar]
- 26.Oliveira S, Li Y-J, Noureddine M, Züchner S, Qin X, Pericak-Vance MA, Vance JM (2005) Identification of risk and age-at-onset genes on chromosome 1p in Parkinson disease. Am J Hum Genet 77:252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dausset J, Cann H, Cohen D, Lathrop M, Lalouel JM, White R (1990) Centre d’etude du polymorphisme humanin (CEPH): collaborative genetic mapping of the human genome. Genomics 6:575–577 10.1016/0888-7543(90)90491-C [DOI] [PubMed] [Google Scholar]
- 28.Xu H, Gregory SG, Hauser ER, Stenger JE, Pericak-Vance MA, Vance JM, Zuchner S, Hauser MA (2005) SNPselector: a Web tool for selecting SNPs for genetic association studies. Bioinformatics 21:4181–4186 10.1093/bioinformatics/bti682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shephard N, John S, Cardon L, McCarthy MI, Zeggini E (2005) Will the real disease gene please stand up? BMC Genet Suppl 6:S66 10.1186/1471-2156-6-S1-S66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skol AD, Scott LJ, Abecasis GR, Boehnke M (2006) Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet 38:209–213 10.1038/ng1706 [DOI] [PubMed] [Google Scholar]
- 31.Martin ER, Bass MP, Hauser ER, Kaplan NL (2003) Accounting for linkage in family-based tests of association with missing parental genotypes. Am J Hum Genet 73:1016–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abecasis GR, Cookson WO (2000) GOLD—graphical overview of linkage disequilibrium. Bioinformatics 16:182–183 10.1093/bioinformatics/16.2.182 [DOI] [PubMed] [Google Scholar]
- 33.Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- 34.Li M, Boehnke M, Abecasis GR (2005) Joint modeling of linkage and association: identifying SNPs responsible for a linkage signal. Am J Hum Genet 76:934–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Connelly JJ, Wang T, Cox JE, Haynes C, Wang L, Shah SH, Crosslin DR, Hale AB, Nelson S, Crossman DC, et al (2006) GATA2 is associated with familial early-onset coronary artery disease. PLoS Genet 2:e139 10.1371/journal.pgen.0020139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knaus UG (2000) Rho GTPase signaling in inflammation and transformation. Immunol Res 21:103–109 10.1385/IR:21:2-3:103 [DOI] [PubMed] [Google Scholar]
- 37.Cernuda-Morollon E, Ridley AJ (2006) Rho GTPases and leukocyte adhesion receptor expression and function in endothelial cells. Circ Res 98:757–767 10.1161/01.RES.0000210579.35304.d3 [DOI] [PubMed] [Google Scholar]
- 38.Wherlock M, Mellor H (2002) The Rho GTPase family: a Racs to Wrchs story. J Cell Sci 115:239–240 [DOI] [PubMed] [Google Scholar]
- 39.Schwartz M (2004) Rho signalling at a glance. J Cell Sci 117:5457–5458 10.1242/jcs.01582 [DOI] [PubMed] [Google Scholar]
- 40.Tzima E (2006) Role of small GTPases in endothelial cytoskeletal dynamics and the shear stress response. Circ Res 98:176–185 10.1161/01.RES.0000200162.94463.d7 [DOI] [PubMed] [Google Scholar]
- 41.Shimokawa H, Takeshita A (2005) Rho-kinase is an important therapeutic target in cardiovascular medicine. Arterioscler Thromb Vasc Biol 25:1767–1775 10.1161/01.ATV.0000176193.83629.c8 [DOI] [PubMed] [Google Scholar]
- 42.Ren J, Fang CX (2005) Small guanine nucleotide-binding protein Rho and myocardial function. Acta Pharmacol Sin 26:279–285 10.1111/j.1745-7254.2005.00059.x [DOI] [PubMed] [Google Scholar]
- 43.Noma K, Oyama N, Liao JK (2006) Physiological role of ROCKs in the cardiovascular system. Am J Physiol Cell Physiol 290:C661–C668 10.1152/ajpcell.00459.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Penzes P, Johnson RC, Alam MR, Kambampati V, Mains RE, Eipper BA (2000) An isoform of kalirin, a brain-specific GDP/GTP exchange factor, is enriched in the postsynaptic density fraction. J Biol Chem 275:6395–6403 10.1074/jbc.275.9.6395 [DOI] [PubMed] [Google Scholar]
- 45.Steven R, Kubiseski TJ, Zheng H, Kulkarni S, Mancillas J, Ruiz MA, Hogue CW, Pawson T, Culotti J (1998) UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell 92:785–795 10.1016/S0092-8674(00)81406-3 [DOI] [PubMed] [Google Scholar]
- 46.Sundaresan M, Yu ZX, Ferrans VJ, Sulciner DJ, Gutkind JS, Irani K, Goldschmidt-Clermont PJ, Finkel T (1996) Regulation of reactive-oxygen-species generation in fibroblasts by Rac1. Biochem J 318:379–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Irani K, Xia Y, Zweier JL, Sollott SJ, Der CJ, Fearon ER, Sundaresan M, Finkel T, Goldschmidt-Clermont PJ (1997) Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts. Science 275:1649–1652 10.1126/science.275.5306.1649 [DOI] [PubMed] [Google Scholar]
- 48.Hordijk PL (2006) Regulation of NADPH oxidases: the role of Rac proteins. Circ Res 98:453–462 10.1161/01.RES.0000204727.46710.5e [DOI] [PubMed] [Google Scholar]
- 49.Alam MR, Johnson RC, Darlington DN, Hand TA, Mains RE, Eipper BA (1997) Kalirin, a cytosolic protein with spectrin-like and GDP/GTP exchange factor-like domains that interacts with peptidylglycine α-amidating monooxygenase, an integral membrane peptide-processing enzyme. J Biol Chem 272:12667–12675 10.1074/jbc.272.19.12667 [DOI] [PubMed] [Google Scholar]
- 50.McPherson CE, Eipper BA, Mains RE (2002) Genomic organization and differential expression of Kalirin isoforms. Gene 284:41–51 10.1016/S0378-1119(02)00386-4 [DOI] [PubMed] [Google Scholar]
- 51.McPherson CE, Eipper BA, Mains RE (2004) Kalirin expression is regulated by multiple promoters. J Mol Neurosci 22:51–62 10.1385/JMN:22:1-2:51 [DOI] [PubMed] [Google Scholar]
- 52.Rabiner CA, Mains RE, Eipper BA (2005) Kalirin: a dual Rho guanine nucleotide exchange factor that is so much more than the sum of its many parts. Neuroscientist 11:148–160 10.1177/1073858404271250 [DOI] [PubMed] [Google Scholar]
- 53.Colomer V, Engelender S, Sharp AH, Duan K, Cooper JK, Lanahan A, Lyford G, Worley P, Ross CA (1997) Huntingtin-associated protein 1 (HAP1) binds to a Trio-like polypeptide, with a rac1 guanine nucleotide exchange factor domain. Hum Mol Genet 6:1519–1525 10.1093/hmg/6.9.1519 [DOI] [PubMed] [Google Scholar]
- 54.Ratovitski EA, Bao C, Quick RA, McMillan A, Kozlovsky C, Lowenstein CJ (1999) An inducible nitric-oxide synthase (NOS)-associated protein inhibits NOS dimerization and activity. J Biol Chem 274:30250–30257 10.1074/jbc.274.42.30250 [DOI] [PubMed] [Google Scholar]
- 55.Cooke JP, Losordo DW (2002) Nitric oxide and angiogenesis. Circulation 105:2133–2135 10.1161/01.CIR.0000014928.45119.73 [DOI] [PubMed] [Google Scholar]
- 56.Ignarro LJ (2002) Nitric oxide as a unique signaling molecule in the vascular system: a historical overview. J Physiol Pharmacol 53:503–514 [PubMed] [Google Scholar]
- 57.Fujita A, Nakamura K, Kato T, Watanabe N, Ishizaki T, Kimura K, Mizoguchi A, Narumiya S (2000) Ropporin, a sperm-specific binding protein of rhophilin, that is localized in the fibrous sheath of sperm flagella. J Cell Sci 113:103–112 [DOI] [PubMed] [Google Scholar]
- 58.Watanabe G, Saito Y, Madaule P, Ishizaki T, Fujisawa K, Morii N, Mukai H, Ono Y, Kakizuka A, Narumiya S (1996) Protein kinase N (PKN) and PKN-related protein rhophilin as targets of small GTPase Rho. Science 271:645–648 10.1126/science.271.5249.645 [DOI] [PubMed] [Google Scholar]