Abstract
The duplication 17p11.2 syndrome, associated with dup(17)(p11.2p11.2), is a recently recognized syndrome of multiple congenital anomalies and mental retardation and is the first predicted reciprocal microduplication syndrome described—the homologous recombination reciprocal of the Smith-Magenis syndrome (SMS) microdeletion (del(17)(p11.2p11.2)). We previously described seven subjects with dup(17)(p11.2p11.2) and noted their relatively mild phenotype compared with that of individuals with SMS. Here, we molecularly analyzed 28 additional patients, using multiple independent assays, and also report the phenotypic characteristics obtained from extensive multidisciplinary clinical study of a subset of these patients. Whereas the majority of subjects (22 of 35) harbor the homologous recombination reciprocal product of the common SMS microdeletion (∼3.7 Mb), 13 subjects (∼37%) have nonrecurrent duplications ranging in size from 1.3 to 15.2 Mb. Molecular studies suggest potential mechanistic differences between nonrecurrent duplications and nonrecurrent genomic deletions. Clinical features observed in patients with the common dup(17)(p11.2p11.2) are distinct from those seen with SMS and include infantile hypotonia, failure to thrive, mental retardation, autistic features, sleep apnea, and structural cardiovascular anomalies. We narrow the critical region to a 1.3-Mb genomic interval that contains the dosage-sensitive RAI1 gene. Our results refine the critical region for Potocki-Lupski syndrome, provide information to assist in clinical diagnosis and management, and lend further support for the concept that genomic architecture incites genomic instability.
Nonallelic homologous recombination (NAHR) between region-specific low-copy repeats (LCRs) (also known as “segmental duplications”) is a major cause of DNA rearrangements associated with many genomic disorders.1,2 The proximal short arm of chromosome 17 is particularly rich in LCRs and is a regional locus for four genomic disorders, including Charcot-Marie-Tooth disease type 1A (CMT1A [MIM 118220]),3 hereditary neuropathies with liabilities to pressure palsies (HNPP [MIM 162500]),4 Smith-Magenis syndrome (SMS [MIM 182290]),5 and the recently recognized duplication 17p11.2 syndrome.6 The proximal and distal SMS LCRs (called “SMS-REPs”)7 mediate the common deletion and reciprocal duplication at meiosis, resulting in SMS and duplication 17p11.2 syndrome, respectively.8,9 The architectural features of the genome in this region also stimulate nonrecurrent constitutional chromosomal rearrangements that yield different-sized deletions and duplications,10,11 translocations,11 marker chromosomes,12–14 and somatic rearrangements involving 17p11.2.15
SMS is a well-characterized syndrome comprising multiple congenital anomalies and mental retardation and is associated with a heterozygous 17p11.2 deletion or point mutation of the retinoic acid inducible 1 gene (RAI1 [MIM 607642]) that maps within 17p11.2.16–22 Of the patients with SMS who harbor the deletion, ∼70%–80% have the recurrent ∼3.7-Mb common deletion mediated by NAHR with the proximal and distal SMS-REPs as recombination substrates,5,18 whereas the remainder have smaller or larger deletions apparently stimulated by other LCRs in the region.11,23
Like SMS, duplication 17p11.2 syndrome is also associated with congenital anomalies and neurodevelopmental and behavioral phenotypes, yet the clinical features of each syndrome are distinct. Cytogenetic and clinical features of persons with partial trisomy of proximal 17p have been described, mostly in isolated case reports or literature reviews, and nonspecific and noncharacterizing findings include developmental delay, mental retardation, and dysmorphic features.12,14,24–30 Many cases of 17p duplication have been defined only by routine G-banded chromosome analyses and FISH, and, in these reports, the duplicated regions are not well characterized at the molecular level. Elsewhere, we described patients with the predicted reciprocal recombination product of the common SMS deletion, dup(17)(p11.2p11.2), and we found, from the limited information available, that the phenotype was mild compared with that of individuals with del(17)(p11.2p11.2).6 Only one of the patients (patient 990) participated in a systematic clinical evaluation in the General Clinical Research Center (GCRC) at Texas Children’s Hospital, and more-detailed findings are described here. Interestingly, patient 990 and the patient described by Moog et al.29 were reported to have features of autism. Patients with duplication of only the RAI1 locus have not been described.
Here, the molecular assays of 35 subjects with dup(17)(p11.2p11.2) are reported. Of these subjects, 22 harbor a “common” duplication (∼3.7 Mb), and 13 harbor nonrecurrent duplications ranging in size from 1.3 to 15.2 Mb, as determined by multiple independent molecular assays. Phenotypic characterization of this microduplication syndrome is achieved, not only by review of the medical literature and available medical records, but also by systemic multidisciplinary clinical evaluations through a clinical protocol in the GCRC of a subset of 10 subjects, including 1 subject who harbors the smallest duplication identified to date. Apart from developmental delay, language impairment, and cognitive impairment, the most frequent clinical features in persons with dup(17)(p11.2p11.2) are hypotonia, poor feeding and failure to thrive in infancy, oral-pharyngeal dysphasia, autistic features, obstructive and central sleep apnea, structural cardiovascular abnormalities, electroencephalogram (EEG) abnormalities, and hypermetropia. Features reported in >50% of patients with the SMS deletion17,18 that either are not observed or are seen only infrequently in the duplication 17p11.2 syndrome include short stature, hearing impairment, otolaryngologic abnormalities, ophthalmic abnormalities such as myopia and iris hamartomata, genitourinary and/or renal anomalies, clinically significant scoliosis, and hypercholesterolemia. Because duplication 17p11.2 syndrome is a distinct clinical entity from its recombination reciprocal and because the cytogenetic nomenclature can be cumbersome when used to refer to affected individuals, we propose that this newly characterized microduplication syndrome be referred to by the eponym “Potocki-Lupski syndrome” (PLS).
Material and Methods
Human Subjects
Thirty-five subjects (table 1) with duplication of the proximal short arm of chromosome 17 were enrolled in a molecular protocol that was approved by the Baylor College of Medicine (BCM) Institutional Review Board (IRB). Of these subjects, 10 (age range 25 mo to 14.5 years; 6 males) also participated in a BCM IRB–approved multidisciplinary clinical study through the GCRC at the Texas Children’s Hospital. Informed consent was obtained from the participants and parents or legal guardians. All but two were ascertained through an abnormal G-banded chromosome analysis, although many subjects had at least one chromosome analysis that was interpreted as normal. The diagnosis for two recently identified patients was confirmed by a commercially available array comparative genomic hybridization (array CGH) performed after a normal G-banded chromosome analysis (patients 2543 and 2555) and subtelomeric FISH analysis (patient 2543).
Table 1. .
Summary of Cytogenetic and Molecular Analyses of Subjects[Note]
| Breakpointc |
|||||
| Subjecta | Karyotype | Duplication Type (Size in Mb) |
Mechanismb | Distal | Proximal |
| 5046,26,31 | 46,XY,dir dup(17)(pter-p11.2:p12-p11.2:p11.2-qter) | Common | P, inter | … | … |
| 52731,32 | 46,XY,dup(17)(p11.2p12) | (10.5) | M, intra | RP11-756K11 | RP11-822E23 |
| 56325,31 | 46,XX,dup(17)(p11.2p12) | (5.5) | P | RP11-726O12 | RP11-98L14 |
| 62131 | 46,XY,inv dup(17)(p13.3p11.2) | Complex | M | Outside | RP11-45M22 |
| 9906 | 46,XY,dup(17)(p11.2p11.2) | Common | P, inter | … | … |
| 10066,33 | 46,XX,del(17)(p12p12)dup(17)(p11.2p11.2) | Common | P, intra | RP11-92B11 | RP11-434D2 |
| 11926 | 46,XX,dup(17)(p11.2p11.2) | Common | P, inter | … | … |
| 1229 | 46,XY,dup(17)(p11.2p11.2) | (6.4) | P, intra | RP11-849N15 | Outside |
| 12516,34 | 46,XX,dup(17)(p11.2p11.2) | Common | P, intra | … | … |
| 13536 | 46,XY,dup(17)(p11.2p11.2) | Common | M, inter | … | … |
| 13646 | 46,XY,dup(17)(p11.2p11.2) | Common | M, intra | … | … |
| 145828 | 46,XY,dup(17)(p11.2p11.2) | (8.2) | M, intra | RP11-590H8 | Outside |
| 1529 | 46,XX,dup(17)(p11.2p11.2) | Common | … | … | … |
| 1579 | 46,XY,dup(17)(p11.2p11.2) | Common | M, inter | … | … |
| 1602 | 46,XY,dup(17)(p11.2p11.2) | Common | … | … | … |
| 1618 | 46,XY,dup(17)(p11.2p11.2) | Common | P, inter | … | … |
| 1632 | 46,XY,dup(17)(p11.2p11.2) | Common | M, inter | … | … |
| 1671 | 46,XX,dup(17)(p11.2p11.2) | Common | M, intra | … | … |
| 1786 | 46,XX,dup(17)(p11.2p11.2) | Common | P, intra | … | … |
| 1789 | 46,XX,dup(17)(p11.2p11.2) | Common | P, intra | RP11-92B11 | RP11-434D2 |
| 1838 | 46,XY,dup(17)(p11.2p11.2) | Common | P, intra | … | … |
| 186130 | 46,XX,dic(17)dup(17)(p10p11.2) | (7.1) | M, intra | RP11-998F8 | Outside |
| 1913 | 46,XX,dup(17)(p11.2p11.2) | Common | P, intra | RP11-92B11 | RP11-434D2 |
| 2153 | 46,XY,dup(17)(p11.2p12) | Complex | P, inter | RP11-92B11 | RP11-434D2 |
| 2167 | 46,XX,dup(17)(p11.2p11.2) | Common | P | RP11-92B11 | RP11-434D2 |
| 2211 | 46,XY,dup(17)(p11.2p12) | (8.2) | M, intra | RP11-590H8 | Outsidee |
| 2306 | 46,XY,dup(17)(p11.2p11.2) | Common | M, inter | … | … |
| 2337d | 46,XY,der(17).ish del(17)(p13.3)dup(17)(p11.2p12) inv(17)(p11.2p13.3) | Complex | M, intra | RP11-131K5 | RP11-311F12 |
| 2362 | 46,XY,dup(17)(p11.2p11.2) | (6.9) | … | RP11-64B12 | RP11-822E23e |
| 2414 | 46,XY,dup(17)(p11.2p11.2) | Common | … | RP11-92B11 | RP11-434D2 |
| 2440 | 46,XX,dup(17)(p11.2p11.2) | (5.0) | P, inter | RP11-385D13 | RP11-434D2 |
| 2488 | 47,XYY,dup(17)(p11.2p11.2) | (7.6) | M, intra | RP11-601N13 | RP11-822E23e |
| 2543 | 46,XY | (1.3) | M, inter | RP1-48J14 | RP11-258F1 |
| 2555 | 46,XX | Common | M, inter | RP11-92B11 | RP11-434D2 |
| 2571 | 46,XX,dup(17)(p11.2p11.2) | Common | P, inter | RP11-92B11 | RP11-434D2 |
Note.— Although initial chromosome analyses may have been normal, the result leading to referral is given. All subjects with a common ∼3.7-Mb duplication had an ∼1.1-Mb junction fragment evidenced by PFGE. No patient with a large or small duplication had this junction fragment. All subjects except 621 were duplicated by FISH analysis performed with FLI1 and/or RAI1.
Laboratory numbers are given, and citations of references are included if the subject was described elsewhere.
M = maternal; P = paternal; inter = interchromosomal; intra = intrachromosomal.
These breakpoint data are from array CGH. “Outside” indicates that the breakpoint is outside array detection.
L. E. L. M. Vissers, P. Stankiewicz, S. A. Yatsenko, E. Crawford, H. Creswick, V. K. Proud, B. B. A. de Vries, R. Pfundt, C. L. M. Marcelis, J. Zackowski, W. Bi, A. Geurts van Kessel, J. R. Lupski, and J. A. Veltman, personal communication.
The proximal breakpoint is located within the pericentromeric region as determined by FISH.
Clinical evaluations performed at the GCRC included physical examination, developmental and cognitive profiles, psychiatric history and diagnostic review, speech and language assessment, ophthalmologic and otolaryngologic examinations, audiologic assessment, swallow-function study, echocardiogram, electrocardiogram, overnight sleep study and multiple sleep latency test that included an EEG, renal ultrasound, scoliosis survey, radiographs of forearms and hands, fasting lipid profile, and thyroid function studies. Available medical records and previously reported data were reviewed for the subjects who did not participate in the multidisciplinary clinical protocol. Clinical information and/or partial molecular analyses have been reported elsewhere for subjects 504,6,26,31 527,31,32 563,25,31 621,31 990,6 1006,6,33 1192,6 1251,6,34 1353,6 1364,6 1458,28 1861,30 and 2337 (L. E. L. M. Vissers, P. Stankiewicz, S. A. Yatsenko, E. Crawford, H. Creswick, V. K. Proud, B. B. A. de Vries, R. Pfundt, C. L. M. Marcelis, J. Zackowski, W. Bi, A. Geurts van Kessel, J. R. Lupski, and J. A. Veltman, personal communication).
Cytogenetic Analysis
Peripheral blood samples for FISH were collected in green-top tubes containing sodium heparin. BAC DNA was labeled by nick translation with digoxigenin (Roche) or biotin (Invitrogen). Dual-color FISH analysis was performed on metaphase and interphase preparations of blood lymphocytes or lymphoblastoid cells as described elsewhere.35 The duplication was confirmed by interphase FISH analysis with probes specific for FLI136 and ZFP179 (ZNF179),37 each of which maps within the SMS common deletion region; for analyses of patients identified more recently, a probe specific for RAI1 was used.38 The probe specific for the peripheral myelin protein 22 gene39 (PMP22 [MIM 601097]), mapping within the commonly duplicated CMT1A region, was the control. Dual-color FISH by use of SMS-REP flanking clones as probes11 was applied to patients 2555 and 2571 to confirm their common duplications. For determination of proximal breakpoints, 17 α-satellite centromeric probe (D17Z1) (Oncor) and BAC RP11-22N2, which maps to 17q11.1, were directly labeled as probes.
Pulsed-Field Gel Electrophoresis (PFGE) Analyses
PFGE was performed on subject samples as described elsewhere.5,6 In brief, high-molecular-weight genomic DNA was digested with the rare cutter restriction endonuclease NotI, was separated by PFGE, and, after Southern blotting, was probed with a 1.1-kb DNA fragment corresponding to CLP1, which maps within both proximal and distal copies of SMS-REPs. The duplication is considered to be common if the two breakpoints map within the proximal and distal SMS-REPs, and it is distinguished from uncommon duplication by a unique de novo ∼1.1-Mb NotI band, corresponding to the rearrangement-specific common junction fragment.
Array CGH
A custom human 17p array was developed with the total of 83 human BAC/PAC clones from 17p, tiling an 11-Mb region around RAI1 that spans the genomic interval from BAC clone RP11-462C21 in 17p13.1 to BAC clone RP11-1109M24 in 17p11.1 and includes 10 representative clones in 17p13. Sixteen control clones were from chromosomes other than chromosome 17. One PAC clone, RP5-836L9, was included in our array but was later excluded from our analysis after invalidation by FISH analysis. Clones CTD-457L16 and RP11-160E2 are in a region with copy-number variations (CNVs).40 Clones were printed in quadruplicate. Array printing and hybridization were performed as described elsewhere.41 Reference genomic DNA was prepared from a sex-matched normal individual. Each patient was examined twice with dye reversal, and the final result was a normalized combination of the two hybridization experiments. The details for data processing were described elsewhere.10
Genotyping
We determined the parental origin of the duplicated chromosomes and recombination mechanisms, using a combination of microsatellite haplotype reconstruction and the segregation of marker genotypes on genomic DNA, as described elsewhere.6 Size and relative intensities of the peaks were calculated with GeneMapper (v.3.7) software (Applied Biosystems). Phases of parental haplotypes were determined on the bases of the most parsimonious explanation for observed genotypes in the patient and under the assumption of no recombination.
Results
Proximal 17p Duplications
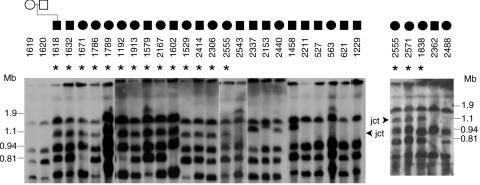
The common duplication in chromosome 17p11.2 is an ∼3.7-Mb interstitial duplication that is mediated by the distal and proximal SMS-REPs and is the reciprocal of the common deletion observed in SMS. A de novo recombination-specific junction fragment of ∼1.1 Mb was detected previously by PFGE in seven patients with the common duplication (patients 504, 990, 1006, 1192, 1251, 1353, and 1364).6 This ∼1.1-Mb junction fragment was also detected in 15 of 28 new subjects (fig. 1). The junction fragment was not observed in the parental samples and thus is deemed to occur de novo in all subjects. Thirteen subjects had no junction fragment or had a junction fragment smaller or larger than 1.1 Mb (fig. 1), indicating an unusual-sized duplication. Thus, 22 (∼63%) of the 35 subjects with duplication 17p11.2 harbor a common recurrent duplication (table 1).
Figure 1. .
Identification of the common duplication and differentiation from unusual-sized duplications by PFGE analysis. An ∼1.1-Mb novel junction (jct) fragment was predicted for the common duplication after the genomic DNA was digested with NotI, was separated by PFGE, and was probed with a genomic fragment from the CLP gene. An asterisk (*) indicates the subjects who harbor this junction fragment. Junction fragment data from subject 1192 was reported elsewhere.6 Subject 2555 is shown in both gel segments. The ∼1.1-Mb junction fragment was not observed in the parents, which is consistent with a de novo event. A junction fragment was not observed in 13 of the 35 subjects, indicating that those 13 have unusual-sized duplications.
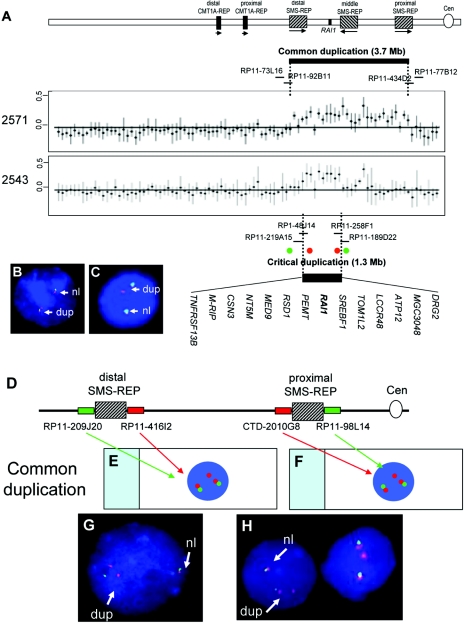
A custom array CGH identified effectively four distinct genomic disorders in proximal 17p.10 To define the duplication breakpoints in subjects with PLS, we extended the previous array to cover almost the entire short arm of chromosome 17, spanning the genomic regions from 17p13 to the centromere, such that the array tiled the genomic regions that correspond to band 17p12 and subbands 17p11.2 and 17p11.1. Array CGH was performed on samples from seven patients with a common duplication (patients 1006,10 1789,10 1913,10 2167, 2414, 2555, and 2571), and the results indicated that each patient harbors a common duplication spanning the genomic region from the BAC clone RP11-92B11, mapping in the distal SMS-REP, to BAC clone RP11-434D2, mapping in the proximal SMS-REP, which independently confirms that the recombination-specific junction fragment identified by PFGE is an indicator of a common duplication (table 1, fig. 2, and data not shown).
Figure 2. .
A, Array CGH analysis of patients 2571 and 2543. A combined result from two dye-swap experiments was presented with normalized log2(Cy3/Cy5) ratios of patient versus control for each individual clone plotted on the Y-axis and represented by dots with SD. Patient 2571 has the common duplication spanning a region from the distal to the proximal SMS-REP. Patient 2543 harbors a duplicated genomic region smaller in size than the common recurrent dup(17)(p11.2p11.2). The size of the duplicated genomic interval is ∼1.3 Mb, spanning from the distal SMS-REP to a site telomeric to the middle SMS-REP. Since patient 2543 exhibits the key phenotypic features of PLS, this ∼1.3-Mb duplicated interval represents the critical region. The 14 genes, including RAI1, contained within this critical interval are listed. The regions of the critical duplication and the common duplication are indicated by thick horizontal lines. The BACs in the vicinity of the breakpoints are indicated by short black lines. Note that one clone, CTD-457L16, located in a CNV region between the middle and the proximal SMS-REPs showed increased copy number. B and C, FISH analyses of interphase nuclei, which detected the breakpoints in subject 2543. BAC probes used in FISH analysis are indicated by red or green circles. In panel A, the probes are differentially labeled to give a green signal for RP11-209J20 and a red signal for RP11-416I2. In panel B, the presence of three red signals (RP11-416I2) and two green signals (RP11-209J20) indicate that the distal breakpoint maps within the distal SMS-REP. In panel C, the presence of three red signals (RP11-258F1) and two green signals (RP11-189D22) indicate that the proximal breakpoint maps distal to the middle SMS-REP. D–H, Common dup(17)(p11.2p11.2) detected by a FISH assay. D, Schematic representation of a dual-color interphase FISH assay developed to screen for common versus unusual duplications. A map of chromosome 17p11.2 with the placement of FISH probes is above. The SMS-REP–flanking clones are differentially labeled and detected with green and red dyes, respectively. The presence of three red signals indicates that the breakpoints map between RP11-209J20 and RP11-416I2 (E) and between CTD-2010G8 and RP11-98L14 (F). G and H, FISH analysis of interphase nuclei of patient 2555 to confirm the common duplication. The presence of three red signals (RP11-416I2 and CTD-2010G8) and two green signals (RP11-209J20 and RP11-98L14) indicates that the breakpoints map within the distal (G) and proximal SMS-REPs (H). Cen = centromere; dup = duplicated chromosome 17; nl = normal chromosome 17.
We previously developed a rapid and reliable FISH assay to distinguish common from nonrecurrent SMS deletions with dual-color interphase FISH with the use of SMS-REP–flanking clones as probes,11,18 which can also distinguish common from nonrecurrent duplications. The distal SMS-REP–flanking BAC clones RP11-416I2 and RP11-209J20 and the proximal SMS-REP–flanking clones RP11-98L14 and CTD-2010G8 were used concurrently in FISH on interphase lymphocytes. We applied this novel FISH test to the analysis of patients with proximal 17p duplications. As predicted, in the common duplications, three signals were observed for the BAC probes RP11-416I2 and CTD-2010G8 within the SMS common deletion interval, but only two signals were observed for the probe telomeric to the distal SMS-REP (RP11-209J20) and the probe centromeric to the proximal SMS-REP (RP11-98L14) (fig. 2D–2H).
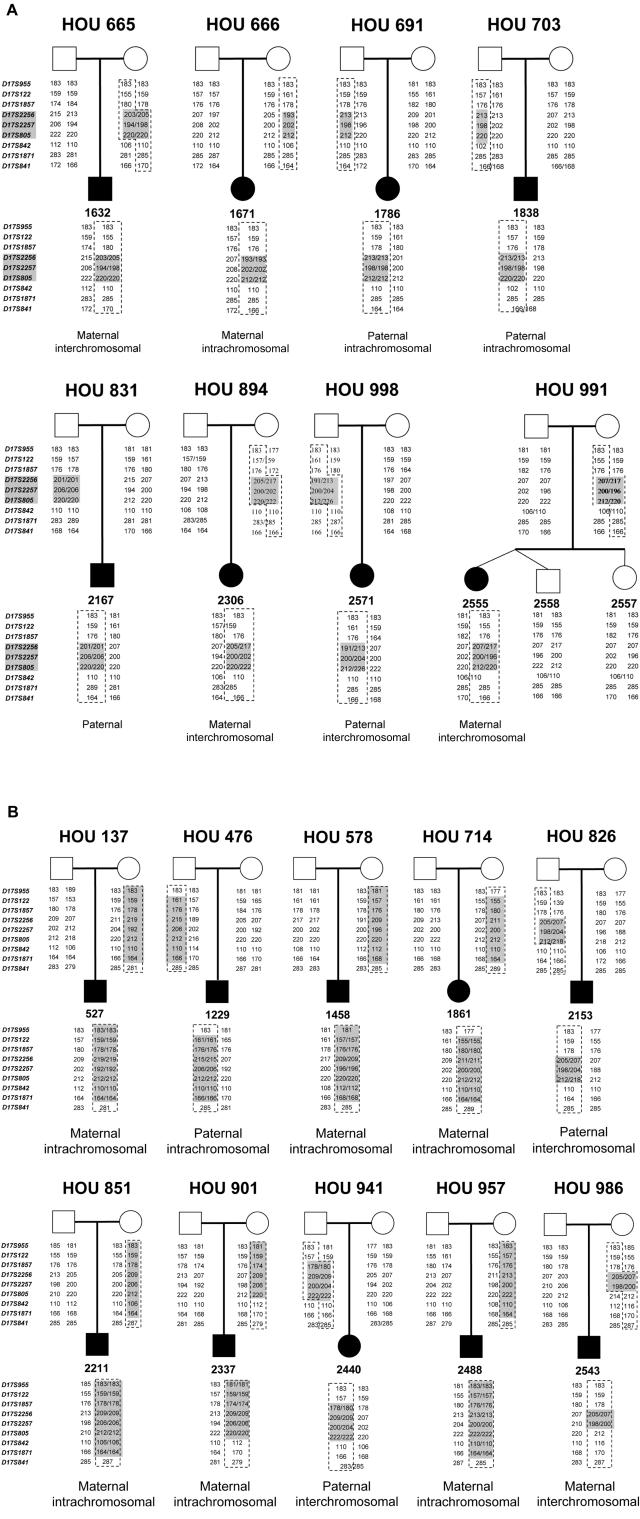
Our previous studies showed that recombination resulting in common recurrent deletions in 17p11.25,42 and uncommon nonrecurrent deletions11 showed no apparent bias in parental origin and that these occurred via both inter- and intrachromosomal mechanisms. To investigate the genetic recombination mechanism for proximal 17p duplication, we reconstructed the haplotypes of eight patients with common duplication dup17p11.2, using 10 microsatellite markers, and we analyzed the segregation of marker genotypes (table 1 and fig. 3). These data are consistent with our previous results for 11 families42 and further confirm that there is no substantial bias for parental origin of the duplication and that the crossover occurs via interchromosomal and intrachromosomal mechanisms at approximately equal frequencies.
Figure 3. .
Pedigrees and haplotypes of 18 Houston families (HOU). Patient numbers are given below the blackened symbols representing the affected children in the pedigree. A, Eight families with common recurrent proximal 17p duplications. B, Ten families with uncommon nonrecurrent proximal 17p duplications. To the left of each pedigree is a list of microsatellite markers used for genotyping. The sizes of each allele are written below each family member. The dotted lines outline alleles inherited by the patients from the parent of origin.
Uncommon Duplications and Their Breakpoints
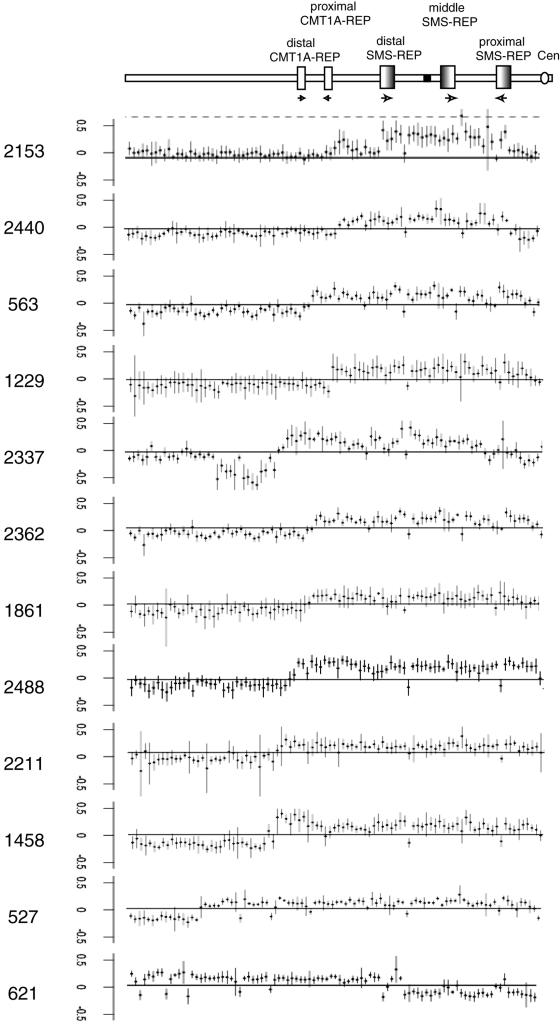
In our cohort, 13 subjects (37%) had 17p duplications that were not mediated by the distal and proximal SMS-REPs and are thus classified as uncommon duplications. Not surprisingly, there is imperfect correlation between G-banded chromosome analyses and breakpoint analyses (table 1). None of these subjects, however, showed the ∼1.1-Mb junction fragment on PFGE (table 1 and fig. 1). The custom array CGH proved to be the most informative and rapid method for determining the breakpoints in the uncommon duplications. The array CGH revealed that the centromeric breakpoints group in the pericentromeric region, with some mapping to a remaining sequencing gap close to the centromere, and it provided evidence that narrowed the critical region for PLS.
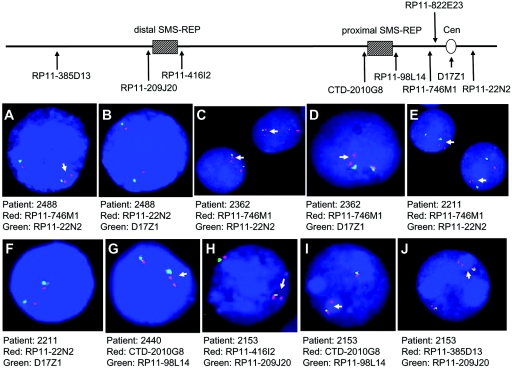
BAC microarray analyses have been previously performed for three patients with duplication (patients 563, 1229, and 1861) on a custom array for proximal 17p.10 Although clones covering the pericentromeric region were lacking, these subjects were found to have proximal breakpoints centromeric to the LCR17pG copy.10 To further refine the proximal breakpoints of the subjects with uncommon duplications, we applied array CGH analysis, using our new extended microarray, and FISH analysis of a subset of subjects (figs. 4 and 5). Analysis by array CGH determined that 3 of the 13 subjects (patients 621, 2153, and 2337) have more than a simple duplication of a single region within 17p and thus are classified as having complex rearrangements (table 1 and fig. 6). Of the 10 subjects with uncommon duplications with simple rather than complex rearrangement, 3 (patients 527, 2362, and 2488) have a proximal breakpoint that maps within the two overlapping BACs in the pericentromeric region, RP11-822E23 and RP11-1109E24. Interestingly, this 17p breakpoint grouping is at the gap at 21.5 Mb that is among the nine euchromatic gaps in chromosome 17.43 It is one of the two gaps that are apparently refractory to cloning and are associated with primate-specific breaks in conserved synteny.44 Four subjects (1229, 1458, 1861, and 2211) have a proximal breakpoint that maps proximal to the BAC RP11-1109E24, indicating that their proximal breakpoints are in the pericentromeric region (2211) (fig. 4) or in the centromere (186130) (fig. 6). Thus, 7 of 10 simple duplication events have breakpoints within the pericentromeric or centromeric region and may have been stimulated by pericentromeric structures. Three simple duplications have a proximal breakpoint distant from the centromere. The breakpoint for subject 2440 mapped within the proximal SMS-REP, which was confirmed by FISH analysis (fig. 4). For subject 563, the centromeric breakpoint is located within the BAC RP11-98L14 that contains a 23-kb genomic fragment homologous to the SMS-REPs, whereas subject 2543 has a proximal breakpoint between the distal and middle SMS-REPs, where no LCR has been identified to date.
Figure 4. .
FISH analyses of interphase nuclei for mapping of breakpoints. Above is a map of proximal 17p, with the locations of FISH probes indicated by arrows. A–F, Centromeric breakpoints in patients 2488 (A and B), 2362 (C and D), and 2211 (E and F) mapped within the pericentromeric region. The probes used are listed below interphase nuclei. The presence of one signal in addition to the normal two signals indicates duplication in that region. Duplication was observed for BAC probe RP11-746M1 in 17p11.2. FISH using centromeric probe D17Z1 and BAC probe RP11-22N2 in 17q11.1 indicated no duplication in either the centromere or 17q. G, FISH with the proximal SMS-REP flanking clones CTD-2010G8 (red) and RP11-98L14 (green), mapping the proximal breakpoint in patient 2440 in the proximal SMS-REP. H–J, Two separate duplications in proximal 17p in patient 2153, confirmed by FISH. Duplicated chromosome 17 is indicated by arrows. Cen = centromere.
Figure 5. .
Determination of duplicated regions by array CGH analysis. A diagram of proximal 17p and important architectural features are shown at the top with the centromere (white circle), SMS-REPs (gradient boxes), and CMT1A-REPs (white boxes) marked. Horizontal arrows depict orientation of the LCRs. Normalized log2(Cy3/Cy5) ratios of patient versus control for each individual clone are plotted on the Y-axis. The patients are arranged by size of duplication.
Figure 6. .
Summary of breakpoint mapping in 13 patients with uncommon duplications of 17p11.2. Top, Schematic representation of the short arm of chromosome 17 with LCRs depicted. The clones used in the array analysis are depicted just below the diagram of the LCRs and are ordered from the telomere (left) to the centromere (right). The positions of some individual large insert clones are indicated by dashed lines. Below the clones are 13 horizontal bars labeled with individual patient numbers. Blue depicts regions with normal dosage, red represents duplicated genomic segments, and green represents deletion. The sizes of the duplicated or deleted segments are labeled just above each subject bar. The asterisk (*) adjacent to patients 621, 2153, and 2337 indicates that these subjects harbor a complex chromosomal rearrangement including the proximal 17p duplication. Cen = centromere.
Although the proximal breakpoints in the uncommon duplications group near the pericentromeric region, the distal breakpoints in the 10 uncommon simple duplications did not cluster into groups (table 1 and fig. 6). Two of the distal breakpoints map within LCR-containing BAC clones; subject 2543 had a breakpoint in the distal SMS-REP, and the distal breakpoint of subject 2440 is in RP11-385D13, the BAC that resides within LCR17pA. This large 383-kb LCR is the progenitor of many repeats in 17p,43,44 encompasses the breakpoints of the evolutionary translocation t(4;19) in Gorilla gorilla,44,45 and contains breakpoints for several different chromosome aberrations, including the uncommon but recurrent deletions defined in a subset of patients with SMS.23 No LCRs or other higher-order sequence structures were identified in the other seven distal breakpoints.
We sought to determine the parental origin of the duplication in the uncommon duplications and analyzed 10 families with nonrecurrent duplication (fig. 3). Parental origin information was previously published for two additional patients (patients 563(pat) and 621(mat)).31 In total, eight duplications are maternal in origin, whereas four are paternal. Seven resulted from intrachromosomal recombination, and three from interchromosomal crossover. Interestingly, in each of the six patients studied (527, 1229, 1458, 1861, 2211, and 2488), the uncommon duplications with a proximal breakpoint close to the centromere resulted from intrachromosomal recombinations, five of which are maternal in origin.
Subjects Who Harbor an Uncommon Duplication and Other Rearrangements Involving 17p
Three of the 13 subjects with uncommon duplications were found to have a complex chromosomal rearrangement (table 1 and fig. 6). Subject 621 has a 15.2-Mb duplication that does not encompass RAI1 (fig. 6), as well as a paracentric inversion of 17p11.2p13.3.31 As observed in array CGH analysis and confirmed by FISH analysis, subject 2153 has two duplicated segments—the common ∼3.7-Mb duplication and a smaller duplication involving two overlapping BAC clones (RP11-385D13 and RP11-640I15) that are ∼1 Mb telomeric from the distal SMS-REP (figs. 4, 5, and 6). Subject 2337 has a complex rearrangement including a 5.9-Mb duplication of 17p11.2, a 1.9-Mb deletion of 17p12 that is ∼400 kb distal to the duplicated region (fig. 6), and an inversion. Extensive additional molecular analyses and breakpoint mapping of the complex chromosomal rearrangements found in patient 2337 will be reported elsewhere (L. E. L. M. Vissers, P. Stankiewicz, S. A. Yatsenko, E. Crawford, H. Creswick, V. K. Proud, B. B. A. de Vries, R. Pfundt, C. L. M. Marcelis, J. Zackowski, W. Bi, A. Geurts van Kessel, J. R. Lupski, and J. A. Veltman, personal communication).
Distinctive Phenotype of Subjects Who Harbor the Common Duplication
The 22 subjects determined to have the common 17p11.2 duplication represent a molecularly defined cohort of patients with a gain in copy number for a discreet genomic segment. The facial features in most patients were not strikingly dysmorphic, yet there were shared similar characteristics (fig. 7A–7J). The patient described by Kozma et al.26 and subject 1913 described here have more-pronounced facial dysmorphisms. Short stature (height <2nd percentile) was observed in five of the seven subjects described by Potocki et al.6; however, only one (patient 1913) of the seven patients with the common duplication in this cohort had short stature for her age. At age 13 years, this patient measured at the 50th percentile for a 7.5-year-old female. Results of growth-hormone-stimulation assays performed before her admission to GCRC were normal.
Figure 7. .
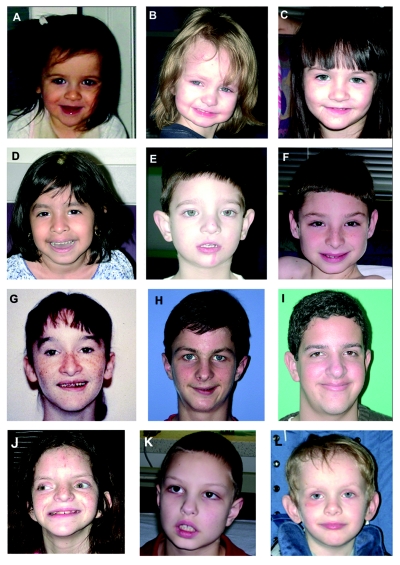
Facial features of PLS. A–J, Patients with the common duplication. A, Patient 2555, at age 22 mo. B, Patient 2306, at age 33 mo. C, Patient 2167, at age 3 years 3 mo. D, Patient 1671, at age 3 years 9 mo. E, Patient 1579, at age 4 years 2 mo. F, Patient 2414, at age 9 years 6 mo. G, Patient 1006, at age 14 years 2 mo. H, Patient 1618, at age 14 years 6 mo. I, Patient 990, at age 16 years 1 mo. J, Patient 1913, at age 13 years. Shared features include a broad forehead, gentle down-slant of the palpebral fissures, and relatively long nasal tip. Younger patients have a triangular face with prominence to the angle of the jaw and micrognathia. A more oval-shaped face and larger chin is seen in older individuals. Patients 1579 (E) and 1913 (J) were microcephalic and hyperteloric. Patient 1913 has more pronounced facial dysmorphisms, including down-slanting palpebral fissures, low-set and posteriorly rotated ears, and broad mouth. An interesting feature shared by most patients is an asymmetric smile that is seen in multiple photographs taken of each patient in the GCRC and in photographs shared by their parents. K, Patient 2543, who harbors a small (∼1.3-Mb) duplication, at age 8 years and 1 mo. L, Patient 2211, who harbors a large (8.2-Mb) duplication, at age 4 years and 10 mo. Physical features seen in these patients are very similar to those of patients with the common duplication.
The key clinical features of PLS are summarized in table 2. A more detailed history was available for the subjects who participated in the multidisciplinary clinical protocol. The history ascertained for all subjects included infantile hypotonia, poor feeding, and developmental delay. A formal diagnosis of failure to thrive was given to five of seven patients with the common duplication, and three of these patients were fed by gastrostomy tube (table 2). Gestational history was also significant in this group. Both failure to progress in labor and subsequent cesarean section were reported for four of the seven subjects. Interestingly, three patients had an abnormal triple analyte screen that suggested an increased risk of Down syndrome. Prenatal chromosome analyses were performed on two subjects, and results were interpreted as normal for both. Four of the seven subjects were small for gestational age. In the non-GCRC group, birth weight was provided for 11 patients, and 5 of them were small for gestational age.
Table 2. .
Key Clinical Features of PLS[Note]
| Patient |
|||||||||||
| Trait | 990 | 1579 | 1618 | 1913 | 2167 | 2306 | 2414 | 2211 | 2440 | 2543 | Non-GCRC Subjectsa |
| Sex | M | M | M | F | F | F | M | M | F | M | |
| Age | 9 y 10 mo | 4 y 2 mo | 14.5 y | 13 y | 3 y 3 mo | 2 y 9 mo | 9.5 y | 4 y 10 mo | 2 y 1mo | 8 y 1 mo | |
| Duplicationb | C | C | C | C | C | C | C | L | L | S | C |
| Obstetrical and birth history: | |||||||||||
| Birth weight low for gestational age | − | + | − | − | + | + | + | + | − | − | 5/11 |
| Failure to progress and/or cesarean section | + | + | − | − | + | − | + | − | + | − | 2/8 |
| Medical and developmental history: | |||||||||||
| Poor feeding as infantc | + | + | + | + | + | + | + | + | + | + | 8/9 |
| Gastroesophageal refluxc | + | − | + | + | + | − | + | + | + | + | 3/5 |
| Failure to thrive in infancy or early childhoodc | − | + | + | + | + | + | − | + | + | + | 5/8 |
| Gastrostomy tube | − | + | + | + | − | − | − | + | + | − | 1/8 |
| Hypotonia as infantc | + | + | + | + | + | + | + | + | + | + | 9/11 |
| Developmental delayc | + | + | + | + | + | + | + | + | + | + | 14/14 |
| Epilepsy | − | − | − | − | − | − | − | − | − | − | 1/10 |
| Subjective sleep disturbance | − | − | − | − | − | − | − | + | − | − | 4/6 |
| Short stature | − | − | − | +d | − | − | − | − | − | − | 4/11 |
| Neuropsychiatric evaluation: | |||||||||||
| Cognitive impairmentc | + | + | + | + | + | + | + | + | + | + | 14/14 |
| Low adaptive functionc | + | + | + | + | + | + | + | + | + | + | 5/5 |
| Autistic featuresc | + | − | + | + | + | + | + | + | + | + | 1/2 |
| Speech and language evaluation: | |||||||||||
| Language impairmentc | + | + | + | + | + | + | + | + | + | + | 9/10 |
| Articulation difficultiesc | + | + | + | + | + | + | + | + | + | + | 8/9 |
| Oral-pharyngeal dysphasiac,e | + | + | + | + | + | − | + | + | + | 1/1 | |
| Central and/or obstructive sleep apneac | + | + | + | + | + | + | − | + | + | ||
| EEG abnormalityc | + | + | + | + | + | + | + | + | − | 3/5 | |
| Epileptiform abnormalities on EEG | + | − | + | + | − | − | + | + | − | 0/3 | |
| Hypermetropia seen on ophthalmic examinationc | + | + | + | + | + | − | + | + | + | − | 1/5 |
| Hearing impairment | −f | − | − | −f | − | − | − | − | − | − | 1/6 |
| CNS abnormality by MRI | − | +g,h | +g,i | +g,h | +g,j | +g,j | − | +g,i | +g,k | − | 0/4 |
| Cardiovascular abnormality | − | − | + | + | + | + | + | − | −l | 1/2 | |
| Structural renal anomaly | − | − | − | − | − | − | − | + | − | − | 1/3 |
| Scoliosis >10 degrees | − | − | − | − | − | − | + | − | + | + | 1/3 |
| Low total cholesterol and low LDL | − | + | − | + | + | − | − | − | − | − | |
Note.— Plus sign (+) = feature present; minus sign (−) = feature not present.
Data for non-GCRC subjects is given as number with trait/number tested. Male:female ratio is 6:9. Age range is 17 mo to 24 years.
Duplication size: C = common; L = large; S = small.
Feature observed in 70%–100% of subjects with the common duplication evaluated at GCRC.
Not growth-hormone deficient.
Swallow-function study with a speech pathologist present.
Mild sensitivity loss at 4,000 Hz.
Magnetic resonance imaging (MRI) not performed at GCRC.
Microcephaly.
Mild attenuation of corpus callosum.
Mild delay in myelination.
Possible Rathke’s cleft remnant and prominence of left semicircular canal and vestibule.
Borderline prolonged QT interval.
Developmental delay, cognitive impairment, and communication disorders were present in all subjects (tables 2 and 3). Among the GCRC common-duplication group, the milestone of walking was attained, on average, at age 31 mo (range 18 mo to 5 years). Speech delay, absent speech, and other speech and language abnormalities, such as immediate and delayed echolalia, were observed in all subjects. Expressive and receptive language impairment was clearly evident when assessed objectively by standard means (Oral and Written Language Scales and Preschool Language Scale–4th Edition). All GCRC subjects demonstrated articulation dysfunction, and five demonstrated difficulties with motor planning and/or sequencing sounds within words that indicate verbal apraxia. Three GCRC subjects used augmentative communication, such as manual signing and the picture-exchange communication system.
Table 3. .
Neuropsychiatric and Communicative Profile of PLS[Note]
| Patient |
|||||||||
| Trait | 990 | 1579 | 1618 | 1913 | 2167 | 2306 | 2414 | Mean Standard Score (Proportion Affected)a | Patient 2543 |
| Sex | M | M | M | F | F | F | M | M | |
| Age | 9 y 10 mo | 4 y 2 mo | 14 y 6 mo | 13 y | 3 y 3 mo | 2 y 9 mo | 9 y 6 mo | 8 y 1 mo | |
| Duplication | C | C | C | C | C | C | C | C | S |
| Communication skills: | |||||||||
| Receptive language | 74 | 65 | 40 | 40 | 51 | 85 | 40 | 56 | 50 |
| Expressive language | 73 | 50 | 50 | b | 61 | 78 | 40 | 59 | 50 |
| Total language | 72 | 50 | 43 | b | 51 | 80 | 40 | 56 | 50 |
| Articulation difficulties | + | + | + | + | + | + | + | (7/7) | + |
| Augmentative communication | − | + | − | + | − | − | + | (3/7) | + |
| Cognitive assessment IQ/DQc: | |||||||||
| Global | 65 | 49 | 40 | 36 | 49 | 69 | 36 | 49 | 42 |
| Verbal | 78 | 46 | 46 | 36 | 37 | 82 | 38 | 52 | |
| Nonverbal | 68 | 62 | 46 | 38 | 46 | 63 | 36 | 51 | |
| Adaptive function: | |||||||||
| Behavior composited | 51 | 39 | 30 | 42 | 73 | 27 | 41 | 28 | |
| Daily livingd | 52 | 50 | 27 | 40 | 67 | 34 | 45 | 20 | |
| Socializationd | 36 | 32 | 39 | 37 | 72 | 26 | 41 | 43 | |
| Behavioral features consistent with autism: | |||||||||
| Decreased eye contact | + | − | − | − | − | + | + | (3/7) | + |
| Repetitive motor mannerisms | + | − | + | − | + | + | − | (4/7) | + |
| Echolalia | + | − | + | + | + | + | + | (6/7) | + |
Note.— Plus sign (+) = feature present; minus sign (−) = feature not present.
Mean values for seven patients (990, 1579, 1618, 1913, 2167, 2306, and 2414).
The complete formal language assessment not performed on this subject at GCRC. She was nearly nonverbal and used augmentative communication.
Global, verbal, and nonverbal IQs and developmental quotients (DQs) were obtained using the Stanford-Binet Intelligence Scale–4th Edition46 and the Mullen Scales of Early Learning,47 respectively.
Vineland Adaptive Behavior Scales.48
Cognitive assessments by Stanford-Binet Intelligence Scale46 or the Mullens Scales of Early Learning47 revealed global intelligence quotients (IQs) within the mild (IQ level 50–55 to ∼70) to moderate (IQ 35–40 to 50–55) range of mental retardation, although two patients with the common duplication scored within the borderline (IQ 70–79) or low-average (IQ 80–89) range in verbal subsets (table 3). Adaptive functioning was uniformly low in all but one subject (2306) across all domains, measured by standardized score <20–69 on the Vineland Adaptive Behavior Scales.48
Regarding behavior, the most frequent history from the non-GCRC group was attention deficit and hyperactivity disorder. These reports differ from the findings for psychometric testing and psychiatric assessment of the subjects assessed at the GCRC, for which most patients demonstrated features of the autistic spectrum disorders (tables 2 and 3). Only one subject (1579) did not exhibit any autistic characteristics on examination or have a history of them. Autistic features gathered by history or established on examination included decreased eye contact (3 of 7 patients), motor mannerisms or posturing (4 of 7), sensory hypersensitivities or preoccupations (5 of 7), repetitive behaviors or preoccupations (5 of 7), lack of appropriate functional or symbolic play (3 of 7), and lack of joint attention (3 of 7). Patient 2414 was evaluated with an Autism Diagnostic Interview–Revised (ADI-R) and Autism Diagnostic Observation Schedule–Generic (ADOS-G), each of which was positive for autism.
Several clinical evaluations were performed as part of the GCRC protocol. Many studies were either not performed and/or were not reported for other molecularly characterized patients. No parents (of the GCRC cohort) reported disturbed sleep in their children, and no patient exhibited substantial signs of airway obstruction by otolaryngologic examination. Thus, the abnormalities on overnight sleep study were unexpected in this cohort, revealing sleep-disordered breathing characterized by obstructive sleep apnea, central sleep apnea, and significant oxygen desaturation and hypercarbia. In addition, the EEG findings were abnormal for all subjects with the common duplication. Six had a slow occipital dominant rhythm (“alpha”), and four had generalized and/or focal epileptiform abnormalities (spikes, sharp waves, and spike and slow-wave discharges); no EEG seizure discharges or clinical seizures were recorded. No subject received anti-seizure medication or had a history of seizures. Abnormalities of the circadian rhythm of melatonin have been established in persons with deletion 17p11.2 (SMS), yet we did not find this to be a feature in one patient with duplication 17p11.2.49
Although organ developmental abnormalities were not recognized as major features of individuals with duplication 17p11.2,6,24,26,27,29,34 through systematic evaluation of the GCRC patients, we found that more than half showed structural cardiovascular anomalies when assessed by echocardiography (table 2). Patient 1913 had a large secundum type atrial septal defect with left-to-right shunting and moderately dilated aortic root; patient 2167 evidenced a small secundum atrial septal defect with left-to-right shunting; patient 2306 manifested bicommissural aortic valve and patent foramen ovale; and patient 2414 had a mildly dilated pulmonary annulus. Balarin et al.28 described a patient with a large duplication 17p11.2 and Alport syndrome, yet the only abnormality seen on renal ultrasound in the GCRC subjects was poor corticomedullar differentiation in one patient (1913). Renal ultrasound was normal in all other subjects. Urinalysis and measures of blood urea nitrogen and creatinine were normal in all subjects studied in the GCRC. Few patients evaluated in the GCRC manifested mild scoliosis (<10 degrees) (2 of 7 subjects), mildly low total cholesterol and low-density lipoproteins (LDL) (3 of 7 subjects), and mildly low thyroxin with normal levels of thyroid-stimulating hormone (2 of 7 subjects).
An Uncommon Duplication That Narrows the Critical Region to a 1.3-Mb Interval That Includes RAI1
The size of duplication in the 13 patients with uncommon nonrecurrent duplication studied ranged from 1.3 Mb to >15.2 Mb. The smallest duplication (in subject 2543) was beyond the limits of resolution of G-band analysis and was detected by array CGH. We determined the breakpoints of the duplicated segment in subject 2543 to be at BACs RP11-92B11 and RP11-258F1 (table 1 and fig. 2A). The telomeric breakpoint maps within the distal SMS-REP, and the centromeric breakpoint is distal to the middle SMS-REP. FISH analysis confirmed the distal breakpoint by use of SMS-REP-flanking BAC clones RP11-209J20 and RP11-416I2 as probes (fig. 2B) and confirmed the proximal breakpoint by use of clones RP11-258F1 and RP11-189D22 as probes (fig. 2C). This duplicated segment contains 14 genes, including RAI1, the major contributing gene for the reciprocal deletion causing SMS.
Genotype-Phenotype Correlation in Patients with Proximal 17p Duplication
The cardinal features of PLS have now been defined by multidisciplinary clinical analysis of a cohort of seven patients with the common duplication and have been supported by review of medical records of subjects with common duplication who did not participate in the clinic protocol (table 2). Variability in the phenotype is observed despite a common genomic imbalance in these subjects. However, it is expected that persons harboring large duplications that encompass the CMT1A region within 17p12 will have a more severe phenotype, including peripheral neuropathy.29,31,32,50 Of the subjects with uncommon duplications, patients 2543, 2440, and 2211 were evaluated at the GCRC (table 2 and figs. 2 and 6).
Patient 2211 (fig. 7L), whose duplication encompasses PMP22, has features typical of PLS, including hypermetropia, speech and language impairment, autistic spectrum disorder, sleep apnea, oral-pharyngeal dysphagia, and EEG abnormalities that include epileptiform changes, such as multifocal spike and wave discharges. Mild scoliosis (<9 degrees) was also seen and was not unexpected, given his peripheral neuropathy. Features of this patient that have not been observed in persons with the common duplication include severe peripheral neuropathy, syrinx of the spinal canal, renal anomaly (malrotation of the left kidney), and severe and hemodynamically significant cardiovascular disease. Whereas a moderate aortic root dilatation and bicommissural aortic valve were observed in subjects with the common duplication (patients 1913 and 2306, respectively), patient 2211 had a massively dilated aortic root and bicommissural aortic valve requiring surgical intervention.
Clinical assessments in conjunction with molecular analyses of subjects with smaller duplications can enable definition of the critical region for PLS. Subject 2543 (fig. 7K), who has the smallest duplication (1.3 Mb) reported to date, was evaluated at the GCRC at age 8 years and 1 mo; his key clinical features are listed in table 2. As an infant, he displayed hypotonia, failure to thrive, poor sucking, vomiting, and recurrent respiratory illnesses. Although a gastrostomy tube was avoided, a prolonged period (nearly 12 h per d) was required to complete his feeding. Developmental milestones were delayed; he sat at age 9 mo, crawled at age 13 mo, and walked at age 20 mo. He was toilet trained at age 6–7 years and began reading single words at age 7 years. He was given a formal diagnosis of autism at age 3 years, was still nonverbal at age 8 years, and uses an augmentative communication device. Chromosome analysis was normal in this patient, yet a diagnosis of autism and mental retardation prompted further analysis with array CGH, thus revealing a duplication involving 17p11.2. Clinical evaluation at the GCRC revealed normal ophthalmic, otolaryngologic, and audiolgic examinations and normal studies of lipid levels and thyroid function. Scoliosis survey revealed a 14-degree curvature of the thoracolumbar spine that was not clinically apparent. Echocardiogram was normal, although an electrocardiogram revealed a borderline-prolonged QT interval (a measure of the time between the start of the Q wave and the end of the T wave in the heart's electrical cycle). The 24-h sleep study was significant because of multiple nocturnal awakenings and mild obstructive sleep apnea associated with oxygen desaturation. Results of the EEG performed concurrently with the sleep study were normal. As observed in persons with the common duplication, subject 2543 exhibited impairment in language, cognition, and adaptive function. The previous diagnosis of autism was confirmed by testing using ADI-R and ADOS-G.
Among this cohort, patient 62131 is the only one who has a large duplication outside the critical region and has a normal dosage of the RAI1 gene. Dysmorphic features, failure to thrive, mental retardation, and CMT1A neuropathy were reported. However, many of the key clinical features of PLS, including autistic features, EEG abnormalities, hypermetropia, and sleep apnea, were not evaluated for this patient; thus, further genotype-phenotype correlation is not possible.
Discussion
Autistic Spectrum Disorder, a Feature of PLS
PLS (dup(17)(p11.2p11.2)) is clinically recognizable and is associated with hypotonia and failure to thrive in infancy and early childhood, developmental delay, mental retardation, severe communication disorder, autistic spectrum disorder, sleep disordered breathing, EEG abnormalities, cardiovascular anomalies, and hypermetropia. We have defined this syndrome on the basis of a shared molecular structural abnormality6 and, by systematic analysis of a larger cohort of subjects, have elucidated further the clinical and behavioral phenotype of this microduplication syndrome. Initially, we hypothesized that patients with duplication 17p11.2 did not come to medical attention because of the milder phenotype.6 However, our findings now reveal that these patients may have substantial medical illness as well as neurobehavioral abnormalities that, except for the developmental delay, may go unrecognized until later infancy or childhood. Most patients likely elude an etiological diagnosis because of the limitations of the current conventional cytogenetic analyses.
In the absence of rearrangement-type–specific selection, the prevalence of dup(17)(p11.2p11.2) should be equal to that of its recombination reciprocal del(17)(p11.12p11.2). However, because of the subtle appearance of the duplicated G-light band, and since interphase FISH is required to resolve visually the duplicated region, many patients escape ascertainment by conventional cytogenetic methods. The availability of array CGH in clinical diagnostic laboratories has revolutionized clinical genetics, because it enables high-resolution genome analysis with highly sensitive and specific detection of deletion or duplication of targeted regions of the human genome. The array CGH assays that are clinically implemented comprise a targeted array of clones with which unbalanced rearrangements of the telomeres and regions associated with genomic disorders can be resolved.41,51,52 We expect that the recognized prevalence of the duplication 17p11.2 syndrome will increase as this method becomes more widely used by physicians.
Another essential factor in the recognition of this disorder is the appreciation of the associated behavioral characteristics.6 Our objective clinical assessment of the duplication 17p11.2 syndrome clearly indicates features of autistic spectrum disorder in the vast majority of patients. Linkage studies document evidence of a locus that confers susceptibility to attention deficit hyperactivity disorder in 17p11.2.53,54 There have been no linkage or association studies reporting a susceptibility locus for autism in 17p11.2, but two studies suggest such a locus in 17q11.2.55,56 We should note that genomic duplications can result in the misinterpretation of marker genotypes because of the triallelic nature of a duplicated locus compared with the normal biallelic locus.50,57,58
Until these clinical features are widely recognized, most patients with dup(17)(p11.2p11.2) will likely receive a diagnosis by a laboratory test before anyone suspects a specific syndrome. Nonetheless, the phenotypic delineation presented here will allow appropriate medical management, clinical care, and anticipatory guidance for persons with PLS. For example, because autistic spectrum disorder is common to PLS, patients should undergo diagnostic analysis specifically for autism (ADI-R and ADOS-G), and, if criteria are met, specific therapy, such as targeted social communication treatment, should be considered.59 Even in the absence of a formal diagnosis of autism, considering the significant communication impairment in persons with PLS, specific speech and language interventions and augmentative communication can be integrated into their education and language-development programs. Abnormalities noted during the sleep studies may have substantial consequences for daytime cognition and behavior; thus, overnight sleep studies and clinical assessments should be considered as part of the evaluation of persons with PLS. Although no subject whom we evaluated had clinically recognized seizures, four had potentially epileptiform abnormalities of generalized and focal character on EEG and may be at an increased risk of future epilepsy. Thus, any medications that decrease the seizure threshold should be prescribed with caution. Although still considered preliminary, our findings also suggest that recommendations for clinical care of persons with PLS should include echocardiography with evaluation of the aortic root. Other evaluations, such as audiologic testing, ophthalmic examination, and routine studies of blood and urine chemistry, tend to be standard recommendations for most patients evaluated for cognitive impairment and behavioral abnormalities.
The Critical Region for PLS
Whereas both SMS due to deletion 17p11.2 and PLS due to duplication 17p11.2 share the common recombination mechanism, our data show that the clinical features are different and, at times, are even divergent between these two genomic disorders. For example, myopia is common in SMS, whereas hypermetropia is common in PLS, and hyperlipidemia is common in SMS, whereas low cholesterol is a feature of PLS. Thus, CNV resulting from deletions and duplications can cause a clinical phenotype, but the manifestation can differ for genomic losses and genomic gains. Although >50 genes map to the commonly deleted SMS interval, only RAI1 has been shown to be mutated in patients with nondeletion SMS.19,20,60,61 Even though haploinsufficiency of RAI1 is necessary and sufficient to cause many of the clinical features of SMS, the full phenotypic spectrum of SMS is evident only in persons harboring the chromosomal deletion.18,62 Since no patient has been identified with a duplication involving RAI1 alone, it remains to be determined whether RAI1 is the major dosage-sensitive gene of consequence in PLS. A single subject (2543) narrows the critical region for PLS to a 1.3-Mb interval spanning from the distal SMS-REP to a site <200 kb telomeric from the middle SMS-REP. This individual exhibits the clinical and behavioral phenotype of PLS, including autistic spectrum disorder. Interestingly, this patient also lacks two frequent features found in persons with the common duplication—namely, hypermetropia and EEG abnormalities. Thus, one can speculate that dosage abnormalities of RAI1 or other genes within this region are not responsible for these findings. Several more patients need to be evaluated to enable appreciation of the clinical variability in persons with identical molecular aberrations.18
It is challenging to determine the dosage-sensitive gene(s) responsible for PLS, the dup(17)(p11.2p11.2) syndrome. Whereas the dosage-sensitive gene for deletion syndromes can often be determined by the identification of rare patients with loss-of-function (e.g., nonsense or frameshift allele) point mutations consistent with haploinsufficiency, duplication syndromes provide greater challenges. Among the PLS candidates, RAI1 may be the culprit, since it represents a dosage-sensitive gene that has been shown to be responsible for the majority of SMS features. Animal models are useful adjuncts to clinical analyses because they allow targeted investigation and controlled observation. Human chromosome 17p11.2 is syntenic to the 32–34 cM region of murine chromosome 11. CNV of RAI1, including decreased or increased dosage, causes distinct neurobehavioral and craniofacial consequences in both humans and mice, indicating that a critical window of gene expression exists for RAI1.63 Interestingly, the deletion and duplication animals recapitulate some of the physical and neurobehavioral features seen in patients.64,65 We have shown recently that both physical and behavioral features observed in Dp(11)17/+ mice can be rescued by restoring the normal Rai1 gene dosage in Dp(11)17/Rai1− compound heterozygous animals.63 Further analyses may help to provide insights that could be applicable to clinical management and therapy.
Stimulation of Chromosome 17p Duplication by Genomic Architecture
Consistent with the finding that the majority of patients with SMS have a common deletion, the common recurrent duplication of proximal 17p also predominates. These observations further support the concept that directly orientated highly homologous LCRs represent a major factor conferring susceptibility to dosage imbalance of genomic material by serving as substrates for NAHR. Uncommon recurrent deletions ∼5 Mb in size, by use of alternative LCRs in proximal 17p as recombination substrates, have been reported in six patients.23 However, none of the uncommon 17p duplications share breakpoints with the uncommon recurrent deletions. Identification of the predicted reciprocal duplication of the uncommon recurrent deletion in proximal 17p requires investigation of additional patients with duplication.
Many nonrecurrent deletion breakpoints in the proximal 17p occurred within LCRs.11 In this study, three proximal and two distal nonrecurrent duplication breakpoints mapped within BACs that contain LCRs. In addition, seven proximal duplication junctions grouped at or near the pericentromeric region. Therefore, genome architecture likely stimulates both nonrecurrent deletions and duplications in proximal 17p.
A wide spectrum of structural genomic aberrations has been reported in proximal 17p, likely because of both enrichment of LCRs in this highly unstable region and ascertainment secondary to the presence of genes (e.g., RAI1 and PMP22) in which copy-number alteration conveys clinical phenotypes.66 Breakpoints in nonrecurrent translocations and marker chromosomes have been frequently mapped at the centromere.11–14 By array CGH, we mapped duplication breakpoints in the 13 subjects with uncommon duplications. Remarkably, 7 of 10 simple duplications have proximal breakpoints mapping to the centromere or the pericentromeric region. In contrast, none of the uncommon deletion breakpoints (n=18) are located within or close to the centromere. This difference is statistically significant (P=.002) by Fisher’s exact test. Interestingly, three of the breakpoints in this 17p breakpoint grouping are at the gap at 21.5 Mb that is among the nine euchromatic gaps in chromosome 17.43 It is one of only two gaps that are apparently refractory to cloning and are associated with primate-specific breaks in conserved synteny. This gap at 21.5 Mb is flanked by AT-rich genomic sequence; fosmid XXfos-82794E5 mapping distal to the gap has a 57% AT content, and XXfos-82794E5 mapping proximal to the gap has a 66% AT content. Regions of several hundred kilobases flanking this gap are enriched with both interchromosomal and intrachromosomal segmental duplications according to the Segmental Duplication Database. The complete sequence of BAC RP11-822E23 shares 92%–96% homology with 10–100-kb segments from chromosomes 2, 4, 7, 8, and 17. Further genome architectural complexity includes a 30-kb fragment in this BAC that was found in an inverted orientation with 97% homology to BAC RP11-1109M24, a clone that maps to the proximal side of the gap that is ∼450 kb from the centromeric heterochromatin. Two structural variations (variations _0497 and _0498) are present in the BAC proximally adjacent to RP11-1109M24 (Database of Genomic Variants Human Genome Assembly build 36). A recent CNV map of the human genome40 showed that the chromosome 17p pericentromeric region is a highly variable CNV region. This 17p pericentromeric region also appears to manifest genomic instability in somatic cells.67 In addition, whereas, for the uncommon duplications with proximal breakpoints close to the centromere, five of six cases studied resulted from maternal intrachromosomal recombination, 12 of the 14 uncommon deletions were of paternal origin.11
Given these data, we propose that the mechanism responsible for proximal 17p duplication may be different from that involved in 17p deletion. An instability factor for duplication occurrence appears to be related to the specific structure and organization at or near the centromere. Pericentromeric regions provide boundaries between the heterochromatic centromeric alpha satellite and the unique euchromatic gene-containing sequences. In the short arm of chromosome 17, the pericentromeric region shows extensive zones of segmental duplication ranging from 500 kb to 5.5 Mb.68 Our data indicate that LCRs near the centromere stimulate nonrecurrent duplications. Pericentromeric and centromeric regions are also the frequent breakpoints for translocations11 and marker chromosomes12–14 involving proximal 17p. Several breakpoints of translocations were also located within LCRs,11 indicating that segmental duplications may play a role in these genomic rearrangements as well as during evolution. Our breakpoint analysis of proximal 17p duplications provide further evidence to support the concept that genomic architecture plays an important role in genome instability.
Grouping of duplication breakpoints in specific regions has also been observed recently in a study of the duplication rearrangements associated with Pelizaeus-Merzbacher disease, a genomic disorder most commonly caused by genomic duplications of the proteolipid protein gene (PLP1 [MIM 300401]).69 PLP1 duplications are different in size and do not cluster to a specific set of LCRs. Breakpoint mapping in patients with multiple duplication showed that most of the breakpoints appear to be in proximity to segmental duplications.70 Nucleotide sequence of recombinant junction reveals that both duplications71 and deletions72 involving PLP1 appear to occur by nonhomologous end joining and not by NAHR. The PLP1 duplication studies and our breakpoint analyses of dup17p11.2 suggest that genomic structure stimulates but does not mediate these nonrecurrent duplication events.
In summary, we phenotypically characterized PLS (dup(17)(p11.2p11.2) syndrome), narrowed the critical region to a 1.3-Mb interval, further documented LCR involvement in duplication rearrangements, again demonstrated NAHR as the predominant recombination mechanism, provided evidence in support of the hypothesis that RAI1 is the dosage-sensitive gene causing this syndrome, and objectively defined a dosage-sensitive locus conferring an autism phenotype that maps within the critical interval for PLS.
Acknowledgments
We thank the patients and their families, for their willing and productive participation; Marianna Hörz, for coordinating the many clinical studies; and Joanne Williams and the nursing staff at the GCRC who provided care, for each patient’s admission to the Texas Children’s Hospital. We acknowledge the generous referral from the many physicians and genetic professionals who shared both information and subjects for these studies. R.A.L. is a Senior Scientific Investigator of Research to Prevent Blindness, New York, which provided unrestricted funding for part of this protocol. This work was supported in part by National Institute of Child Health and Human Development grants P01 HD39420 (to J.R.L.) and K08 HD01149 (to L.P.), Baylor College of Medicine Mental Retardation Research Center grant HD2406407, and Texas Children’s Hospital GCRC grant M01RR00188.
Web Resources
The URLs for data presented herein are as follows:
- Database of Genomic Variants, http://projects.tcag.ca/variation/ (for Human Genome Assembly build 36)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CMT1A, HNPP, SMS, RAI1, PMP22, and PLP1)
- Segmental Duplication Database, http://humanparalogy.gs.washington.edu/
References
- 1.Lupski JR (1998) Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 14:417–422 10.1016/S0168-9525(98)01555-8 [DOI] [PubMed] [Google Scholar]
- 2.Stankiewicz P, Lupski JR (2002) Genome architecture, rearrangements and genomic disorders. Trends Genet 18:74–82 10.1016/S0168-9525(02)02592-1 [DOI] [PubMed] [Google Scholar]
- 3.Pentao L, Wise CA, Chinault AC, Patel PI, Lupski JR (1992) Charcot-Marie-Tooth type 1A duplication appears to arise from recombination at repeat sequences flanking the 1.5 Mb monomer unit. Nat Genet 2:292–300 10.1038/ng1292-292 [DOI] [PubMed] [Google Scholar]
- 4.Reiter LT, Murakami T, Koeuth T, Pentao L, Muzny DM, Gibbs RA, Lupski JR (1996) A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nat Genet 12:288–297 10.1038/ng0396-288 [DOI] [PubMed] [Google Scholar]
- 5.Chen K-S, Manian P, Koeuth T, Potocki L, Zhao Q, Chinault AC, Lee CC, Lupski JR (1997) Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nat Genet 17:154–163 10.1038/ng1097-154 [DOI] [PubMed] [Google Scholar]
- 6.Potocki L, Chen K-S, Park S-S, Osterholm DE, Withers MA, Kimonis V, Summers AM, Meschino WS, Anyane-Yeboa K, Kashork CD, et al (2000) Molecular mechanism for duplication 17p11.2—the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat Genet 24:84–87 10.1038/71743 [DOI] [PubMed] [Google Scholar]
- 7.Park S-S, Stankiewicz P, Bi W, Shaw C, Lehoczky J, Dewar K, Birren B, Lupski JR (2002) Structure and evolution of the Smith-Magenis syndrome repeat gene clusters, SMS-REPs. Genome Res 12:729–738 10.1101/gr.82802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw CJ, Bi W, Lupski JR (2002) Genetic proof of unequal meiotic crossovers in reciprocal deletion and duplication of 17p11.2. Am J Hum Genet 71:1072–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bi W, Park S-S, Shaw CJ, Withers MA, Patel PI, Lupski JR (2003) Reciprocal crossovers and a positional preference for strand exchange in recombination events resulting in deletion or duplication of chromosome 17p11.2. Am J Hum Genet 73:1302–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shaw CJ, Shaw CA, Yu W, Stankiewicz P, White LD, Beaudet AL, Lupski JR (2004) Comparative genomic hybridisation using a proximal 17p BAC/PAC array detects rearrangements responsible for four genomic disorders. J Med Genet 41:113–119 10.1136/jmg.2003.012831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stankiewicz P, Shaw CJ, Dapper JD, Wakui K, Shaffer LG, Withers M, Elizondo L, Park S-S, Lupski JR (2003) Genome architecture catalyzes nonrecurrent chromosomal rearrangements. Am J Hum Genet 72:1101–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stankiewicz P, Parka S-S, Holder SE, Waters CS, Palmer RW, Berend SA, Shaffer LG, Potocki L, Lupski JR (2001) Trisomy 17p10-p12 resulting from a supernumerary marker chromosome derived from chromosome 17: molecular analysis and delineation of the phenotype. Clin Genet 60:336–344 10.1034/j.1399-0004.2001.600503.x [DOI] [PubMed] [Google Scholar]
- 13.Shaw CJ, Stankiewicz P, Bien-Willner G, Bello SC, Shaw CA, Carrera M, Perez Jurado L, Estivill X, Lupski JR (2004) Small marker chromosomes in two patients with segmental aneusomy for proximal 17p. Hum Genet 115:1–7 10.1007/s00439-004-1119-5 [DOI] [PubMed] [Google Scholar]
- 14.Yatsenko SA, Treadwell-Deering D, Krull K, Lewis RA, Glaze D, Stankiewicz P, Lupski JR, Potocki L (2005) Trisomy 17p10-p12 due to mosaic supernumerary marker chromosome: delineation of molecular breakpoints and clinical phenotype, and comparison to other proximal 17p segmental duplications. Am J Med Genet A 138:175–180 [DOI] [PubMed] [Google Scholar]
- 15.Barbouti A, Stankiewicz P, Nusbaum C, Cuomo C, Cook A, Hoglund M, Johansson B, Hagemeijer A, Park S-S, Mitelman F, et al (2004) The breakpoint region of the most common isochromosome, i(17q), in human neoplasia is characterized by a complex genomic architecture with large, palindromic, low-copy repeats. Am J Hum Genet 74:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenberg F, Guzzetta V, Montes de Oca-Luna R, Magenis RE, Smith AC, Richter SF, Kondo I, Dobyns WB, Patel PI, Lupski JR (1991) Molecular analysis of the Smith-Magenis syndrome: a possible contiguous-gene syndrome associated with del(17)(p11.2). Am J Hum Genet 49:1207–1218 [PMC free article] [PubMed] [Google Scholar]
- 17.Greenberg F, Lewis RA, Potocki L, Glaze D, Parke J, Killian J, Murphy MA, Williamson D, Brown F, Dutton R, et al (1996) Multi-disciplinary clinical study of Smith-Magenis syndrome (deletion 17p11.2). Am J Med Genet 62:247–254 [DOI] [PubMed] [Google Scholar]
- 18.Potocki L, Shaw CJ, Stankiewicz P, Lupski JR (2003) Variability in clinical phenotype despite common chromosomal deletion in Smith-Magenis syndrome [del(17)(p11.2p11.2)]. Genet Med 5:430–434 [DOI] [PubMed] [Google Scholar]
- 19.Slager RE, Newton TL, Vlangos CN, Finucane B, Elsea SE (2003) Mutations in RAI1 associated with Smith-Magenis syndrome. Nat Genet 33:466–468 10.1038/ng1126 [DOI] [PubMed] [Google Scholar]
- 20.Bi W, Saifi GM, Shaw CJ, Walz K, Fonseca P, Wilson M, Potocki L, Lupski JR (2004) Mutations of RAI1, a PHD-containing protein, in nondeletion patients with Smith-Magenis syndrome. Hum Genet 115:515–524 10.1007/s00439-004-1187-6 [DOI] [PubMed] [Google Scholar]
- 21.Madduri N, Peters SU, Voigt RG, Llorente AM, Lupski JR, Potocki L (2006) Cognitive and adaptive behavior profiles in Smith-Magenis syndrome. J Dev Behav Pediatr 27:188–192 10.1097/00004703-200606000-00002 [DOI] [PubMed] [Google Scholar]
- 22.Goldman AM, Potocki L, Walz K, Lynch JK, Glaze DG, Lupski JR, Noebels JL (2006) Epilepsy and chromosomal rearrangements in Smith-Magenis syndrome [del(17)(p11.2p11.2)]. J Child Neurol 21:93–98 [DOI] [PubMed] [Google Scholar]
- 23.Shaw CJ, Withers MA, Lupski JR (2004) Uncommon deletions of the Smith-Magenis syndrome region can be recurrent when alternate low-copy repeats act as homologous recombination substrates. Am J Hum Genet 75:75–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Docherty Z, Hulten MA, Honeyman MM (1983) De novo tandem duplication 17p11→cen. J Med Genet 20:138–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magenis RE, Brown MG, Allen L, Reiss J (1986) De novo partial duplication of 17p [dup(17)(p12→p11.2)]: clinical report. Am J Med Genet 24:415–420 10.1002/ajmg.1320240304 [DOI] [PubMed] [Google Scholar]
- 26.Kozma C, Meck JM, Loomis KJ, Galindo HC (1991) De novo duplication of 17p [dup(17)(p12→p11.2)]: report of an additional case with confirmation of the cytogenetic, phenotypic, and developmental aspects. Am J Med Genet 41:446–450 10.1002/ajmg.1320410413 [DOI] [PubMed] [Google Scholar]
- 27.Brown A, Phelan MC, Patil S, Crawford E, Rogers RC, Schwartz C (1996) Two patients with duplication of 17p11.2: the reciprocal of the Smith-Magenis syndrome deletion? Am J Med Genet 63:373–377 [DOI] [PubMed] [Google Scholar]
- 28.Balarin MA, da Silva Lopes VL, Varella-Garcia M (1999) A dup(17)(p11.2p11.2) detected by fluorescence in situ hybridization in a boy with Alport syndrome. Am J Med Genet 82:183–186 [DOI] [PubMed] [Google Scholar]
- 29.Moog U, Engelen JJ, Weber BW, Van Gelderen M, Steyaert J, Baas F, Sijstermans HM, Fryns JP (2004) Hereditary motor and sensory neuropathy (HMSN) IA, developmental delay and autism related disorder in a boy with duplication (17)(p11.2p12). Genet Couns 15:73–80 [PubMed] [Google Scholar]
- 30.Shaw CJ, Stankiewicz P, Christodoulou J, Smith E, Jones K, Lupski JR (2004) A girl with duplication 17p10-p12 associated with a dicentric chromosome. Am J Med Genet A 124:173–178 10.1002/ajmg.a.20355 [DOI] [PubMed] [Google Scholar]
- 31.Roa BB, Greenberg F, Gunaratne P, Sauer CM, Lubinsky MS, Kozma C, Meck JM, Magenis RE, Shaffer LG, Lupski JR (1996) Duplication of the PMP22 gene in 17p partial trisomy patients with Charcot-Marie-Tooth type-1A neuropathy. Hum Genet 97:642–649 [PubMed] [Google Scholar]
- 32.Lupski JR, Wise CA, Kuwano A, Pentao L, Parke JT, Glaze DG, Ledbetter DH, Greenberg F, Patel PI (1992) Gene dosage is a mechanism for Charcot-Marie-Tooth disease type 1A. Nat Genet 1:29–33 10.1038/ng0492-29 [DOI] [PubMed] [Google Scholar]
- 33.Potocki L, Chen K-S, Koeuth T, Killian J, Iannaccone ST, Shapira SK, Kashork CD, Spikes AS, Shaffer LG, Lupski JR (1999) DNA rearrangements on both homologues of chromosome 17 in a mildly delayed individual with a family history of autosomal dominant carpal tunnel syndrome. Am J Hum Genet 64:471–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider MC, Hughes CR, Forrester S, Kimonis V (2000) Mild phenotype due to tandem duplication of 17p11.2. Am J Med Genet 94:296–299 [DOI] [PubMed] [Google Scholar]
- 35.Shaffer LG, Kennedy GM, Spikes AS, Lupski JR (1997) Diagnosis of CMT1A duplications and HNPP deletions by interphase FISH: implications for testing in the cytogenetics laboratory. Am J Med Genet 69:325–331 [DOI] [PubMed] [Google Scholar]
- 36.Chen K-S, Gunaratne PH, Hoheisel JD, Young IG, Miklos GL, Greenberg F, Shaffer LG, Campbell HD, Lupski JR (1995) The human homologue of the Drosophila melanogaster flightless-I gene (flil) maps within the Smith-Magenis microdeletion critical region in 17p11.2. Am J Hum Genet 56:175–182 [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao Q, Chen K-S, Bejjani BA, Lupski JR (1998) Cloning, genomic structure, and expression of mouse ring finger protein gene Znf179. Genomics 49:394–400 10.1006/geno.1998.5285 [DOI] [PubMed] [Google Scholar]
- 38.Vlangos CN, Wilson M, Blancato J, Smith AC, Elsea SH (2005) Diagnostic FISH probes for del(17)(p11.2p11.2) associated with Smith-Magenis syndrome should contain the RAI1 gene. Am J Med Genet A 132:278–282 [DOI] [PubMed] [Google Scholar]
- 39.Patel PI, Roa BB, Welcher AA, Schoener-Scott R, Trask BJ, Pentao L, Snipes GJ, Garcia CA, Francke U, Shooter EM, et al (1992) The gene for the peripheral myelin protein PMP-22 is a candidate for Charcot-Marie-Tooth disease type 1A. Nat Genet 1:159–165 10.1038/ng0692-159 [DOI] [PubMed] [Google Scholar]
- 40.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al (2006) Global variation in copy number in the human genome. Nature 444:444–454 10.1038/nature05329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheung SW, Shaw CA, Yu W, Li J, Ou Z, Patel A, Yatsenko SA, Cooper ML, Furman P, Stankiewicz P, et al (2005) Development and validation of a CGH microarray for clinical cytogenetic diagnosis. Genet Med 7:422–432 [DOI] [PubMed] [Google Scholar]
- 42.Shaw CJ, Bi W, Lupski JR (2002) Genetic proof of unequal meiotic crossovers in reciprocal deletion and duplication of 17p11.2. Am J Hum Genet 71:1072–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zody MC, Garber M, Adams DJ, Sharpe T, Harrow J, Lupski JR, Nicholson C, Searle SM, Wilming L, Young SK, et al (2006) DNA sequence of human chromosome 17 and analysis of rearrangement in the human lineage. Nature 440:1045–1049 10.1038/nature04689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stankiewicz P, Shaw CJ, Withers M, Inoue K, Lupski JR (2004) Serial segmental duplications during primate evolution result in complex human genome architecture. Genome Res 14:2209–2220 10.1101/gr.2746604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stankiewicz P, Park S-S, Inoue K, Lupski JR (2001) The evolutionary chromosome translocation 4;19 in Gorilla gorilla is associated with microduplication of the chromosome fragment syntenic to sequences surrounding the human proximal CMT1A-REP. Genome Res 11:1205–1210 10.1101/gr.181101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thorndike RL, Hagen EP, Sattler JM (1986) The Stanford-Binet Intelligence Scale, 4th edition: guide for administering and scoring. Riverside Publishing, Itasca, IL [Google Scholar]
- 47.Mullen E (1995) Mullen Scales of Early Llearning. American Guidance Services, Circle Pines, MN [Google Scholar]
- 48.Sparrow SS, Balla DA, Cicchetti DV (1984) Vineland Adaptive Behavior Scales, interview edition, survey form manual. American Guidance Services, Circle Pines, MN [Google Scholar]
- 49.Potocki L, Glaze D, Tan D-X, Park S-S, Kashork CD, Shaffer LG, Reiter RJ, Lupski JR (2000) Circadian rhythm abnormalities of melatonin in Smith-Magenis syndrome. J Med Genet 37:428–433 10.1136/jmg.37.6.428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lupski JR, de Oca-Luna RM, Slaugenhaupt S, Pentao L, Guzzetta V, Trask BJ, Saucedo-Cardenas O, Barker DF, Killian JM, Garcia CA, et al (1991) DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell 66:219–232 10.1016/0092-8674(91)90613-4 [DOI] [PubMed] [Google Scholar]
- 51.Trilochan S, Cheung SW, Ward P, Darilek S, Patel A, Del Gaudio D, Kang SH, Lalani S, Li J, McAdoo S, et al (2006) Prenatal diagnosis of chromosomal abnormalities using array-based comparative genomic hybridization. Genet Med 8:719–727 [DOI] [PubMed] [Google Scholar]
- 52.Shaffer LG, Kashork CD, Saleki R, Rorem E, Sundin K, Ballif BC, Bejjani BA (2006) Targeted genomic microarray analysis for identification of chromosome abnormalities in 1500 consecutive clinical cases. J Pediatr 149:98–102 10.1016/j.jpeds.2006.02.006 [DOI] [PubMed] [Google Scholar]
- 53.Arcos-Burgos M, Castellanos FX, Pineda D, Lopera F, Palacio JD, Palacio LG, Rapoport JL, Berg K, Bailey-Wilson JE, Muenke M (2004) Attention-deficit/hyperactivity disorder in a population isolate: linkage to loci at 4q13.2, 5q33.3, 11q22, and 17p11. Am J Hum Genet 75:998–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ogdie MN, Fisher SE, Yang M, Ishii J, Francks C, Loo SK, Cantor RM, McCracken JT, McGough JJ, Smalley SL, et al (2004) Attention deficit hyperactivity disorder: fine mapping supports linkage to 5p13, 6q12, 16p13, and 17p11. Am J Hum Genet 75:661–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bartlett CW, Goedken R, Vieland VJ (2005) Effects of updating linkage evidence across subsets of data: reanalysis of the autism genetic resource exchange data set. Am J Hum Genet 76:688–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sutcliffe JS, Delahanty RJ, Prasad HC, McCauley JL, Han Q, Jiang L, Li C, Folstein SE, Blakely RD (2005) Allelic heterogeneity at the serotonin transporter locus (SLC6A4) confers susceptibility to autism and rigid-compulsive behaviors. Am J Hum Genet 77:265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matise TC, Chakravarti A, Patel PI, Lupski JR, Nelis E, Timmerman V, Van Broeckhoven C, Weeks DE (1994) Detection of tandem duplications and implications for linkage analysis. Am J Hum Genet 54:1110–1121 [PMC free article] [PubMed] [Google Scholar]
- 58.Lupski JR (2003) 2002 Curt Stern Award address: genomic disorders recombination-based disease resulting from genomic architecture. Am J Hum Genet 72:246–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aldred C, Green J, Adams C (2004) A new social communication intervention for children with autism: pilot randomised controlled treatment study suggesting effectiveness. J Child Psychol Psychiatry 45:1420–1430 10.1111/j.1469-7610.2004.00338.x [DOI] [PubMed] [Google Scholar]
- 60.Girirajan S, Elsas LJ 2nd, Devriendt K, Elsea SH (2005) RAI1 variations in Smith-Magenis syndrome patients without 17p11.2 deletions. J Med Genet 42:820–828 10.1136/jmg.2005.031211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bi W, Saifi GM, Girirajan S, Xin S, Szomju BB, Firth H, Magenis E, Potocki L, Elsea SH, Lupski JR (2006) RAI1 point mutations, CAG repeat variations, and SNP analysis in nondeletion Smith-Magenis syndrome. Am J Med Genet A 140:2454–2463 [DOI] [PubMed] [Google Scholar]
- 62.Girirajan S, Vlangos CN, Szomju BB, Edelman E, Trevors CD, Dupuis L, Nezarati M, Bunyan DJ, Elsea SH (2006) Genotype-phenotype correlation in Smith-Magenis syndrome: evidence that multiple genes in 17p11.2 contribute to the clinical spectrum. Genet Med 8:417–427 [DOI] [PubMed] [Google Scholar]
- 63.Walz K, Paylor R, Yan J, Bi W, Lupski JR (2006) Rai1 duplication causes physical and behavioral phenotypes in a mouse model for dup(17)(p11.2p11.2). J Clin Invest 116:3035–3041 10.1172/JCI28953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walz K, Caratini-Rivera S, Bi W, Fonseca P, Mansouri DL, Lynch J, Vogel H, Noebels JL, Bradley A, Lupski JR (2003) Modeling del(17)(p11.2p11.2) and dup(17)(p11.2p11.2) contiguous gene syndromes by chromosome engineering in mice: phenotypic consequences of gene dosage imbalance. Mol Cell Biol 23:3646–3655 10.1128/MCB.23.10.3646-3655.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walz K, Spencer C, Kaasik K, Lee CC, Lupski JR, Paylor R (2004) Behavioral characterization of mouse models for Smith-Magenis syndrome and dup(17)(p11.2p11.2). Hum Mol Genet 13:367–378 10.1093/hmg/ddh044 [DOI] [PubMed] [Google Scholar]
- 66.Lupski JR, Stankiewicz P (2005) Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet 1:e49 10.1371/journal.pgen.0010049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mendrzyk F, Korshunov A, Toedt G, Schwarz F, Korn B, Joos S, Hochhaus A, Schoch C, Lichter P, Radlwimmer B (2006) Isochromosome breakpoints on 17p in medulloblastoma are flanked by different classes of DNA sequence repeats. Genes Chromosomes Cancer 45:401–410 10.1002/gcc.20304 [DOI] [PubMed] [Google Scholar]
- 68.She X, Horvath JE, Jiang Z, Liu G, Furey TS, Christ L, Clark R, Graves T, Gulden CL, Alkan C, et al (2004) The structure and evolution of centromeric transition regions within the human genome. Nature 430:857–864 10.1038/nature02806 [DOI] [PubMed] [Google Scholar]
- 69.Inoue K, Osaka H, Imaizumi K, Nezu A, Takanashi J, Arii J, Murayama K, Ono J, Kikawa Y, Mito T, et al (1999) Proteolipid protein gene duplications causing Pelizaeus-Merzbacher disease: molecular mechanism and phenotypic manifestations. Ann Neurol 45:624–632 [DOI] [PubMed] [Google Scholar]
- 70.Lee JA, Inoue K, Cheung SW, Shaw CA, Stankiewicz P, Lupski JR (2006) Role of genomic architecture in PLP1 duplication causing Pelizaeus-Merzbacher disease. Hum Mol Genet 15:2250–2265 10.1093/hmg/ddl150 [DOI] [PubMed] [Google Scholar]
- 71.Woodward KJ, Cundall M, Sperle K, Sistermans EA, Ross M, Howell G, Gribble SM, Burford DC, Carter NP, Hobson DL, et al (2005) Heterogeneous duplications in patients with Pelizaeus-Merzbacher disease suggest a mechanism of coupled homologous and nonhomologous recombination. Am J Hum Genet 77:966–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Inoue K, Osaka H, Thurston VC, Clarke JT, Yoneyama A, Rosenbarker L, Bird TD, Hodes ME, Shaffer LG, Lupski JR (2002) Genomic rearrangements resulting in PLP1 deletion occur by nonhomologous end joining and cause different dysmyelinating phenotypes in males and females. Am J Hum Genet 71:838–853 [DOI] [PMC free article] [PubMed] [Google Scholar]