Abstract
Congenital anomalies of the kidney and urinary tract (CAKUT) include vesicoureteral reflux (VUR). VUR is a complex, genetically heterogeneous developmental disorder characterized by the retrograde flow of urine from the bladder into the ureter and is associated with reflux nephropathy, the cause of 15% of end-stage renal disease in children and young adults. We investigated a man with a de novo translocation, 46,X,t(Y;3)(p11;p12)dn, who exhibits multiple congenital abnormalities, including severe bilateral VUR with ureterovesical junction defects. This translocation disrupts ROBO2, which encodes a transmembrane receptor for SLIT ligand, and produces dominant-negative ROBO2 proteins that abrogate SLIT-ROBO signaling in vitro. In addition, we identified two novel ROBO2 intracellular missense variants that segregate with CAKUT and VUR in two unrelated families. Adult heterozygous and mosaic mutant mice with reduced Robo2 gene dosage also exhibit striking CAKUT-VUR phenotypes. Collectively, these results implicate the SLIT-ROBO signaling pathway in the pathogenesis of a subset of human VUR.
Congenital anomalies of the kidney and urinary tract (CAKUT) make up a family of diseases with a diverse anatomical spectrum, including kidney anomalies (e.g., renal dysplasia, duplex kidney, and hydronephrosis) and ureter anomalies (e.g., vesicoureteral reflux [VUR], megaureter, and ureterovesical junction [UVJ] obstruction).1,2 In particular, VUR (MIM %193000), a polygenic genetic disorder with an incidence of ∼1 in 100 infants,3,4 is one of the most common clinical manifestations of CAKUT. VUR is characterized by the reflux of urine from the bladder into the ureters and sometimes into the kidneys and is a risk factor for urinary tract infection (UTI).5 In combination with intrarenal reflux, the resulting inflammatory reaction may result in renal injury or scarring, also called “reflux nephropathy.”6 Extensive renal scarring impairs renal function and may predispose patients to hypertension, proteinuria, and renal insufficiency. Reflux nephropathy accounts for as much as 15% of end-stage renal disease in children and young adults.7 Primary VUR results from a developmental defect of the UVJ8 and is known to occur in multiple members of families. In siblings and offspring of affected patients, the prevalence is as high as 50%.9,10 Despite its high incidence in the pediatric population, the genetic basis of VUR remains to be elucidated.
Human ROBO1–4 encode homologs of Drosophila Roundabout (Robo), a transmembrane receptor that binds SLIT ligand and transduces a signal to prevent axons from recrossing the CNS midline.11 On the basis of the mouse Robo1 mutant phenotype, ROBO1 is a candidate gene for pulmonary hypoplasia and adenocarcinoma,12,13 as well as for dyslexia (DYX5).14 Mutations in ROBO3 result in horizontal-gaze palsy with progressive scoliosis,15 whereas zebrafish Robo4 is implicated in angiogenesis.16 Robo2 loss-of-function mutations in zebrafish and mice result in retinal and commissural pathfinding defects, respectively.17,18 Interestingly, Robo2 and Slit2 mouse mutants reveal an additional key role for SLIT-ROBO signaling in regulating the metanephric expression of glial cell derived neurotrophic factor (Gdnf), which in turn induces ureteric bud outgrowth from the nephric duct and restricts it to a single site.19 However, a role for ROBO2 in human disease has not been identified elsewhere.
Material and Methods
FISH and Array Comparative Genomic Hybridization Analyses
Metaphase FISH was performed according to standard methods. RP11 BAC clones were obtained from BACPAC Resources, were labeled as FISH probes, and were hybridized to metaphase chromosomes prepared from a t(Y;3)(p11;p12)dn lymphoblastoid cell line established from patient DGAP107. Array comparative genomic hybridization (CGH) experiments were performed by Spectral Genomics (SpectralChip 2006 array) and Agilent Technologies (Human Genome CGH Microarray Kit 44A).
Southern and RT-PCR Analyses
Southern and RT-PCR analyses were performed according to routine protocols. RT-PCR primers used to amplify the 2.8-kb ROBO2 cDNA (GenBank accession number NM_002942) were ROBO2-F1 and ROBO2-R1; those used to amplify ROBO2-PCDH11Y fusion transcripts were ROBO2-F2 (same for all transcripts) and PCDH11Y-R1, PCDH11Y-R2, and PCDH11Y-R3; those used to amplify wild-type ROBO2 transcripts were ROBO2-F2 and ROBO2-qR; and those used to amplify PCDH11Y cDNA (GenBank accession number NM_032971) were PCDH11Y-rtF1 and PCDH11Y-rtR1 (appendix C). All primer sequences are listed in appendix G.
Quantitative Real-Time PCR Analyses
PCR primers and TaqMan fluorogenic probes for analysis of the ROBO2 nontranslocated allele and the Fu-129 and Fu-153 fusion transcripts were designed using Primer Express software (Applied Biosystems). TaqMan primers and probes for Gapdh and β-actin were used for normalization. Probe melting temperatures were ∼7°C–10°C higher than those for the matching primer pair. High-pressure liquid chromatography–purified fluorogenic probes contained covalently attached 5′-FAM reporter and 3′-BHQ1 quencher dyes. Sequences of TaqMan primer and probe sets are listed in appendix G. RT-PCR reactions were performed using an iCycler IQ Real-Time Detection System (Bio-Rad). SuperScript One-Step RT-PCR with Platinum Taq kits (Invitrogen) were used for triplicate RT-PCR amplifications, with each 50-μl reaction containing 200 ng total RNA, 5 mM MgSO4, 500 nM forward and reverse primers, and 200 nM fluorogenic probes. Controls included either no reverse transcriptase or the substitution of H2O for RNA for each primer and probe set. The one-step RT-PCR protocol was 15 min at 50°C and 5 min at 95°C, followed by 45 cycles, each consisting of 15 s at 95°C and 1 min at 60°C. IQ Supermix reagent for real-time PCR (Bio-Rad) was used for two-step RT-PCR. Relative gene expression was analyzed using standard curve and comparative threshold cycle methods.
Fusion Proteins
cDNA sequences for Fu-129 and Fu-153 were amplified by PCR with the use of forward primer 5′-hR2(E1) and reverse primers 3′-LEVA(X1) and 3′-SRSC(X1) (appendix G) cloned into EcoRI and XhoI sites in pcDNA3, under control of the cytomegalovirus promoter. To express yellow fluorescent protein (YFP)–ROBO2-PCDH11Y fusions, the YFP (Venus) coding sequence was cloned into BamHI and EcoRI sites in pCS, whereas ROBO2-PCDH11Y coding sequences were inserted inframe with YFP at EcoRI and XhoI sites. YFP-ROBO2-PCDH11Y fusion proteins were expressed under the control of the simian cytomegalovirus IE94 promoter. Myc-SLIT and hemagglutinin (HA)–RoboN (Robo1-N) constructs have been described elsewhere.20
Neuronal Migration Assay
The in vitro neuronal migration assay that uses postnatal anterior subventricular zone (SVZa) cells was described elsewhere.21 In brief, P1-6 Sprague-Dawley rat brains devoid of meninges were placed in 10% fetal calf serum in Dulbecco's modified Eagle medium (DMEM) and were embedded. Coronal sections of 300 μm were prepared by vibratome, and tissues within the SVZa borders were dissected to make SVZa explants 200–300 μm in diameter. Explants were embedded, together with human embryonic kidney (HEK) cell aggregates in collagen and matrigel (3:2:1 ratio of collagen:matrigel:medium), and were cultured in DMEM with 5% CO2 at 37°C for 24 h. Cocultured cells were washed in PBS for 10 min and were fixed in 4% paraformaldehyde at 4°C overnight.
To make cell aggregates, HEK cells were transiently transfected to express mouse Slit2 or RoboN, DGAP107 Fu-129 or Fu-153, or pcDNA3 or Semaphorin 3A expression vectors as negative controls, with the use of Effectene Transfection Kit (Qiagen). After 24 h, transfected HEK cells were detached and were collected by brief centrifugation, and cell pellets were resuspended in an equal volume of DMEM. Ten microliters of suspended cells were hung from the dish cover at 37°C, in 5% CO2 for 1–2 h, to form aggregates. Aggregated cells were washed in DMEM and were squared with a needle.
Western Blot
HEK cells were transfected to express YFP-ROBO2-PCDH11Y-Fu-129, YFP-ROBO2-PCDH11Y-Fu-153, Myc-Slit, and HA-RoboN. Cells transfected with pCS2 vector were used as a negative control. To remove cell debris 48 h after transfection, media were collected and were centrifuged for 20 min at 4°C. Supernatants were diluted with sixfold protein loading buffer and were heated at 95°C for 20 min. Cells were lysed (1× PBS, 0.5% Triton X-100, and 1× protease inhibitor), were diluted with sixfold protein loading buffer, and were heated at 95°C for 20 min. Proteins were resolved by 12% SDS-PAGE, with YFP (Venus) fusion proteins detected by monoclonal anti–green fluorescent protein (GFP) antibody that detects YFP (Clontech), and Myc-Slit2 and HA-RoboN were blotted with monoclonal antibodies against Myc and HA, respectively.
Mutation Analysis
ROBO2 mutation screening employed PCR amplification of each of the 26 human ROBO2 exons and intron-exon boundaries, followed by purification and bidirectional DNA sequencing. Sequences of the ROBO2 PCR primer sets are listed in appendix G. Sequence data were analyzed using Lasergene (DNAStar) sequence analysis software. DNA samples with sequence changes were confirmed by resequencing. National Center for Biotechnology Information RefSeq ROBO2 cDNA sequence (GenInfo identifier [GI] 109254774) and protein sequence (Entrez Protein accession number NP_002933) (GI 61888896) were used to calculate the nucleotide and amino acid positions.
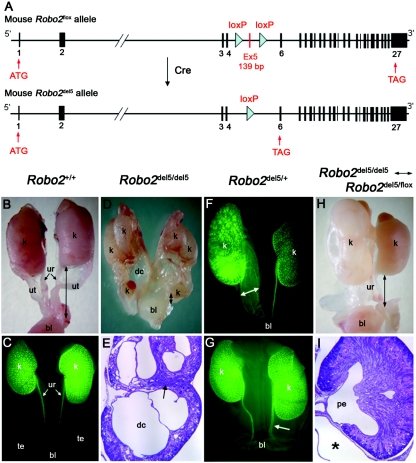
Preparation of Robo2flox and Robo2del5 Alleles
Robo2flox mice were produced using homologous recombination in 129 embryotic stem cells and blastocyst injection. After germline transmission, mice were backcrossed to C57BL/6 and were analyzed thereafter in a mixed C57BL/6-129/Sv background. The Robo2flox allele was genotyped by PCR amplification, followed by SpeI restriction digestion with the use of PCR primers Ro2-MEBAC15F and Ro2-MEBAC15R (appendix G), which amplify a 1,100-bp fragment for both wild-type and Robo2flox alleles. After SpeI digestion, the Robo2flox amplicon remains uncut, whereas the wild-type amplicon yields 750-bp and 350-bp products. Robo2flox/+ mice were bred with TgEIIa-Cre (stock number 003724 [Jackson Laboratory]) to produce the Robo2del5 allele. The Robo2del5 allele was amplified by primers Robo2koF and Robo2R, which produce a 1,100-bp fragment. The wild-type allele was amplified by primers Robo2wtF and Robo2R, which yield a 1,390-bp fragment. F2 Robo2del5/del5↔Robo2del5/flox mosaic mice were prepared as described in appendix F and were analyzed for the presence of urinary tract phenotypes. To examine the ureter and kidney defects, Hoxb7-GFP transgenic mice (gift from Dr. Frank Costantini, Columbia University) were bred with Robo2 mutants. GFP fluorescence was monitored and photographed using a Nikon SMZ-1500 epifluorescence stereomicroscope.
Human and Animal Studies
All human studies were performed under informed consent protocols approved by the Partners HealthCare System Human Research Committee (Boston), the Human Research Ethics Committee (Institute of Child Health, University College London), or the University Medical Center (Utrecht). Mouse protocols were approved by the Institutional Animal Care and Use Committee at Harvard Medical School or Boston University Medical Center, with additional approval from King's College, London.
Results
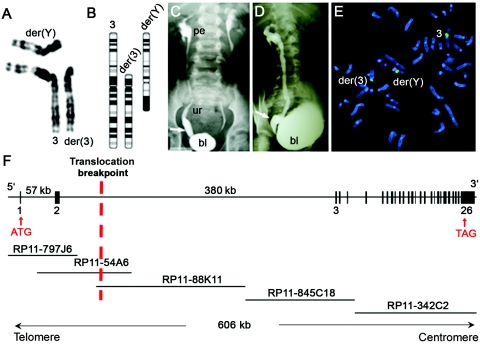
The Developmental Genome Anatomy Project (DGAP) is a collaborative effort to use chromosomal rearrangements associated with developmental disorders to identify the underlying genetic etiology. DGAP107 is a man aged 18 years with a 46,X,t(Y;3)(p11;p12)dn translocation, whose phenotype includes bilateral high-grade VUR and right megaureter at the UVJ (fig. 1A–1D and appendix A). He required ureteral reimplantation surgery at age 9 years and was found to have wide-open right and left ureteral orifices due to bilateral absence of intravesical ureteral segments. Normally, these submucosal ureteral segments obliquely traverse the muscular layers of the bladder to prevent retrograde flow of urine by a flap-valve mechanism. By metaphase FISH, we identified a BAC clone (RP11-54A6) that crosses the 3p12 breakpoint, which disrupts intron 2 of ROBO2, which is composed of 26 exons and spans ∼606 kb of genomic DNA (fig. 1E and 1F).
Figure 1. .
ROBO2 disrupted in DGAP107. Partial karyogram (A) and idiogram (B) for 46,X,t(Y;3)(p11;p12)dn is shown. VCUG of DGAP107 shows anterior-posterior (C) and lateral (D) views of bilateral grade IV VUR and megaureter at the right UVJ (arrows). bl = Bladder; pe = renal pelvis; ur = ureter. E, FISH analysis showing BAC RP11-54A6 (green), which hybridizes to normal chromosome 3, der(3), and der(Y) and crosses the 3p12 breakpoint. F, Intron-exon structure of ROBO2, with select exons numbered and the relevant BAC contig. The location of the 3p12 translocation breakpoint is indicated by a red dotted vertical line.
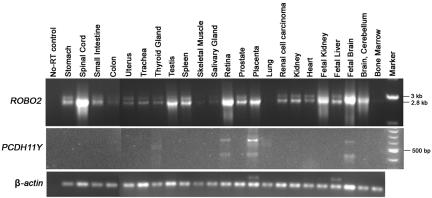
We cloned and sequenced the breakpoints on the der(3) and der(Y) chromosomes (appendix B). In addition to disruption of ROBO2 at 3p12, the protocadherin gene PCDH11Y at Yp11 was also disrupted by the translocation. A contribution of PCDH11Y disruption to the VUR phenotype in DGAP107 is unlikely, however, since PCDH11Y expression has been detected only in placenta, brain, retina, and testis22 and has not been detected in embryonic kidney (appendix C). By array CGH and FISH, we also identified a 3.4-Mb interstitial deletion at 17p11.2 in DGAP107. This region is within the common microdeletion region pathogenetic in Smith-Magenis syndrome (SMS [MIM #182290]), a mental retardation syndrome associated with behavioral and sleep disturbances and craniofacial and skeletal anomalies.23 Thus, a role for del(17)(p11.2) in the sleep, behavioral, and cognitive deficits of DGAP107 seems likely. The del(17)(p11.2) microdeletion could also contribute to the pathogenesis of VUR in DGAP107. However, for reasons described below and in appendix D, we conclude that ROBO2 disruption alone is sufficient to account for the VUR phenotype observed in DGAP107.
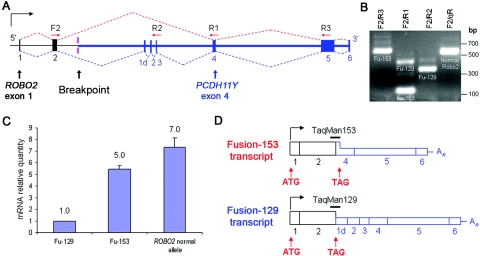
The t(Y;3) translocation in DGAP107 juxtaposes ROBO2 and PCDH11Y in the same transcriptional orientation. On the der(Y), the promoter and the first two exons of ROBO2 reside upstream of exons 1d–6 of PCDH11Y (fig. 2A). From RT-PCR experiments that used DGAP107 lymphoblast RNA, we identified two ROBO2-PCDH11Y fusion transcripts driven by the ROBO2 promoter (fig. 2B). Each transcript contains the first two exons of ROBO2 spliced out of frame to PCDH11Y downstream exons, resulting in premature stop codons shortly after ROBO2 exon 2. When assayed by real-time RT-PCR, these fusion transcripts, denoted Fu-129 and Fu-153, are expressed at somewhat reduced levels compared with the wild-type ROBO2 transcripts derived from the nontranslocated allele (fig. 2C); the wild-type transcripts contained no detectable mutations. Fu-129 and Fu-153 encode 129- and 153-residue polypeptides, respectively, containing the first ROBO2 extracellular immunoglobulin (Ig) domain, but they are truncated before the transmembrane and cytoplasmic domains required for SLIT-ROBO signal transduction (fig. 2D).
Figure 2. .
The t(Y;3) translocation in DGAP107, which generates novel ROBO2 fusion transcripts. A, ROBO2 and PCDH11Y intron-exon structure surrounding the der(Y) breakpoint. The forward primer F2 in ROBO2 exon 2 (black bar) was used in RT-PCR with three reverse primers—R2, R1, and R3—in PCDH11Y exons 3, 4, and 5, respectively (blue bars). Dotted lines indicate the observed splicing patterns of the two fusion transcripts. The red splicing pattern generates Fu-153, which encodes 153 aa, and the blue pattern generates Fu-129, which encodes 129 aa. B, RT-PCR fusion transcript amplification. Lane 1, F2/R3 primers amplify Fu-153 (641 bp) and Fu-129 transcripts; only the shorter Fu-153 amplicon is shown. Lane 2, F2/R1 primers amplify transcripts for both Fu-129 (456 bp) and Fu-153 (122 bp). Lane 3, F2/R2 primers amplify only Fu-129 transcripts (347 bp). Lane 4, F2/qR primers amplify only transcripts from the wild-type nontranslocated ROBO2 allele (606 bp). qR primer is located in exon 7 of ROBO2. C, Real-time RT-PCR quantitation of ROBO2 fusion transcripts Fu-129 and Fu-153 (detected by TaqMan probes shown in panel D) and of ROBO2 nontranslocated allele transcripts (detected by TaqMan probe across ROBO2 exons 2 and 3) in DGAP107 lymphoblast RNA. D, Exon structure of Fu-153 and Fu-129. Horizontal bars indicate TaqMan probes used to quantify fusion transcripts. Black boxes indicate ROBO2 exons; blue boxes, PCDH11Y exons; full-height boxes, coding exons; and half-height boxes, noncoding exons.
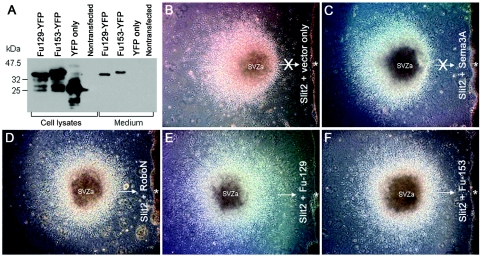
When expressed without transmembrane and cytoplasmic domains, the soluble extracellular Ig domains of ROBO, denoted as RoboN, are able to bind SLIT ligand.24 Moreover, the first ROBO Ig domain is necessary and potentially sufficient for SLIT binding.24 RoboN isoforms can thus inhibit SLIT-ROBO signaling by competing with wild-type ROBO for SLIT binding.25 We therefore hypothesized that the truncated proteins encoded by the ROBO2-PCDH11Y fusions might act in a dominant-negative manner to block endogenous SLIT-ROBO signaling. To test this hypothesis, we performed an in vitro neuronal migration assay21 in which SVZa explants were cultured in proximity to HEK cell aggregates secreting Slit2 or Slit2 and either RoboN, Semaphorin 3A, Fu-129, or Fu-153 (fig. 3A). We then examined the directionality of neuronal migration away from the SVZa explants (fig. 3B–3F). HEK cell aggregates alone have no effect on SVZa explants, resulting in a radially symmetric pattern of neuronal outgrowth.25
Figure 3. .
ROBO2 fusion proteins inhibiting SLIT chemorepulsion. A, YFP-tagged ROBO2 fusion proteins (Fu129-YFP [40 kDa] and Fu153-YFP [42 kDa]) detected by an anti-YFP antibody, expressed in HEK cell lysates, and secreted into the medium. In the presence of aggregated cells transfected with Slit2 plus empty vector (B) or Slit2 plus Sema3A (Semaphorin 3A, with no effect on Slit2 repulsive activity) (C), cells migrate out of SVZa explants and away from the Slit2-expressing cell aggregate (asterisk). In the presence of aggregated cells transfected with Slit2 plus RoboN (the Robo extracellular domain, which inhibits Slit repulsive activity), cells migrate out of SVZa explants symmetrically in all directions (D) including toward (arrow) the Slit2 and RoboN-expressing cell aggregate (asterisk). Fu-129 and Fu-153 also effectively block Slit2 repulsive activity (E and F), allowing symmetrical neuronal migration out of SVZa explants and toward (arrows) Slit2 and Fu-129 or Slit2– and Fu-153–expressing cell aggregates (asterisks).
When they were cultured with HEK cell aggregates transfected with vectors expressing Slit2 only or Slit2 and Semaphorin 3A (a molecule having no effect on Slit function, as a control), the Slit2-expressing cell aggregates acted upon the SVZa explants to repel SVZa neuronal outgrowth (fig. 3B and 3C). In contrast, in aggregates coexpressing Slit2 and either RoboN, Fu-129, or Fu-153, the latter molecules abrogated the chemorepulsive effect of Slit2 on the SVZa explant and significantly increased the number of neurons able to migrate towards the Slit2 source, resulting in a radially symmetric pattern of neuronal outgrowth (fig. 3B–3F). These results indicate that the ROBO2-PCDH11Y fusion proteins that result from t(Y;3) can act as dominant-negative molecules to block SLIT-mediated chemorepulsive function. The fusion proteins could further compromise ROBO2 function in DGAP107, which retains only hemizygous ROBO2 expression from the nontranslocated allele.
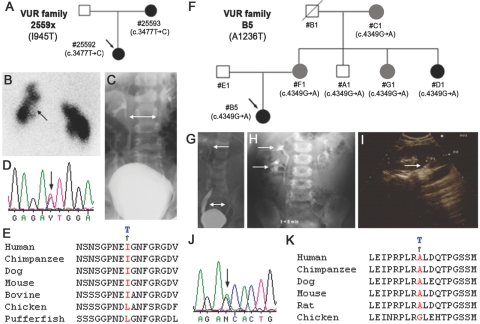
Family studies indicate that primary VUR frequently segregates with autosomal dominant inheritance and incomplete penetrance.9,26 To test whether mutations in ROBO2 are associated with CAKUT and VUR in the general population, we sequenced the 26 exons and intron-exon boundaries of ROBO2 in 124 families with VUR with potential autosomal dominant inheritance. One sequence change—c.2436T→C, or I598T—was observed in exon 12 in the ROBO2 extracellular domain but was also identified in three control DNA samples (see below). It therefore most likely represents a sequence polymorphism and was discounted from further study. In contrast, two novel ROBO2 intracellular coding sequence changes were identified that were not found in 276 controls (see below). These produce nonconservative amino acid substitutions in two independent families with CAKUT-VUR (fig. 4 and appendix E). In family 2559x with CAKUT-VUR, the affected daughter, 25592, has bilateral VUR, hypoplastic kidneys, and nephropathy, whereas her mother, 25593, required ureteral reimplantation because of severe VUR (fig. 4A–4C). Both individuals have a heterozygous T→C change at position 3477 in coding exon 19 (c.3477T→C) that would cause a nonconservative missense I945T substitution in the ROBO2 intracellular domain (ICD) (fig. 4D and 4E).
Figure 4. .
ROBO2 missense mutations in familial CAKUT and VUR. A, Family 2559x with CAKUT-VUR and exon 19 (c.3477T→C) mutation. Arrow indicates the proband. Blackened and gray symbols indicate patients with CAKUT-VUR and family members with urinary tract symptoms and radiological evidence of CAKUT. Nucleotide changes are shown under each individual. B, 99mTc-dimercaptosuccinic acid renogram of proband 25592 showing bilateral renal parenchymal defects (arrow). C, VCUG of proband 25592 showing bilateral reflux (bidirectional arrow). Chromatograms show T→C change (arrow) in exon 19 of family 2559x (D) and amino acid conservation across species (E). F, Family B5 with CAKUT-VUR and exon 23 (c.4349G→A) mutation. G, VCUG showing bilateral VUR (bidirectional arrow) and right duplex kidney (arrow) in proband B5. H, IVP detecting right duplex kidney (arrows) in proband B5. I, US showing suspected duplex (arrow) in upper pole of the right kidney in D1, an asymptomatic aunt of proband B5. Chromatograms show G→A change (arrow) in exon 23 of family B5 (J) and amino acid conservation across species (K).
In family B5 with CAKUT-VUR, the proband has bilateral VUR and a right duplex collecting system and kidney. Her mother and two aunts have urinary tract symptoms and ultrasonographical evidence of CAKUT, whereas her grandmother has a unilateral small kidney (fig. 4F–4I). All five family members carry a heterozygous G→A sequence alteration at position 4349 in coding exon 23 (c.4349G→A) that would cause a nonconservative missense amino acid substitution, A1236T, in the ROBO2 ICD (fig. 4J and 4K). An additional family member, uncle A1, also has this alteration but did not exhibit an ultrasonographically detectable renal phenotype; however, nonpenetrance of VUR is common.9 Both I945 and A1236 are evolutionarily conserved in all mammals and are only slightly divergent in birds and fish, organisms that lack a urinary bladder and UVJ (fig. 4E and 4K).
To further assess the likelihood that these sequence changes represent functional missense variants, as opposed to rare neutral variants found in the general population, we sequenced ROBO2 exons 12, 19, and 23 in 180 unrelated, clinically unaffected controls of ethnic backgrounds similar to those of the affected individuals. Two occurrences of c.2436T→C in exon 12 were detected, but no nucleotide changes were identified in exons 19 or 23. In addition, to determine the full spectrum of ROBO2 sequence variation, we resequenced the 26 ROBO2 exons and intron-exon boundaries in an additional 96 controls. We found only one reoccurrence of c.2436T→C. Of several nonvalidated putative synonymous and nonsynonymous ROBO2 coding SNPs listed in Ensembl v39, we detected only one—c.737C→A, or R32R—in our own sequencing efforts. Moreover, this apparent change was found to represent a sequencing artifact. Thus, in sum, these results suggest that the two sequence changes in the ICD identified in familial VUR are deleterious missense variants that contribute to the CAKUT-VUR phenotype.
Both I945T and A1236T could alter the function of the ROBO ICD, which regulates actin polymerization and cellular migration,27 by creating novel threonine phosphorylation sites or by influencing the binding of proteins that interact with the ROBO ICD.28 For example, the SH3 domain of srGAP1 binds to the ROBO1 ICD CC3 subdomain,27,28 which is partly conserved in ROBO2. The ROBO2 CC3 subdomain (residues 1193–1201) resides close to A1236, and an extended ROBO1 CC3 peptide binds the srGAP1 SH3 domain much more strongly than does the isolated CC3,27 suggesting that residues outside CC3 also mediate srGAP binding. I945T and A1236T may act as either dominant gain- or loss-of-function mutations that influence protein binding to the ROBO2 ICD.
To establish further the involvement of ROBO2 in the pathogenesis of the CAKUT-VUR phenotype postnatally, we next generated and analyzed a conditional Robo2 mouse mutant. A homozygous Robo2-null mouse, described elsewhere,19 with a targeted deletion of exon 1 exhibits a multiple ureter phenotype and fails to survive after birth. To determine whether heterozygosity for Robo2 loss of function could produce an abnormal urinary tract phenotype and recapitulate human CAKUT-VUR, we prepared a mouse Robo2 floxed allele, Robo2flox, containing loxP sites flanking Robo2 exon 5 (fig. 5A). Robo2flox/flox homozygotes are viable and fertile and lack urinary tract abnormalities. We then produced Robo2del5/+ mice that lack exon 5, by crossing the floxed allele with the TgEIIa-Cre deletor strain.29 The deletion of Robo2 exon 5 causes a reading frameshift. RT-PCR and in situ hybridization experiments showed that Robo2del5 transcripts were expressed at markedly reduced levels compared with transcripts from the wild-type allele. We thus conclude that Robo2del5 is, effectively, a null allele. Consistent with results reported for the Robo2 null allele described elsewhere,19 Robo2del5/del5 homozygotes uniformly died shortly after birth with multiplex, dysplastic kidneys and short ureters (fig. 5B–5E). However, whereas no heterozygous phenotype was described for the existing Robo2 null allele,19 when a Hoxb7-GFP reporter transgene that specifically identifies the ureteric epithelium30 was introduced into the Robo2del5/+ background, 4 (15%) of 26 Robo2del5/+ heterozygous newborns exhibited a unilateral CAKUT-VUR phenotype (fig. 5F and fig. 5G). This heterozygous phenotype included both massive and lesser degrees of megaureter and a wide-open UVJ, similar to the pathology identified in DGAP107.
Figure 5. .
Robo2del5/del5 homozygous, Robo2del5/+ heterozygous, and Robo2del5/del5↔Robo2del5/flox mosaic newborn mice expressing striking CAKUT phenotypes. A, Structures of the mouse Robo2flox and Robo2del5 alleles. The Robo2flox allele encodes a wild-type, full-length 1,470-aa Robo2 protein but contains two loxP sites flanking exon 5. The Robo2del5 allele is generated from Robo2flox by Cre, which deletes Robo2 exon 5 to produce an aberrant transcript expressed only at low levels. B and C, Wild-type female (B) and male (C) newborn mouse excretory system. The male excretory system in panel C is illuminated by the Hoxb7-GFP transgene. k = kidney; bl = bladder; ur = ureter; ut = uterus; te = testis. Black bidirectional arrows indicate ureter length in panels B and D. Robo2del5/del5 newborn homozygotes display multiplex dysplastic kidneys (D) and, at 25× magnification (E), reveal dysplastic cysts (dc) in the calyces and an internalized nephrogenic zone (arrow). Hoxb7-GFP transgene–positive Robo2del5/+ heterozygous newborns show megaureter dilation (F) (bidirectional arrow) and early ureter dilatation (G) (arrow). Robo2del5/del5↔Robo2del5/flox mosaic newborns show hydronephrosis in the left kidney (H). At 25× magnification (I), they show megaureter (asterisk). Black bidirectional arrows indicate ureter length in panel H. pe = pelvis.
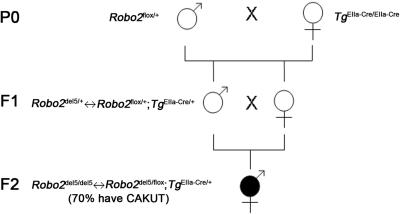
To test whether further reductions in Robo2 gene dosage could increase CAKUT-VUR penetrance, we took advantage of the variable expression of the EIIa-directed Cre recombinase in the early preimplantation embryo,31 to generate mosaic progeny that consisted of admixtures of Robo2del5/del5 (null) and Robo2del5/flox (haploinsufficient) cells. This mosaicism originates from the incomplete, stochastic action of the EIIa-Cre transgene on the Robo2flox allele in the early embryo before implantation.31 Remarkably, 4 (40%) of 10 Robo2del5/del5↔Robo2del5/flox mosaic newborns (resulting from the union of Robo2del5;TgEIIa-Cre and Robo2flox gametes) exhibited unilateral urinary tract defects, including short ureter, megaureter, and hydronephrosis (fig. 5H and 5I).
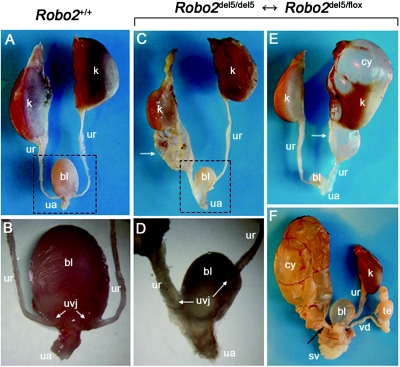
To determine whether Robo2del5/del5↔Robo2del5/flox mosaic newborns with urinary tract defects could survive after prolonged reflux and obstruction, we followed another cohort of these mice to adulthood. Seven (70%) of 10 Robo2del5/del5↔Robo2del5/flox adult mosaics, ranging in age from 45 d to 77 d, manifested defects involving the UVJ (fig. 6). These UVJ defects were bilateral and especially notable, in that one UVJ was typically located laterally and cephalad in the bladder (fig. 6A–6D), a location commonly associated with reflux in humans,32 whereas the contralateral UVJ was located caudad in the bladder or even ectopically in the urethra. The caudal UVJ location was associated with obstruction, resulting in megaureter and severe hydronephrosis (fig. 6C and 6E). In some male Robo2del5/del5↔Robo2del5/flox mice, the ureter was connected to the vas deferens, resulting in massive hydronephrosis (fig. 6F). The Robo2del5/del5↔Robo2del5/flox mouse model is thus consistent with the frequent coexistence of reflux and obstruction in the same patient with VUR.33 During embryonic development, the nephric duct undergoes apoptosis, transposing the ureter orifice from the nephric duct to the urogenital sinus epithelium, to form the UVJ.34 Mutation in mouse Robo2 causes abnormal sites of ureteric bud outgrowth,19 which provides a developmental explanation for the ectopic UVJ sites frequently observed in VUR.
Figure 6. .
Adult Robo2del5/del5↔Robo2del5/flox mosaics exhibiting megaureter, hydronephrosis, and UVJ defects. A–D, Ventral views. k = kidney; ur = ureter; bl = bladder; ua = urethra. A, Urinary tract in a wild-type mouse aged 87 d. B, Higher magnification of boxed region in panel A, indicating normal position of the UVJ. C, Right megaureter (arrow) in a Robo2del5/del5↔Robo2del5/flox mosaic aged 45 d. D, Higher magnification of boxed region in panel C, demonstrating abnormal bilateral UVJ. The obstructed right UVJ connects to a caudal site in the bladder close to the urethra, causing megaureter. The left UVJ is located laterally in the bladder, a site commonly associated with human VUR. E and F, Dorsal views. E, Right megaureter (arrow) and hydronephrosis in a mosaic aged 45 d. Hydronephrosis replaces the normal renal parenchyma (k), causing an upper pole cyst (cy). F, Left ureter of a male mosaic mouse aged 77 d that remains connected to the vas deferens (vd) (arrow), resulting in obstruction and severe hydronephrosis. The left kidney has lost all parenchyma and is replaced by a large cyst. The right kidney, ureter, and vas deferens are normal in appearance. sv = seminal vesicle; te = testis.
Discussion
Collectively, these results demonstrate that reduced Robo2 gene dosage can contribute to the pathogenesis of CAKUT-VUR (table 1 and appendix F). In human primary VUR, linkage studies have produced inconsistent results,35 underscoring the need for other methods to identify the responsible genes. This problem is especially acute for VUR, since the manifestations may vary during life, progressing or resolving spontaneously.36 Furthermore, because asymptomatic individuals cannot be classified as “unaffected,” linkage studies may be inconclusive or yield false-negative results unless confined to affected individuals.36
Table 1. .
Penetrance of CAKUT in Robo2del5 Mutant Mice
| Genotype | Robo2del5/+Newborn |
Robo2del5/flox Mosaic Newborn |
Robo2del5/flox Mosaic Adult | Robo2del5/del5 Newborn |
| CAKUT penetrance (%) | 15 | 40 | 70 | 100 |
| Total number observed | 26 | 10 | 10 | 20 |
Lastly, intrafamilial phenotypic variability and genetic heterogeneity3,35 also exist. Since we identified only two coding-region changes segregating with VUR in 124 families with VUR, alterations in ROBO2 itself are likely to account for a small subset of VUR. Interestingly, however, recent studies indicate that ROBO2 resides in an inherently unstable genomic region, 3p12.3, that is prone to evolutionary chromosomal rearrangements and to loss of heterozygosity in human cancers.37 This raises the yet-untested possibility that ROBO2 may be subject to frequent rearrangement or microdeletion and duplication at either the organismal or cellular level, which could be missed by direct sequencing. In addition, variants in other genes whose products function in the ROBO2 signal-transduction pathway may be implicated in the molecular pathogenesis of renal dysplasia and VUR, which may coexist because of interrelated pathophysiology, common underlying genetic abnormality, or both mechanisms.
Acknowledgments
We thank Roxana Peters, Robert Eisenman, Diana Donovan, Annick Turbe-Doan, and Juan Liu, for technical support; Yiping Shen, Anne Higgins, and Fowzan Alkuraya, for assistance with array CGH; Chantal Farra, for referral of the DGAP107 subject; Frank Costantini, for providing Hoxb7-GFP transgenic mice; Wellington Cardoso and Jining Lu, for help with fluorescence stereomicroscopy; and Natalia Leach, Irfan Saadi, Kate Ackerman, Azra Ligon, David Harris, Gail Bruns, Grigoriy Kryukov, Shamil Sunyaev, Monica Banerjee, Maria Bitner-Glindzicz, Sue Malcolm, Dagan Jenkins, Ramon Bonegio, and David Salant, for helpful suggestions. This work was supported by National Institutes of Health grants PO1GM061354 (to C.C.M.) and RO1DK063316 (to R.L.M.); the Hilda Gershon Sugarman Young Investigator grant from the National Kidney Foundation, a Department of Medicine Pilot Project grant, and an Evans Medical Foundation grant (to W.L.); Dutch Kidney Foundation grant C02.2009 (to J.C.G. and A.M.v.E.); a Health Research Council grant (to M.R.E.); a Kids Kidney Appeal grant, a Kidney Research UK grant, and Wellcome Trust grant 066647 (to A.S.W. and S.A.F.); and a Medical Research Council Career Establishment grant (to W.A. and V.S.).
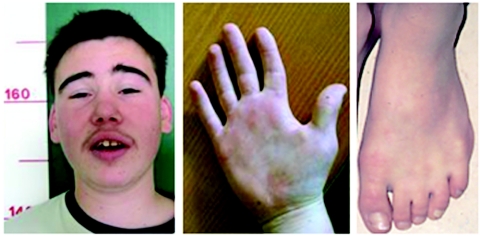
Appendix A: DGAP107 Phenotype
Figure A1. .
Facial and limb abnormalities of the DGAP107 proband. Note the low-set, dysplastic ears and subtle membranous syndactyly and clinodactyly. Blepharophimosis is also present.
Table A1. .
Clinical Findings in the DGAP107 Subject at Age 14 Years
| Characteristic | Patient Phenotype |
| Weight (kg) | 40 (10th percentile) |
| Height (cm) | 146 (<5th percentile) |
| Visual disorders | Daltonism, strabismus, hypermetropia |
| Limb defects | Mild syndactyly, clinodactyly, brachymetacarpia |
| Urinary tract defects | VUR grade IV, bilateral UVJ defects, unilateral megaureter |
| Learning disabilities | Global verbal retardation, verbal IQ 69, performance IQ 46a |
| Facial features | Blepharophimosis, low-set and dysplastic ears |
| Dental anomalies | Malformed lower incisor (“T” shape) |
| Genital anomalies | Complete left testicular agenesis |
| Neurological defects | Seizures, hyperactivity, sleep disorder |
| Orthopedic abnormality | Hyperlordosis |
| Growth retardation | Delayed puberty |
| Other abnormalities | Bilateral inguinal hernia |
IQ = intelligence quotient.
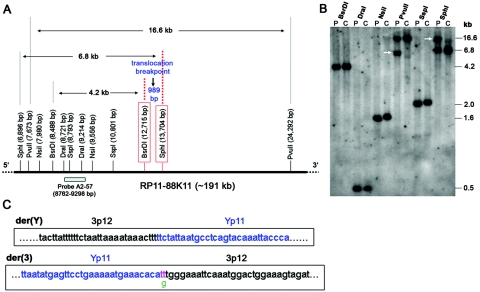
Appendix B: The DGAP107 3p12 Breakpoint Disrupts ROBO2
Figure B1. .
ROBO2 disrupted in DGAP107, with the breakpoint lying within intron 2. A, Restriction map surrounding the 3p12 breakpoint. The base-pair position of BAC RP11-88K11 (AC131005, within intron 2 of ROBO2 [see BAC contig in fig. 1F]) was used to calculate the distance between restriction enzyme sites. RP11-88K11 overlaps with BAC clone RP11-54A6 used in FISH and also contains the breakpoint, which is between boxed BsrDI and SphI sites, on the basis of the aberrant bands detected by Southern blot analysis. B, Southern blot analysis of DGAP107 (P) and unaffected control (C) genomic DNA, with use of the designated restriction enzymes and the probe A2-57 shown in panel A. Aberrant bands (white arrows) are present only in DGAP107 DNA digested with SphI and PvuII. C, Breakpoint cloning showing the sequence of the junction fragment from der(3) with a 1-bp deletion (g [green]) and a 2-bp insertion (tt [pink]). There is no gain or loss of nucleotides at the der(Y) breakpoint.
Appendix C: ROBO2 and PCDH11Y Expression in Human Adult and Fetal Tissues
Figure C1. .
Expression of ROBO2 and PCDH11Y in human tissues. RT-PCR amplified 2.8-kb ROBO2 cDNAs with the use of primers ROBO2-F1 and ROBO2-R1. The 3-kb ROBO2 cDNA product (upper band of doublet) contains an alternatively spliced exon 24B. RT-PCR amplification of 404-bp and 620-bp PCDH11Y cDNAs used primers PCDH11Y-F1 and PCDH11Y-R1. The intensity of the fragments indicates the approximate expression level of ROBO2 and PCDH11Y in these tissues. Notably, there is no expression of PCDH11Y in the fetal kidney. β-actin was used as a cDNA loading control.
Appendix D: Analysis of the DGAP107 del(17)(p11.2) Microdeletion
The pleiotropic nature of the DGAP107 phenotype suggests that both the t(Y;3)(p11;p12)dn translocation and the del(17)(p11.2) microdeletion may contribute to the overall DGAP107 phenotype. However, we parse the contribution of del(17)(p11.2) to the VUR phenotype as follows. Point mutations in RAI1, which resides in the SMS critical deletion region, suggest that RAI1 haploinsufficiency accounts for many features of SMS.38 Less frequently observed cardiac, renal, and other defects may reflect hemizygosity for other genes in the SMS common deletion region.39 Of note, RAI1 is included in the 3.4-Mb del(17)(p11.2) in DGAP107, the boundaries of which were defined by FISH experiments (not shown). It seems likely that the del(17)(p11.2) microdeletion contributes to some aspects of the sleep, behavioral, and cognitive deficits in DGAP107, because similar phenotypes are observed in patients with SMS who exhibit 17p11.2 deletions or RAI1 haploinsufficiency. Thus, ROBO2 disruption, 17p11.2 microdeletion, or both could theoretically account for VUR in DGAP107. Our human and mouse experimental results indicate that ROBO2 disruption can contribute substantially to the VUR phenotype. Conversely, our analyses also suggest that, whereas the 17p11.2 microdeletion in DGAP107 may contribute to the pathogenesis of VUR in DGAP107, it is not likely to play a primary role. There are three bases for this conclusion.
First, although we note an isolated case report describing VUR in SMS,40 reexamination of the frequency of renal defects in SMS indicates that these are relatively infrequent and less frequent than originally believed.23,41
Second, in the context of this study, we examined and observed no kidney or collecting system defects in three age-matched genetically engineered mouse models of SMS: SMSdf(11)17/+ (Δ2 Mb), SMSdf(11)17−l/+ (Δ500 kb), and Rai1−/−.42–44 The first two models represent deletion alleles that eliminate 2 Mb and 500 kb, respectively, from mouse chromosome 11. These regions are homologous to and share conservation of synteny with the 17p11.2 region that is involved in SMS. Although SMSdf(11)17 and SMSdf(11)17−l homozygotes die before nephrogenesis (9.5 embryonic d), examination of 20 SMSdf(11)17−l/+ mice in the context of this study revealed no UVJ or other urinary tract defects. In addition, Rai1−/− are not reported to exhibit any evidence for renal or ureteral defects,42 and our own analysis of these mice confirms this finding.
Third, analysis of the urinary tract in five SMSdf(11)17−l/+;Robo2del5/+ transheterozygotes, the genotype of which nominally approximates the DGAP107 genotype, revealed no observable phenotype compared with genetic background and age-matched littermate controls. Thus, whereas the 17p11.2 microdeletion could be a contributory factor, it seems unlikely to play a major role in the pathogenesis of CAKUT and VUR phenotypes in DGAP107.
Appendix E: Clinical Data for Families 2559x and B5
Clinical Data for Family 2559x
2559x is a white British family. The index patient (25592) has bilateral VUR and bilateral nephropathy. She presented at age 3 years with a symptomatic, documented Escherichia coli UTI, at which time ultrasound (US) revealed two small kidneys that were each 6.0 cm long (normal mean for age 7.0 cm). An indirect isotope cystogram with mercaptoacetyltriglycine (MAG3) showed bilateral VUR. The patient was treated long term with antibiotics. At age 4 years, her plasma creatinine was 107 μmol/liter (normal mean for age 56 μmol/liter). Her formal EDTA-glomerular filtration rate was 31 ml/min/1.73 m2 (normal mean >90 ml/min/1.73 m2), and a formal voiding cystourethrogram (VCUG) confirmed bilateral VUR. A repeat US confirmed two small kidneys with dimensions of 4.7 cm (left) and 6.0 cm (right) (50th percentile for age 7.2 cm), both with scarred upper poles. At age 9 years, she had progressive renal failure with a plasma creatinine of 306 μmol/liter. By age 12 years, her plasma creatinine had risen to 407 μmol/liter. Her mother (25593) has a history of VUR that required ureteral reimplantation surgery.
Clinical Data for Family B5
B5 is a white Dutch family. By VCUG and intravenous pyelogram (IVP), we determined that the proband B5 had bilateral VUR and a right kidney duplex system. US investigations further documented the proband’s double collecting system on the right side and a single system on the left, with a slightly dilated upper pole system. Renal US studies were also performed in all other family members except grandfather B1, who is deceased. The US findings for family B5 include the following: A1 (uncle) had normal kidneys, with an 11.0-cm right kidney and a 12.5-cm left kidney (normal [±SD] 11.5 ± 1.0 cm); C1 (grandmother) had a small, 9.6-cm right kidney, an 11.4-cm left kidney, a slim collecting system on both sides, a normal corticomedullary ratio, no signs of a duplex system, and normal flow in the right renal artery; D1 (aunt) had an 11.8-cm right kidney with an upper pole duplex system (column of Bertini), an 11.0-cm left kidney, a normal corticomedullary ratio, and no dilatation or urologic complaints; E1 (father) had a normal kidneys, a slim collecting system on both sides, a small peripelvic cyst on the right side, and no urologic complaints; F1 (mother) had a left kidney with fetal lobulation on the right side, no visible column from the renal parenchyma to the hilus, a duplex system that can neither be proven nor excluded, a slim collecting system on both sides, and febrile UTI history; and G1 (aunt) had no dilatation on either kidney, a cyst (4.8×3.5 cm) in the upper pole of the left kidney, and pyelonephritis at age 8 years.
Controls
All controls for genetic studies of families 2559x and B5 were unrelated. Control samples were from 180 Americans of white European descent (CEPH) and from 96 whites in the same geographic region as family B5. These latter 96 samples were subjected both to targeted sequencing of exons 12, 19, and 23 and to complete resequencing of all 26 coding ROBO2 exons.
Appendix F: Analysis of Robodel5 Mutant Mice
Figure F1. .
Generation of F2 Robo2del5/del5↔Robo2del5/flox mosaics
Table F1. .
Phenotype-Genotype Correlation in F2 Robo2del5/del5↔Robo2del5/flox Mosaics[Note]
| Identification Number |
Age (d) |
Sex | Phenotypea |
| 3235 | 45 | F | L and R: megaureter, hydronephrosis |
| 3239 | 45 | F | R: megaureter, hydronephrosis; L: normal |
| 404 | 62 | M | L: megaureter, hydronephrosis with complete loss of renal parenchyma; R: normal |
| 3218 | 77 | M | R: megaureter, hydronephrosis with complete loss of renal parenchyma; L: normal |
| 3236 | 77 | F | No discernible gross phenotype |
| 403 | 62 | M | No discernible gross phenotype |
| 3372 | 27 | M | No discernible gross phenotype |
| 3964 | 20 | F | L: megaureter, hydronephrosis; R: normal |
| 3963 | 20 | F | L: megaureter, hydronephrosis; R: normal |
| 3962 | 20 | F | L and R: megaureter, hydronephrosis |
Note.— Genotyping was performed using PCR of mouse tail DNA. All mosaics were positive for Robo2del5, Robo2flox, and TgEIIa-Cre.
L = left; R = right.
Notes on the Breeding Scheme
In the P0 generation, Robo2flox/+ heterozygotes were crossed with TgEIIa-cre/EIIa-cre homozygotes. The 50% of the progeny that receive TgEIIa-cre and Robo2flox gametes constitute the F1 generation shown (fig. F1). When transmitted from males, TgEIIa-cre is variably expressed in the F1 embryos after fertilization and before implantation,29,31 resulting in recombination in some embryonic cells but not others. Thus, the resulting F1 progeny are mosaic and contain mixtures of Robo2flox/+ (no recombination) and Robo2del5/+ (recombination) cells. These mosaics are genotypically denoted Robo2del5/+↔Robo2flox/+;TgEIIa-cre/+. PCR experiments demonstrated variable ratios of the Robo2flox and the Robo2del5 alleles, confirming the mosaicism of the F1 mice (not shown). As expected, 10 F1 mosaic mice examined exhibit no observable phenotype, because heterozygous Robo2del5/+ mice exhibit a phenotype at a low percentage (i.e., 15%), and, even at the most extreme degree of mosaicism (100% Robo2del5/+ and 0% Robo2flox/+), the overall reduction in Robo2 gene dosage would not surpass that in Robo2del5/+ heterozygotes.
The F2 generation was generated by intercross of the subset of F1 germline mosaics that also carried the TgE11a-cre/+ transgene. The F2 generation thus also consisted of mosaic progeny because of the union of Robo2del5;TgEIIa-cre and Robo2flox gametes. This particular combination of gametes results in a subset of F2 mosaics that contains both Robo2del5/del5 and Robo2del5/flox cells—that is, a mixture of heterozygous and homozygous cells. Cells carrying the Robo2del5/del5 genotype in these Robo2del5/del5↔Robo2del5/flox mosaics derive from the action of Cre in cells that commence embryogenesis with the Robo2del5/flox genotype; Robo2del5/flox cells result when Cre activity in those cells is insufficient to cause recombination. As described in the text and in table F1, 70% of these mosaics exhibit striking CAKUT phenotypes, whereas 30% exhibit no observable phenotype. The presence of a CAKUT phenotype presumably correlates with the percentage of Robo2del5/del5 cells that make up the urinary tracts of these mosaics.
The mosaic nature of the F2 mice (at least in the germline) was established by further intercross of a subset of F2 Robo2del5/del5↔Robo2del5/flox;TgEIIa-cre mosaics to generate an F3 generation. In this case, a high percentage of Robo2del5/del5 cells in the F2 germline result in a preponderance of gametes carrying the Robo2del5 mutant allele. The union of these Robo2del5 gametes results in an increased proportion of Robo2del5/del5 mice (90% of progeny) in the F3 generation that die at birth from CAKUT phenotypes (not shown). The remaining 10% retain the Robo2del5/flox allele and survive. The non-Mendelian ratio of these resulting F3 genotypes confirms germline mosaicism in the F2 mice.
Appendix G: PCR Primers
Table G1. .
PCR Primers and Probes
| Primer or Probe Useda | Type | Location | Sequence |
| RT-PCR analyses: | |||
| 2.8-kb ROBO2 cDNAb: | |||
| ROBO2-F1 | Forward | Exon 9 of ROBO2 | 5′-GAGCAAGGCACACTGCAGATTA-3′ |
| ROBO2-R1 | Reverse | Exon 26 of ROBO2 | 5′-AGTCATCACTTCCATGAGTCCG-3′ |
| 404-bp and 620-bp PCDH11Y cDNAb: | |||
| PCDH11Y-F1 | Forward | Exon 1 of PCDH11Y | 5′-CAGAAACAACCTCAGCGACTCC-3′ |
| PCDH11Y-R1 | Reverse | Exon 4 of PCDH11Y | 5′-GAACACCACGCATACTAGCAGG-3′ |
| ROBO2-PCDH11Y fusion transcriptsc: | |||
| ROBO2-F2 | Forward | Exon 2 of ROBO2 | 5′-GCAGTGAGTCGAAATGCGTC-3′ |
| PCDH11Y-R1 | Reverse | Exon 4 of PCDH11Y | 5′-GAACACCACGCATACTAGCAGG-3′ |
| PCDH11Y-R2 | Reverse | Exon 3 of PCDH11Y | 5′-CCCACTGTCATTCAGTCCTCAT-3′ |
| PCDH11Y-R3 | Reverse | Exon 5 of PCDH11Y | 5′-TGTTTCAATGACATCGAGGCC-3′ |
| 606-bp ROBO2 nontranslocated allelec: | |||
| ROBO2-F2 | Forward | Exon 2 of ROBO2 | 5′-GCAGTGAGTCGAAATGCGTC-3′ |
| ROBO2-qR | Reverse | Exon 7 of ROBO2 | 5′-TGGCCGAACCACAAACTGT-3′ |
| Quantitative real-time PCR analyses: | |||
| ROBO2 nontranslocated alleled: | |||
| ROBO2-F2 | Forward | Exon 2 of ROBO2 | 5′-GCAGTGAGTCGAAATGCGTC-3′ |
| ROBO2-qR2 | Reverse | Exon 3 of ROBO2 | 5′-GCTCTCCAGCTGCCACTACAA-3′ |
| ROBO2-FAM2 | TaqMan probe | ROBO2 exon 2–exon 3 junction | 5′-CTGGAAGTGGCATTGTTACGAGATGACTTCC-3′ |
| Fu-129 transcriptd,e: | |||
| ROBO2-F2 | Forward | Exon 2 of ROBO2 | 5′-GCAGTGAGTCGAAATGCGTC-3′ |
| PCDH11Y-qR1 | Reverse | Exon 1d of PCDH11Y | 5′-TATTCCATCCTCTTCCATCCATTC-3′ |
| TaqMan 129 | TaqMan probe | ROBO2 exon 2 and PCDH11Y exon 1d junction | 5′-CTGGAAGTGGCATAGGGTGCTTAAAAAGTACAGA-3′ |
| Fu-153 transcriptd,e: | |||
| ROBO2-F2 | Forward | Exon 2 of ROBO2 | 5′-GCAGTGAGTCGAAATGCGTC-3′ |
| PCDH11Y-qR2 | Reverse | Exon 4 of PCDH11Y | 5′-ATGTACGTCCCGGACAACAAA-3′ |
| TaqMan 153 | TaqMan probe | ROBO2 exon 2 and PCDH11Y exon 4 junction | 5′-TGGAAGTGGCATTGTTGTGCGGGT-3′ |
| Genotyping of Robo2flox and Robo2del5 mice: | |||
| Robo2flox allelef: | |||
| Ro2-MEBAC15F | Forward | Intron 5 of Robo2 | 5′-CCAATCATAGTCTCTCCACG-3′ |
| Ro2-MEBAC15R | Reverse | Intron 5 of Robo2 | 5′-CCTCTGATTCAATGAGATGC-3′ |
| 1,180-bp fragment from Robo2del5 allele: | |||
| Robo2koF | Forward | Intron 4 of Robo2 | 5′-CCACTATGCTGGCTCTGTCTCACAC-3′ |
| Robo2R | Reverse | Intron 5 of Robo2 | 5′-GGTTTTGGAGGTCTTACTACGTAGC-3′ |
| 1,390-bp fragment from Robo2+ wild-type allele: | |||
| Robo2wtF | Forward | Intron 4 of Robo2 | 5′-CAACTTTTCCTTTTCCGGGAGG-3′ |
| Robo2R | Reverse | Intron 5 of Robo2 | 5′-GGTTTTGGAGGTCTTACTACGTAGC-3′ |
| Synthesis of fusion protein constructs: | |||
| Fu-129 ROBO2-PCDH11Y fusion cDNA (129-aa fusion protein): | |||
| 5′-hR2(E1) | Forward | Exon 1 of ROBO2 with 5′ EcoRI linker | 5′-ATCGAATTCATGAGTCTGCTGATGTTTACACAACTACTG-3′ |
| 3′-LEVA(X1) | Reverse | Intron 1d of PCDH11Y with 5′ XhoI linker | 5′-TTACTCGAGCTATGCCACTTCCAGAGACGCATTTCG-3′ |
| Fu-153 ROBO2-PCDH11Y fusion cDNA (153-aa fusion protein): | |||
| 5′-hR2(E1) | Forward | Exon 1 of ROBO2 with 5′ EcoRI linker | 5′-ATCGAATTCATGAGTCTGCTGATGTTTACACAACTACTG-3′ |
| 3′-SRSC(X1) | Reverse | Intron 4 of PCDH11Y with 5′ XhoI linker | 5′-CCACTCGAGCTAGCAGGACCGCGAAAATGTACGTCC-3′ |
Primers and probes were used for amplification and quantitation.
As shown in figure C1.
As shown in figure 2B.
As shown in figure 2C.
As shown in figure 2D.
PCR amplifies a 1,100-bp fragment from both wild-type and Robo2flox alleles. After SpeI digestion, Robo2flox allele will not cleave (1,100-bp product), whereas the wild-type allele will be cleaved by SpeI to yield two smaller products of 750 bp and 350 bp.
Table G2. .
PCR Primer Sets Used to Amplify 26 Human ROBO2 Exons and Intron-Exon Boundaries in Mutation Analysis
| Exon and Primer | Amplicon Size (bp) |
Location in ROBO2 | Sequence |
| 1: | 286 | ||
| Forward | 5′-UTR | 5′-TTTGCTCTTCTTGACTTTAATTAGTATCTAGG-3′ | |
| Reverse | Introns 1–2 | 5′-TATAACCCACATCAAATTCAAAAAGAAAT-3′ | |
| 2: | 501 | ||
| Forward | Introns 1–2 | 5′-CGAAGAGTTTAATTTCCCCATCA-3′ | |
| Reverse | Introns 2–3 | 5′-GCGTCTATGGGAACACATCAAAA-3′ | |
| 3: | 264 | ||
| Forward | Introns 2–3 | 5′-TTGTACAACAAAAAGCCTAAGTTACTGTC-3′ | |
| Reverse | Introns 3–4 | 5′-AAAATTCAATCTCTCTGGGCCAT-3′ | |
| 4: | 280 | ||
| Forward | Introns 3–4 | 5′-TAATGACCTTTATTTTCTATTCTGTTCCTTT-3′ | |
| Reverse | Introns 4–5 | 5′-TTATATGGCCCAGTTTTTAATGTTAGTAATACT-3′ | |
| 5: | 291 | ||
| Forward | Introns 4–5 | 5′-CTTTTTTCATAATGTACTTAAAGCATGCA-3′ | |
| Reverse | Introns 5–6 | 5′-GTGGCATTGTAGCTGTCCTTTTATT-3′ | |
| 6: | 427 | ||
| Forward | Introns 5–6 | 5′-TTGCACTTTGTGGCTGATTTG-3′ | |
| Reverse | Introns 6–7 | 5′-TAATTTTATTTCAACTAATGATAGAGAGGACAC-3′ | |
| 7: | 289 | ||
| Forward | Introns 6–7 | 5′-CAACATAGTACCATATTTTCTCCTTGACATA-3′ | |
| Reverse | Introns 7–8 | 5′-AAGCAAGGCAAGCTTTCAGG-3′ | |
| 8: | 336 | ||
| Forward | Introns 7–8 | 5′-CCCACTGTATTCCTTAATTGTAGTAGCTT-3′ | |
| Reverse | Introns 8–9 | 5′-TCCACATGGTTAACGTGTATCTAGAAA-3′ | |
| 9: | 351 | ||
| Forward | Introns 8–9 | 5′-TTCAGTGTCAATATATCAAGCCTACTGA-3′ | |
| Reverse | Introns 9–10 | 5′-CACTATGCAATTTTTCCATAGAGCAG-3′ | |
| 10: | 231 | ||
| Forward | Introns 9–10 | 5′-TGGCTGTCATTGAGTAATTATTCTGC-3′ | |
| Reverse | Introns 10–11 | 5′-TCCCCCCTTAACTTATTATTTGATATTG-3′ | |
| 11: | 343 | ||
| Forward | Introns 10–11 | 5′-CTGTCTAGGTCAGGTCCTTTAGTAGACTG-3′ | |
| Reverse | Introns 11–12 | 5′-CAGCAGGATAGTTCAGGTGACATT-3′ | |
| 12: | 324 | ||
| Forward | Introns 11–12 | 5′-AACCTTTGTCATTGATACCCAACTC-3′ | |
| Reverse | Introns 12–13 | 5′-TCCTCATCAAGCCCCTCGT-3′ | |
| 13: | 298 | ||
| Forward | Introns 12–13 | 5′-AGTTCTAAAGACATGAGGTTGATTTACATAA-3′ | |
| Reverse | Introns 13–14 | 5′-CACTCTTTGTTCATTTGCATTTTCC-3′ | |
| 14: | 409 | ||
| Forward | Intron 13–14 | 5′-AGGACAGAAATGGGACAAATGAA-3′ | |
| Reverse | Intron 14–15 | 5′-TTCTAAGGAAGATAACAAATAGGTACTGTAACA-3′ | |
| 15: | 294 | ||
| Forward | Introns 14–15 | 5′-AGTCTCCTGCAACTTGTCTTTATACTCAT-3′ | |
| Reverse | Introns 15–16 | 5′-TCATTCGTGAGACACTGAGATTTCT-3′ | |
| 16: | 321 | ||
| Forward | Introns 15–16 | 5′-AATATTTGATCAGTTACAGTAGTCTCGTTACC-3′ | |
| Reverse | Introns 16–17 | 5′-TGCAAAATCATCATCCACCTTG-3′ | |
| 17: | 338 | ||
| Forward | Introns 16–17 | 5′-TCTTCATTTTTGATGCACCATGT-3′ | |
| Reverse | Introns 17–18 | 5′-TTTCTGTTCCTTCCATTCATTTCAT-3′ | |
| 18: | 211 | ||
| Forward | Introns 17–18 | 5′-CCTCAGCTCTAAACTAAGGGCCA-3′ | |
| Reverse | Introns 18–19 | 5′-TCTTACTATAGAGTTTCCCCAGTCCTG-3′ | |
| 19: | 240 | ||
| Forward | Introns 18–19 | 5′-AAATCTTCCATTTCTTAACGCTTTATATTG-3′ | |
| Reverse | Introns 19–20 | 5′-AAAAACACAACTTACCTCCACGG-3′ | |
| 20: | 432 | ||
| (continued) | |||
| Forward | Introns 19–20 | 5′-GATAGTTTTGGGCTTCCGGTG-3′ | |
| Reverse | Introns 20–21 | 5′-TGAATCACTAAGTCAAACAACAAATACTAATT-3′ | |
| 21: | 306 | ||
| Forward | Introns 20–21 | 5′-CATAAATACACCTTGCCATCTGATG-3′ | |
| Reverse | Introns 21–22 | 5′-TGGCAAAAATGAACAACAGAGAG-3′ | |
| 22: | 411 | ||
| Forward | Introns 21–22 | 5′-TGCATGTATGTGTATATGTATTTGTGTCA-3′ | |
| Reverse | Introns 22–23 | 5′-TGTAGTTCTATCAGAATCTCTTGTGAATTTATT-3′ | |
| 23: | 379 | ||
| Forward | Introns 22–23 | 5′-AAGACAGTATGAGTTACTATAGCATGCATTT-3′ | |
| Reverse | Introns 23–24 | 5′-GGAAGTAGTTGACTTTTGATGCATTTTA-3′ | |
| 24: | 325 | ||
| Forward | Introns 23–24 | 5′-AGGTAGATTTACAGGTTAGTCATAGTGCA-3′ | |
| Reverse | Introns 24–25 | 5′-CATGGAGCACGTCTCTTCAGC-3′ | |
| 25: | 351 | ||
| Forward | Introns 24–25 | 5′-TGGTAAAGTAGGCCATTCACAGTG-3′ | |
| Reverse | Introns 25–26 | 5′-CAAGATTCTTTCTGAATCACGATAGC-3′ | |
| 26: | 574 | ||
| Forward | Introns 25–26 | 5′-TCACAAACTCATCTATCTGAAGACCTTAT-3′ | |
| Reverse | 3′-UTR | 5′-AAAATTGCAGTGCAAAATTTAAACA-3′ | |
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- BACPAC Resources, http://bacpac.chori.org/
- DGAP, http://dgap.harvard.edu/
- Ensembl, http://www.ensembl.org/Homo_sapiens/index.html
- Entrez Protein, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Protein (for ROBO2 [accession number NP_002933])
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for ROBO2 [accession number NM_002942] and PCDH11Y [accession number NM_032971])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for VUR and SMS)
References
- 1.Pope JC 4th, Brock JW 3rd, Adams MC, Stephens FD, Ichikawa I (1999) How they begin and how they end: classic and new theories for the development and deterioration of congenital anomalies of the kidney and urinary tract, CAKUT. J Am Soc Nephrol 10:2018–2028 [DOI] [PubMed] [Google Scholar]
- 2.Nakanishi K, Yoshikawa N (2003) Genetic disorders of human congenital anomalies of the kidney and urinary tract (CAKUT). Pediatr Int 45:610–616 10.1046/j.1442-200X.2003.01779.x [DOI] [PubMed] [Google Scholar]
- 3.Feather SA, Malcolm S, Woolf AS, Wright V, Blaydon D, Reid CJ, Flinter FA, Proesmans W, Devriendt K, Carter J, et al (2000) Primary, nonsyndromic vesicoureteric reflux and its nephropathy is genetically heterogeneous, with a locus on chromosome 1. Am J Hum Genet 66:1420–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sargent MA (2000) What is the normal prevalence of vesicoureteral reflux? Pediatr Radiol 30:587–593 10.1007/s002470000263 [DOI] [PubMed] [Google Scholar]
- 5.Bailey RR (1973) The relationship of vesico-ureteric reflux to urinary tract infection and chronic pyelonephritis-reflux nephropathy. Clin Nephrol 1:132–141 [PubMed] [Google Scholar]
- 6.Dillon MJ, Goonasekera CD (1998) Reflux nephropathy. J Am Soc Nephrol 9:2377–2383 [DOI] [PubMed] [Google Scholar]
- 7.Kincaid-Smith PS, Bastos MG, Becker GJ (1984) Reflux nephropathy in the adult. Contrib Nephrol 39:94–101 [DOI] [PubMed] [Google Scholar]
- 8.Mackie GG, Awang H, Stephens FD (1975) The ureteric orifice: the embryologic key to radiologic status of duplex kidneys. J Pediatr Surg 10:473–481 10.1016/0022-3468(75)90187-6 [DOI] [PubMed] [Google Scholar]
- 9.Noe HN, Wyatt RJ, Peeden JN Jr, Rivas ML (1992) The transmission of vesicoureteral reflux from parent to child. J Urol 148:1869–1871 [DOI] [PubMed] [Google Scholar]
- 10.Van den Abbeele AD, Treves ST, Lebowitz RL, Bauer S, Davis RT, Retik A, Colodny A (1987) Vesicoureteral reflux in asymptomatic siblings of patients with known reflux: radionuclide cystography. Pediatrics 79:147–153 [PubMed] [Google Scholar]
- 11.Kidd T, Brose K, Mitchell KJ, Fetter RD, Tessier-Lavigne M, Goodman CS, Tear G (1998) Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell 92:205–215 10.1016/S0092-8674(00)80915-0 [DOI] [PubMed] [Google Scholar]
- 12.Xian J, Aitchison A, Bobrow L, Corbett G, Pannell R, Rabbitts T, Rabbitts P (2004) Targeted disruption of the 3p12 gene, Dutt1/Robo1, predisposes mice to lung adenocarcinomas and lymphomas with methylation of the gene promoter. Cancer Res 64:6432–6437 10.1158/0008-5472.CAN-04-2561 [DOI] [PubMed] [Google Scholar]
- 13.Xian J, Clark KJ, Fordham R, Pannell R, Rabbitts TH, Rabbitts PH (2001) Inadequate lung development and bronchial hyperplasia in mice with a targeted deletion in the Dutt1/Robo1 gene. Proc Natl Acad Sci USA 98:15062–15066 10.1073/pnas.251407098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hannula-Jouppi K, Kaminen-Ahola N, Taipale M, Eklund R, Nopola-Hemmi J, Kaariainen H, Kere J (2005) The axon guidance receptor gene ROBO1 is a candidate gene for developmental dyslexia. PLoS Genet 1:e50 10.1371/journal.pgen.0010050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jen JC, Chan WM, Bosley TM, Wan J, Carr JR, Rub U, Shattuck D, Salamon G, Kudo LC, Ou J, et al (2004) Mutations in a human ROBO gene disrupt hindbrain axon pathway crossing and morphogenesis. Science 304:1509–1513 10.1126/science.1096437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bedell VM, Yeo SY, Park KW, Chung J, Seth P, Shivalingappa V, Zhao J, Obara T, Sukhatme VP, Drummond IA, et al (2005) Roundabout4 is essential for angiogenesis in vivo. Proc Natl Acad Sci U S A 102:6373–6378 10.1073/pnas.0408318102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fricke C, Lee JS, Geiger-Rudolph S, Bonhoeffer F, Chien CB (2001) Astray, a zebrafish roundabout homolog required for retinal axon guidance. Science 292:507–510 [DOI] [PubMed] [Google Scholar]
- 18.Long H, Sabatier C, Ma L, Plump A, Yuan W, Ornitz DM, Tamada A, Murakami F, Goodman CS, Tessier-Lavigne M (2004) Conserved roles for Slit and Robo proteins in midline commissural axon guidance. Neuron 42:213–223 10.1016/S0896-6273(04)00179-5 [DOI] [PubMed] [Google Scholar]
- 19.Grieshammer U, Le M, Plump AS, Wang F, Tessier-Lavigne M, Martin GR (2004) SLIT2-mediated ROBO2 signaling restricts kidney induction to a single site. Dev Cell 6:709–717 10.1016/S1534-5807(04)00108-X [DOI] [PubMed] [Google Scholar]
- 20.Li HS, Chen JH, Wu W, Fagaly T, Zhou L, Yuan W, Dupuis S, Jiang ZH, Nash W, Gick C, et al (1999) Vertebrate slit, a secreted ligand for the transmembrane protein roundabout, is a repellent for olfactory bulb axons. Cell 96:807–818 10.1016/S0092-8674(00)80591-7 [DOI] [PubMed] [Google Scholar]
- 21.Ward ME, Rao Y (2005) Investigations of neuronal migration in the central nervous system. Methods Mol Biol 294:137–156 [DOI] [PubMed] [Google Scholar]
- 22.Blanco P, Sargent CA, Boucher CA, Mitchell M, Affara NA (2000) Conservation of PCDHX in mammals; expression of human X/Y genes predominantly in brain. Mamm Genome 11:906–914 10.1007/s003350010177 [DOI] [PubMed] [Google Scholar]
- 23.Potocki L, Shaw CJ, Stankiewicz P, Lupski JR (2003) Variability in clinical phenotype despite common chromosomal deletion in Smith-Magenis syndrome [del(17)(p11.2p11.2)]. Genet Med 5:430–434 [DOI] [PubMed] [Google Scholar]
- 24.Liu Z, Patel K, Schmidt H, Andrews W, Pini A, Sundaresan V (2004) Extracellular Ig domains 1 and 2 of Robo are important for ligand (Slit) binding. Mol Cell Neurosci 26:232–240 10.1016/j.mcn.2004.01.002 [DOI] [PubMed] [Google Scholar]
- 25.Wu W, Wong K, Chen J, Jiang Z, Dupuis S, Wu JY, Rao Y (1999) Directional guidance of neuronal migration in the olfactory system by the protein Slit. Nature 400:331–336 10.1038/22477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chapman CJ, Bailey RR, Janus ED, Abbott GD, Lynn KL (1985) Vesicoureteric reflux: segregation analysis. Am J Med Genet 20:577–584 10.1002/ajmg.1320200403 [DOI] [PubMed] [Google Scholar]
- 27.Li X, Chen Y, Liu Y, Gao J, Gao F, Bartlam M, Wu JY, Rao Z (2006) Structural basis of Robo proline-rich motif recognition by the srGAP1 Src homology 3 domain in the Slit-Robo signaling pathway. J Biol Chem 281:28430–28437 10.1074/jbc.M604135200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong K, Ren XR, Huang YZ, Xie Y, Liu G, Saito H, Tang H, Wen L, Brady-Kalnay SM, Mei L, et al (2001) Signal transduction in neuronal migration: roles of GTPase activating proteins and the small GTPase Cdc42 in the Slit-Robo pathway. Cell 107:209–221 10.1016/S0092-8674(01)00530-X [DOI] [PubMed] [Google Scholar]
- 29.Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H (1996) Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA 93:5860–5865 10.1073/pnas.93.12.5860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srinivas S, Goldberg MR, Watanabe T, D'Agati V, al-Awqati Q, Costantini F (1999) Expression of green fluorescent protein in the ureteric bud of transgenic mice: a new tool for the analysis of ureteric bud morphogenesis. Dev Genet 24:241–251 [DOI] [PubMed] [Google Scholar]
- 31.Holzenberger M, Lenzner C, Leneuve P, Zaoui R, Hamard G, Vaulont S, Bouc YL (2000) Cre-mediated germline mosaicism: a method allowing rapid generation of several alleles of a target gene. Nucleic Acids Res 28:E92 10.1093/nar/28.21.e92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanagho EA, Guthrie TH, Lyon RP (1969) The intravesical ureter in primary reflux. J Urol 101:824–832 [DOI] [PubMed] [Google Scholar]
- 33.Stauss J, Connolly LP, Connolly SA, Zurakowski D, Treves ST, Peters CA (2003) Dynamic renal scintigraphy in children with vesicoureteral reflux and suspected coexisting ureteropelvic junction obstruction. J Urol 170:1966–1970 10.1097/01.ju.0000092163.08445.e5 [DOI] [PubMed] [Google Scholar]
- 34.Batourina E, Tsai S, Lambert S, Sprenkle P, Viana R, Dutta S, Hensle T, Wang F, Niederreither K, McMahon AP, et al (2005) Apoptosis induced by vitamin A signaling is crucial for connecting the ureters to the bladder. Nat Genet 37:1082–1089 10.1038/ng1645 [DOI] [PubMed] [Google Scholar]
- 35.Sanna-Cherchi S, Reese A, Hensle T, Caridi G, Izzi C, Kim YY, Konka A, Murer L, Scolari F, Ravazzolo R, et al (2005) Familial vesicoureteral reflux: testing replication of linkage in seven new multigenerational kindreds. J Am Soc Nephrol 16:1781–1787 10.1681/ASN.2004121034 [DOI] [PubMed] [Google Scholar]
- 36.Eccles MR, Bailey RR, Abbott GD, Sullivan MJ (1996) Unravelling the genetics of vesicoureteric reflux: a common familial disorder. Hum Mol Genet 5:1425–1429 [DOI] [PubMed] [Google Scholar]
- 37.Yue Y, Grossmann B, Galetzka D, Zechner U, Haaf T (2006) Isolation and differential expression of two isoforms of the ROBO2/Robo2 axon guidance receptor gene in humans and mice. Genomics 88:772–778 10.1016/j.ygeno.2006.05.011 [DOI] [PubMed] [Google Scholar]
- 38.Slager RE, Newton TL, Vlangos CN, Finucane B, Elsea SH (2003) Mutations in RAI1 associated with Smith-Magenis syndrome. Nat Genet 33:466–468 10.1038/ng1126 [DOI] [PubMed] [Google Scholar]
- 39.Bi W, Yan J, Stankiewicz P, Park SS, Walz K, Boerkoel CF, Potocki L, Shaffer LG, Devriendt K, Nowaczyk MJ, et al (2002) Genes in a refined Smith-Magenis syndrome critical deletion interval on chromosome 17p11.2 and the syntenic region of the mouse. Genome Res 12:713–728 10.1101/gr.73702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chou IC, Tsai FJ, Yu MT, Tsai CH (2002) Smith-Magenis syndrome with bilateral vesicoureteral reflux: a case report. J Formos Med Assoc 101:726–728 [PubMed] [Google Scholar]
- 41.Greenberg F, Lewis RA, Potocki L, Glaze D, Parke J, Killian J, Murphy MA, Williamson D, Brown F, Dutton R, et al (1996) Multi-disciplinary clinical study of Smith-Magenis syndrome (deletion 17p11.2). Am J Med Genet 62:247–254 [DOI] [PubMed] [Google Scholar]
- 42.Bi W, Ohyama T, Nakamura H, Yan J, Visvanathan J, Justice MJ, Lupski JR (2005) Inactivation of Rai1 in mice recapitulates phenotypes observed in chromosome engineered mouse models for Smith-Magenis syndrome. Hum Mol Genet 14:983–995 10.1093/hmg/ddi085 [DOI] [PubMed] [Google Scholar]
- 43.Walz K, Caratini-Rivera S, Bi W, Fonseca P, Mansouri DL, Lynch J, Vogel H, Noebels JL, Bradley A, Lupski JR (2003) Modeling del(17)(p11.2p11.2) and dup(17)(p11.2p11.2) contiguous gene syndromes by chromosome engineering in mice: phenotypic consequences of gene dosage imbalance. Mol Cell Biol 23:3646–3655 10.1128/MCB.23.10.3646-3655.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan J, Keener VW, Bi W, Walz K, Bradley A, Justice MJ, Lupski JR (2004) Reduced penetrance of craniofacial anomalies as a function of deletion size and genetic background in a chromosome engineered partial mouse model for Smith-Magenis syndrome. Hum Mol Genet 13:2613–2624 10.1093/hmg/ddh288 [DOI] [PubMed] [Google Scholar]