Abstract
We report a female patient with a de novo balanced translocation, 46,X,t(X;2)(p11.2;q37)dn, who exhibits seizures, capillary abnormality, developmental delay, infantile hypotonia, and obesity. The 2q37 breakpoint observed in association with the seizure phenotype is of particular interest, because it lies near loci implicated in epilepsy in humans and mice. Fluorescence in situ hybridization mapping of the translocation breakpoints showed that no known genes are disrupted at Xp11.2, whereas diacylglycerol kinase delta (DGKD) is disrupted at 2q37. Expression studies in Drosophila and mouse suggest that DGKD is involved in central nervous system development and function. Electroencephalographic assessment of Dgkd mutant mice revealed abnormal epileptic discharges and electrographic seizures in three of six homozygotes. These findings implicate DGKD disruption by the t(X;2)(p11.2;q37)dn in the observed phenotype and support a more general role for DGKD in the etiology of seizures.
Epilepsy is a relatively common multifactorial neurological disorder characterized by recurrent unprovoked seizures, affecting ∼3% of the population during their life span.1 Despite the evidence of a genetic component, especially in idiopathic epilepsy,2,3 and the frequency of seizure disorders in humans, the genetic basis is difficult to determine because of trait heterogeneity and complexity. Not surprisingly, most genes identified as causative for epilepsy syndromes encode ion channels, including subunits of sodium channels in generalized epilepsy with febrile seizures (GEFS+ [MIM 604233]),4,5 potassium channels in neonatal human epilepsy (EBN1 [MIM 121200], EBN2 [MIM 121201]),6 ligand gated ion channels (e.g., GABAA receptor subunits) in childhood absence epilepsy (ECA2 [MIM 607681]),7 and neuronal nicotinic acetylcholine receptors in autosomal dominant nocturnal frontal lobe epilepsy (ENFL1 [MIM 600513] and ENFL3 [MIM 605375]).8 However, there are several examples of epilepsy genes that do not fall into the channelopathy category, including the Aristaless-related homeobox gene (ARX) in human X-linked epilepsy (ISSX [MIM 308350]),9 the ethanolamine kinase gene (eas) in the Drosophila “easily shocked” seizure mutant,10 the leucine-rich glioma inactivated 1 gene (LGI1) in a temporal lobe epilepsy (ADLTE [MIM 600512]),11 the very large G-protein coupled receptor 1 (VLGR1/MASS1) in febrile seizures (FEB4 [MIM 604352]),12 and the EF-hand domain–containing protein 1 (EFHC1) in juvenile myoclonic epilepsy (EJM1 [MIM 254770].13 Such functional diversity makes gene discovery in epilepsy especially challenging.
A successful gene-discovery approach in the setting of functional diversity is analysis of balanced chromosomal rearrangements in humans with developmental disorders. Phenotypic abnormalities observed in association with balanced chromosomal rearrangements can be explained by alteration of expression of a gene(s) residing at or near the chromosomal breakpoint(s). This principle serves as the rationale for the Developmental Genome Anatomy Project (DGAP), a study from which this case originated. DGAP is an ongoing research endeavor approved by the Partners HealthCare System Human Research Committee. In this study, the finding of an apparently balanced t(X;2)(p11.2;q37)dn in an individual with epilepsy (designated “DGAP095”) provided an opportunity to identify a gene liable for the phenotype. We found that the 2q37 breakpoint disrupts diacylglycerol kinase delta (DGKD), and our expression studies support a role for DGKD in CNS development and function. Electroencephalographic (EEG) assessment of Dgkd knockdown mice revealed abnormal epileptic discharges in half of the mutant mice evaluated, indicating DGKD involvement in the seizure phenotype observed in DGAP095.
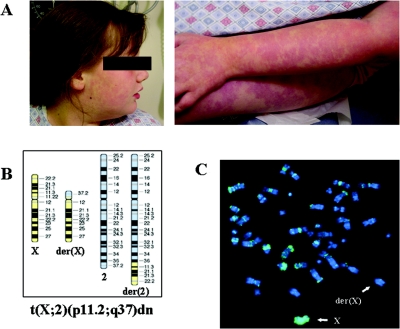
At the time of assessment, DGAP095 was a mildly obese 12-year-old female with epilepsy, congenital capillary abnormalities, mild hypotonia, and a history of developmental delay (fig. 1A). Her weight was at the 95th percentile, her height at the 10th percentile, and her head circumference at the 90th percentile. She was the first child of healthy, nonconsanguineous parents who also have a healthy younger son. She was delivered vaginally after a normal pregnancy (birth weight 8 pounds 10 ounces; Apgar scores 8 and 9). She crawled and walked at ages 2 and 3.5 years, respectively. At 3.5 years of age, she received the diagnosis of generalized tonic clonic seizures, which evolved into drop attacks at age 5 years that eventually subsided; at 11 years of age, she developed staring events with an occasional myoclonic component, and currently she has 1–2 generalized tonic clonic seizures per week. DGAP095 first exhibited cognitive delay at the age of 18 mo and verbal/auditory processing difficulties characterized by delayed responses and perseveration. She also experienced self-stimulatory behavior, including hair pulling, leg slapping, hand flapping, and echolalia. Seizures in conjunction with behavioral findings are consistent with the Lennox-Gastaut syndrome form of epilepsy.14 Brain magnetic resonance imaging did not show any abnormalities, and comprehensive metabolic testing showed results within normal limits. A skin biopsy did not reveal any pathological changes. Chromosomal analysis revealed an apparently balanced de novo translocation between chromosomes X and 2, designated 46,X,t(X;2)(p11.2;q37)dn (fig. 1B). To exclude the possibility that her clinical phenotype reflected some form of genomic imbalance, nonrandom inactivation of the normal X chromosome was confirmed by BrdU-based replication studies (fig. 1C), and no copy number alterations were detected by whole-genome array comparative genome hybridization (CGH) at a resolution of ∼1 Mb (GenomeChip V1.2 Human BAC Arrays [Spectral Genomics]). The potential etiology of the phenotype due to the t(X;2) was supported by phenotypic overlap between DGAP095 and patients with del(2)(q37),15–19 genetic linkage studies implicating 2q36 as a seizure-susceptibility locus,20,21 and the fact that the mouse chromosomal region with homologous synteny to human 2q37 is reported to contain a seizure susceptibility locus.22 Thus, the cytogenetic anomaly in DGAP095, in conjunction with the published linkage data, provided a clue to the location of a potential epilepsy gene at 2q37.
Figure 1. .
A, DGAP095 capillary abnormalities on face and limbs. B, Ideogram of t(X;2)(p11.2;q37)dn present in DGAP095. C, Metaphase spread following staining with DAPI and fluorescein-conjugated BrdU antibodies. Note that the normal chromosome X is BrdU-stained, indicating its late-replication status.
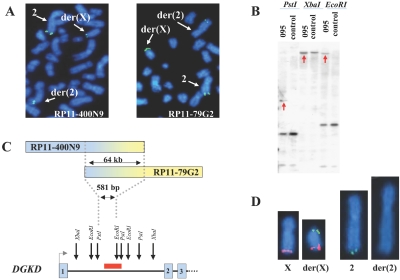
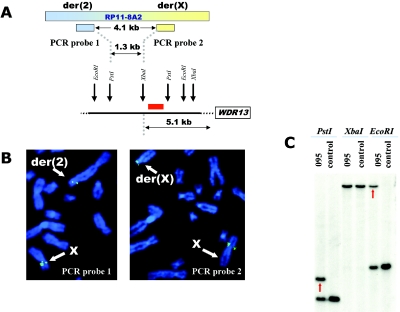
Consecutive FISH experiments with BACs, either directly labeled with SpectrumOrange and SpectrumGreen (Abbott Molecular/Vysis) or indirectly labeled with biotin, were employed for initial narrowing of the breakpoint regions. On chromosome 2, BAC clone RP11-400N9 showed hybridization signals localized to both the der(2) and der(X) chromosomes, demonstrating that it spanned the breakpoint (fig. 2A). FISH with another BAC clone, RP11-79G2, which overlaps clone RP11-400N9, revealed that it also hybridized to both derivative chromosomes, thus placing the breakpoint within a 64-kb region of overlap (fig. 2A and 2C). Analyses of National Center for Biotechnology Information and University of California Santa Cruz gene maps indicated that the breakpoint on chromosome 2 disrupted DGKD. For further breakpoint refinement, Southern blot analyses hybridized with radioactive PCR probes from the overlapping area were employed and narrowed the breakpoint region to 581 bp within intron 1 (fig. 2B and 2C). Indirect labeling with biotin and subsequent amplification of the signal by use of avidin and anti-avidin conjugated with FITC (Roche) were used for FISH, with a cDNA for DGKD (KIAA0145, clone ha00914 [Kazusa DNA Research Institute]). The DGKD cDNA showed expected hybridization signals on both the normal chromosome 2 and der(X) (fig. 2D). No DGKD hybridization was observed on the der(2), because of a very small target sequence (a single exon), confirming translocation of the majority of DGKD to the der(X). The breakpoint on the X chromosome was found to reside within BAC clone RP11-8A2 (data not shown). At the time of mapping, this BAC represented a draft clone; however, it overlapped several fully sequenced clones. On the basis of the sequence of these overlapping clones, PCR probes were designed and used for FISH to narrow the breakpoint region to 4.1 kb (fig. 3A and 3B). Southern blot analyses further localized the X chromosome breakpoint to a 1,393-bp region (figs. 3A and 3C). This chromosomal break did not occur within any known or putative genes, with the closest gene residing at a distance of 5.1 kb. Expression of this gene, WDR13, appeared to be unaffected by the translocation by RT-PCR analysis (data not shown). Interestingly, both breakpoint regions contained Alu-Sx repetitive elements, which could provide a molecular mechanism for the formation of the translocation by illegitimate recombination between chromosomes X and 2.
Figure 2. .
Mapping of the breakpoint on chromosome 2. A, Representative metaphase spread following FISH with clone RP11-400N9 (left) and RP11-79G2 (right); both probes are biotin labeled and detected by FITC-conjugated avidin. FISH signals are detected on the normal chromosome 2, der(X), and der(2), indicating that the breakpoint is contained within the same genomic sequence on RP11-400N9 and RP11-79G2. B, Refinement of the breakpoint region, performed using Southern blot hybridization with a 1,363-bp radioactive PCR probe from the chromosome 2 breakpoint region. Red arrows indicate novel aberrant restriction fragments in DGAP095 DNA caused by chromosomal rearrangement. C, Orientation of the RP11-400N9 and RP11-79G2 clones relative to each other (and reflective of uneven split signals on the derivative chromosomes). The defined breakpoint region falls within the first intron of DAG kinase delta (DGKD), as shown on the DGKD structural diagram, where numbered boxes represent exons. Restriction sites of the enzymes used for the Southern blot are indicated by arrows, and the location of the PCR probe is represented by a red rectangle. D, Representative metaphase chromosomes X, 2, der(2), and der(X) after hybridization with DGKD cDNA labeled with FITC. Note the absence of DGKD hybridization signal on the der(2). (Chromosome X subtelomeric probe labeled with SpectrumOrange was used to facilitate identification of the der(X).)
Figure 3. .
Mapping of the breakpoint on chromosome X. A, Graphic representation of orientation of PCR probes used for FISH and map of restriction sites of the enzymes used for the Southern blot, with the probe location represented by a red rectangle. B, Metaphase chromosomes following FISH with PCR probe 1 (left) and PCR probe 2 (right); both probes are labeled by biotin and detected by FITC-conjugated avidin. Probe 1 hybridizes to the normal chromosome X and to the der(2), whereas probe 2 hybridizes to the normal X and der(X), indicating that the breakpoint is located within the sequence flanked by the probes. C, Southern blot hybridized to the 371-bp radioactive PCR probe from the chromosome X breakpoint region (originally defined by FISH). (No aberrant fragment is present in the XbaI digest because the breakpoint occurs outside of the area flanked by XbaI sites.)
3′-RACE analysis (Qiagen) performed on RNA isolated from DGAP095 lymphoblastoid cells showed the presence of two alternatively spliced DGKD transcripts but no evidence of chimeric transcripts. This result is expected, because of the absence of an ORF close to the reciprocal breakpoint on the X chromosome. Also, to exclude hidden heterozygosity in DGAP095, the coding region of DGKD was analyzed, with the exception of a highly CG-rich repetitive region encompassing exon 1 that was refractory to sequence analysis, and no mutations were found in the regions assessed. The absence of novel transcripts and of any sequence mutations in the second DGKD allele implies that DGAP095's abnormal phenotype is due to DGKD haploinsufficiency.
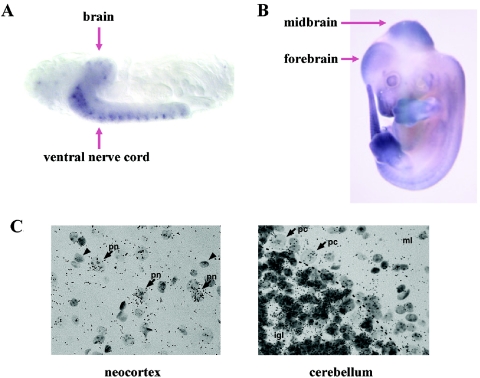
Whole-mount in situ hybridization experiments were performed to investigate Dgkd developmental expression in both mouse and fly. Embryos were fixed in 4% paraformaldehyde/PBS, permeabilized with proteinase K, and then hybridized to a digoxigenin-labeled probe antisense to a unique sequence within the respective Dgkd orthologs. Probes were prepared by RT-PCR followed by subcloning into the pDrive cloning vector (Qiagen) and labeled using a digoxigenin RNA labeling kit (Boehringer Mannheim). Expression of the presumptive Dgkd fly ortholog (CG31187), identified by two-directional BLAST analysis, was confined to the developing brain and ventral nerve cord of late-stage (stage 16) fly embryos (fig. 4A). Murine Dgkd, identified by homologous synteny, displayed prominent expression in the developing midbrain and forebrain of E12.5 embryos (fig. 4B). Radioactive in situ hybridization was used to determine Dgkd expression patterns in adult mouse brain. A [35S]-labeled Dgkd riboprobe specific to the unique 3′ UTR region of Dgkd was generated by PCR followed by subcloning into the pCR 2.1-TOPO vector (Invitrogen) and was synthesized and labeled using an in vitro transcription kit (Roche). Sagittal sections of mouse brain were hybridized to 106 counts/min of riboprobe and were exposed at 4°C for 2 wk in Kodak NTB emulsion. After exposure, slides were developed and counterstained with hematoxylin. The study revealed a broad neuronal expression pattern with particular prominence in pyramidal neurons of the neocortex and hippocampus, and also within internal granule cell neurons of the cerebellum (fig. 4C). The observed Dgkd expression patterns suggest the relevance of Dgkd to neural development and brain pathophysiology.
Figure 4. .
Whole-mount in situ hybridization showing expression of Dgkd orthologs. A, Stage 16 fly embryo hybridized to the Drosophila Dgkd antisense probe. Note expression in brain and ventral nerve cord. B, E12.5 mouse embryo hybridized to the mouse Dgkd antisense probe. Note prominent expression in forebrain and midbrain. C, In situ hybridization performed using a 3′ UTR probe to the Dgkd transcript on adult wild-type mouse neocortex and cerebellum. Dark granules represent Dgkd transcripts. Neocortex: moderate to high Dgkd expression in large pyramidal neurons (pn and arrows) and relatively absent to low in small cells with the morphology of glia (arrowheads). In the cerebellum, expression is relatively low in glia and neurons of the molecular layer (ml), intermediate within the Purkinje cell and Bergmann glial cell layer (pc and arrows point to Purkinje cells), and highest in granule neurons within the internal granule cell layer (igl).
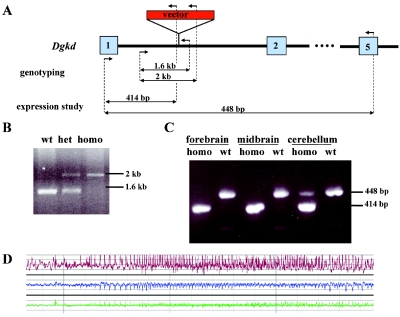
To evaluate further the role of DGKD disruption in the DGAP095 phenotype, a Dgkd mutant mouse was generated. Mouse protocols were approved by the Institutional Animal Care and Use Committee at Harvard Medical School. A mouse embryonic stem cell line (RRT600, strain Ola/129) developed using gene-trap methodology was identified in the BayGenomics database by use of BLAST with human DGKD cDNA sequence. RT-PCR of the RNA pool isolated from the mouse embryos and the brain of adult mice showed that the RRT600 sequence tag belonged to the first exon of murine Dgkd; thus, the ES cell line was designated as DgkdGt(pGTlxf)Byg. The precise location of the gene-trap vector pGTlxf insertion site was determined to be within the first intron of Dgkd (fig. 5A and 5B). The gene-trap vector possesses a strong splice-site–acceptor sequence (i.e., Engrailed-2 [En2] exon), and, as a result of its integration, the first exon splices to the vector sequences resulting in generation of a fusion transcript between Dgkd exon 1 and the gene-trap vector. Because Dgkd is expressed in adult wild-type mouse brain, RT-PCR studies were performed on total RNA isolated from mutant mouse brain (RNeasy kit [Qiagen]) to validate that the gene-trap insertion affected Dgkd expression. Homozygous mutant mice showed absent Dgkd expression in forebrain and midbrain tissues; however, faint expression was detected in cerebellum (fig. 5C). Therefore, the DgkdGt(pGTlxf)Byg allele appears to represent a knockdown or hypomorphic allele rather than a categoric null. In addition, Dgkd encodes two alternative transcripts. Insertion of the gene-trap vector affected expression of the most abundant transcript, but expression of a second, low-abundance transcript that initiates in exon 2 was comparatively unaffected (data not shown). Of note, the site of the gene-trap insertion is analogous to the DGKD disruption in DGAP095, in which the translocation breakpoint falls within the first intron. Also, the knockdown nature of the introduced mutation approximates the haploinsufficient state for DGKD in DGAP095. Thus, the knockdown mutant represents a directly relevant model for assessing DGAP095's genetic defect.
Figure 5. .
A, Graphic representation of the gene-trap vector insertion within Dgkd intron 1. Numbered boxes represent exons, arrows represent primers used for genotyping and expression assays, and double-sided arrows represent expected amplicons. B, Representative genotyping assay performed on wild-type, heterozygous (het), and homozygous (homo) mice. C, Dgkd expression analysis in brain tissues from homozygous and wild-type mice. D, EEG recording showing epileptiform activity in a Dgkd knockdown mutant mouse. A sudden onset of ictal electrographic discharge is recorded from both hemispheres (upper channel [red] and middle channels [blue] represent left and right hemispheres, respectively), with higher amplitude on the left. The frequency and amplitude of the discharge evolves in a manner typical of seizure activity. Simultaneous EMG recording (lower channel [green]) demonstrates fast, low-voltage frequencies that might reflect fine-motor activity associated with electrographic seizure discharge. (It is possible that the EMG channel is contaminated with EEG activity, as the recording electrode is placed close to the head.) No gross movements are observed, excluding the artifact as a cause of the EEG changes. Time scale is 5 s per major division. Amplitude scale is 800 microvolts per major division.
Dgkd homozygous mutant mice were viable and fertile. Twenty-four mutant mice of different ages (14 homozygotes and 10 heterozygotes) were assessed for gross anatomical and histopathological abnormalities by complete necropsy, and no consistent morphological abnormalities were noted. Nevertheless, these analyses do not exclude more-subtle physical abnormalities or those requiring additional physiological studies. A potential seizure phenotype in mutant mice was initially investigated by a seizure-susceptibility study using a subthreshold dose of pentylenetetrazol (PTZ) (40 mg/kg) in 10 homozygous mutants at the age of 2 mo (5 females and 5 males) and in 10 wild-type littermate controls (5 females and 5 males). The latency and severity of seizures were evaluated as described elsewhere,22 and scores were compared between wild-type and knockdown littermates. No significant difference in susceptibility to PTZ-induced seizures was noted.
To examine potential electroencephalic abnormalities, EEG/electromyography (EMG) assessments were performed on six homozygotes (three females and three males) and two wild-type male littermate controls at 12 wk of age. Animals were implanted with two EMG electrodes inserted into neck muscles and four EEG electrodes on the skull at the following coordinates: 2 mm lateral to and 1 mm anterior to the Bregma for the two front electrodes, and 2 mm lateral to and 3 mm posterior to the Bregma for the two rear electrodes. On the 4th d after surgery, animals were wired to an MP150 physiological signal recording device (Biopac Systems). Two channels of EEG, one from each side of the brain, and one channel of EMG data were recorded simultaneously from each animal. High- and low-pass filters were 0.1 Hz and 100 Hz for EEG and 1.0 Hz and 500 Hz for EMG, respectively. Continuous EEG/EMG recording was allowed for all mice, provided they had good signals, until the experiment was discontinued on the 24th d after surgery. For each animal, the continuous EEG was visually inspected to identify electrographic seizure activity. Epileptiform activity was recognized on the basis of commonly used principles of EEG analysis, including the occurrence of epileptiform spikes distinguished on the basis of morphology, electrical field and amplitude, and changes in baseline activity with an evolving pattern of rhythmic activity of higher amplitude than the baseline. The pattern of activity and the specific morphology of the EEG activity were assessed by an experienced electroencepalographer (A.J.C.). For each animal, two channels of EEG recording were available along with a single channel of EMG recording to allow the assessment of associated muscle activity. In three mutant mice (two males and one female), discrete seizures were identified in association with rhythmic EMG activities, whereas no seizures were noted in the wild-type mice. One mutant mouse had a bilateral ictal discharge (fig. 5D), whereas, in the other two mutants, the ictal EEG activity was confined to a single hemisphere, suggesting a focal origin. Analysis of contemporaneous EMG activity excluded the possibility that the abnormal EEG activity represented movement artifact. Interestingly, one of the mutant animals had frequent, high-amplitude (800–1000 microvolts), interictal epileptiform spikes occurring once every 10–120 s. In that animal, direct observation was made during the occurrence of an electrographic seizure; a long spike train was observed when the mouse was motionless in a normal sleeping posture, and only a fine jittering motion of the head was noted. Postmortem analyses excluded the possibility of artifacts introduced by improper electrode implantation. The fact that only half of the mutant mice exhibited seizures could be due to the mixed C57BL/6 X Ola/129 genetic background in which the stock was maintained; the nature of the hypomorphic mutation, which could attenuate robust expression of the phenotype; or the limited observation period.
In respect to the other DGAP095 clinical features, the potential vascular and obesity phenotypes were not exhaustively investigated in mutant mice. The absence of apparent capillary abnormalities in mutant mice does not exclude the potential presence of a physiological abnormality that has not been observed. Also, the mutant mice tended to be overweight, but the comprehensive analysis of the obesity trait was hindered by the propensity of C57BL/6 mice to become obese. It must also be noted that mouse models do not always fully recapitulate human phenotypes.
Interestingly, 2 (9%) of 21 homozygous mutants and 2 (7%) of 28 heterozygotes (monitored for at least 5 mo) developed tumors. One homozygous 11-mo-old male mouse received a diagnosis of squamous cell carcinoma of the tongue (a rare murine cancer), one 15-mo-old homozygous male mouse developed hepatoma, and two heterozygous mice 5.5 mo and 16 mo of age had histiocytic sarcoma. In contrast, 0 of 7 wild-type littermates, monitored until 18 mo of age, succumbed to cancer. The finding of tumors in 4 (8%) of 49 of mutant mice could be related potentially to altered DAG signaling.
Recently, a Dgkd null knockout mouse model has been described.23 These mice developed respiratory difficulties and were invariably dead within the first 24 h after birth. Besides small size, the newborn pups were also noted to have open eyelids. A dramatically reduced viability of the Dgkd homozygous null mice demonstrates the critical function of Dgkd expression. The drastic difference in the phenotype of our knockdown mutants in comparison with the knockout null mutants is interpreted to be due to their hypomorphic nature and potentially to the fact that expression of only one isoform is affected.
As in mice, DGKD in humans has two alternatively spliced isoforms with the low-abundance transcript initiating in exon 2 and being present in spleen, ovary, and some tumor-derived cells.24 Both isoforms possess kinase activity and can form homo- and hetero-oligomers but are expressed under different regulatory mechanisms and have different intracellular localization in response to phorbol ester stimulation.24 The t(X;2) in DGAP095 causes abolishment of expression of the abundant DGKD isoform as it separates exon 1 from the rest of the gene. It is possible that expression of the other isoform also could be affected if its transcription requires an upstream regulatory element. The function of the DGKD gene product supports its involvement in the seizure phenotype. It belongs to a family of diacylglycerol (DAG) kinases that phosphorylate DAG and convert it to phosphatidic acid (PA). Both DAG and PA are important second messengers in a pathway of lipid signaling that has been implicated in epilepsy and other neurological diseases, such as depression and Alzheimer disease.25–27 Moreover, mice deficient in the gene encoding DAG kinase epsilon, Dgke, display an altered susceptibility to seizures.28 Therefore, the patient’s phenotype could be explained by an altered balance between DAG and PA caused by DGKD haploinsufficiency. Of special interest, in this respect, are DAG-gated transient receptor potential cation channels (e.g., TRPC3, -6, and -7), in which DAG binding directly opens the channel.29,30 Existence of these channels in the brain places DAG kinases in proximity to the category of neuronal channelopathies. It is of note that the neurological and capillary abnormalities in DGAP095 could potentially have a common etiology, because vascular and neural development share many common genetic pathways and signaling mechanisms.31 For example, the angiogenic factor VEGF is also involved in the stimulation of neurogenesis.32 Intriguingly, the downstream targets of DAG kinases are implicated in the regulation of angiogenesis and VEGF-induced signaling,33–35 and DAG-gated channels are believed to play a critical role in vasoconstrictor-activated cation influx and regulation of arterial myogenic tone.36
In summary, the collective evidence, including direct disruption of DGKD by the t(X;2) in an individual with epilepsy, expression of DGKD orthologs in the developing nervous system in Drosophila and mouse embryos, broad expression in regions of the mouse brain known to produce seizures, and the occurrence of seizures in Dgkd mutant mice provide strong support for DGKD’s involvement in epilepsy disorders in humans and mice.
Acknowledgments
We express our gratitude to the patient and her parents for participating in DGAP; Dr. Wayne Frankel for discussions and advice related to epilepsy assessment in mice; Dr. George F. Murphy for dermatopathological assessment of mice; Dr. Carolien Panhuysen for her help with interpretation of linkage data; Heather Ferguson and Chantal Kelly for their assistance as study coordinators; and Qiongchao Xi, Lori Arbeitman, and Diana Donovan for their assistance with murine analyses. This study was supported by National Institutes of Health grants PO1 GM061354 (to C.C.M.), F32 HD43627 (to N.T.L.), and K08 GM077357 (to M.B.K.) and was assisted by the Cytogenetics and Rodent Histopathology Core Facilities of the Dana Farber Harvard Cancer Center (P30 CA06516).
Web Resources
The URLs for data presented herein are as follows:
- BayGenomics, http://baygenomics.ucsf.edu/
- Developmental Genome Anatomy Project (DGAP), http://www.bwhpathology.org/dgap/
- Kazusa DNA Research Institute, http://www.kazusa.or.jp/
- National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/
- University of California Santa Cruz, http://genome.ucsc.edu/
References
- 1.Hauser WA, Annegers JF, Kurland LT (1993) Incidence of epilepsy and unprovoked seizures in Rochester, Minnesota: 1935–1984. Epilepsia 34:453–468 10.1111/j.1528-1157.1993.tb02586.x [DOI] [PubMed] [Google Scholar]
- 2.Hirose S, Mitsudome A, Okada M, Kaneko S (2005) Genetics of idiopathic epilepsies. Epilepsia Suppl 1 46:38–43 10.1111/j.0013-9580.2005.461011.x [DOI] [PubMed] [Google Scholar]
- 3.Jallon P, Latour P (2005) Epidemiology of idiopathic generalized epilepsies. Epilepsia Suppl 9 46:10–14 10.1111/j.1528-1167.2005.00309.x [DOI] [PubMed] [Google Scholar]
- 4.Wallace RH, Wang DW, Singh R, Scheffer IE, George AL Jr, Phillips HA, Saar K, Reis A, Johnson EW, Sutherland GR, et al (1998) Febrile seizures and generalized epilepsy associated with a mutation in the Na+-channel β1 subunit gene SCN1B. Nat Genet 19:366–370 10.1038/448 [DOI] [PubMed] [Google Scholar]
- 5.Escayg A, Heils A, MacDonald BT, Haug K, Sander T, Meisler MH (2001) A novel SCN1A mutation associated with generalized epilepsy with febrile seizures plus--and prevalence of variants in patients with epilepsy. Am J Hum Genet 68:866–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biervert C, Schroeder BC, Kubisch C, Berkovic SF, Propping P, Jentsch TJ, Steinlein OK (1998) A potassium channel mutation in neonatal human epilepsy. Science 279:403–406 10.1126/science.279.5349.403 [DOI] [PubMed] [Google Scholar]
- 7.Wallace RH, Marini C, Petrou S, Harkin LA, Bowser DN, Panchal RG, Williams DA, Sutherland GR, Mulley JC, Scheffer IE, et al (2001) Mutant GABAA receptor γ2-subunit in childhood absence epilepsy and febrile seizures. Nat Genet 28:49–52 10.1038/88259 [DOI] [PubMed] [Google Scholar]
- 8.Bertrand D, Picard F, Le Hellard S, Weiland S, Favre I, Phillips H, Bertrand S, Berkovic SF, Malafosse A, Mulley J (2002) How mutations in the nAChRs can cause ADNFLE epilepsy. Epilepsia Suppl 5 43:112–122 10.1046/j.1528-1157.43.s.5.16.x [DOI] [PubMed] [Google Scholar]
- 9.Stromme P, Mangelsdorf ME, Shaw MA, Lower KM, Lewis SM, Bruyere H, Lutcherath V, Gedeon AK, Wallace RH, Scheffer IE, et al (2002) Mutations in the human ortholog of Aristaless cause X-linked mental retardation and epilepsy. Nat Genet 30:441–445 10.1038/ng862 [DOI] [PubMed] [Google Scholar]
- 10.Pavlidis P, Ramaswami M, Tanouye MA (1994) The Drosophila easily shocked gene: a mutation in a phospholipid synthetic pathway causes seizure, neuronal failure, and paralysis. Cell 79:23–33 10.1016/0092-8674(94)90397-2 [DOI] [PubMed] [Google Scholar]
- 11.Kalachikov S, Evgrafov O, Ross B, Winawer M, Barker-Cummings C, Martinelli Boneschi F, Choi C, Morozov P, Das K, Teplitskaya E, et al (2002) Mutations in LGI1 cause autosomal-dominant partial epilepsy with auditory features. Nat Genet 30:335–341 10.1038/ng832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakayama J, Fu YH, Clark AM, Nakahara S, Hamano K, Iwasaki N, Matsui A, Arinami T, Ptacek LJ (2002) A nonsense mutation of the MASS1 gene in a family with febrile and afebrile seizures. Ann Neurol 52:654–657 10.1002/ana.10347 [DOI] [PubMed] [Google Scholar]
- 13.Suzuki T, Delgado-Escueta AV, Aguan K, Alonso ME, Shi J, Hara Y, Nishida M, Numata T, Medina MT, Takeuchi T, et al (2004) Mutations in EFHC1 cause juvenile myoclonic epilepsy. Nat Genet 36:842–849 10.1038/ng1393 [DOI] [PubMed] [Google Scholar]
- 14.Commission on Classification and Terminology of the International League Against Epilepsy (1989) Proposal for revised classification of epilepsies and epileptic syndromes. Epilepsia 30:389–399 [DOI] [PubMed] [Google Scholar]
- 15.Young RS, Shapiro SD, Hansen KL, Hine LK, Rainosek DE, Guerra FA (1983) Deletion 2q: two new cases with karyotypes 46,XY,del(2)(q31q33) and 46,XX,del(2)(q36). J Med Genet 20:199–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher AM, Ellis KH, Browne CE, Barber JC, Barker M, Kennedy CR, Foley H, Patton MA (1994) Small terminal deletions of the long arm of chromosome 2: two new cases. Am J Med Genet 53:366–369 10.1002/ajmg.1320530412 [DOI] [PubMed] [Google Scholar]
- 17.Conrad B, Dewald G, Christensen E, Lopez M, Higgins J, Pierpont ME (1995) Clinical phenotype associated with terminal 2q37 deletion. Clin Genet 48:134–139 [DOI] [PubMed] [Google Scholar]
- 18.Wenger SL, Boone LY, Surti U, Steele MW (1997) Terminal 2q deletion--a recognizable syndrome. Clin Genet 51:290 [DOI] [PubMed] [Google Scholar]
- 19.Bijlsma EK, Aalfs CM, Sluitjer S, Oude Luttikhuis ME, Trembath RC, Hoovers JM, Hennekam RC (1999) Familial cryptic translocation between chromosomes 2qter and 8qter: further delineation of the Albright hereditary osteodystrophy-like phenotype. J Med Genet 36:604–609 [PMC free article] [PubMed] [Google Scholar]
- 20.Scheffer IE, Phillips HA, O’Brien CE, Saling MM, Wrennall JA, Wallace RH, Mulley JC, Berkovic SF (1998) Familial partial epilepsy with variable foci: a new partial epilepsy syndrome with suggestion of linkage to chromosome 2. Ann Neurol 44:890–899 10.1002/ana.410440607 [DOI] [PubMed] [Google Scholar]
- 21.Sander T, Schulz H, Saar K, Gennaro E, Riggio MC, Bianchi A, Zara F, Luna D, Bulteau C, Kaminska A, et al (2000) Genome search for susceptibility loci of common idiopathic generalised epilepsies. Hum Mol Genet 9:1465–1472 10.1093/hmg/9.10.1465 [DOI] [PubMed] [Google Scholar]
- 22.Ferraro TN, Golden GT, Smith GG, St Jean P, Schork NJ, Mulholland N, Ballas C, Schill J, Buono RJ, Berrettini WH (1999) Mapping loci for pentylenetetrazol-induced seizure susceptibility in mice. J Neurosci 19:6733–6739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crotty T, Cai J, Sakane F, Taketomi A, Prescott SM, Topham MK (2006) Diacylglycerol kinase δ regulates protein kinase C and epidermal growth factor receptor signaling. Proc Natl Acad Sci USA 103:15485–15490 10.1073/pnas.0604104103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakane F, Imai S, Yamada K, Murakami T, Tsushima S, Kanoh H (2002) Alternative splicing of the human diacylglycerol kinase δ gene generates two isoforms differing in their expression patterns and in regulatory functions. J Biol Chem 277:43519–43526 10.1074/jbc.M206895200 [DOI] [PubMed] [Google Scholar]
- 25.Pacheco MA, Jope RS (1996) Phosphoinositide signaling in human brain. Prog Neurobiol 50:255–273 10.1016/S0301-0082(96)00035-4 [DOI] [PubMed] [Google Scholar]
- 26.Bordi F, Ugolini A (1999) Group I metabotropic glutamate receptors: implications for brain diseases. Prog Neurobiol 59:55–79 10.1016/S0301-0082(98)00095-1 [DOI] [PubMed] [Google Scholar]
- 27.van Blitterswijk WJ, Houssa B (2000) Properties and functions of diacylglycerol kinases. Cell Signal 12:595–605 10.1016/S0898-6568(00)00113-3 [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez de Turco EB, Tang W, Topham MK, Sakane F, Marcheselli VL, Chen C, Taketomi A, Prescott SM, Bazan NG (2001) Diacylglycerol kinase ε regulates seizure susceptibility and long-term potentiation through arachidonoyl- inositol lipid signaling. Proc Natl Acad Sci USA 98:4740–4745 10.1073/pnas.081536298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hofmann T, Obukhov AG, Schaefer M, Harteneck C, Gudermann T, Schultz G (1999) Direct activation of human TRPC6 and TRPC3 channels by diacylglycerol. Nature 397:259–263 10.1038/16711 [DOI] [PubMed] [Google Scholar]
- 30.Okada T, Inoue R, Yamazaki K, Maeda A, Kurosaki T, Yamakuni T, Tanaka I, Shimizu S, Ikenaka K, Imoto K, et al (1999) Molecular and functional characterization of a novel mouse transient receptor potential protein homologue TRP7: Ca(2+)-permeable cation channel that is constitutively activated and enhanced by stimulation of G protein-coupled receptor. J Biol Chem 274:27359–27370 10.1074/jbc.274.39.27359 [DOI] [PubMed] [Google Scholar]
- 31.Carmeliet P (2003) Blood vessels and nerves: common signals, pathways and diseases. Nat Rev Genet 4:710–720 10.1038/nrg1158 [DOI] [PubMed] [Google Scholar]
- 32.Jin K, Zhu Y, Sun Y, Mao XO, Xie L, Greenberg DA (2002) Vascular endothelial growth factor (VEGF) stimulates neurogenesis in vitro and in vivo. Proc Natl Acad Sci USA 99:11946–11950 10.1073/pnas.182296499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eerola I, Boon LM, Mulliken JB, Burrows PE, Dompmartin A, Watanabe S, Vanwijck R, Vikkula M (2003) Capillary malformation-arteriovenous malformation, a new clinical and genetic disorder caused by RASA1 mutations. Am J Hum Genet 73:1240–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baldanzi G, Mitola S, Cutrupi S, Filigheddu N, van Blitterswijk WJ, Sinigaglia F, Bussolino F, Graziani A (2004) Activation of diacylglycerol kinase α is required for VEGF-induced angiogenic signaling in vitro. Oncogene 23:4828–4838 10.1038/sj.onc.1207633 [DOI] [PubMed] [Google Scholar]
- 35.Kranenburg O, Gebbink MF, Voest EE (2004) Stimulation of angiogenesis by Ras proteins. Biochim Biophys Acta 1654:23–37 [DOI] [PubMed] [Google Scholar]
- 36.Dietrich A, Kalwa H, Rost BR, Gudermann T (2005) The diacylgylcerol-sensitive TRPC3/6/7 subfamily of cation channels: functional characterization and physiological relevance. Pflugers Arch 451:72–80 10.1007/s00424-005-1460-0 [DOI] [PubMed] [Google Scholar]