Abstract
Branchio-oto-renal syndrome (BOR) is an autosomal dominant developmental disorder characterized by the association of branchial arch defects, hearing loss, and renal anomalies. Mutations in EYA1 are known to cause BOR. More recently, mutations in SIX1, which interacts with EYA1, were identified as an additional cause of BOR. A second member of the SIX family of proteins, unc-39 (SIX5), has also been reported to directly interact with eya-1 in Caenorhabditis elegans. We hypothesized that this interaction would be conserved in humans and that interactors of EYA1 represent good candidate genes for BOR. We therefore screened a cohort of 95 patients with BOR for mutations in SIX5. Four different heterozygous missense mutations were identified in five individuals. Functional analyses of these mutations demonstrated that two mutations affect EYA1-SIX5 binding and the ability of SIX5 or the EYA1-SIX5 complex to activate gene transcription. We thereby identified heterozygous mutations in SIX5 as a novel cause of BOR.
Branchio-oto-renal syndrome (BOR [MIM 113650]) is a clinically heterogeneous autosomal dominant disorder comprising branchial arch defects, hearing loss, and renal anomalies. The syndrome has an estimated prevalence of 1 in 40,000 and accounts for ∼2% of profound deafness.1 Abnormal pinnae, cervical fistulas, pits, and/or cysts are the most common branchial arch defects seen in patients, whereas hearing loss can be conductive, sensorineural, or mixed and often is associated with preauricular tags or pits.2 Renal defects, which are estimated to be severe in only 6% of patients, can include collecting system duplication, hypoplasia, cystic dysplasia, hydronephrosis, and agenesis.3 The penetrance of the syndrome is highly variable both between and within families. Heimler and Lieber4 published a report on a large, four-generation kindred containing 16 individuals with confirmed manifestations of the syndrome. Of these, only four had abnormalities of all three systems, whereas seven had branchial arch and/or hearing defects but no reported renal abnormalities. However, because only one of these individuals had undergone a detailed renal evaluation, it is possible that the incidence of renal defects in this kindred is higher than reported.
In addition to being clinically heterogeneous, BOR also displays genetic heterogeneity. Mutations in the EYA1 gene, the human ortholog of the eyes absent gene in Drosophila, were the first cause of BOR to be identified after the disease was linked to chromosome 8q13.3.5,6 Loss-of-function alleles, including nonsense, frameshift, and splice-site mutations, were found in 7 of 42 patients screened by sequencing. In addition, deletions estimated to span 6, 7, and 20 kb were identified in 3 of 21 patients screened by Southern blot analysis. Although it was initially predicted that causative mutations would cluster in the eyes absent homologous region (eyaHR), also known as the EYA domain (ED) of EYA1,7 a large number of alterations distributed throughout the gene have since been reported, including complex rearrangements involving each of the 16 exons of the gene and multiple loss-of-function and missense mutations in all exons, with the exception of exons 1–3.8–11
EYA proteins are characterized by a divergent N-terminal transactivation domain and the conserved C-terminal ED, which mediates interactions with DNA-binding proteins such as SIX1 and SIX2 (human orthologs of Drosophila “sine oculis” [so]), thereby forming a potent transactivation complex.12,13 To determine whether mutation of residues within the ED affects the ability of Eya1 to directly interact with Six1 or Six2, Buller et al.12 performed GST pull-down and yeast two-hybrid assays on five mutations. Three ED constructs, each containing a single substituted residue (E330K, S454P, and L472R), showed a reduction in Eya1-Six binding in both assays, suggesting that these residues are essential for direct Eya1-Six interaction. Mutsuddi et al.14 used in vitro biochemical assays, in addition to in vivo studies of Drosophila eye, to also test the loss of function due to the S454P and L472R EYA1 mutations. (Because a different isoform of EYA1 was used for amino acid numbering by Mutsuddi et al., S454P and L472R are described as S487P and L505R, respectively.) Both mutations showed an approximately fivefold reduction in the ability of the eya-so complex to transactivate. However, coimmunoprecipitation studies showed that this reduction was not due to an inability of the complex to form, as both mutant constructs were able to complex efficiently with so.
Following the identification of EYA1 as the first BOR gene, a number of reports on the exclusion of the 8q13 locus and mapping of additional BOR loci, such as the BOS2 (branchiootic syndrome 2 [MIM %120502]) locus on 1q31, were published.15–17 Most recently, we used a large BOR kindred with >40 affected individuals to map a third BOR locus (branchiootic syndrome-3 [MIM 608389]) to 14q21.3.18 Located within the 33-Mb critical interval were three members of the SIX family of genes (SIX1, SIX4, and SIX6). Mutation screening of these genes in the family used to map the locus revealed a heterozygous Y129C missense mutation in SIX1, with two further mutations (delE133 and R110W) in three additional patients, confirming SIX1 as a second gene for BOR.19 Functional analysis of these mutations demonstrated that the R110W mutation, located in the SIX domain (SD), interferes with formation of the Eya1-Six1 complex, whereas the Y129C and delE133 mutations affect not only this protein-protein interaction but also the ability of Six1 to bind DNA.19
In 2004, the Caenorhabditis elegans interactome was published, containing data on >4,000 direct protein-protein interactions in the nematode identified by high-throughput, yeast two-hybrid analysis.20 At the center of one of the subnetworks made up of >75 interactions was C49A1.4 (eya-1), the ortholog of EYA1. In addition to the ortholog of SIX1 (ceh-33), a second member of the SIX family of proteins, SIX5 (unc-39/ceh-35), was also shown to directly interact with eya-1 in the nematode. Because SIX5 has a high degree of homology to SIX1 and directly interacts with eya-1 in C. elegans, we considered it an excellent candidate for mutation analysis in a cohort of patients with BOR who did not have mutations in either EYA1 or SIX1.
Patients.—The diagnosis of BOR was based on published criteria.10 All samples were negative for EYA1 or SIX1 mutations. This study was approved by the institutional review boards of the University of Freiburg (303102) and the University of Michigan Medical School (2004-0322, HUM00003330). Patient samples were obtained after informed consent was given.
Mutation analysis.—Primers were designed to amplify each of the three exons and surrounding intronic sequence of SIX5. Standard PCR were performed, and sequencing was performed on an ABI capillary sequencer. Sequence traces were analyzed using the Sequencher software program (Gene Codes). Absence of sequence changes in 150 European white control individuals was performed by sequencing or, where possible, by digestion of the PCR product with a restriction enzyme.
Generation of SIX5 constructs.—The human SIX5 full-length cDNA was cloned by RT-PCR into either pGBKT7 vector for yeast two-hybrid assays or pcDNA3 vector for cell-culture analysis. To introduce the same amino acid substitutions found in patients with BOR into the human SIX5 protein, PCR-based mutagenesis was performed using the QuickChange mutagenesis kit (Stratagene). Once mutant colonies were identified, the plasmid DNA was isolated and sequenced through the mutation-containing region. Four mutant cDNAs (A158T, A296T, G365R, and T552M) were constructed in the same manner in either the pGBKT7 or the pcDNA3 vector.
Yeast two-hybrid assay.—The Matchmaker Two-Hybrid System 3 (Clontech) was used for yeast interaction assays. SIX5 was fused with the GAL4 DNA-binding domain of pGBKT7 (GAL4 BD-SIX5). The Eya1 domain (Eya1D) region of Eya1 was fused with the GAL4 activation domain of pGAD424 (GAL4 AD-Eya1D). Small scale LiAc cotransformations of the plasmids into AH109 cells, colony-lift filter assays, and liquid culture β-galactosidase assays were performed as described in the Clontech protocols.
Luciferase assay.—The human embryonic kidney HEK293 cell line and Cos-7 cells were cultured as described elsewhere.12 The myogenin luciferase reporter pGL3-6×MEF3 was used as a reporter. One microgram of pGL3-6×MEF3 plasmid was cotransfected with 1 μg of pcDNA3-SIX5 wild type (wtSIX5) or its mutant plasmids, or 1 μg Eya1 alone, or both SIX5 and Eya1 together with use of FuGENE 6 Transfection Reagent (Roche), according to the manufacturer’s instructions. Cells were grown in dishes 6 cm in diameter and were additionally cotransfected with 0.25 μg of the cytomegalovirus (CMV) promoter-β-galactosidase (pCMVβ-gal) as an internal control. Forty-eight hours after transfection, cell extracts were prepared and assayed for luciferase activities.
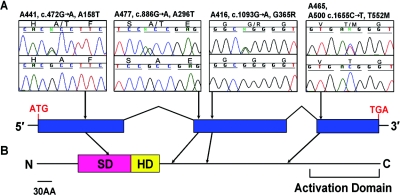
Mutation screening of SIX5 in 95 unrelated patients with BOR revealed four different heterozygous missense mutations in five individuals (table 1 and fig. 1). Individual A441, who has bilateral dysplastic kidneys and a right preauricular tag but normal hearing, was found to carry a c.472G→A substitution (amino acid change A158T). Individual A477 has bilateral cervical fistulae; hemifacial microsomia; a preauricular sinus and pinna malformation on the right side; hearing loss in both ears, with the right ear more affected than the left; and bilateral renal dysplasia with diminished renal function. This patient was found to have a c.886G→A transition (amino acid change A296T). In individual A416, a c.1093G→A substitution (amino acid change G365R) was found. In addition, a c.1655C→T transition (amino acid change T552M) was found in two individuals: A465, who has cervical fistulae with hypoplastic kidneys, and A500, who has cervical fistulae, moderate to severe hearing loss in both ears, and left renal agenesis with a hypoplastic right kidney. All sequence changes were absent in 150 healthy control individuals. The phenotypes of the patients presented here show a similarly high degree of variability, as seen in patients with BOR who had mutations in EYA1 or SIX1.10,19 Although the cohort of patients is small, there is no evidence of a genotype-phenotype correlation associated with SIX5 mutations.
Table 1. .
Clinical and Genetic Data on Five Patients with SIX5 Mutations[Note]
| Patient | Branchial Defect | Hearing Loss | Renal Defect | Nucleotide Change | Amino Acid Change |
| A441 | PTR | - | DKB | c.472G→A | A158T |
| A477 | CFB, HMR, PSR, PMR | +L, ++R | DKB, ↓ function | c.886G→A | A296T |
| A416 | ND | ND | ND | c.1093G→A | G365R |
| A465 | CFB | - | HKB | c.1655C→T | T552M |
| A500 | CFB | ++L, +R | AL, HKR | c.1655C→T | T552M |
Note.— − = absent, + = affected, ++ = more affected; L = left, R = right, B = bilateral; PT = preauricular tag, DK = dysplastic kidneys, CF = cervical fistulae, HM = hemifacial microsomia, PS = preauricular sinus, PM = pinna malformation, ↓ function = diminished renal function, HK = hypoplastic kidneys, A = agenesis; ND = no data available.
Figure 1. .
Four missense mutations in SIX5 identified in patients with BOR. A, A158T, A296T, and G365R mutations, present in heterozygous form in a single patient. The T552M mutation was detected in patients A465 and A500. Sequences from the affected individual are shown above sequences from healthy control individuals. Patient identifiers, nucleotide changes, and amino acid changes are shown above the traces. B, SIX5 has an SD and a homeodomain (HD) characteristic of the SIX family of proteins at the N-terminus. An activation domain has also been mapped to the C-terminus.21 The A158T mutation is located in the SIX domain; the remaining three mutations are located between the HD and the activation domain.
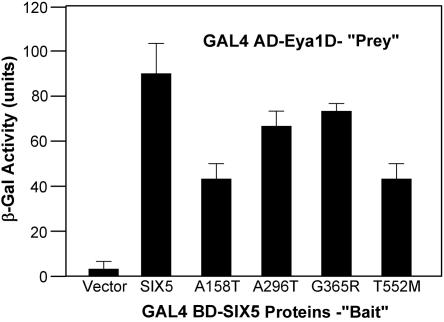
To test whether any of the mutations identified in patients affect the ability of SIX5 to directly interact with Eya1, yeast two-hybrid liquid β-galactosidase assays were performed. Cotransformation of yeast with GAL4 BD-SIX5 and GAL4 AD-Eya1D constructs led to strong lacZ expression as a result of interaction between the two fusion proteins (fig. 2). Introduction of mutations into the SIX5 construct showed varying levels of reduction in lacZ expression. A296T and G365R showed only a slight reduction in expression, whereas both A158T and T552M showed a >2-fold reduction, indicating that these two residues of SIX5 may be required for efficient binding with EYA1.
Figure 2. .
Mutations in SIX5 affect SIX5-Eya1 binding. Yeast two-hybrid assays were performed to test the effect of the mutations on the ability of SIX5 to bind to Eya1. Cotransformation of GAL4BD-SIX5 and GAL4AD-Eya1D leads to strong lacZ expression. An >2-fold reduction in LacZ expression is seen when SIX5 proteins containing an A158T or a T552M mutation are cotransformed with Eya1D fusion proteins.
To investigate what effect these mutations may have on the ability of SIX5 or the Eya1-SIX5 complex to activate transcription, the myogenin luciferase reporter pGL3-6×MEF3 was used. This construct contains six MEF3 sites to which SIX proteins are known to bind.19,21 Transfection of HEK293 cells with wtSIX5 resulted in an ∼7-fold increase in luciferase activity compared with vector alone (fig. 3). Transfection of the A296T and G365R mutant constructs alone did not result in a significant decrease in luciferase reporter activity. However, the A158T and T552M mutations resulted in significant decreases in luciferase activity when compared with wtSIX5. Decreases were 50% for A158T and 25% for T552M. Coexpression of wtSIX5 with Eya1 resulted in a 12-fold increase in luciferase activity. However, when each of the mutant SIX5 proteins was coexpressed with Eya1, the increase in activity was reduced to 52%, 80%, 70%, and 56% of that seen for wtSIX5 with Eya1 for the A158T, A296T, G365R, and T552M mutants, respectively. Although the reduction in transcriptional activity is small for the A296T and G365R mutants, these reductions are statistically significant when compared with the activity seen on coexpression of wtSIX5 with Eya1 (fig. 3). The results were reproducible in three independent experiments, each performed in duplicate.
Figure 3. .
Mutations in SIX5 reduce its ability to activate transcription of the MEF3 promoter. The myogenin luciferase reporter pGL3-6×MEF3 was cotransfected with pcDNA3 vector containing SIX5 or its mutants (A158T, A296T, G365R, and T552M), pFlag-Eya1, or both the SIX5 and Eya1 plasmids together in HEK293 cells. Luciferase activity in the cell lysate was normalized with β-galactosidase activity of pCMVβ-gal as an internal control. The activity at each data point is relative to that obtained by the control pCMV vector. The mean fold activation from three independent experiments (each performed in duplicate) is shown with the standard deviation. A filled circle (•) indicates P<.002, compared with SIX5; an asterisk (*) indicates P<.002, compared with SIX5/Eya1 (one-way analysis of variance [ANOVA]).
In this study, we have identified heterozygous mutations in SIX5 as a novel cause of BOR. SIX5 was considered to be a good candidate gene for BOR, owing to its similarity to SIX1, a known BOR gene,19 and data on C. elegans on its direct interaction with eya-1,20 mutations of which also cause BOR.6 Four different missense mutations were identified in five patients from a cohort of 95 unrelated individuals. Although it was not possible to perform segregation analyses on these mutations, because DNA samples from family members were not available, the absence of each mutation in a cohort of 150 healthy control individuals strongly suggests that these changes are disease-causing mutations rather than nonpathogenic polymorphisms. This is confirmed by the functional studies of Eya1-SIX5 interaction and reduced function in transcriptional activation assays.
Unlike SIX1, SIX5 possesses an additional activation domain (AD) at the C-terminus, which retains its transactivation function when fused to a GAL4 DNA-binding domain.22 Consistent with this, our data show that expression of SIX5 alone is able to transactivate MEF3-reporter transcription in cultured cells (fig. 3). SIX5 is also able to synergistically activate MEF3 reporter gene expression when coexpressed with Eya proteins21 (fig. 3). Among the four mutations identified in the SIX5 protein, the A158T mutation is located in the SD domain, while the other three mutations are located between the HD and AD domains (fig. 1B). Our results show that the SD mutation A158T resulted in a reduction (50%) in reporter activity in a yeast two-hybrid assay (fig. 2) and that this reduction can be explained by a decrease in the Eya1-SIX5 complex formation. Interestingly, the luciferase assay showed that the SD mutation (A158T) decreased the reporter activity in the absence of Eya1. Because the SD is known to mediate not only protein-protein interaction but also protein-DNA binding affinity through dimerization,21,23,24 the reduction in reporter activity observed in cultured cells without addition of Eya1 is likely caused by the SD mutation affecting the formation of the SIX5-DNA binding complex.
In contrast to A158T, the two mutations A296T and G365R have no effect on the ability of SIX5 alone to activate transcription but do affect the transcriptional activity of the Eya1-SIX5 complex, to varying degrees. The largest effect on coexpression was again seen for A158T and the second largest for the T552M mutation. Because the mutated T552M residue is near the C-terminal activation domain, it may affect SIX5 transactivation function.
It should be noted that the effect of SIX1 mutations19 was greater than that seen with the SIX5 mutations presented here. This may be in part because SIX5 itself is transcriptionally active but also because, when transfected together with Eya1, Eya2, or Eya3, the combination of Six5 with Eya3 showed the highest levels of activity on a myogenin luciferase reporter: 8-fold activation compared with the 4-fold activation seen with Six5 and Eya1.21 Therefore, although EYA1 and SIX5 do interact, there may be some additional transcriptional activation through EYA3.
We have identified mutations in SIX5 as a new cause of BOR. In addition, we provided evidence to support the interaction between Eya1 and SIX5 and showed that mutations of SIX5 compromise the interaction between the two proteins, decreasing the transcriptional activity of the Eya1-SIX5 complex.
Acknowledgments
We thank the patients and their families for their contribution. We also thank Shrawan Kumar, Harold Bass, and Cor Cremers for ascertainment of patient samples. This work was supported by grants from the National Institutes of Health to F.H. (R01-DK045345-01), P-X.X. (RO1 DC005824 and DK064640), and C.H.C. (T32DK0655517-01). F.H. is the Frederick G. L. Huetwell Professor of the Cure and Prevention of Birth Defects and a Doris Duke Distinguished Clinical Scientist.
Web Resource
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for BOR, BOS2, and branchiootic syndrome-3 locus)
References
- 1.Fraser FC, Sproule JR, Halal F (1980) Frequency of the branchio-oto-renal (BOR) syndrome in children with profound hearing loss. Am J Med Genet 7:341–349 10.1002/ajmg.1320070316 [DOI] [PubMed] [Google Scholar]
- 2.Weber KM, Kousseff BG (1999) “New” manifestations of BOR syndrome. Clin Genet 56:306–312 10.1034/j.1399-0004.1999.560408.x [DOI] [PubMed] [Google Scholar]
- 3.Izzedine H, Tankere F, Launay-Vacher V, Deray G (2004) Ear and kidney syndromes: molecular versus clinical approach. Kidney Int 65:369–385 10.1111/j.1523-1755.2004.00390.x [DOI] [PubMed] [Google Scholar]
- 4.Heimler A, Lieber E (1986) Branchio-oto-renal syndrome: reduced penetrance and variable expressivity in four generations of a large kindred. Am J Med Genet 25:15–27 10.1002/ajmg.1320250104 [DOI] [PubMed] [Google Scholar]
- 5.Kalatzis V, Abdelhak S, Compain S, Vincent C, Petit C (1996) Characterization of a translocation-associated deletion defines the candidate region for the gene responsible for branchio-oto-renal syndrome. Genomics 34:422–425 10.1006/geno.1996.0307 [DOI] [PubMed] [Google Scholar]
- 6.Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Weil D, Cruaud C, Sahly I, Leibovici M, et al (1997) A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat Genet 15:157–164 10.1038/ng0297-157 [DOI] [PubMed] [Google Scholar]
- 7.Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Levi-Acobas F, Cruaud C, Le Merrer M, Mathieu M, et al (1997) Clustering of mutations responsible for branchio-oto-renal (BOR) syndrome in the eyes absent homologous region (eyaHR) of EYA1. Hum Mol Genet 6:2247–2255 10.1093/hmg/6.13.2247 [DOI] [PubMed] [Google Scholar]
- 8.Vincent C, Kalatzis V, Abdelhak S, Chaib H, Compain S, Helias J, Vaneecloo FM, Petit C (1997) BOR and BO syndromes are allelic defects of EYA1. Eur J Hum Genet 5:242–246 [PubMed] [Google Scholar]
- 9.Vervoort VS, Smith RJ, O’Brien J, Schroer R, Abbott A, Stevenson RE, Schwartz CE (2002) Genomic rearrangements of EYA1 account for a large fraction of families with BOR syndrome. Eur J Hum Genet 10:757–766 10.1038/sj.ejhg.5200877 [DOI] [PubMed] [Google Scholar]
- 10.Chang EH, Menezes M, Meyer NC, Cucci RA, Vervoort VS, Schwartz CE, Smith RJ (2004) Branchio-oto-renal syndrome: the mutation spectrum in EYA1 and its phenotypic consequences. Hum Mutat 23:582–589 10.1002/humu.20048 [DOI] [PubMed] [Google Scholar]
- 11.Clarke JC, Honey EM, Bekker E, Snyman LC, Raymond RM Jr, Lord C, Brophy PD (2006) A novel nonsense mutation in the EYA1 gene associated with branchio-oto-renal/branchiootic syndrome in an Afrikaner kindred. Clin Genet 70:63–67 10.1111/j.1399-0004.2006.00642.x [DOI] [PubMed] [Google Scholar]
- 12.Buller C, Xu X, Marquis V, Schwanke R, Xu PX (2001) Molecular effects of Eya1 domain mutations causing organ defects in BOR syndrome. Hum Mol Genet 10:2775–281 10.1093/hmg/10.24.2775 [DOI] [PubMed] [Google Scholar]
- 13.Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, Zipursky SL (1997) The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 91:881–891 10.1016/S0092-8674(00)80480-8 [DOI] [PubMed] [Google Scholar]
- 14.Mutsuddi M, Chaffee B, Cassidy J, Silver SJ, Tootle TL, Rebay I (2005) Using Drosophila to decipher how mutations associated with human branchio-oto-renal syndrome and optical defects compromise the protein tyrosine phosphatase and transcriptional functions of eyes absent. Genetics 170:687–695 10.1534/genetics.104.039156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar S, Deffenbacher K, Marres HA, Cremers CW, Kimberling WJ (2000) Genomewide search and genetic localization of a second gene associated with autosomal dominant branchio-oto-renal syndrome: clinical and genetic implications. Am J Hum Genet 66:1715–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar S, Marres HA, Cremers CW, Kimberling WJ (1998) Autosomal-dominant branchio-otic (BO) syndrome is not allelic to the branchio-oto-renal (BOR) gene at 8q13. Am J Med Genet 76:395–401 [DOI] [PubMed] [Google Scholar]
- 17.Stratakis CA, Lin JP, Rennert OM (1998) Description of a large kindred with autosomal dominant inheritance of branchial arch anomalies, hearing loss, and ear pits, and exclusion of the branchio-oto-renal (BOR) syndrome gene locus (chromosome 8q13.3). Am J Med Genet 79:209–214 [DOI] [PubMed] [Google Scholar]
- 18.Ruf RG, Berkman J, Wolf MT, Nurnberg P, Gattas M, Ruf EM, Hyland V, Kromberg J, Glass I, Macmillan J, et al (2003) A gene locus for branchio-otic syndrome maps to chromosome 14q21.3-q24.3. J Med Genet 40:515–519 10.1136/jmg.40.7.515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruf RG, Xu PX, Silvius D, Otto EA, Beekmann F, Muerb UT, Kumar S, Neuhaus TJ, Kemper MJ, Raymond RM Jr, et al (2004) SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc Natl Acad Sci USA 101:8090–8095 10.1073/pnas.0308475101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, et al (2004) A map of the interactome network of the metazoan C. elegans. Science 303:540–543 10.1126/science.1091403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohto H, Kamada S, Tago K, Tominaga SI, Ozaki H, Sato S, Kawakami K (1999) Cooperation of Six and Eya in activation of their target genes through nuclear translocation of Eya. Mol Cell Biol 19:6815–6824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawakami K, Sato S, Ozaki H, Ikeda K (2000) Six family genes—structure and function as transcription factors and their roles in development. Bioessays 22:616–626 [DOI] [PubMed] [Google Scholar]
- 23.Pham YC, Man N, Holt I, Sewry CA, Pall G, Johnson K, Morris GE (2005) Characterisation of the transcription factor, SIX5, using a new panel of monoclonal antibodies. J Cell Biochem 95:990–1001 10.1002/jcb.20454 [DOI] [PubMed] [Google Scholar]
- 24.Harris SE, Winchester CL, Johnson KJ (2000) Functional analysis of the homeodomain protein SIX5. Nucleic Acids Res 28:1871–1878 10.1093/nar/28.9.1871 [DOI] [PMC free article] [PubMed] [Google Scholar]