Abstract
We have identified one frameshift mutation, one splice-site mutation, and two missense mutations in highly conserved residues in ZDHHC9 at Xq26.1 in 4 of 250 families with X-linked mental retardation (XLMR). In three of the families, the mental retardation phenotype is associated with a Marfanoid habitus, although none of the affected individuals meets the Ghent criteria for Marfan syndrome. ZDHHC9 is a palmitoyltransferase that catalyzes the posttranslational modification of NRAS and HRAS. The degree of palmitoylation determines the temporal and spatial location of these proteins in the plasma membrane and Golgi complex. The finding of mutations in ZDHHC9 suggests that alterations in the concentrations and cellular distribution of target proteins are sufficient to cause disease. This is the first XLMR gene to be reported that encodes a posttranslational modification enzyme, palmitoyltransferase. Furthermore, now that the first palmitoyltransferase that causes mental retardation has been identified, defects in other palmitoylation transferases become good candidates for causing other mental retardation syndromes.
Mental retardation is the clinical description of difficulties with intellectual processing. The underlying cellular abnormalities that cause this phenotype are enormously variable, and the term “mental retardation” reflects a final, common description of multiple, complex processes in brain development and sustained function. Mental retardation may occur if neuronal numbers are reduced early in embryogenesis due to a failure of appropriate cell division, as in microcephaly,1,2 or if there is failure of neural migration.3,4 Also, abnormalities of the structural components of the synapse will affect the speed and accuracy of sustained neural processing,5,6 and several transcription factors7,8 and members of the guanosine triphosphastase protein signaling cascades have also been identified as causing mental retardation.9,10
To identify novel genes that cause X-linked mental retardation (XLMR), we have embarked on a systematic screen for mutations in genes on the X chromosome. We have collected and characterized a cohort of 250 families in which at least two males in the family have mental retardation as the predominant clinical phenotype and in which there was no known molecular diagnosis. All families have been examined and have no abnormalities by conventional karyotype analysis at 500 G-banding resolution. DNA from probands from the families have been screened and are negative for mutations in 61 known syndromic and nonsyndromic XLMR genes by sequence analysis of the coding exons and splice junctions. Expansions of the FMR1 trinucleotide repeat were also excluded. Although all families have mental retardation as the predominant feature, some families also had both consistent and inconsistent additional clinical abnormalities, such as macrocephaly, short stature, and spasticity. These features were recorded but were not used as selection criteria for screening. DNA from an affected individual from each family was screened for variants in the coding exons and splice junctions of the remaining 676 Vega-annotated X-chromosome genes (Vega Genome Browser) by bidirectional, PCR-based direct sequencing.
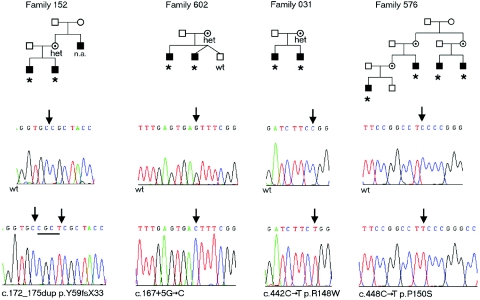
Two truncating and two missense mutations were found in the zinc finger, DHHC-type containing 9 gene (ZDHHC9 [GenBank accession number NM_016032]), located in Xq26.1. In family 152, there is a 4-bp duplication, c.172_175dup (p.Y59fsX33), present in two brothers and their mother (fig. 1). The maternal uncle of the brothers, who was also affected, was unavailable for analysis. In family 602, a splice-site mutation, c.167+5G→C, was detected in two affected brothers and their mother and was absent from an unaffected brother. In silico analysis of the splice-site variant c.167+5G→C by use of NNSPLICE (BDGP: Splice Site Prediction by Neural Network) predicts the reduction of the strength of the splice-donor site from 76.46% to 62.23% . This was confirmed by sequence analysis of ZDHHC9 cDNA, prepared from lymphocytes from the proband and amplified using PCR primers situated within exons 2 and 4 (table 1). The transcript identified was abnormal and used an upstream splice-donor site (consensus strength 76.82%) within exon 2, resulting in a frameshift and protein truncation, T11fsX33 (fig. 2). In families 031 and 576, missense variants were identified. In family 031, c.442C→T (p.R148W) was detected in two brothers and their mother. In family 576, the variant c.448C→T (p.P150S) was found in four affected males (fig. 1). We evaluated the potential effects of these missense variants on ZDHHC9 function by comparing amino acid conservation at these positions in paralogs and orthologs within and across species (fig. 3). Both R148 and P150 are conserved from man to yeast in the context of only 31% total amino acid identity in ZDHHC9 between these two species (blastp [NCBI BLAST Web site]). Moreover, they reside within a larger block of 52 aa residues that are highly conserved and form the active site of the palmitoyltransferase enzyme (figs. 3 and 4). Comparison of paralogs of ZDHHC9 in humans show that the residues equivalent to R148 and P150 in ZDHHC14, ZDHHC5, ZDHHC18, ZDHHC8, and ZDHHC19 are also conserved (fig. 3). In the remaining 18 human paralogs, these residues are not universally conserved, but these paralogs are the more distantly related proteins. Conservation across species shows that these residues are conserved in ZDHHC14 and ZDHHC5 in D. rerio and in the only two paralogs of the ZDHHC family in C. elegans (fig. 3). The conservation of these amino acids and their position within a known functional domain of the protein indicate that they significantly influence ZDHHC9 function and, therefore, that these missense variants are likely to be disease causing (fig. 4). No other variants in ZDHHC9 were detected in the 250-case series. The four sequence variants were not detected in DNA from 445 normal control samples, which represent 667 X chromosomes. Furthermore, no additional common or rare coding sequence variants were detected in the control series, demonstrating that ZDHHC9 shows little variation in unaffected individuals. Details of PCR primers and sequencing temperatures are given in table 1.
Figure 1. .
Pedigrees of families with XLMR and mutations in ZDHHC9. Individuals marked with an asterisk (*) carry the mutant allele in each family. Carrier females who have been shown to be heterozygous for the wild-type allele and the mutation are labeled “het.” Representative sequence chromatograms of wild-type and mutant sequences are shown below each pedigree. n.a. = no sample was available for testing; wt = wild-type sequence.
Table 1. .
Primers Used for PCR Amplification of ZDHHC9
| Primer Sequence(5′→3′) |
||
| Segment | Forward | Reverse |
| Exon 2 | ACCCAGCGGCAGGAATAG | CAGCCCCAAATCAGTACTACAAA |
| Exon 3 | TGTATGTTATAGGTGGCAGCAGT | CAGGATCTCAGAGAGAAAAGGC |
| Exon 4 | TGCATGCACTTAGTGGAGTTTT | GATTTGCAGTTTTTCAGGAGC |
| Exon 5 | TTACAACTGAAATAGACAATGATCCTG | ACCAGTAATTATGGCCCCAGT |
| Exon 6 | GATGAGGCTCTTCACCCTTG | ATGAAGAACTGGTGGGGGA |
| Exon 7 | ATGGAAACATGGCTTTTTGTG | GAGGCACCAGCTAGCCTTTA |
| Exon 8a | GCAGATCAAAGGATCATGGA | AGAGAGTTAACAAAGAACTGGGC |
| Exon 9 | GACATTTCCTGGTTTGTGGA | AAATCTTAATCTACATCAAGGACACA |
| Exon 10 | TTCAGTTGTGTTCTGCCGAG | CTTCTGCCACTGCCTTGAGT |
| cDNA 2–4 | ACCCAGCGGCAGGAATAG | GGAAGATCTTGCATGTGTAACAG |
Primer was placed in the coding sequence because of flanking intronic repeats.
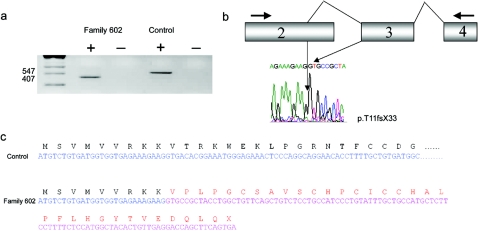
Figure 2. .
a, RT-PCR analysis of the ZDHHC9 gene by use of lymphocyte RNA from an affected male individual in family 602 and an unaffected control. A 547-bp PCR product was identified from cDNA made from control cells containing sequence from exons 2–4, by use of the predicted splice junctions. In family 602, only a 407-bp PCR product was identified. b, Diagram of the cDNA detected in family 602, which included an upstream splice-donor site, resulting in the loss of 140 bp at the terminus of exon 2, and the sequence chromatogram of the novel splicing event, leading to the mutation. c, Effect of the upstream splice-donor site within exon 2 spliced to exon 3. This results in a frameshift and truncated protein p.T11fsX33.
Figure 3. .
Sequence alignment of the conserved palmitoyltransferase active site of proteins ZDHHC9, ZDHHC14, and ZDHHC5 in multiple species (Homo sapiens, Canis familiaris, Mus musculus, Gallus gallus, Danio rerio, Xenopus laevis, Tetraodon nigroviridis, Drosophila melanogaster, Caenorhabditis elegans, Neurospora crassa, and Saccharomyces cerevisiae). Sequence alignment was performed using ClustalW. Conserved amino acid residues are highlighted in yellow, and the two missense mutations are identified in red.
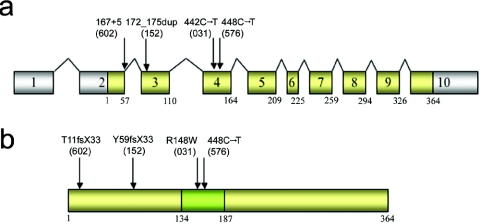
Figure 4. .
a, Diagrammatic representation of the exon structure of ZDHHC9, with positions of mutations found in families with XLMR. Exons that are translated are indicated by yellow shading. b, Diagrammatic representation of the protein ZDHHC9, with the palmitoyltransferase domain indicated by green shading. The location of the amino acid changes in the protein due to mutations in ZDHHC9 are indicated by arrows. The family numbers are in parentheses.
The presenting phenotype in the four families was moderate mental retardation in two or more males. In three families (602, 031, and 576) a diagnosis of XLMR with a Marfanoid habitus or Marfan syndrome was considered. Although the physique of the affected individuals was generally a long and thin habitus, none met the Ghent criteria for Marfan syndrome.11 In family 152, FG syndrome was briefly considered for the diagnosis but was dismissed. The physique was not Marfanoid in this family either.
In family 152, the uncle was said to be profoundly retarded and unable to walk or talk and needed full-time care. His problems were attributed to birth injury, because the umbilical cord was around his neck at birth. No other details are available. The two sibling nephews were referred when they were 6 and 4 years old, respectively, because both boys had developmental delay and moderate learning disability. The first child was born at term by elective cesarean delivery due to failure of progression during labor, but he did not require admission to a special-care baby unit after delivery. His birth weight was 3.4 kg. A solitary juvenile xanthogranuloma appeared at age 4 wk and disappeared at age 15 mo. Developmental delay was noted at age 8 mo, since the child was floppy and not sitting. He was not walking at age 18 mo, and speech and language were delayed. At age 6 years, his height was 111.8 cm (50th percentile), and his weight was 20.3 kg (25th percentile). His head circumference was 52.5 cm (75th percentile). A clinical diagnosis of FG syndrome was considered because of the presence of hypotonia, a cowlick, and a high forehead but, in the absence of macrocephaly, severe constipation, and hypertelorism, this clinical diagnosis was deemed unlikely. The clinical features of the second child were similar to those of the older brother, but the developmental progression was slower. His birth weight was 3.2 kg, and his growth parameters at age 4 years were the 25th percentile for height, the 75th percentile for weight, and the 75th percentile for head circumference. Results of metabolic investigations, including white blood cell enzymes, urine organic acids, very-long-chain fatty acids, and creatine kinase levels, were all normal.
In family 602, two brothers presented with developmental delay. The older brother was delivered at term after an elective cesarean delivery at 42 wk gestation, and his birth weight was 4.2 kg. He walked at age 3 years 6 mo and developed limited speech at age 4 years 6 mo. At age 13 years, a diagnosis of Marfan syndrome was considered. His head circumference was 58 cm (>97th percentile), height was 178 cm (90th–97th percentile), arm span was 190 cm (span:height ratio 1.06), and upper:lower segment ratio was 0.93. Additional clinical features were pectus carinatum, pes planus, and arachnodactyly. Although he had at least two major skeletal features of Marfan syndrome, the results of eye examination and cardiac echocardiogram were normal. Additional investigations excluded homocystinuria and other metabolic diseases. The younger affected brother is a DZ twin and had similar features to the older affected brother, including pectus carinatum, pes planus, and thin facial features. The younger brother's height was in the 75th–90th percentile, his arm span:height ratio was 1.0, and his upper:lower segment ratio was 0.96. In adulthood, the younger affected brother has developed major behavioral problems, and a diagnosis of schizophrenia has been made. He receives medication in a supported environment. The unaffected twin is intellectually normal and does not have a Marfanoid habitus.
In family 031, two brothers presented with developmental delay and mental retardation with unremarkable birth histories. They were described as having Marfanoid features. The older brother’s birth weight was 3.2 kg, and he was a slow feeder, sat at age 13 mo, walked at age 3 years, and talked at age 4 years. He attended a special school and lives semi-independently in a supervised home. As an adult, his head circumference is 55.5 cm (9th–25th percentile), his height is 169.5 cm (9th–25th percentile), and his arm span is 173.5 cm (span:height ratio 1.02), with mild fixed flexion deformity at the elbows. The younger brother has similar facial features, with large ears, long fingers and toes, and pes planus, and has similar growth and developmental parameters. His birth weight was 3.5 kg, and, at age 16 years, his height was 172.4 cm (25th percentile), his arm span was 176 cm (span:height ratio 1.02), and his head circumference was 57.6 cm (50th percentile).
In family 576, the proband was referred to a genetics service because of joint hypermobility, pectus excavatum, long mobile digits, and delayed sitting at age 12 mo. Pregnancy and delivery were uncomplicated, but the child presented with pyloric stenosis at age 6 wk. Adducted thumbs were noted shortly after birth but gradually resolved spontaneously by age 14 mo. At age 14 mo, the child had a long face, strabismus, and prominent ears that were not crumpled. His palate was normal and not high, and examination of the spine showed that it was normal, with no evidence of kyphoscoliosis. Long, thin limbs were noted, with long digits, 5th-finger camptodactyly, and long toes with camptodactyly. His height was in the 90th–98th percentile, his weight was <50th percentile, and his head circumference was in the 50th–75th percentile. A clinical diagnosis of XLMR and Marfanoid habitus was made on the basis of the family history. The mother of this boy reported that her brother had a similar physique, was tall and thin, was double-jointed, and had similarly unusual hands. He also has significant learning difficulties and has managed little employment. She also has two maternal cousins who are affected. In summary, the clinical phenotype in the four families is one of hypotonia, significant delay in walking disproportionate to the intellectual disability, and a Marfanoid habitus in some individuals.
ZDHHC9 (GenBank accession number NP_057116) is an integral membrane protein of 364 aa containing four predicted transmembrane spans, a proline-rich domain, and a zinc-finger DHHC (aspartic acid–histidine-histidine-cysteine) domain in the cytoplasmic loop. It is a palmitoyltransferase that catalyzes the posttranslational modification of RAS.12 The ZDHHC9 gene is expressed highly in kidney, skeletal muscle, brain, lung, and liver; to a lesser extent in placenta, heart, colon, and small intestine; and at low levels in peripheral blood cells, thymus, and spleen.12 ZDHHC9 forms a complex with GCP16 and colocalizes in the Golgi apparatus when overexpressed in mammalian cells. This complex palmitoylates HRAS and NRAS and not GAP-43 or Gαi1.12 There may be additional target proteins modified by ZDHHC9, but they have not been identified thus far. In mammalian cells, palmitoylation determines both the temporal and spatial localization of NRAS and HRAS.13 KRAS is not subject to posttranslational modification by palmitoyltransferase and is mainly identified at the plasma membrane. Monopalmitoylated NRAS displays a more pronounced Golgi localization, a faster retrograde plasma membrane to Golgi trafficking, and a several-fold shorter time at the plasma membrane, compared with the dually palmitoylated HRAS species.13,14
Recently abnormalities in HRAS and KRAS have been identified in Costello syndrome (MIM 218040), cardiofaciocutaneous syndrome (MIM 115150), and Noonan syndrome (MIM 163950), suggesting that changes in these and other molecules in the RAS–mitogen-activated protein (MAP) kinase pathway can cause significant developmental abnormalities.15–18 Interestingly, these syndromes are associated with a mental retardation phenotype, providing further evidence of the role of this pathway in intellectual processing. The mechanism by which loss-of-function mutations in ZDHHC9 cause mental retardation is unclear, but it may be through alteration of the relative proportion of the RAS proteins within the different compartments of nerve cells. This may result in alterations in the growth and development characteristics of neurons similar to those seen by alteration of palmitoylation of GAP-43 and PSD-95.19,20 Palmitoylation of PSD-95, a major component of the postsynaptic density, is required for postsynaptic targeting and clustering of glutamate receptors and palmitoylation of GAP43 is sufficient to alter the branching of dendrites and axons in hippocampal neurons. The finding of mutations in ZDHHC9 therefore adds a further layer of complexity to our understanding of the mental retardation phenotype, as it suggests that alterations in the concentrations and cellular distribution of normal proteins is sufficient to cause disease.
Our findings provide further support for the importance of RAS signaling in the development of intellectual processing. Furthermore, now that the first palmitoyltransferase that causes mental retardation has been identified, defects in other palmitoylation transferases become good candidates for causing other forms of X-linked or autosomal forms of mental retardation.19,20
Acknowledgments
We thank the families, for their long-term cooperation in this work, and Luciannne Vandeleur, for tissue-culture support. This work was supported by Australian National Health and Medical Research Council program grant 400121; the State of New South Wales (NSW) Health Department, through their support of the NSW GOLD Service; National Institute of Child Health and Human Development grant HD26202; a grant from the South Carolina Department of Disabilities and Special Needs; and the Wellcome Trust.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- BDGP: Splice Site Prediction by Neural Network, http://www.fruitfly.org/seq_tools/splice.html (for NNSPLICE)
- ClustalW, http://www.ebi.ac.uk/clustalw/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for ZDHHC9 [accession number NM_016032] and ZDHHC9 [accession number NP_057116])
- NCBI BLAST, http://www.ncbi.nlm.nih.gov/blast/ (for blastp)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Costello, Noonan, and cardiofaciocutaneous syndromes)
- Vega Genome Browser, http://vega.sanger.ac.uk/index.html
References
- 1.Bond J, Roberts E, Springell K, Lizarraga SB, Scott S, Higgins J, Hampshire DJ, Morrison EE, Leal GF, Silva EO, et al (2005) A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet 37:353–355 10.1038/ng1539 [DOI] [PubMed] [Google Scholar]
- 2.Bond J, Scott S, Hampshire DJ, Springell K, Corry P, Abramowicz MJ, Mochida GH, Hennekam RC, Maher ER, Fryns JP, et al (2003) Protein-truncating mutations in ASPM cause variable reduction in brain size. Am J Hum Genet 73:1170–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.des Portes V, Pinard JM, Billuart P, Vinet MC, Koulakoff A, Carrie A, Gelot A, Dupuis E, Motte J, Berwald-Netter Y, et al (1998) A novel CNS gene required for neuronal migration and involved in X-linked subcortical laminar heterotopia and lissencephaly syndrome. Cell 92:51–61 10.1016/S0092-8674(00)80898-3 [DOI] [PubMed] [Google Scholar]
- 4.Gleeson JG, Allen KM, Fox JW, Lamperti ED, Berkovic S, Scheffer I, Cooper EC, Dobyns WB, Minnerath SR, Ross ME, et al (1998) Doublecortin, a brain-specific gene mutated in human X-linked lissencephaly and double cortex syndrome, encodes a putative signaling protein. Cell 92:63–72 10.1016/S0092-8674(00)80899-5 [DOI] [PubMed] [Google Scholar]
- 5.Tarpey P, Parnau J, Blow M, Woffendin H, Bignell G, Cox C, Cox J, Davies H, Edkins S, Holden S, et al (2004) Mutations in the DLG3 gene cause non-syndromic X-linked mental retardation. Am J Hum Genet 75:318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, et al (2003) Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet 34:27–29 10.1038/ng1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stromme P, Mangelsdorf ME, Shaw MA, Lower KM, Lewis SM, Bruyere H, Lutcherath V, Gedeon AK, Wallace RH, Scheffer IE, et al (2002) Mutations in the human ortholog of Aristaless cause X-linked mental retardation and epilepsy. Nat Genet 30:441–445 10.1038/ng862 [DOI] [PubMed] [Google Scholar]
- 8.Gibbons RJ, Picketts DJ, Villard L, Higgs DR (1995) Mutations in a putative global transcriptional regulator cause X-linked mental retardation with α-thalassemia (ATR-X syndrome). Cell 80:837–845 10.1016/0092-8674(95)90287-2 [DOI] [PubMed] [Google Scholar]
- 9.D’Adamo P, Menegon A, Lo Nigro C, Grasso M, Gulisano M, Tamanini F, Bienvenu T, Gedeon AK, Oostra B, Wu SK, et al (1998) Mutations in GDI1 are responsible for X-linked non-specific mental retardation. Nat Genet 19:134–139 10.1038/487 [DOI] [PubMed] [Google Scholar]
- 10.Kutsche K, Yntema H, Brandt A, Jantke I, Nothwang HG, Orth U, Boavida MG, David D, Chelly J, Fryns JP, et al (2000) Mutations in ARHGEF6, encoding a guanine nucleotide exchange factor for Rho GTPases, in patients with X-linked mental retardation. Nat Genet 26:247–250 10.1038/80002 [DOI] [PubMed] [Google Scholar]
- 11.De Paepe A, Devereux RB, Dietz HC, Hennekam RC, Pyeritz RE (1996) Revised diagnostic criteria for the Marfan syndrome. Am J Med Genet 62:417–426 [DOI] [PubMed] [Google Scholar]
- 12.Swarthout JT, Lobo S, Farh L, Croke MR, Greentree WK, Deschenes RJ, Linder ME (2005) DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J Biol Chem 280:31141–31148 10.1074/jbc.M504113200 [DOI] [PubMed] [Google Scholar]
- 13.Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PI (2005) An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 307:1746–1752 10.1126/science.1105654 [DOI] [PubMed] [Google Scholar]
- 14.Rocks O, Peyker A, Bastiaens PI (2006) Spatio-temporal segregation of Ras signals: one ship, three anchors, many harbors. Curr Opin Cell Biol 18:351–357 10.1016/j.ceb.2006.06.007 [DOI] [PubMed] [Google Scholar]
- 15.Aoki Y, Niihori T, Kawame H, Kurosawa K, Ohashi H, Tanaka Y, Filocamo M, Kato K, Suzuki Y, Kure S, et al (2005) Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat Genet 37:1038–1040 10.1038/ng1641 [DOI] [PubMed] [Google Scholar]
- 16.Schubbert S, Zenker M, Rowe SL, Boll S, Klein C, Bollag G, van der Burgt I, Musante L, Kalscheuer V, Wehner LE, et al (2006) Germline KRAS mutations cause Noonan syndrome. Nat Genet 38:331–336 10.1038/ng1748 [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez-Viciana P, Tetsu O, Tidyman WE, Estep AL, Conger BA, Cruz MS, McCormick F, Rauen KA (2006) Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science 311:1287–1290 10.1126/science.1124642 [DOI] [PubMed] [Google Scholar]
- 18.Niihori T, Aoki Y, Narumi Y, Neri G, Cave H, Verloes A, Okamoto N, Hennekam RC, Gillessen-Kaesbach G, Wieczorek D, et al (2006) Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat Genet 38:294–296 10.1038/ng1749 [DOI] [PubMed] [Google Scholar]
- 19.Gauthier-Campbell C, Bredt DS, Murphy TH, El-Husseini AE-D (2004) Regulation of dendritic branching and filopodia formation in hippocampal neurons by specific acylated protein motifs. Mol Biol Cell 15:2205–2217 10.1091/mbc.E03-07-0493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Husseini AE-D, Schnell E, Dakoji S, Sweeney N, Zhou Q, Prange O, Gauthier-Campbell C, Aguilera-Moreno A, Nicoll RA, Bredt DS (2002) Synaptic strength regulated by palmitate cycling on PSD-95. Cell 108:849–863 10.1016/S0092-8674(02)00683-9 [DOI] [PubMed] [Google Scholar]