Abstract
Congenital diaphragmatic hernia (CDH) is a relatively common birth defect associated with high mortality and morbidity. Although the exact etiology of most cases of CDH remains unknown, there is a growing body of evidence that genetic factors play an important role in the development of CDH. In this review, we examine key findings that are likely to form the basis for future research in this field. Specific topics include a short overview of normal and abnormal diaphragm development, a discussion of syndromic forms of CDH, a detailed review of chromosomal regions recurrently altered in CDH, a description of the retinoid hypothesis of CDH, and evidence of the roles of specific genes in the development of CDH.
Congenital diaphragmatic hernia (CDH [MIM 142340, 222400, 610187, and 306950]) is defined as a protrusion of abdominal viscera into the thorax through an abnormal opening or defect that is present at birth. In some cases, this protrusion is covered by a membranous sac. In contrast, diaphragmatic eventrations are extreme elevations, rather than protrusions, of part of the diaphragm that is often atrophic and abnormally thin. CDH is a relatively common birth defect, with an incidence of ∼1 in every 3,000 live births.1,2 CDH is often associated with potentially lethal lung hypoplasia and pulmonary hypertension. Despite advances in therapy, mortality remains high, especially among severely affected infants, and long-term morbidity among survivors is common.3
The most common type of CDH is the posterolateral, or Bochdalek-type, hernia, which accounts for 90%–95% of CDH cases.1 Other types of CDH include anterior retrosternal or peristernal Morgagni hernias, central (septum transversum) hernias, and pars sternalis hernias, which are found in the pentalogy of Cantrell—a rare association involving abnormalities of the anterior diaphragm, sternum, heart, and abdominal wall.
Although there are multiple examples of familial cases of CDH in the literature, the recurrence risk for isolated cases of CDH is often reported to be <2% on the basis of a mathematical model of multifactorial inheritance risk.1,4,5 Empiric data also suggest a relatively low recurrence risk for CDH.6–8 Although multifactorial inheritance may best explain most cases of CDH in humans, much has been learned about the genetic factors that play a role in the development of CDH by studies of patients with CDH caused by specific genetic syndromes and chromosome anomalies. Our understanding of CDH has also been aided by basic research with the use of dietary, teratogen-induced, and knockout models of CDH.
Overview of Normal and Abnormal Diaphragm Development
The development of the human diaphragm occurs between the 4th and 12th wk of gestation. Traditional views of diaphragm development suggest that the diaphragm arises from four different structures.9 The septum transversum gives rise to the central portion of the diaphragm, the pleuroperitoneal folds (PPFs) give rise to the posterolateral section of the diaphragm, the dorsal (esophageal) mesentery gives rise to a portion of the diaphragm posterior to the esophagus, and elements from the thoracic body wall contribute to a rim of musculature around the diaphragm’s periphery. In contrast to this traditional view, systematic examinations of diaphragm development in rodents have failed to identify contributions to the diaphragm musculature from the lateral body wall, the septum transversum, or the esophageal mesenchyme.10 Rather, myogenic cells and axons were shown to coalesce within the PPF and then to expand to form the neuromuscular component of the diaphragm.10 If further investigation shows that this model provides an accurate depiction of diaphragm development in humans, the classic view of diaphragm development will need to be revised.11
Several theories have been proposed concerning the primary embryologic events that lead to the development of CDH. Events implicated in these theories have included (1) abnormalities in (ipsilateral) lung development, (2) failure of closure of the pleuroperitoneal canals, (3) defective myoblast formation, and (4) abnormal phrenic nerve innervation.12–14
Although it is possible that each of these abnormalities may play a role in the development of some cases of CDH, there is growing evidence from animal models that CDH arises from malformation of the amuscular mesenchymal substratum of the PPF before pleuroperitoneal canal closure.10,15,16 Critical findings that support this model include the normal formation of the primordial diaphragm in Fgf10−/− mouse embryos that have complete lung agenesis and the ability to induce defects characteristic of CDH in c-met−/− mouse embryos that do not form diaphragm muscle fibers because of a defect in muscle precursor migration.16
Pulmonary hypoplasia is one of the most serious clinical complications accompanying CDH. The role of physical compression on the development of pulmonary hypoplasia in CDH was effectively demonstrated in studies of surgically produced CDH in fetal lambs and is consistent with the observation that pulmonary hypoplasia is usually more severe on the side of the diaphragmatic defect.17,18 However, studies of lung development in rodents with CDH caused by in utero exposure to the herbicide nitrofen suggested that pulmonary hypoplasia was present before development of a diaphragmatic defect.12,19 This observation led to the development of the dual-hit hypothesis, which states that pulmonary hypoplasia can be caused by the combined effect of an early insult that directly affects lung development followed by further restriction in lung growth, later in gestation, secondary to diminished fetal breathing movements and competition for space as a result of the herniation of the abdominal contents into the thoracic cavity.19
It is possible that these two hits may be caused by defects within a single gene that affects both lung and diaphragm development. Examples of genes that are known to affect both lung and diaphragm development include Friend of GATA2 (FOG2 [MIM 603693]) and GATA-binding protein 4 (GATA4 [MIM 600576]).20–22 In the future, it may be possible to formally test the dual-hit hypothesis by generating conditional knockout mice in which the lungs and the primordial diaphragm are targeted separately. These studies may also provide another means of testing whether diaphragmatic defects can be induced or altered by a primary pulmonary insult.
Syndromic Forms of CDH
CDH may occur either as an isolated birth defect or in association with other non–hernia-related anomalies (known as “nonisolated CDH” or “CDH+”). Some anomalies—including lung hypoplasia, abnormalities in cardiac position, intestinal malrotation, and patent ductus arteriosus—are typically considered secondary effects of CDH and are not considered grounds for classification as nonisolated CDH. Common findings associated with CDH include cardiovascular abnormalities, abnormalities of the CNS, and genitourinary and/or renal anomalies.
Some individuals with nonisolated CDH have patterns of anomalies that are strongly suggestive of a specific genetic syndrome. In patients with CDH for whom a syndromic diagnosis can be provided, the most frequently diagnosed syndrome is Fryns syndrome (MIM 229850).23–25 However, reports of individuals with Fryns-like phenotypes associated with chromosomal anomalies—including duplication of 1q24-q31.2; deletion of the terminal portion of 6q, 8p23.1, and 15q26; and partial trisomy 22—suggests that some cases of CDH attributed to this autosomal recessive syndrome are likely to represent genocopies of this disorder.24,26–29
Many of the syndromes associated with CDH have specific Mendelian inheritance patterns and, in some cases, the location and/or the identity of the causative gene(s) is known. Examples of CDH syndromes associated with a particular chromosomal locus or causative gene(s) are shown in table 1. CDH is a cardinal feature of some of these syndromes, whereas, for others, the rates of CDH are lower but probably exceed the level seen in the general population.
Table 1. .
Examples of Genetic Syndromes Associated with CDH
| Syndrome Name | Chromosome(s) | Gene(s) | Brief Description |
| Beckwith-Wiedemann (MIM 130650) | 11p15, 5q35 | CDKN1C (MIM 600856), NSD1 (MIM 606681) | Autosomal dominant inheritance, macroglossia, hypoglycemia, visceromegaly, abdominal-wall defects, and overgrowth |
| CHARGE (MIM 214800) | 8q12.1 | CHD7 (MIM 608892) | Autosomal dominant inheritance, coloboma, cardiac abnormalities, choanal atresia, growth retardation, genital abnormalities, ear abnormalities, and hearing loss |
| Cornelia de Lange (MIM 122470 and 300590) | 5p13.1, Xp11.22-p11.21 | NIPBL (MIM 608667), SMC1A (MIM 300040) | Autosomal dominant inheritance, distinctive facial features, microcephaly, hirsutism, malformations of the upper limbs, and growth retardation |
| Craniofrontonasal (MIM 304110) | Xq12 | EFNB1 (MIM 300035) | X-linked dominant inheritance, females more severely affected, craniosynostosis, hypertelorism, broad nasal tip, grooved nails of the hallux and thumb, syndactyly, and skeletal abnormalities |
| Denys-Drash (MIM 194080) | 11p13 | WT1 (MIM 607102) | Autosomal dominant inheritance, male pseudohermaphroditism, genital abnormalities, and increased risk of Wilms tumor |
| Donnai-Barrow (MIM 222448) | 2q23-q3130 | … | Autosomal recessive inheritance, CDH, omphalocele, agenesis of the corpus callosum, hypertelorism, and hearing loss |
| Fryns (MIM 229850) | Fryns-like phenotype has been seen with duplication of 1q24-q31.2; deletion of the terminal portion of 6q, 8p23.1, and 15q26; and partial trisomy 2224,26–29 | … | Autosomal recessive inheritance, CDH, coarse facial features, cleft lip/palate, cardiac malformations, cerebral abnormalities, and hypoplastic finger/toenails |
| Pallister-Killian (MIM 601803) | Mosaic tetrasomy 12p | … | Coarse facial features with broad forehead and hypertelorism, sparse temporal hair, hypopigmentations, and mental retardation |
| Simpson-Golabi-Behmel (MIM 312870 and 300209) | Xq26, Xp22.3-p22.2 | GPC3 (MIM 300037), CXORF5 (MIM 300170) | X-linked recessive inheritance, macrosomia, coarse facial features, hypertelorism, macroglossia, skeletal abnormalities, abdominal-wall defects, and renal abnormalities |
| Thoracoabdominal (MIM 313850) | Xq25-q26.1 | … | X-linked dominant inheritance, diaphragmatic and ventral hernias, hypoplastic lungs, and cardiac anomalies |
| Wolf-Hirschhorn (MIM 194190) | 4p16 | … | “Greek helmet” facial appearance, mental and growth retardation, cleft lip/palate, cardiac defects, and epilepsy |
The existence of genetic syndromes associated with CDH provides one of the strongest lines of evidence that genetic factors play a role in the development of CDH. It is likely that much of our understanding of CDH will be shaped by studies that focus on understanding the molecular mechanisms by which changes in these genes result in diaphragmatic defects. These studies may, in turn, help us identify interacting genes that are involved in the development of other forms of CDH.
Chromosomal Abnormalities Described in Patients with CDH
Chromosomal anomalies have been identified as an important etiology for nonisolated CDH.31 In the majority of published cases, chromosome anomalies were identified using a combination of G-banded chromosome analysis and/or FISH. The use of new genomic technologies—like array-based comparative genomic hybridization (array CGH)—is likely to increase the number of chromosomal anomalies identified in individuals with CDH and may aid in the identification of CDH-related genes.23,24,32
Trisomy 13, 18, and 21 and 45,X are the most common aneuploidies described in association with CDH.31 Structural abnormalities—including deletions, duplications, inversions, and translocations—of nearly all chromosomes have also been described in association with CDH.33,34 Both Lurie33 and Enns et al.34 have published useful reviews of chromosomal anomalies associated with CDH. Using these reviews as a foundation, we have compiled an updated list of the CDH-associated chromosomal anomalies (table 2).
Table 2. .
Structural Chromosomal Anomalies Described in Patients with CDH[Note]
| Chromosome, Type of Anomaly, and Patient Karyotype | Study Author(s) |
| Chromosome 1: | |
| Balanced translocation: | |
| 46,X,t(X;1)(q26;q12) | Punnett35 |
| 46,XY,t(1;15)(q41;q21.2) de novo | Smith et al.36 |
| 46,XY,t(1;21)(q32;q22)pat | Howe et al.37 |
| 46,XY,t(1;14)(p22;q13),inv(6)(p25q22),del(15)(q26.1q26.2) | Klaassens et al.38 |
| Inversion: | |
| 46,XY,inv(1)(q41q44)mat | Tonks et al.39 |
| Duplication: | |
| 46,XY/46,X,der(Y)t(Y;1)(q12;q12) | Ahn et al.40 |
| 46,XY[9]/46,X,der(Y),t(Y;1)(q12;q12)[12] | Zeng et al.41 |
| 46,XX,der(22)t(1;22)(q12;p12)[11]/46,XX[9] | Ahmed et al.42 |
| der(9)t(1;9)(q32.3;p24.1) | Kousseff43 |
| dup(1)(q22q32) | Schneider et al.44 |
| dup(1)(q22q32)mosaicism | van Dooren45 |
| 46,XY/46,XY,dup(1)(q24.2q31.2) | Clark and Fenner-Gonzalez26 |
| dup(1)(q25q31.2) | Mehraein et al.46 |
| Deletion: | |
| der(1)t(1;21)mosaicism | Philip et al.47 |
| 46,XX,del(1)(p) | Benjamin et al.48 |
| 46,XX,del(1)(q32.3q42.3) | Youssoufian et al.49 |
| 46,XY,del(1)(q41q42.12) | Kantarci et al.23 |
| 46,XY,del(1)(q32.3q42.2) | Slavotinek et al.50 |
| 46,XX,del(1)(q42.11q42.3) | Rogers et al.51 |
| Chromosome 2: | |
| Duplication: | |
| 46,XX/47,XX,der(2)del(2)(p13)del(2)(q12) | Grevengood et al.52 |
| der(X)t(X;2)(q27;p13)mat | Sarda et al.53 |
| dup(2)(p13p25) | van Dooren45 |
| dup(2)(p21p25) | van Dooren45 |
| der(6)t(2;6)(p23;p25) | Bender et al.54 |
| 46,XY,dup(2)(p21p25) | Heathcote et al.55 |
| 46,XY,der(7)t(2;7)(p25.3;q34)mat | Enns et al.34 |
| dup(2)(q33q37) | Johnson et al.56 |
| 46,XY,der(15)t(2;15)(q37.2;q26.2) | Scott et al.57 |
| Deletion: | |
| 46,XX,der(2)t(2;7)(q36;q37)pat | Brackley et al.58 |
| 46,XY,del(2)(q33q35 or q35q37) de novo | Tonks et al.39 |
| 46,XY,der(2)t(2;8)(q37;p11.2)pat | Tonks et al.39 |
| 46,XX,der(2)t(2;14)(q37.1;q31.2) | van Dooren45 |
| der(2)t(2;14)(q37;q31.2) | De La Fuente et al.59 |
| 46,XX,del(2)(q37.1) | Casas et al.60 |
| 46,XY,del(2)(q37.3) | Reddy et al.61 |
| Chromosome 3: | |
| Balanced translocation: | |
| 46,XY,t(3;12)(p21.1;p13.3) de novo | Tonks et al.39 |
| Duplication: | |
| der(21)t(3;21)(p24.3;q11.2)mat | Pettigrew62 |
| 46,XX,der(15)t(3;15)(q29;q26.1)mat | Rosenberg et al.63 |
| Deletion: | |
| del(3)(p) | Steinhorn et al.64 |
| del(3)(p12p21) | Pfeiffer et al.65 |
| 46,XY,del(3)(q11.1q13.2)/47,XY,del(3)(q11.2q13.2),+r(3) | Brennan et al.66 |
| 46,XY,del(3)(q21q23) | Wolstenholme et al.67 |
| del(3)(q22); two patients | Dillon et al.68 |
| der(3)t(3;5)(q27;q31) | Kristeshavilli et al.69 |
| Miscellaneous: | |
| 46,XY,der,t(3;8)(p23;p23.1) | Tibboel and Gaag31 |
| Chromosome 4: | |
| Ring chromosomea: | |
| 45,XX,−4/46,XX,r(4)(p1?6;q3?3) | Kocks et al.70 |
| Duplication: | |
| 46,XY,rec(4),dup(4)(q),inv(4)(p15.2q25)pat | Kobori et al.71 |
| 46,XY,inv dup(4)(q32q26),del(4)(q32) | Frints et al.72 |
| 46,XX,der(22)t(4;22)(q28.3;p13) | Celle et al.73 |
| 46,XY,der(18)t(4;18)(q31;q23) | Yunis et al.74 |
| dup(4)(q25q31) | van Dooren45 |
| Deletion: | |
| 46,XY,del(4)(p16) | van Dooren et al.75 |
| del(4)(p16); two patients | Howe et al.37 |
| 46,XY,del(4)(p16) | Tachdjian et al.76 |
| del(4)(p16) | Pober et al.8 |
| del(4)(p16.3) | Casaccia et al.77 |
| 46,XY,rec(4),dup(4)(q),inv(4)(p15.2q25)pat | Kobori et al.71 |
| del(4)(p16); two patients | Laziuk et al.78 |
| 46,XX,del(4)(p13) | Sergi et al.79 |
| del(4)(p16.3) | Van Buggenhout et al.80 |
| del(4)(p16.3) | Slavotinek et al.50 |
| 46,XX,der(4)t(4;13)(p16;q32) | Tapper et al.81 |
| del(4)(q31.3) | Del Campo et al.82 |
| 46,XX,del(4)(qter) | Park et al.83 |
| del(4)(q31) | van Dooren45 |
| del(4)(q31.1q31.3 or q31.3q32.2) | Wakui et al.84 |
| del(4)(q31) | Young et al.85 |
| 46,XY,inv dup(4)(q32q26),del(4)(q32) | Frints et al.72 |
| der(4)t(4;20)(q34.2;q13.1)pat | Pober et al.8 |
| der(4)t(4;20)(q34.2;q13.1)pat | Reiss et al.86 |
| Chromosome 5: | |
| Duplication: | |
| Partial trisomy 5 | Bollmann et al.87 |
| dup(5)(q33) | Korner et al.88 |
| 46,XY,−9,+t(5q;9p) | Torfs et al.1 |
| der(15)t(5;15)(p15.3;q24), two cases | Aviram-Goldring et al.89 |
| der(9)t(5;9)(p13;p22) | Liberfarb et al.90 |
| 47,XY,t(5;13)(p15;q21)+der(13)t(5;13)(p15;q21)mat | Masuno et al.91 |
| der(3)t(3;5)(q27;q31) | Kristeshavilli et al.69 |
| Deletion: | |
| del(5)(q13q22) | Kousseff43 |
| Chromosome 6: | |
| Balanced translocation: | |
| 46,XY,t(6;8)(q24;q23) | Howe et al.37 |
| Inversion: | |
| 46,XY,t(1;14)(p22;q13),inv(6)(p25q22),del(15)(q26.1q26.2) | Klaassens et al.38 |
| Duplication: | |
| 47,XY,+der(22)t(6;22)(p25;q11.2) | Scarbrough et al.92 |
| 46,XY,der(15)t(6;15)(p25;q24)mat | Kristofferson et al.93 |
| der(15)t(6;15)(p25;q24)mat | Kristofferson et al.93 |
| 46,XX, inv dup(6)(p25.2p22.2) | Scott et al.57 |
| Deletion: | |
| der(6)t(2;6)(p23;p25) | Bender et al.54 |
| 46,XY,der(6)t(X;6)(p21.2;p25) | Batanian et al.94 |
| 46,XY,der(6)t(6;8)(p25.1;q24.23) | Baruch and Erickson95 |
| del(6)(q15q21) | Yu and Bock96 |
| 46,XY,del(6)(q23) | Shen-Schwarz et al.97 |
| del(6)(q23) | van Dooren45 |
| 46,XX,del(6)(q25.3) | Krassikoff and Sekhon27 |
| del(6)(qter)mat | Le Caignec et al.32 |
| Miscellaneous: | |
| 46,XX,add(6)(q23 or q25) | Tonks et al.39 |
| Chromosome 7: | |
| Duplication: | |
| dup(7)(p15p22) | Herrmann et al.98 |
| 46,XX,der(2)t(2;7)(q36;q37)pat | Brackley et al.58 |
| 46,XY,der(18)t(7;18)(qter;p11.1) | Habedank and Trost-Binkhues99 |
| Deletion: | |
| del(7)(p21) | van Dooren45 |
| del(7)(q) | Fauza and Wilson100 |
| del(7)(q11q22) | Klep-de Pater et al.101 |
| 46,XY,del(7)(q32) | Torfs et al.1 |
| 46,XX,del(7)(q32) | Dott et al.102 |
| der(7)t(7;20)(q33.2;p13) | Kjaer et al.103 |
| 46,XY,der(7)t(2;7)(p25.3;q34)mat | Enns et al.34 |
| Miscellaneous: | |
| chtb(7)(q31.1) | Bonneau et al.104 |
| Chromosome 8: | |
| Balanced translocation: | |
| 46,XY,t(6;8)(q24;q23) | Howe et al.37 |
| t(8;14)(q24;q21) | Philip et al.47 |
| 46,XX,t(8;13)(q22.3q22)mat | Temple et al.105 |
| 46,XX,t(8;15)(q22.3q15) de novo | Temple et al.105 |
| Duplication: | |
| Trisomy 8 mosaicism | Pober et al.8 |
| 46,XY,der(2)t(2;8)(q37;p11.2)pat | Tonks et al.39 |
| 46,XY, inv dup(8)(p23.1p11.22) | Ringer et al.106 |
| dup(8)(p21) | van Dooren45 |
| 46,XY,der(12)t(8;12)(p21;p13) | Moreno Fuenmayor et al.107 |
| 46,XX,der(15)t(8;15)(q24.1;q26.1) | Chen et al.108 |
| 46,XY,dup(8)(q) | Hilfiker et al.109 |
| 46,XY,der(6)t(6;8)(p25.1;q24.23) | Baruch and Erickson95 |
| Deletion: | |
| 46,XY,del(8) | Thorpe-Beeston et al.110 |
| del(8)(p) | Pober et al.8 |
| del(8)(p22) | Kousseff43 |
| 46,XY,del(8)(p23.1) | Howe et al.37 |
| del(8)(p23.1) | Faivre et al.111 |
| 46,XY,del(8)(p23.1p23.1) | Shimokawa et al.112 |
| 46,XX,del(8)(p23.1) | Borys and Taxy113 |
| 46,XY,del(8)(p23.1) | Lopez et al.114 |
| 46,XX,del(8)(p23.1) | Pecile et al.115 |
| 46,XY,del(8)(p23.1) | Fraer et al.116 |
| 46,XY,del(8)(p23.1:p23.1) | Slavotinek et al.24 |
| del(8)(q21.2q22) | Maerzke et al.117 |
| del(8)(q22q24.1) | Harnsberger et al.118 |
| del(8)(q22q24.1) | Capellini et al.119 |
| Miscellaneous: | |
| 46,XX,add(8)(p?) | Betremieux et al.120 |
| 46,XY,der,t(3;8)(p23;p23.1) | Tibboel and Gaag31 |
| Chromosome 9: | |
| Ring chromosomea: | |
| r(9) | Dillon et al.68 |
| Duplication: | |
| 47,XX,+9 | Chen et al.121 |
| 47,XY,+9 | Suzumori et al.122 |
| 47,XX,+9 | Sepulveda et al.123 |
| Trisomy 9 | Frohlich124 |
| Trisomy 9 | Robert et al.125 |
| Trisomy 9 | Dott et al.102 |
| 47,XX,+i(9p) | Henriques-Coelho et al.126 |
| Deletion: | |
| 46,XX,der(9)t(9;16)(p22;q24) | Alfi et al.127,128 |
| der(9)t(5;9)(p13;p22); two patients | Liberfarb et al.90 |
| 46,XY,der(9)t(9;11)(p24;p13)pat | Donnenfeld et al.129 |
| 46,XY,−9,+t(5q;9p) | Torfs et al.1 |
| 46,XY,der(9)t(9;16)(q34.3;q24.3) | Ferrero et al.130 |
| der(9)t(1;9)(q32.3;p24.1) | Kousseff43 |
| Chromosome 10: | |
| Balanced translocation: | |
| t(X,10) de novo | Cunniff et al.131 |
| Duplication: | |
| 46,XY,der(21)t(10;21)(p11;p11) | Yunis et al.132 |
| 46,XY,der(20)t(10;20)(p12;p12) | Lurie et al.133 |
| Miscellaneous: | |
| 46,XY,add(10)(q?q24) de novo | Tonks et al.39 |
| Chromosome 11: | |
| Duplication: | |
| 46,XY,der(9)t(9;11)(p24;p13)pat | Donnenfeld et al.129 |
| 47,XX or XY,+der(22)t(11;22)(q23;q11) | Iselius et al.,134 Fraccaro et al.,135 Phelan et al.,136 Azancot et al.,137 de Beaufort et al.,138 Aurias et al.,139 Noel et al.,140 Dean et al.,29 Kousseff,43 Hickmann et al.,141 van Dooren,45 Tonks et al.,39 Dott et al.,102 Borys and Taxy,113 and Kadir et al.142 |
| 47,XY,+der(13)t(11;13)(q21;q14) | Park et al.143 |
| 46,XY,der(12)t(11;12)(q23.3;q24.3)mat | Klaassens et al.144 |
| Deletion: | |
| 46,XY,del(11)(p12p15.1) | Scott et al.145 |
| del(11)(p13) | Gustavson et al.146 |
| 46,XY,?del(11)(q23),9qh+ | Dott et al.102 |
| 46,XX,der(11)t(11;12)(q24;p11.2) | Decker-Philips et al.147 |
| Chromosome 12: | |
| Balanced translocation: | |
| t(12;15) | Fauza and Wilson100 |
| 46,XY,t(3;12)(q21.1;p13.3) de novo | Tonks et al.39 |
| Duplication: | |
| Mosaic tetrasomy 12p | Bergoffen et al.,148 Corning et al.,149 Rodriguez et al.,150 Donnenfeld et al.,129,151 Dott et al.,102 Betremieux et al.,152 Veldman et al.,153 Witters et al.,154 Tonks et al.,39 Borys and Taxy,113 Takakuwa et al.,155 and Pober et al.8 |
| 46,XX,der(11)t(11;12)(q24;p11.2) | Decker-Philips et al.147 |
| der(15)t(12;15) | Pober et al.8 |
| Deletion: | |
| 46,XY,der(12)t(8;12)(p21;p13) | Moreno Fuenmayor et al.107 |
| 46,XY,del(12) | Howe et al.37 |
| 46,XY,der(12)t(11;12)(q23.3;q24.3)mat | Klaassens et al.144 |
| Chromosome 13: | |
| Balanced translocation: | |
| 46,XX,t(8;13)(q22.3q22)mat | Temple et al.105 |
| Ring chromosomea: | |
| r(13) | van Dooren45 |
| Duplication: | |
| 47,XY,+der(13)(qter→q31::q31→neo→qter) | Warburton et al.156 and Tohma et al.157 |
| 46,XX,der(4)t(4;13)(p16;q32) | Tapper et al.81 |
| 47,XY,+der(13)t(11;13)(q21;q14) | Park et al.143 |
| 47,XY,t(5;13)(p15;q21)+der(13)t(5;13)(p15;q21)mat | Masuno et al.91 |
| Deletion: | |
| 46,XX,13q- | Benjamin et al.48 |
| Chromosome 14: | |
| Balanced translocation: | |
| t(8;14)(q24;q21) | Philip et al.47 |
| 46,XY,t(1;14)(p22;q13),inv(6)(p25q22),del(15)(q26.1q26.2) | Klaassens et al.38 |
| Duplication: | |
| dup(14)(q24q32) | van Dooren45 |
| 46,XX,dup(14)(q32.1) | Masada et al.158 |
| der(2)t(2;14)(q37;q31.2) | De La Fuente et al.59 |
| 46,XY/47,XY,+14 | Howe et al.37 |
| 46,XX/46,XX,i(14)(q10) | Scott et al.57 |
| Deletion: | |
| 46,XY,del(14)(q32.11qter), bilateral eventration | Masada et al.158 |
| Chromosome 15: | |
| Balanced translocation: | |
| 46,XY,t(1;15)(q41;q21.2) de novo | Smith et al.36 |
| 46,XX,t(8;15)(q22.3q15) de novo | Temple et al.105 |
| t(12;15) | Fauza and Wilson100 |
| Duplication: | |
| inv dup(15) | van Dooren45 |
| 46,XY,dup(15)(q11q13)mat | Boyar et al.159 |
| 47,XX,+dic(15)(q11.2) | Howe et al.37 |
| dup(15)(q15q26) | van Dooren45 |
| dup(15)(q15q26) + del(X)(p22) | van Dooren45 |
| der(X)t(X;15)(p22;q15)mat | Zabel and Baumann160 |
| Deletion: | |
| 46,XY,r(15)(p11q26) | de Jong et al.28 |
| 46,XY,r(15)(p11q26.1), two cases | Klaassens et al.38 |
| r(15)(q25.3) | Elghezal et al.161 |
| 46,XY,der(15)t(6;15)(p25;q24)mat | Kristofferson et al.93 |
| der(15)t(6;15)(p25;q24)mat | Kristofferson et al.93 |
| 46,XX,der(15)t(3;15)(q29;q26.1)mat | Rosenberg et al.63 |
| 46,XX,der(15)t(15;17)(q24.3;q23.3) | Howe et al.37 |
| 46,XY,del(15)(q24) 46,XX,del(15)(q24) | Bettelheim et al.162 |
| 46,XX,der(15)t(8;15)(q24.1;q26.1) | Chen et al.108 |
| 46,XY,der(15)t(15;20)(q26.3;q13.1) | Reiss et al.86 |
| der(15)t(5;15)(p15.3;q24), two cases | Aviram-Goldring et al.89 |
| 46,XX,del(15)(q25q26.2) | Schlembach et al.163 |
| 46,XX,del(15)(q26.1) | Biggio et al.164 |
| 46,XX,del(15)(q26.1) de novo | Hengstschlager et al.165 |
| 46,XY,del(15)(q26.1) de novo | Tonks et al.39 |
| 46,XY,r(15)(q26.2) | Tumer et al.166 |
| 46,XY,t(1;14)(p22;q13),inv(6)(p25q22),del(15)(q26.1q26.2) | Klaassens et al.38 |
| del(15)(q26) | Pober et al.8 |
| der(15)t(12;15) | Pober et al.8 |
| 46,XX,del(15)(q26.2) | Slavotinek et al.24 |
| 46,XX,del(15)(q26.2;26.2) | Slavotinek et al.24 |
| 46,XY,der(15)t(8;15)(q24.2;q26.2) | Slavotinek et al.50 |
| del(15)(q26.1) | Lopez et al.114 |
| 46,XY,der(15)t(2;15)(q37.2;q26.2) | Scott et al.57 |
| Chromosome 16: | |
| Duplication: | |
| 47,XY+ mar 16 | Howe et al.37 |
| 46,XX,der(9)t(9;16)(p22;q24) | Alfi et al.127,128 |
| 46,XY,der(9)t(9;16)(q34.3;q24.3) | Ferrero et al.130 |
| 47,XX,+16[3]/46,XX[15] | Chen et al.167 |
| 47,XX,+16 | Johnson et al.168 |
| Chromosome 17: | |
| Ring chromosomea: | |
| 46,XX,r(17)/45,XX,−17 | Baldermann et al.169 |
| Duplication: | |
| 45,XX,der(15)t(15;17)(q24.3;q23.3) | Howe et al.37 |
| Chromosome 18: | |
| Duplication: | |
| 46,XX/46,XX,del(18)(ptel)/46,XX,−18, +i(18q) | Le Caignec et al.32 |
| 46,XY,idic(18)(p11)[15]/45,XY,−18[6]/46,XY,del(18)(p11.7)[6]/spurious cells[3] | Dott et al.102 |
| iso(18)(q) | Hayashi et al.170 |
| Deletion: | |
| 46,XX/46,XX,del(18)(ptel)/46,XX,−18, +i(18q) | Le Caignec et al.32 |
| 46,XY,idic(18)(p11)[15]/45,XY,−18[6]/46,XY,del(18)(p11.7)[6]/spurious cells[3] | Dott et al.102 |
| iso(18)q | Hayashi et al.170 |
| 46,XY,der(18)t(7;18)(qter;p11.1) | Habedank and Trost-Binkhues99 |
| 46,XY,der(18)t(4;18)(q31;q23) | Yunis et al.74 |
| 45,XX,der(18)t(18;22)(qter;q11),−22 | Geneix et al.171 |
| Chromosome 20: | |
| Duplication: | |
| der(4)t(4;20)(q34.2;q13.1)pat | Reiss et al.86 |
| der(7)t(7;20)(q33.2;p13) | Kjaer et al.103 |
| 46,XY,der(15)t(15;20)(q26.3;q13.1) | Reiss et al.86 |
| der(4)t(4;20)(q34.2;q13.1)pat | Pober et al.8 |
| Deletion: | |
| 46,XY,der(20)t(10;20)(p12;p12) | Lurie et al.133 |
| Chromosome 21: | |
| Translocation: | |
| 46,XY,t(1;21)(q32;q22)pat | Howe et al.37 |
| Duplication: | |
| der(1)t(1;21)mosaicism | Philip et al.47 |
| Tetrasomy 21 | Pober et al.8 |
| Deletion: | |
| 46,XY,der(21)t(10;21)(p11;p11) | Yunis et al.132 |
| 46,XY,+X,dic(X;21)(p11.1;p11.1) | Smith et al.172 |
| der(21)t(3;21)(p24.3;q11.2)mat | Pettigrew62 |
| Chromosome 22: | |
| Duplication: | |
| 47,XY,+der(22)t(6;22)(6p25;q11.2) | Scarbrough et al.92 |
| 47,XX or XY,+der(22)t(11;22)(q23;q11) | Iselius et al.,134 Fraccaro et al.,135 Phelan et al.,136 Azancot et al.,137 de Beaufort et al.,138 Aurias et al.,139 Noel et al.,140 Dean et al.,29 Kousseff,43 Hickmann et al.,141 van Dooren,45 Tonks et al.,39 Dott et al.,102 Borys and Taxy,113 and Kadir et al.142 |
| Trisomy 22 | Kim et al.,173 Ladonne et al.,174 Phillipson et al.,175 Dean et al.,29 Golombek and Shaw,176 Ramsing et al.,177 and Van Voss et al.178 |
| Deletion: | |
| 45,XX,der(18)t(18;22)(qter;q11),−22 | Geneix et al.171 |
| 46,XX,der(22)t(1;22)(q12;p12)[11]/46,XX[9] | Ahmed et al.42 |
| del(22)(q11q11) | Betremieux et al.152 |
| 46,XX,der(22)t(4;22)(q28.3;p13) | Celle et al.73 |
| Chromosome X: | |
| Balanced translocation: | |
| 46,X,t(X;1)(q26;q12) | Punnett35 |
| Monosomy: | |
| 45,X | David and Illingworth,6 Benjamin et al.,48 Bollmann et al.,87 Tibboel and Gaag,31 Cunniff et al.,131 Robert et al.,125 Dawani et al.,180 and Scott et al.57 |
| Diploid/tetraploid mosaicism: | |
| 92,XXXX/46,XX | Witters et al.154 |
| Duplication: | |
| 46,XY,der(6)t(X;6)(p21.2;p25) | Batanian et al.94 |
| Deletion: | |
| 46,X,del(X)(p22.1) | Plaja et al.179 |
| der(X)t(X;2)(q27;p13)mat | Sarda et al.53 |
| der(X)t(X;15)(p22;q15)mat | Zabel and Baumann160 |
| dup(15)(q15q26) + del(X)(p22) | van Dooren45 |
| der(X)t(X;Y)(p22.3;q11.2) | Pober et al.8 |
| 46,XY,+X,dic(X;21)(p11.1;p11.1) | Smith et al.172 |
| Chromosome Y: | |
| Duplication: | |
| der(X)t(X;Y)(p22.3;q11.2) | Pober et al.8 |
| Deletion: | |
| 46,XY/46,X,der(Y)t(Y;1)(q12;q12) | Ahn et al.40 |
| 46,XY[9]/46,X,der(Y),t(Y;1)(q12;q12)[12] | Zeng et al.41 |
Note.— No abnormalities in chromosome 19 have been described in patients with CDH.
Always with deletion.
Chromosomal regions that are involved in balanced translocation or are recurrently deleted or duplicated in patients with CDH are of particular interest to researchers, because they are more likely to harbor genes that cause or predispose to the development of CDH than are less commonly affected regions of the genome. It is important to note that many of the deletions and duplications described in the literature are the product of unbalanced translocations, and it is possible that the diaphragmatic defects seen in these cases are caused by two or more genes located in nonadjacent chromosomal regions. It should also be noted that, in most instances, CDH occurs in only a fraction of individuals with a particular chromosomal abnormality. This suggests that genetic background, environmental factors, and/or stochastic events may also play a role in determining whether an individual develops CDH.
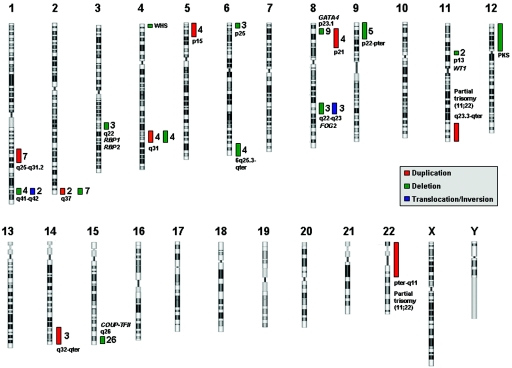
Chromosomal regions that have been associated with CDH in three or more individuals are shown in figure 1 and are described individually below. Several of these intervals overlap the position of genes that are involved in the retinoic-acid signaling pathway—which has been hypothesized to play a role in the development of CDH—or genes that have been implicated in the development of CDH because of studies involving animal models and/or human subjects. In most cases, the chromosomal region described represents a minimally affected region defined by G-banded chromosome analysis and/or FISH. In instances where the minimal affected region has been defined using high-resolution techniques, such as array CGH, we have made specific mention of these results.
Figure 1. .
Chromosomal regions and selected candidate genes for CDH. Recurrent chromosomal abnormalities associated with patients with CDH are represented by colored bars. For each region, the number of patients described with that duplication (red bar), deletion (green bar), or translocation/inversion (blue bar) is given. Selected candidate genes and genetic syndromes are included beside their respective regions. PKS = Pallister-Killian syndrome; WHS = Wolf-Hirschorn syndrome.
Duplication of 1q25q31.2
Duplication of this region has been described in at least seven patients with CDH.26,40–42,44–46 At least three of these cases of CDH were also associated with cleft palate.
Deletion of 1q41-q42
Deletions of this region have been reported in four cases of CDH.23,49–51 Three cases involve a larger deletion, identified by standard cytogenetics techniques. The smallest deletion was determined by Kantarci et al.23 using high-resolution array CGH that refined the interval to an ∼5-Mb region bounded by BACs RP11-553F10 and RP11-275O4. One individual with balanced translocation and one individual with an inversion involving 1q41 have also been described.36,39
Deletion or Duplication of 2q37
CDH has been described in seven patients with deletions of 2q37and in two patients with 2q37 duplications.39,45,56–61 Interestingly, in almost all these patients, the duplication or deletion starts at band q37. Of the patients in whom this region is deleted, two also have duplications of the distal portion of 14q, which is discussed below.45,59
Deletion of 3q22
Deletions of this region have been described in three individuals with CDH.67,68 Two of these patients had blepharophimosis and facial dysmorphism most likely attributable to deletions of FOXL2, which is known to cause blepharophimosis, ptosis, and epicanthus inversus syndrome (BPES [MIM 110100]).67,68 The most-promising CDH candidate genes located in this region are the genes for cellular retinol binding protein 1 (RBP1 [MIM 180260]) and cellular retinol binding protein 2 (RBP2 [MIM 180280]). These genes are part of the retinol signaling pathway and have been shown to play a role in vitamin A homeostasis and lung maturation in mice.181,182 No mutations in RBP1 or RBP2 have been described in patients with CDH to date.
Deletion of 4p16
Wolf-Hirschhorn syndrome (MIM 194190) is associated with deletions of 4p16 and is characterized by a “Greek helmet” facial appearance, growth retardation, mental retardation, seizures and/or epilepsy, cleft lip and/or palate, and cardiac abnormalities. Although not a common finding in Wolf-Hirschhorn syndrome, CDH has been described in association with at least 14 cases of 4p16 deletion.8,37,50,71,75–77,79–81,183 The patient described by Casaccia et al.77 has the smallest known deletion of 4q16 associated with CDH (2.6 Mb), with the deletion extending from locus D4S43 to the telomere.
Duplication or Deletion of 4q31
CDH has been described in four individuals with 4q31 duplications.45,71,73,74 Deletions of this region have also been seen in four individuals with CDH.45,82,84,85
Duplication of 5p15
Duplications of 5p15 have been described in at least four patients with CDH.89–91 All these cases were accompanied by an additional chromosomal anomaly, such as deletion 9p22-pter or deletion 15q26-qter, both of which are discussed below.
Deletion of 6p25
Deletions of this region have been seen in at least three individuals with CDH, all of whom have an additional chromosomal duplication.54,94,95
Deletion of 6q25.3-qter
Deletions involving this region have been seen in four cases of CDH.27,32,45,97 Le Caignec et al.32 used array CGH to identify a <5-Mb subtelomeric deletion of 6q in a patient with CDH and other anomalies. It should be noted, however, that this same deletion was found in the patient’s mother, who presented with only learning disabilities.
Deletion of 8p23.1
Deletions involving 8p23.1 have been described in >30 individuals with abnormal phenotypes, including nine patients with CDH.24,37,43,111–116 More-distal deletions of 8p23.1-p23.2 have also been found in unaffected individuals, suggesting that more-telomeric deletions may be a normal variant in the white population.184 Shimokawa et al. used array CGH to define an ∼6-Mb deletion of 8p23.1 in a patient with CDH.112 This deletion was flanked by low-copy repeats and was bounded by BACs RP11-143D15 and RP11-252C15. GATA4 resides within this region and has been proposed as a candidate gene for CDH. Of note, deletions and loss-of-function mutations of GATA4 have been seen in individuals with cardiac defects involving the cardiac septum, and the majority of patients with CDH with deletion of 8p23.1 also have cardiac anomalies (atrial, ventricular, or atrioventricular septal defect).115,184–187 Gata4 heterozygous-null mice also display diaphragm defects in association with pulmonary and cardiac abnormalities.22 This animal model is discussed in greater detail below.
Duplication of 8p21-p23.1
Duplication of 8p21-p23.1 has been described four times in patients with CDH.39,45,106,107 The patient described by Moreno Fuenmayor et al.107 had a phenotype consistent with that of other patients with duplication 8p21.188 The patient described by Ringer et al.106 had an inverted duplication of 8p11.22-p23.1. In some instances, patients with an inverted duplication of 8p also have a small deletion of 8p23.1, a region recurrently deleted in CDH. Unfortunately, it is unclear whether the patient described by Ringer et al.106 also carried this deletion.
Deletion of 8q22-q23
Three patients with CDH with 8q deletions have been described.117–119 Each of these deletions included bands 8q22-q23, and all these patients had dysmorphic features similar to those of other patients with similar deletions.189 There are also three affected individuals with balanced translocations that involve this region.37,105 FOG2 resides within this region, and data supporting its role in diaphragm development are described below.
Deletion of 9p24-pter
Deletions of this region have been described in five patients with nonisolated CDH.43,90,127,129 All these deletions were terminal deletions as part of unbalanced translocation with another autosome.
Deletion of 11p13
Although only two patients with CDH have been described with a deletion of 11p13, this region is of particular interest because it harbors the Wilms tumor 1 gene (WT1).145,146 Data supporting a role for WT1 in the development of CDH is described below.
Duplication of 11q23.3-qter
This duplication has been described numerous times in patients with CDH. In most cases, this duplication is the result of the more common chromosomal anomaly 47,XX, or XY,+der(22)t(11;22), resulting from 3:1 meiotic segregation.144 Two patients have been described in whom the duplication of 11q23-qter is the result of an unbalanced translocation with another autosome.143,144
Duplication of 12p
Mosaic tetrasomy 12p, or Pallister-Killian syndrome, is characterized by coarse facial features, sparse temporal hair, skin abnormalities, mental retardation, and a high rate of CDH.190 This syndrome usually results from mosaicism for an isochromosome: i(12)(p10).191 Also, one patient with CDH and a balanced translocation involving 12p13.1 has been described.39
Duplication of 14q32
Mosaic trisomy 14 has been described in at least two patients with nonisolated CDH, and duplications of 14q32 have been described three times in association with CDH.37,45,57,59,158
Deletion of 15q26
Deletions of the distal part of the long arm of chromosome 15 have been described in at least 26 patients with nonisolated CDH, making this anomaly one of the most reported structural chromosomal anomalies in CDH.33,38 The majority of patients with deletions of the long arm of 15q have a severe phenotype that can include cardiac abnormalities, limb abnormalities, and dysmorphic features. Chick ovalbumin upstream promoter-transcription factor II (COUP-TFII) resides within this region, and data supporting its role in the development of CDH is described below.
Duplication of 22pter-q11
Duplications of this region have been described numerous times in patients with CDH. This duplication usually is seen as part of the common chromosomal anomaly 47,XX, or XY,+der(22)t(11;22), resulting from 3:1 meiotic segregation.144 Although no patients with isolated duplications of this region have been described, CHD is also a recurrent finding in individuals with trisomy 22.33
Candidate Pathways and Genes
Although the etiology of most cases of CDH remains unknown, there is increasing evidence that specific pathways and genes play a role in the development of CDH. These data are derived from the identification of candidate genes in regions commonly deleted and/or duplicated in CDH and from several genetic animal models. In this section, we review evidence for involvement of the retinoid signaling pathway and genes COUP-TFII, FOG2, GATA4, WT1, and SLIT3 in the development of CDH.
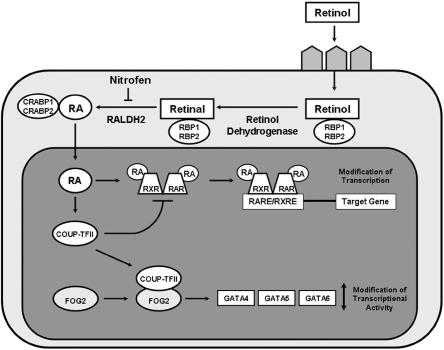
Retinoid Signaling Pathway
Vitamin A (retinol) and its derivatives (retinoids) are essential for embryonic development. Abnormalities in the retinoid signaling pathway and its downstream targets have long been hypothesized to lead to the development of CDH.192 The first connection between retinoids and CDH resulted from the observation that 25%–40% of the offspring of rat dams that were fed a diet deficient in vitamin A developed CDH and that the proportion of affected pups diminished when vitamin A was reintroduced into the diet in midgestation.193–195
Subsequently, in utero exposure to the herbicide nitrofen, bisdiamine (a spermatogenesis inhibitor), SB-210661 (a 5-lipoxygenase inhibitor), and BPCA (a thromboxane-A2 receptor antagonist) was shown to cause CDH in rodents.196 The diaphragmatic defects caused by these substances closely mimicked the characteristics of human posterolateral CDH, including the intermittent incidence of associated cardiac anomalies.197 The connection between these defects and the retinoid signaling pathway became clear when vitamin A was found to decrease the incidence and severity of nitrofen-induced CDH.13 Later, it was shown that nitrofen, bisdiamine, SB210661, and BPCA inhibit RALDH2, a key enzyme responsible for the conversion of retinal to retinoic acid.196
Two knockout mouse models also suggest a role for retinoid signaling in the development of CDH. A proportion of RARα/RARβ receptor double-knockout mice have posterolateral diaphragmatic defects similar to those seen both in humans and in teratogen-induced mouse models of CDH.198 Targeted ablation of Coup-TFII, a gene encoding a transcription factor regulated by the retinoid signaling pathway, has also been shown to cause posterolateral CDH similar to Bochdalek-type CDH seen in humans.199
Preliminary evidence that retinoids may play a role in the development of CDH in human comes from a small study in which the levels of plasma retinol and retinol-binding protein in the cord blood of infants with CDH was found to be 50% lower than those in age-matched controls.200
COUP-TFII
COUP-TFII (also known as NR2F2) is a transcription factor in the steroid/thyroid hormone receptor superfamily. The COUP-TFII gene is located on chromosome 15q26 in a region recurrently deleted in individuals with CDH.33,38 Klaassens et al.38 defined a minimally deleted region for CDH on chromosome 15q26 by use of FISH and array CGH data from patients with nonisolated CDH. Of the genes within this region, COUP-TFII was thought to be the strongest candidate because its expression had been shown previously to be regulated by retinoids and because COUP-TFII regulates gene transcription by influencing retinoic acid receptor or retinoid X receptor heterodimerization (fig. 2).201,202 This region has since been reduced to include COUP-TFII and only eight other known genes.57 As mentioned above in the discussion of the retinoid signaling pathway, homozygous tissue-specific ablation of Coup-TFII in mice causes posterolateral CDH similar to Bochdalek-type CDH seen most commonly in humans.199
Figure 2. .
Retinoic acid (RA) signaling pathway and CDH candidate genes. Retinol travels to target cells via the blood and is taken up by receptors on the cell surface. Once in the cytoplasm, retinol is converted to retinal by retinol dehydrogenases and then to RA by retinal dehydrogenases, of which RALDH2 is the predominant enzyme. The action of RALDH2 can be inhibited by teratogens, such as nitrofen. Several binding proteins are present in the cytoplasm, including retinol-binding proteins 1 and 2 (RBP1 and RBP2), which bind retinol and retinal, and cellular RA-binding proteins 1 and 2 (CRABP1 and CRABP2). When RA enters the nucleus, it mediates its effects by binding to RA receptors (RARs) and retinoid X receptors (RXRs). RARs and RXRs dimerize and regulate gene expression by binding to short DNA sequences—RA-responsive elements (RAREs) and retinoid X–responsive elements (RXREs)—located in the vicinity of target genes. COUP-TFII expression is upregulated by RA. COUP-TFII can act as a repressor of this pathway by directly sequestering RXR, thereby preventing heterodimerization to RAR and inhibiting gene transcription. This process may be a negative feedback system that precisely balances the transcription of certain genes during diaphragm development. COUP-TFII has been shown to interact physically with FOG2, which, in turn, modulates the transcriptional activity of GATA4, GATA5, and GATA6.
Together, these data suggest that deletion of COUP-TFII is likely to play a key role in the development of CDH in individuals with 15q26 deletions. It has not yet been determined whether abnormalities in COUP-TFII are responsible for cases of CDH not associated with 15q26 deletions. Although several research groups are actively screening COUP-TFII in patient cohorts, to date, no CDH-causing mutations in this gene have been published.
FOG2
FOG2 (also known as ZFPM2) is a zinc finger–containing protein that modulates the transcriptional activity of GATA proteins, which, in turn, play important roles in early embryogenesis. The first indication that FOG2 might play a role in normal diaphragm development came with the discovery of an N-ethyl-N-nitrosourea mouse mutant with pulmonary hypoplasia and an abnormal diaphragm that lacked muscularization of the posterolateral and peripheral regions. Sequencing of the Fog2 gene in this mouse revealed a hypomorphic splice-donor mutation.21 A de novo R112X heterozygous mutation was subsequently found in an infant who died shortly after birth with diaphragmatic eventration and severe pulmonary hypoplasia.21
Although no mutations in FOG2 have been found in individuals with CDH, it is interesting to note that FOG2 is located on chromosome 8q23 in a region commonly deleted in individuals with CDH and that FOG2 interacts physically with COUP-TFII.203 It is possible that these proteins work together to regulate downstream target genes that play a role in the development of CDH.
GATA4
GATA4 is a member of a family of DNA-binding proteins that recognize a consensus sequence (the GATA motif), which is found in the promotor regions of many genes.204 GATA4 encodes a transcription factor that interacts with FOG2 during the morphogenesis of the heart.205 GATA4 is located on chromosome 8p23.1, a region recurrently deleted in individuals with CDH.
Recently, Jay et al.22 showed that 70% of heterozygous Gata4+/Δex2 mice on a C57BL/6 background displayed cardiac, lung, or diaphragm defects. The diaphragmatic defects, which affected ∼30% of mice, were located in the ventral midline and were covered by a sac that was continuous with the diaphragm. Together with the occurrence of 8p23.1 deletions in human patients, this research provides additional evidence that GATA4 is important for lung and diaphragm development in humans. To date, no CDH-causing mutations in GATA4 have been identified.
WT1
WT1 is located on chromosome 11p13, a region recurrently deleted in individuals with CDH, and encodes a zinc-finger transcription factor that is expressed in the pleural and abdominal mesothelium that help to form the diaphragm.145,146,206 Mutations in WT1 associated with CDH have been described in two patients with Denys-Drash syndrome (MIM 194080)—characterized by male pseudohermaphroditism, nephropathy, and Wilms tumor—and one patient with Frasier syndrome (MIM 136680)—characterized by focal and segmental glomerulosclerosis, male pseudohermaphroditism, and gonadoblastoma.207–209 A child with Meacham syndrome (MIM 608978)—characterized by CDH, double vagina, sex reversal, and cardiac malformations—was also found to have a de novo WT1 mutation.210 Further evidence of the role of WT1 in CDH comes from homozygous Wt1-null mouse embryos that develop diaphragmatic hernias.211
Recently, Clugston et al.11 compared the Wt1−/− mutant with other CDH animal models—namely, the nitrofen rat model and the vitamin A–deficient rat model. They found that the Wt1 null mutants have defects in the PPF as do the two other models, suggesting that there is a common pathogenic mechanism in dietary, teratogenic, and genetic models of CDH.
Homolog of Drosophila Slit 3 (SLIT3)
SLIT3 is located on chromosome 5q35.1 and is one of three human homologs of the Drosophila Slit gene. In mice, Slit3 is expressed predominantly in the mesothelium of the diaphragm during embryonic development.212 Homozygous Slit3-deficient mice have CDH on or near the ventral midline portion of the central tendon that is similar to the central (septum transversum) type of diaphragmatic hernia seen in humans.212,213 Although SLIT3 seems to be a strong candidate gene for this relatively rare type of CDH, no SLIT3 mutations have been identified in humans with CDH to date.
Discussion
The existence of specific CDH-associated genetic syndromes, recurrently deleted and/or duplicated chromosomal regions, and transgenic mouse models of CDH provide evidence of the important role that genetic factors play in the development of CDH. Future research efforts in each of these areas will provide information that will help us to better understand the etiology of many cases of CDH. Although the genes for several CDH-related syndromes are known, many have not yet been discovered. Additional efforts must also be made to determine the role that these genes play in diaphragm development. The increased use of high-resolution cytogenetic techniques—such as array CGH—in both the clinical and research settings are likely to aid in the discovery of new CDH-related genes as new chromosomal regions associated with CDH are identified and as previously identified regions are refined. Transgenic models have proven to be a valuable resource not only as a way to begin to understand the role that specific genes play in diaphragm development but also as a tool for the discovery of new CDH-related genes. The current emphasis on development of improved resources for transgenic mouse studies will make it easier for researchers to rapidly test hypotheses regarding the involvement of particular genes or gene combinations in diaphragm development. The increasing availability of new technologies, such as micro–magnetic resonance imaging scanners, may also make it easier to screen existing mouse strains for diaphragm defects.
Although several genes have been clearly shown to underlie abnormal diaphragm development in mice, few CDH-related mutations have been identified in corresponding genes in humans. One possible explanation is that the genes and pathways that underlie CDH development in mice are different than those that commonly cause CDH in humans. This, however, seems less likely when one considers that many of these genes are located in chromosomal regions recurrently deleted in individuals with CDH and, therefore, represent excellent candidates for CDH in humans.
Another possibility is that de novo mutations in individual genes are responsible for only a fraction of human CDH cases. The chance of identifying such an event may be particularly low when one considers that this fraction would likely represent a heterogeneous population in which de novo mutations in many different genes can result in the same basic phenotype. If this is the case, identifying de novo mutations in individual genes may require both the recruitment and screening of relatively large numbers of patients with CDH. Such efforts may still be worthwhile because the identification of de novo changes provides valuable evidence that a particular gene is involved in the development of human CDH. Such discoveries could also prove clinically significant if phenotype and/or genotype analysis suggests that a particular subgroup of patients with CDH is more likely to carry de novo mutations in a particular gene. It is important, however, that such screening efforts do not overlook subtle inherited changes that may be important for understanding the complex inheritance pattern that likely underlies the majority of CDH cases.
The assumption that the majority of CDH cases results from a complex inheritance pattern, in which a combination of genetic and environmental factors affect the final phenotype, is consistent with the sporadic nature of the disease and the relatively few instances of familial cases described in the literature.1,4,5 Indeed, it seems reasonable to hypothesize that relatively small inherited changes in the function of two or more genes within the same CDH-related pathway could cause diaphragmatic defects in the offspring of otherwise-normal carrier parents. An additional level of variation may also be added by environmental stressors—such as toxins or nutritional factors such as vitamin A—acting on genetically susceptible individuals. The combined effects of several genes and the environment may also underlie the association of CDH with some chromosomal abnormalities.
Research into the underlying causes of CDH has the potential to positively effect the clinical management of CDH in affected individuals and their families. The description of multiple genetic syndromes associated with CDH highlights the importance of a careful evaluation of patients with CDH. In cases in which CDH is diagnosed prenatally, such an evaluation may have an influence on medical decision making, including decisions made about the possible termination of the pregnancy. It has also become clear that a significant proportion of nonisolated CDH cases are attributable to chromosomal anomalies.30 Since recent studies suggest that some causal chromosomal anomalies can be missed on routine G-banded chromosome analysis, it seems prudent to consider obtaining a higher-resolution cytogenetic study, such array CGH, to look for cryptic deletions and duplications in patients with nonisolated CDH with normal chromosome analyses.23,24,61 Storage of DNA samples from patients with CDH and their parents should also be considered becauase access to such material may ultimately allow a diagnosis to be made, which, in turn, would form the foundation for improved genetics counseling for all family members.
Our understanding of the genetic factors associated with CDH may make it possible to devise preventative strategies or to improve therapeutic interventions for patients with CDH. It is important to keep in mind that measures aimed at improving clinical outcome may not require the prevention or correction of the diaphragmatic defect itself. Instead, these strategies may focus on improvement in postnatal lung function, and, eventually, prenatal modulation (such as tracheal occlusion procedures), since pulmonary hypoplasia and pulmonary hypertension are major contributors to both the morbidity and the mortality associated with CDH. With this in mind, it will be important to identify which CDH-related genes and pathways have direct affects on normal diaphragm and lung development, because they may be particularly good therapeutic targets.
Addendum
After submission of this manuscript, Pasutto et al.214 reported that homozygous mutations in the stimulated by retinoic acid gene 6 homolog (STRA6 [MIM 610745]) cause a broad spectrum of malformations, including CDH, anophthalmia, congenital heart defects, alveolar capillary dysplasia, lung hypoplasia, and mental retardation. In a separate report, Kawaguchi et al.215 showed that STRA6 acts as a membrane receptor for retinol binding protein and mediates cellular uptake of vitamin A.
Acknowledgments
This research was supported by the Sophia Foundation for Scientific Research, Rotterdam, the Netherlands (SSWO 441); the Howard Hughes Medical Institute; the Baylor College of Medicine’s Child Health Research Center (through National Institutes of Health [NIH] grant HD41648); and NIH grant HD-050583.
Web Resource
The URL for data presented herein is as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CDH, FOG2, GATA4, Fryns syndrome, Beckwith-Wiedemann syndrome, CDKN1C, NDS1, CHARGE syndrome, CHD7, Cornelia de Lange syndrome, NIPBL, SMC1A, craniofrontonasal syndrome, EFNB1, Denys-Drash syndrome, WT1, Donnai-Barrow syndrome, Palister-Killian syndrome, Simpson-Golabi-Behmel syndrome, GPC3, CXORF5, thoracoabdominal syndrome, Wolf-Hirschhorn syndrome, BPES, RBP1, RBP2, Frasier syndrome, Meacham syndrome, and STRA6)
References
- 1.Torfs CP, Curry CJ, Bateson TF, Honore LH (1992) A population-based study of congenital diaphragmatic hernia. Teratology 46:555–565 10.1002/tera.1420460605 [DOI] [PubMed] [Google Scholar]
- 2.Skari H, Bjornland K, Haugen G, Egeland T, Emblem R (2000) Congenital diaphragmatic hernia: a meta-analysis of mortality factors. J Pediatr Surg 35:1187–1197 10.1053/jpsu.2000.8725 [DOI] [PubMed] [Google Scholar]
- 3.Harrison MR, Bjordal RI, Langmark F, Knutrud O (1978) Congenital diaphragmatic hernia: the hidden mortality. J Pediatr Surg 13:227–230 10.1016/S0022-3468(78)80391-1 [DOI] [PubMed] [Google Scholar]
- 4.Edwards JH (1960) The simulation of Mendelism. Acta Genet Stat Med 10:63–70 [DOI] [PubMed] [Google Scholar]
- 5.Norio R, Kaariainen H, Rapola J, Herva R, Kekomaki M (1984) Familial congenital diaphragmatic defects: aspects of etiology, prenatal diagnosis and treatment. Am J Med Genet 17:471–483 10.1002/ajmg.1320170210 [DOI] [PubMed] [Google Scholar]
- 6.David TJ, Illingworth CA (1976) Diaphragmatic hernia in the south-west of England. J Med Genet 13:253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czeizel A, Kovacs M (1985) A family study of congenital diaphragmatic defects. Am J Med Genet 21:105–117 10.1002/ajmg.1320210115 [DOI] [PubMed] [Google Scholar]
- 8.Pober BR, Lin A, Russell M, Ackerman KG, Chakravorty S, Strauss B, Westgate MN, Wilson J, Donahoe PK, Holmes LB (2005) Infants with Bochdalek diaphragmatic hernia: sibling precurrence and monozygotic twin discordance in a hospital-based malformation surveillance program. Am J Med Genet A 138:81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rottier R, Tibboel D (2005) Fetal lung and diaphragm development in congenital diaphragmatic hernia. Semin Perinatol 29:86–93 10.1053/j.semperi.2005.04.004 [DOI] [PubMed] [Google Scholar]
- 10.Babiuk RP, Zhang W, Clugston R, Allan DW, Greer JJ (2003) Embryological origins and development of the rat diaphragm. J Comp Neurol 455:477–487 10.1002/cne.10503 [DOI] [PubMed] [Google Scholar]
- 11.Clugston RD, Klattig J, Englert C, Clagett-Dame M, Martinovic J, Benachi A, Greer JJ (2006) Teratogen-induced, dietary and genetic models of congenital diaphragmatic hernia share a common mechanism of pathogenesis. Am J Pathol 169:1541–1549 10.2353/ajpath.2006.060445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iritani I (1984) Experimental study on embryogenesis of congenital diaphragmatic hernia. Anat Embryol (Berl) 169:133–139 [DOI] [PubMed] [Google Scholar]
- 13.Thebaud B, Tibboel D, Rambaud C, Mercier JC, Bourbon JR, Dinh-Xuan AT, Archer SL (1999) Vitamin A decreases the incidence and severity of nitrofen-induced congenital diaphragmatic hernia in rats. Am J Physiol 277:L423–L429 [DOI] [PubMed] [Google Scholar]
- 14.Skandalakis JE, Gray SW, Symbas P (1994) The trachea and lungs. In: Skandalakis JE, Gray SW (eds) Embryology for surgeons. Williams and Wilkins, Baltimore, pp 414–450 [Google Scholar]
- 15.Allan DW, Greer JJ (1997) Pathogenesis of nitrofen-induced congenital diaphragmatic hernia in fetal rats. J Appl Physiol 83:338–347 [DOI] [PubMed] [Google Scholar]
- 16.Babiuk RP, Greer JJ (2002) Diaphragm defects occur in a CDH hernia model independently of myogenesis and lung formation. Am J Physiol Lung Cell Mol Physiol 283:L1310–L1314 [DOI] [PubMed] [Google Scholar]
- 17.de Lorimer AA, Tierney DF, Parker HR (1967) Hypoplastic lungs in fetal lambs with surgically produced congenital diaphragmatic hernia. Surgery 62:12–17 [Google Scholar]
- 18.Okuyama H, Kubota A, Kawahara H, Oue T, Kitayama Y, Yagi M (2006) Correlation between lung scintigraphy and long-term outcome in survivors of congenital diaphragmatic hernia. Pediatr Pulmonol 41:882–886 10.1002/ppul.20466 [DOI] [PubMed] [Google Scholar]
- 19.Keijzer R, Liu J, Deimling J, Tibboel D, Post M (2000) Dual-hit hypothesis explains pulmonary hypoplasia in the nitrofen model of congenital diaphragmatic hernia. Am J Pathol 156:1299–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ackerman KG, Wang J, Luo L, Fujiwara Y, Orkin SH, Beier DR (2006) Gata4 is necessary for normal pulmonary lobar development. Am J Respir Cell Mol Biol (http://ajrcmb.atsjournals.org/cgi/reprint/2006-0211RCv1) (electronically published December 1, 2006; accessed February 28, 2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ackerman KG, Herron BJ, Vargas SO, Huang H, Tevosian SG, Kochilas L, Rao C, Pober BR, Babiuk RP, Epstein JA, et al (2005) Fog2 is required for normal diaphragm and lung development in mice and humans. PloS Genet 1:58–65 10.1371/journal.pgen.0010010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jay PY, Bielinska M, Erlich JM, Mannisto S, Pu WT, Heikinheimo M, Wilson DB (2007) Impaired mesenchymal cell function in Gata4 mutant mice leads to diaphragmatic hernias and primary lung defects. Dev Biol 301:602–614 10.1016/j.ydbio.2006.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kantarci S, Casavant D, Prada C, Russell M, Byrne J, Haug LW, Jennings R, Manning S, Blaise F, Boyd TK, et al (2006) Findings from aCGH in patients with congenital diaphragmatic hernia (CDH): a possible locus for Fryns syndrome. Am J Med Genet A 140:17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slavotinek A, Lee SS, Davis R, Shrit A, Leppig KA, Rhim J, Jasnosz K, Albertson D, Pinkel D (2005) Fryns syndrome phenotype caused by chromosome microdeletions at 15q26.2 and 8p23.1. J Med Genet 42:730–736 10.1136/jmg.2004.028787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slavotinek A (2004) Fryns syndrome: a review of the phenotype and diagnostic guidelines. Am J Med Genet A 124:427–433 10.1002/ajmg.a.20381 [DOI] [PubMed] [Google Scholar]
- 26.Clark RD, Fenner-Gonzales M (1989) Apparent Fryns syndrome in a boy with a tandem duplication of 1q24-31.2. Am J Med Genet 34:422–426 10.1002/ajmg.1320340319 [DOI] [PubMed] [Google Scholar]
- 27.Krassikoff N, Sekhon GS (1990) Terminal deletion of 6q and Fryns syndrome: a microdeletion/syndrome pair? Am J Med Genet 36:363–364 10.1002/ajmg.1320360327 [DOI] [PubMed] [Google Scholar]
- 28.de Jong G, Rossouw RA, Retief AE (1989) Ring chromosome 15 in a patient with features of Fryns’ syndrome. J Med Genet 26:469–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dean JC, Couzin DA, Gray ES, Lloyd DJ, Stephen GS (1991) Apparent Fryns’ syndrome and aneuploidy: evidence for a disturbance of the midline developmental field. Clin Genet 40:349–352 [DOI] [PubMed] [Google Scholar]
- 30.Kantarci S, Donahoe PK, Hill RS, Al-Gazali L, Lacombe D, Chassaing N, Bieth E, Black G, Donnai D, Walsh C, et al (2006) Identification of a genetic locus for Donnai-Barrow syndrome. Paper presented at the Annual Meeting of the American Society of Human Genetics, New Orleans, October 9–13 [Google Scholar]
- 31.Tibboel D, Gaag AV (1996) Etiologic and genetic factors in congenital diaphragmatic hernia. Clin Perinatol 23:689–699 [PubMed] [Google Scholar]
- 32.Le Caignec C, Boceno M, Saugier-Veber P, Jacquemont S, Joubert M, David A, Frebourg T, Rival JM (2005) Detection of genomic imbalances by array-based comparative genomic hybridization in fetuses with multiple malformations. J Med Genet 42:121–128 10.1136/jmg.2004.025478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lurie IW (2003) Where to look for the genes related to diaphragmatic hernia? Genet Couns 14:75–93 [PubMed] [Google Scholar]
- 34.Enns GM, Cox VA, Goldstein RB, Gibbs DL, Harrison MR, Golabi M (1998) Congenital diaphragmatic defects and associated syndromes, malformations and chromosome anomalies: a retrospective study of 60 patients and literature review. Am J Med Genet 79:215–255 [DOI] [PubMed] [Google Scholar]
- 35.Punnett HH (1994) Simpson-Golabi-Behmel syndrome (SGBS) in a female with an X-autosome translocation. Am J Med Genet 50:391–393 10.1002/ajmg.1320500424 [DOI] [PubMed] [Google Scholar]
- 36.Smith SA, Martin KE, Dodd KL, Young ID (1994) Severe microphthalmia, diaphragmatic hernia and Fallot’s tetralogy associated with a chromosome 1;15 translocation. Clin Dysmorphol 3:287–291 [PubMed] [Google Scholar]
- 37.Howe DT, Kilby MD, Sirry H, Berker GM, Roberts E, Davison EV, McHugo J, Whittle MJ (1996) Structural chromosome anomalies in congenital diaphragmatic hernia. Prenat Diagn 16:1003–1009 [DOI] [PubMed] [Google Scholar]
- 38.Klaassens M, Van Dooren MF, Eussen HJ, Douben H, Den Dekker AT, Lee C, Donahoe PK, Galjaard RJ, Goemaere N, De Krijger RR, et al (2005) Congenital diaphragmatic hernia and chromsome 15q26: determination of a candidate region by use of fluorescent in situ hybridization and array-based comparative genomic hybridization. Am J Hum Genet 76:877–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tonks A, Wyldes M, Somerset DA, Dent K, Abhyankar A, Bagchi I, Lander A, Roberts E, Kilby MD (2004) Congenital malformations of the diaphragm: findings of the West Midlands Congenital Anomaly Register 1995 to 2000. Prenat Diagn 24:596–604 10.1002/pd.908 [DOI] [PubMed] [Google Scholar]
- 40.Ahn HY, Shin JC, Kim YH, Ko HS, Park IY, Kim SJ, Rha JG, Kim SP (2005) Prenatal diagnosis of congenital diaphragmatic hernia in a fetus with 46,XY/46,X,−Y,+der(Y)t(Y;1)(q12;q12) mosaicism: a case report. J Korean Med Sci 20:895–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng S, Patil SR, Yankowitz J (2003) Prenatal detection of mosaic trisomy 1q due to an unbalanced translocation in one fetus of a twin pregnancy following in vitro fertilization: a postzygotic error. Am J Med Genet A 120:464–469 10.1002/ajmg.a.20189 [DOI] [PubMed] [Google Scholar]
- 42.Ahmed AA, Gilbert-Barness E (2004) A Fryns syndrome-like phenotype with mosaic t(1;22)(q12;p12) chromosomal translocation. Clin Dysmorphol 13:111–112 10.1097/00019605-200404000-00012 [DOI] [PubMed] [Google Scholar]
- 43.Kousseff BG (2000) Congenital diaphragmatic hernia in genetics. Proc Greenwood Genet Center 19:130–131 [Google Scholar]
- 44.Schneider N, Suhr L, Hawkins H, Hughes M (1991) A second case of Fryns syndrome associated with a duplication of 1q22-32: a karyotype association for congenital diaphragmatic hernia. Am J Hum Genet Suppl 49:275 [Google Scholar]
- 45.van Dooren MF (2004) Congenital diaphragmatic hernia: the importance of genetic and environmental factors. PhD thesis, Erasmus University, Rotterdam, The Netherlands [Google Scholar]
- 46.Mehraein Y, Morlot S, Miller K (2000) De novo duplication of a band fragment of the long arm of chromosome 1. Med Genet 12:96 [Google Scholar]
- 47.Philip N, Gambarelli D, Guys JM, Camboulives J, Ayme S (1991) Epidemiological study of congenital diaphragmatic defects with special reference to aetiology. Eur J Pediatr 150:726–729 10.1007/BF01958765 [DOI] [PubMed] [Google Scholar]
- 48.Benjamin DR, Juul S, Siebert JR (1988) Congenital posterolateral diaphragmatic hernia: associated malformations. J Ped Surg 23:899–903 10.1016/S0022-3468(88)80380-4 [DOI] [PubMed] [Google Scholar]
- 49.Youssoufian H, Chance P, Tuck-Muller CM, Jabs EW, (1988) Association of a new chromosomal deletion [del(1)(q32q42)] with diaphragmatic hernia: assignment of a human ferritin gene. Hum Genet 78:267–270 10.1007/BF00291674 [DOI] [PubMed] [Google Scholar]
- 50.Slavotinek AM, Moshrefi A, Davis R, Leeth E, Schaeffer GB, Burchard GE, Shaw GM, James B, Ptacek L, Pennacchio LA (2006) Array comparative genomic hybridization in patients with congenital diaphragmatic hernia: mapping of four CDH-critical regions and sequencing of candidate genes at 15q26.1-15q26.2. Eur J Hum Genet 14:999–1008 10.1038/sj.ejhg.5201652 [DOI] [PubMed] [Google Scholar]
- 51.Rogers J, Harris D, Pasztor L (1995) Interstitial deletion of the long arm of chromosome 1: del(1)(pter→42.11:q42.3→qter). Am J Hum Genet Suppl 57:A125 10.1002/ajmg.1320570202 [DOI] [Google Scholar]
- 52.Grevengood C, Dalton JD, Dungan JS, Park VM, Tharapel AT, Martens P, Ward JC, Shulman LP, Simpson JL, Elias S (1993) Prenatal detection of a de novo supernumerary marker chromosome as der(2)(p13q12) in a fetus with abnormal facies, single umbilical artery and diaphragmatic hernia. Am J Hum Genet Suppl 53:1796 [Google Scholar]
- 53.Sarda P, Lefort G, Devaux P, Humeau C, Rieu D (1992) Multiple congenital anomalies due to partial 2p13—2pter duplication resulting from an unbalanced X;2 translocation. Ann Genet 35:117–120 [PubMed] [Google Scholar]
- 54.Bender K, Reinwein H, Gorman L, Wolf U (1969) Familial 2/C-translocation: 46,XY,t(2p−;Cp+) and 46,XX,Cp+. Humangenetik 8:94–104 10.1007/BF00295832 [DOI] [PubMed] [Google Scholar]
- 55.Heathcote JG, Sholdice J, Walton JC, Willis NR, Sergovich FR (1991) Anterior segment mesenchymal dysgenesis associated with partial duplication of the short arm of chromosome 2. Can J Ophthalmol 26:35–43 [PubMed] [Google Scholar]
- 56.Johnson J, Beere K, Gunwardene RI, Abassi I (1992) Newborn female with partial trisomy 2q33-2q37 presenting with diaphragmatic hernia and mild dysmorphic features. Am J Hum Genet Suppl 51:A290 [Google Scholar]
- 57.Scott DA, Klaassens M, Holder AM, Lally KP, Fernandes CJ, Galjaard RJ, Tibboel D, de Klein A, Lee B (2007) Genome-wide oligonucleotide-based array comparative genome hybridization analysis of non-isolated congenital diaphragmatic hernia. Hum Mol Genet 16:424–430 10.1093/hmg/ddl475 [DOI] [PubMed] [Google Scholar]
- 58.Brackley KJ, Kilby MD, Morton J, Whittle MJ, Knight SJ, Flint J (1999) A case of recurrent congential fetal anomalies associated with a familial subtelomeric translocation. Prenat Diagn 19:570–574 [DOI] [PubMed] [Google Scholar]
- 59.De La Fuente AA, Gerssen-Schoorl KB, Breed AS (1988) Partial duplication 14q/deletion 2q in two sibs due to t(2;14)(q37.1;q31.2)pat. Ann Genet 31:254–257 [PubMed] [Google Scholar]
- 60.Casas KA, Mononen TK, Mikail CN, Hassed SJ, Li S, Mulvihill JJ, Liin HJ, Falk RE (2004) Chromsome 2q terminal deletion: report of 6 new patients and review of phenotype-breakpoint correlations in 66 individuals. Am J Med Genet A 130:331–339 10.1002/ajmg.a.30156 [DOI] [PubMed] [Google Scholar]
- 61.Reddy KS, Flannery D, Farrer RJ (1999) Microdeletion of chromosome sub-band 2q37.3 in two patients with abnormal situs viscerum. Am J Med Genet 84:460–468 [DOI] [PubMed] [Google Scholar]
- 62.Pettigrew AL (1992) Trisomy 3p: two new cases and a review of the literature. Am J Hum Genet Suppl 51:A86 [Google Scholar]
- 63.Rosenberg C, Blakemore KJ, Kearns WG, Giraldez RA, Escallon CS, Pearson PL, Stetten G (1992) Analysis of reciprocal translocations by chromosome painting: applications and limitations of the technique. Am J Hum Genet 50:700–705 [PMC free article] [PubMed] [Google Scholar]
- 64.Steinhorn RH, Kriesmer PJ, Green TP, McKay CJ, Payne NR (1994) Congenital diaphragmatic hernia in Minnesota: impact of antenatal diagnosis on survival. Arch Ped Adolesc Med 148:626–631 [DOI] [PubMed] [Google Scholar]
- 65.Pfeiffer RA, Rauch A, Ulmer R, Beinder E, Trautmann U (1998) Interstitial deletion del(3)(p12p21) in a malformed child subsequent to paternal paracentric insertion (or intraarm shift) 46,XY,ins(3)(p24.1p12.1p21.31). Ann Genet 41:17–21 [PubMed] [Google Scholar]
- 66.Brennan P, Croaker GD, Heath M (2001) Congenital diaphragmatic hernia and interstitial deletion of chromosome 3. J Med Genet 38:556–558 10.1136/jmg.38.8.556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wolstenholme J, Brown J, Masters KG, Wright C, English CJ (1994) Blepharophimosis sequence and diaphragmatic hernia associated with interstitial deletion of chromosome 3 [46,XY,del(3)(q21q23)]. J Med Genet 31:647–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dillon E, Renwick M, Wright C (2000) Congenital diaphragmatic herniation: antenatal detection and outcome. Br J Radiol 73:360–365 [DOI] [PubMed] [Google Scholar]
- 69.Kristeshavilli JI, Gorgadze IS, Khharabadze KM (1984) A case of partial trisomy for a long arm of chromosome 5 as a result of a balanced translocation t(3;5)(q27;q31) in a father. In: The register of chromosomal disorders in man. Moscow, pp 100–101 [Google Scholar]
- 70.Kocks A, Endele S, Heller R, Schroder B, Schafer HJ, Stadtler C, Makrigeorgi-Butera M, Winterpacht A (2002) Partial deletion of 4p and 4q in a fetus with ring chromosome 4: phenotype and molecular mapping of the breakpoints. J Med Genet 39:E23 10.1136/jmg.39.5.e23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kobori J, Seto-Donlon S, Gregory T, Bangs DD, Hsieh C-L (1993) A case of monosomy 4p and trisomy 4q derived from a meiotic recombination. Am J Hum Genet Suppl 55:1578 [Google Scholar]
- 72.Frints SG, Schrander-Stumpel CT, Engelen JJ, Da Costa AJ, Fryns JP (1996) Partial trisomy and partial monosomy of the distal long arm of chromosome 4: patient report and literature review. Genet Couns 7:135–142 [PubMed] [Google Scholar]
- 73.Celle L, Lee L, Rintoul N, Savani RC, Long W, Mennuti MT, Krantz ID (2000) Duplication of chromosome region 4q28.3-qter in monozygotic twins with discordant phenotypes. Am J Med Genet 94:125–140 [DOI] [PubMed] [Google Scholar]
- 74.Yunis E, Giraldo A, Zuniga R, Egel H, Ramirez E (1977) Partial trisomy 4q. Ann Genet 20:243–248 [PubMed] [Google Scholar]
- 75.Van Dooren MF, Brooks AS, Hoogeboom AJ, Van den Hoonaard TL, De Klein JE, Wouters CH, Tibboel D (2004) Early diagnosis of Wolf-Hirschorn syndrome triggered by a life-threatening event: congenital diaphragmatic hernia. Am J Med Genet A 127:194–196 10.1002/ajmg.a.20613 [DOI] [PubMed] [Google Scholar]
- 76.Tachdjian G, Fondacci C, Tapia S, Huten Y, Blot P, Nessmann C (1992) The Wolf-Hirschorn syndrome in fetuses. Clin Genet 42:281–287 [DOI] [PubMed] [Google Scholar]
- 77.Casaccia G, Mobili L, Braguglia A, Santoro F, Bagolan P (2006) Distal 4p microdeletion in a case of Wolf-Hirschorn syndrome with diaphragmatic hernia. Birth Defects Res A Clin Mol Teratol 76:210–213 10.1002/bdra.20235 [DOI] [PubMed] [Google Scholar]
- 78.Laziuk GI, Cherstvoi ED (1986) The main etiologic groups of congenital developmental defects and the problems of diagnosis and thanatogenesis. Arkh Patol 48:20–25 [PubMed] [Google Scholar]
- 79.Sergi C, Schulze BR, Hager HD, Beedgen B, Zilow E, Linderkamp O, Otto HF, Tariverdian G (1998) Wolf-Hirschorn syndrome: case-report and review of the chromosomal aberrations associated with diaphragmatic defects. Pathologica 90:285–293 [PubMed] [Google Scholar]
- 80.Van Buggenhout G, Melotte C, Dutta B, Froyen G, Van Hummelen P, Marynen P, Matthijs G, de Ravel T, Devriendt K, Fryns JP, et al (2004) Mild Wolf-Hirschhorn syndrome: micro-array CGH analysis of atypical 4p16.3 deletions enables refinement of the genotype-phenotype map. J Med Genet 41:691–698 10.1136/jmg.2003.016865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tapper JK, Zhang S, Harirah HM, Panova NI, Merryman LS, Hawkins JC, Lockhart LH, Gei AB, Velagaleti GV (2002) Prenatal diagnosis of a fetus with unbalanced translocation (4;13)(p16;q32) with overlapping features of Patau and Wolf-Hirschorn syndromes. Fetal Diagn Ther 17:347–351 10.1159/000065383 [DOI] [PubMed] [Google Scholar]
- 82.Del Campo M, De Frutos C, Delicado A, Garcia P, Cabanas F, Quero J (1997) The 4p-syndrome associated with congenital diaphragmatic hernia and dysgenesis of the corpus callosum. Proc Greenwood Genet Center 16:217–218 [Google Scholar]
- 83.Park Y, Gong G, Choe G, Yu E, Kim KS, Lee I (1993) Jarcho-Levin syndrome—a report of an autopsy case with cytogenetic analysis. J Korean Med Sci 8:471–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wakui K, Yamagishi A, Ito T, Imaizumi S (1996) Characterization of an interstitial deletion of chromosome 4 [del(4)(q31.1q31.3 or q31.3q32.3)] in a mother and son by chromosome painting. Jpn J Hum Genet 41:69 [Google Scholar]
- 85.Young RS, Palmer CG, Bender HA, Weaver DD, Hodes ME (1982) Brief cytogenetic case report: a 4.5-year-old girl with deletion 4q syndrome—de novo, 46,XX,del(4)(pter-q31). Am J Med Genet 12:103–107 10.1002/ajmg.1320120114 [DOI] [PubMed] [Google Scholar]
- 86.Reiss RE, Wilkins-Haug L, Quackenbush E, Korf B, Sandstrom M, Weremowicz S, Genest D (1999) Fryns syndrome: association with unbalanced translocations trisomic for 20q13.1-qter in two unrelated families. Am J Hum Genet Suppl 65:A340 [Google Scholar]
- 87.Bollmann R, Kalache K, Mau H, Chaoui R, Tennstedt C (1995) Associated malformations and chromosomal defects in congenital diaphragmatic hernia. Fetal Diagn Ther 10:52–59 [DOI] [PubMed] [Google Scholar]
- 88.Korner H, Tinschert S, Siebke K, Bollmann R, Chaoui R, Wilke T (1991) Pranatale diagnose einer partiellen Trisomie 5q mit Zwerchfelldefekt. Z Klin Med 46:427–429 [Google Scholar]
- 89.Aviram-Goldring A, Daniely M, Frydman M, Shneyour Y, Cohen H, Barkai G (2000) Congenital diaphragmatic hernia in a family segregating a reciprocal translocation t(5;15)(p15.3;q24). Am J Med Genet 90:120–122 [DOI] [PubMed] [Google Scholar]
- 90.Liberfarb RM, Atkins L, Holmes LB (1980) A clinical syndrome associated with 5p duplication and 9p deletion. Ann Genet 23:26–30 [PubMed] [Google Scholar]
- 91.Masuno M, Cholsong Y, Kuwahara T, Shimizu N, Yamaguchi S, Kawabata I, Tamaya T, Morishita Y, Yoshimi N, Orii T (1991) Second meiotic nondisjunction of the rearranged chromosome in a familial reciprocal 5/13 translocation. Am J Med Genet 41:32–34 10.1002/ajmg.1320410110 [DOI] [PubMed] [Google Scholar]
- 92.Scarbrough PR, Carroll AJ, Finley SC, Hamerick K (1986) Partial trisomy 6p and partial trisomy 22 resulting from 3:1 meiotic disjunction of maternal (6p;22q) translocation. J Med Genet 23:185–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kristofferson U, Heim S, Mandahl N, Sundkvist L, Szelest J, Hagerstrand I (1987) Monosomy and trisomy of 15q24→qter in a family with a translocation t(6;15)(p25;q24). Clin Genet 32:169–171 [DOI] [PubMed] [Google Scholar]
- 94.Batanian JR, Grange DK, Fleming R, Gadre B, Wetzel J (2001) Two unbalanced translocations involving a common 6p25 region in two XY female patients. Clin Genet 59:52–57 10.1034/j.1399-0004.2001.590109.x [DOI] [PubMed] [Google Scholar]
- 95.Baruch AC, Erickson RP (2001) Axenfeld-Rieger anomaly, hypertelorism, clinodactyly and cardiac anomalies in sibs with an unbalanced translocation der(6)t(6;8). Am J Med Genet 100:187–190 10.1002/ajmg.1256 [DOI] [PubMed] [Google Scholar]
- 96.Yu CW, Bock HGO (1997) Interstitial deletion of the long arm of chromosome 6: report of a case and review of the literature. Proc Greenwood Genet Center 16:283 [Google Scholar]
- 97.Shen-Schwarz S, Hill LM, Surti U, Marchese S (1989) Deletion of terminal portion of 6q: report of a case with unusual malformations. Am J Med Genet 32:81–86 10.1002/ajmg.1320320117 [DOI] [PubMed] [Google Scholar]
- 98.Herrmann M, Wittwer B, Exeler J, Fabritz L, Horst J (1999) De novo duplication 7(p15p22) in a child with a diaphragmatic hernia. Med Genet 11:166 [Google Scholar]
- 99.Habedank M, Trost-Binkhues G (1983) Monosomy 18p and pure trisomy 18p in a family with translocation (7;18). J Med Genet 20:377–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fauza DO, Wilson JM (1994) Congenital diaphragmatic hernia and associated anomalies: their incidence, identification, and impact on prognosis. J Pediatr Surg 29:1113–1117 10.1016/0022-3468(94)90290-9 [DOI] [PubMed] [Google Scholar]
- 101.Klep-de Pater JM, Bijlsma JB, Bleecker-Wagemakers EM, de France HF, de Vries-Ekkers CM (1979) Two cases with different deletions of the long arm of chromsome 7. J Med Genet 16:151–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dott MM, Wong LY, Rasmussen SA (2003) Population-based study of congenital diaphragmatic hernia: risk factors and survival in Metropolitan Atlanta, 1968-1999. Birth Defects Res A Clin Mol Teratol 67:261–267 10.1002/bdra.10039 [DOI] [PubMed] [Google Scholar]
- 103.Kjaer I, Keeling JW, Graem N (1991) The midline craniofacial skeleton in holoprosencephalic fetuses. J Med Genet 28:846–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bonneau D, Huret JL, Godeau G, Couet D, Putterman M, Tanzer J, Babin P, Larregue M (1991) Recurrent ctb(7)(q31.3) and possible laminin involvement in a neonatal cutis laxa with a Marfan phenotype. Hum Genet 87:317–319 10.1007/BF00200911 [DOI] [PubMed] [Google Scholar]
- 105.Temple IK, Barber JC, James RS, Burge D (1994) Diaphragmatic herniae and translocations involving 8q22 in two patients. J Med Genet 31:735–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ringer K, Rogers J, Pasztor LM (1995) Inversion duplication of chromosome 8 with diaphragmatic hernia. Am J Hum Genet Suppl 57:A124 [Google Scholar]
- 107.Moreno Fuenmayor HM, Meilinger KL, Rucknagel DL, Mohrenweiser HL, Chu EH (1980) Duplication 8p syndrome: studies in a family with a reciprocal translocation between chromosome 8 and 12. Am J Med Genet 7:361–368 10.1002/ajmg.1320070318 [DOI] [PubMed] [Google Scholar]
- 108.Chen CP, Lee CC, Pan CW, Kir TY, Chen BF (1998) Partial trisomy 8q and partial monosomy 15q associated with congenital hydrocephalus, diaphragmatic hernia, urinary tract anomalies, congenital heart defect and kyphoscoliosis. Prenat Diagn 18:1289–1293 [DOI] [PubMed] [Google Scholar]
- 109.Hilfiker ML, Karamanoukian HL, Hudak M, Fisher J, Glick PL (1998) Congenital diaphragmatic hernia and chromosomal abnormalities: report of a lethal association. Pediatr Surg Int 13:550–552 10.1007/s003830050400 [DOI] [PubMed] [Google Scholar]
- 110.Thorpe-Beeston JG, Gosden CM, Nicolaides KH (1989) Prenatal diagnosis of congenital diaphragmatic hernia: associated malformations and chromosomal defects. Fetal Ther 4:21–28 [DOI] [PubMed] [Google Scholar]
- 111.Faivre L, Morichon-Delvallez N, Viot G, Narcy F, Loison S, Mandelbrot L, Aubry MC, Raclin V, Edery P, Munnich A, et al (1998) Prenatal diagnosis of an 8p23.1 deletion in a fetus with a diaphragmatic hernia and review of the literature. Prenat Diagn 18:1055–1060 [DOI] [PubMed] [Google Scholar]
- 112.Shimokawa O, Miyake N, Yoshimura T, Sosonkina N, Harada N, Mizuguchi T, Kondoh S, Kishino T, Ohta T, Remco V, et al (2005) Molecular characterization of del(8)(p23.1p23.1) in a case of congenital diaphragmatic hernia. Am J Med Genet A 136:49–51 [DOI] [PubMed] [Google Scholar]
- 113.Borys D, Taxy JB (2004) Congenital diaphragmatic hernia and chromosomal anomalies: autopsy study. Pediatr Dev Pathol 7:35–38 10.1007/s10024-003-2133-7 [DOI] [PubMed] [Google Scholar]
- 114.Lopez I, Bafalliu JA, Bernabe MC, Garcia F, Costa M, Guillen-Navarro E (2006) Prenatal diagnosis of de novo deletions of 8p23.1 or 15q26.1 in two fetuses with diaphragmatic hernia and congenital heart defects. Prenat Diagn 26:577–580 10.1002/pd.1468 [DOI] [PubMed] [Google Scholar]
- 115.Pecile V, Petroni MG, Fertz MC, Filippi G (1990) Deficiency of distal 8p—report of two cases and review of the literature. Clin Genet 37:271–278 [DOI] [PubMed] [Google Scholar]
- 116.Fraer L, Marchese S, Juda S, Surti U, Huff D, Sherman F, Martin J, Hill LM (1992) Prenatal diagnosis of a de novo 8p23.1 distal deletion. Am J Hum Genet Suppl 51:A408 [Google Scholar]
- 117.Maerzke S, Neumann LM, Hofstaetter C, Plieth M, Reis A (1993) A novel partial monosomy 8q acertained by sonographic abnormalities. Med Genet 5:121 [Google Scholar]
- 118.Harnsberger J, Carey JC, Morgan M (1982) Interstitial deletion of the long arm of the number 8 chromosome and the Langer-Giedion syndrome. Paper presented at the Birth Defects Conference, Birmingham, June 13–16 [Google Scholar]
- 119.Capellini A, Sala E, Colombo D, Villa N, Mariani S (1996) Monosomy 8q and features of Fryns’ syndrome. Eur J Hum Genet Suppl 4:29 [Google Scholar]
- 120.Betremieux P, Lionnais S, Beuchee A, Pladys P, Le Bouar G, Pasquier L, Loeuillet-Olivo L, Azzis O, Milon J, Wodey E, et al (2002) Perinatal management and outcome of prenatally diagnosed congenital diaphragmatic hernia: a 1995-2000 series in Rennes University Hospital. Prenat Diagn 22:988–994 10.1002/pd.454 [DOI] [PubMed] [Google Scholar]
- 121.Chen CP, Chern SR, Cheng SJ, Chang TY, Yeh LF, Lee CC, Pan CW, Wang W, Tzen CY (2004) Second-trimester diagnosis of complete trisomy 9 associated with abnormal maternal serum screen results, open sacral spina bifida and congenital diaphragmatic hernia, and review of the literature. Prenat Diagn 24:455–462 10.1002/pd.900 [DOI] [PubMed] [Google Scholar]
- 122.Suzumori N, Sato T, Okada J, Nakanishi T, Shirai K, Tanemura M, Suzuki Y, Suzumori K (2003) Prenatal findings for complete trisomy 9. Prenat Diagn 23:866–868 10.1002/pd.704 [DOI] [PubMed] [Google Scholar]
- 123.Sepulveda W, Wimalasundera RC, Taylor MJ, Blunt S, Be C, De La Fuente S (2003) Prenatal ultrasound findings in complete trisomy 9. Ultrasound Obstet Gynecol 22:479–483 10.1002/uog.233 [DOI] [PubMed] [Google Scholar]
- 124.Frohlich GS (1982) Delineation of trisomy 9. J Med Genet 19:316–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Robert E, Kallen B, Harris J (1997) The epidemiology of diaphragmatic hernia. Eur J Epidem 13:665–673 10.1023/A:1007395727819 [DOI] [PubMed] [Google Scholar]
- 126.Henriques-Coelho T, Oliva-Teles N, Fonseca-Silva ML, Tibboel D, Guimaraes H, Correia-Pinto J (2005) Congenital diaphragmatic hernia in a patient with tetrasomy 9p. Pediatr Surg 40:e29–e31 10.1016/j.jpedsurg.2005.06.032 [DOI] [PubMed] [Google Scholar]
- 127.Alfi OS, Donnell GN, Allderdice PW, Derencsenyi A (1976) The 9p-syndrome. Ann Genet 19:11–16 [PubMed] [Google Scholar]
- 128.Alfi O, Donnell GH, Crandall BF, Derencsenyi A, Menon R (1973) Deletion of the short arm of chromsome no. 9 (46,9p-): a new deletion syndrome. Ann Genet 16:17–22 [PubMed] [Google Scholar]
- 129.Donnenfeld AE, Campbell TJ, Byers J, Librizzi RJ, Weiner S (1993) Tissue-specific mosaicism among fetuses with prenatally diagnosed diaphragmatic hernia. Am J Obstet Gynecol 169:1017–1021 [DOI] [PubMed] [Google Scholar]
- 130.Ferrero GB, Belligni E, Sorasio L, Delmonaco AG, Oggero R, Faravelli F, Pierluigi M, Silengo M (2006) Phenotype resembling Donnai-Barrow syndrome in a patient with 9qter;16qter unbalanced translocation. Am J Med Genet A 140:892–894 [DOI] [PubMed] [Google Scholar]
- 131.Cunniff C, Jones KL, Jones MC (1990) Patterns of malformation in children with congenital diaphragmatic defects. J Pediatr 116:258–261 10.1016/S0022-3476(05)82884-7 [DOI] [PubMed] [Google Scholar]
- 132.Yunis E, Silva R, Giraldo A (1976) Trisomy 10p. Ann Genet 19:57–60 [PubMed] [Google Scholar]
- 133.Lurie IW, Lazjuk GI, Gurevich DB, Kravtzoa GI, Nedzved MK, Shved IA (1978) Partial trisomy 10p in two generations. Hum Genet 41:235–241 10.1007/BF00273107 [DOI] [PubMed] [Google Scholar]
- 134.Iselius L, Lindsten J, Aurias A, Fraccaro M, Bastard C, Bottelli AM, Bui TH, Caufin D, Dalpra L, Delendi N, et al (1983) The 11q;22q translocation: a collaborative study of 20 new cases and analysis of 110 families. Hum Genet 64:343–355 10.1007/BF00292366 [DOI] [PubMed] [Google Scholar]
- 135.Fraccaro M, Lindsten J, Ford CE, Iselius L (1980) The 11q;22q translocation: a European collaborative analysis of 43 cases. Hum Genet 56:21–51 [DOI] [PubMed] [Google Scholar]
- 136.Phelan MC, Rogers RC, Flannery DB, Albiez K, Byrd JR (1987) An 11q;22q translocation in two families. Proc Greenwood Genet Center 6:22–26 [Google Scholar]
- 137.Azancot A, Eydoux P, Vuillard E, Cusin V, Baumann C, Blot P (2000) Clinical spectrum of prenatal tetralogy of Fallot. Arch Mal Coeur Vaiss 93:587–593 [PubMed] [Google Scholar]
- 138.de Beaufort C, Schneider F, Chafai R, Colette JM, Delneste D, Peirquin G (2000) Diaphragmatic hernia and Fryns syndrome phenotype in partial trisomy 22. Genet Couns 11:181-182 [PubMed] [Google Scholar]
- 139.Aurias A, Turc C, Michiels Y, Sinet PM, Graveleau D, Lejeune J (1975) 2 cases of trisomy 11q(q231→qter) by translocation t(11;22)(q231;q111) in 2 different families. Ann Genet 18:185–188 [PubMed] [Google Scholar]
- 140.Noel B, Levy M, Rethore MO (1976) Partial trisomy of the long arm of the chromosome 11 by malsegregation of a maternal translocation t(11;22)(q23 1q1 11). Ann Genet 19:137 [PubMed] [Google Scholar]
- 141.Hickmann G, Mazauric M, Weik S, Bartsch O, Kozlowski P (2001) Prenatal characterization of 27 autosomal marker chromosomes and outcomes of pregnancies. Eur J Hum Genet Suppl 9:230 [Google Scholar]
- 142.Kadir RA, Hastings R, Economides DL (1997) Prenatal diagnosis of supernumerary chromosome derivative (22) due to maternal balanced translocation in association with diaphragmatic hernia: a case report. Prenat Diagn 17:761–764 [DOI] [PubMed] [Google Scholar]
- 143.Park JP, McDermet MK, Doody AM, Marin-Padilla JM, Moeschler JB, Wurster-Hill DH (1993) Familial t(11;13)(q21;q14) and the duplication 11q,13q phenotype. Am J Med Genet 45:46–48 10.1002/ajmg.1320450113 [DOI] [PubMed] [Google Scholar]
- 144.Klaassens M, Scott DA, Van Dooren MF, Hochstenbach R, Eussen HJ, Cai WW, Galjaard RJ, Wouters C, Poot M, Laudy J, et al (2006) Congenital diaphragmatic hernia and duplication of chromosome 11q23-qter. Am J Med Genet A 140:1580–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Scott DA, Cooper ML, Stankiewicz P, Patel A, Potocki L, Cheung SW (2005) Congenital diaphragmatic hernia in WAGR syndrome. Am J Med Genet A 134:430–433 [DOI] [PubMed] [Google Scholar]
- 146.Gustavson K-H, Anneren G, Wranne L (1984) Two cases of 11p13 interstitial deletion and unusual clinical features. Clin Genet 26:247–249 [Google Scholar]
- 147.Decker-Philips M, McConkie-Rosell A, Qumsiyeh MB, Iafolla AK (1995) Novel unbalanced translocation: 46,XX,der(11)t(11:12)(q24-p11.2). Am J Hum Genet Suppl 57:A309 [Google Scholar]
- 148.Bergoffen J, Punnett H, Campbell TJ, Ross AJ 3rd, Ruchelli E, Zackai EH (1993) Diaphragmatic hernia in tetrasomy 12p mosaicism. J Pediatr 122:603–606 10.1016/S0022-3476(05)83546-2 [DOI] [PubMed] [Google Scholar]
- 149.Corning K, Toburen A, Clarkson K (1999) Lethal Palister-Killian syndrome masquerading as Fryns syndrome. Proc Greenwood Genet Center 19:24–26 [Google Scholar]
- 150.Rodriguez JI, Garcia I, Alvarez J, Delicado A, Palacios J (1994) Lethal Pallister-Killian syndrome: phenotypic similarity with Fryns syndrome. Am J Med Genet 53:176–181 10.1002/ajmg.1320530211 [DOI] [PubMed] [Google Scholar]
- 151.Donnenfeld AE, Campbell TJ, Byers J, Librizzi RJ, Weiner S (1993) Tissue-specific mosaicism among fetuses with prenatally diagnosed diaphragmatic hernia. Am J Obstet Gynecol 169:1017–1021 [DOI] [PubMed] [Google Scholar]
- 152.Betremieux P, Gaillot T, de la Pintiere A, Beuchee A, Pasquier L, Habonimana E, Le Bouar G, Branger B, Milon J, Fremond B, et al (2004) Congenital diaphragmatic hernia: prenatal diagnosis permits immediate intensive care with high survival rate in isolated cases: a population-based study. Prenat Diagn 24:487–493 10.1002/pd.909 [DOI] [PubMed] [Google Scholar]
- 153.Veldman A, Schlosser R, Allendorf A, Fischer D, Heller K, Schaeff B, Fuchs S (2002) Bilateral congenital diaphragmatic hernia: differentiation between Pallister-Killian and Fryns syndromes. Am J Med Genet 111:86–87 10.1002/ajmg.10438 [DOI] [PubMed] [Google Scholar]
- 154.Witters I, Legius E, Moerman P, Deprest J, Van Schoubroeck D, Timmerman D, Van Assche FA, Fryns JP (2001) Associated malformations and chromsomal anomalies in 42 cases of prenatally diagnosed diaphragmatic hernia. Am J Med Genet 103:278–282 10.1002/ajmg.1564 [DOI] [PubMed] [Google Scholar]
- 155.Takakuwa K, Hataya I, Arakawa M, Tamura M, Sekizuka N, Tanaka K (1997) A case of mosaic tetrasomy 12p (Pallister-Killian syndrome) diagnosed prenatally: comparison of chromosome analyses of various cells obtained from the patient. Am J Perinatol 14:641–643 [DOI] [PubMed] [Google Scholar]
- 156.Warburton PE, Dolled M, Mahmood R, Alonso A, Li S, Naritomi K, Tohma T, Nagai T, Hasegawa T, Ohashi H, et al (2000) Molecular cytogenetic analysis of eight inversion duplications of human chromosome 13q that each contain a neocentromere. Am J Hum Genet 66:1796–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tohma T, Ohashi H, Hasegawa T, Nagai T, Fukushima Y, Naritomi K (1998) Two cases of mosaic partial tetrasomy 13q associated with an acentric marker chromosome. Am J Hum Genet Suppl 64:A862 [Google Scholar]
- 158.Masada CT, Olney AH, Fordyce R, Sanger WG (1989) Partial deletion of 14q and partial duplication of 14q in sibs: testicular mosaicism for t(14q;14q) as a common mechanism. Am J Med Genet 34:528–534 10.1002/ajmg.1320340415 [DOI] [PubMed] [Google Scholar]
- 159.Boyar FZ, Whitney MM, Lossie AC, Gray BA, Keller KL, Stalker HJ, Zori RT, Geffken G, Mutch J, Edge PJ, et al (2001) A family with a grand-maternally derived interstitial duplication of proximal 15q. Clin Genet 60:421–430 10.1034/j.1399-0004.2001.600604.x [DOI] [PubMed] [Google Scholar]
- 160.Zabel B, Baumann W (1977) Partial trisomy for the distal part of the long arm of chromosome 15 due to a balanced maternal X/15 translocation. Ann Genet 20:285–289 [PubMed] [Google Scholar]
- 161.Elghezal H, Ben Reguaya M, Denguezli W, Mougou S, Saad A (2006) Prenatal diagnosis of 15q26.1→qter deletion due to a ring chromosome 15. Poster presented at the European Society of Human Genetics Conference, Amsterdam, May 8 [poster 477] [Google Scholar]
- 162.Bettelheim D, Hengstschlager M, Drahonsky R, Eppel W, Bernaschek G (1998) Two cases of prenatally diagnosed diaphragmatic hernia accompanied by the same undescribed chromosomal deletion (15q24 de novo). Clin Genet 53:319–320 [DOI] [PubMed] [Google Scholar]
- 163.Schlembach D, Zenker M, Trautmann U, Ulmer R, Beinder E (2001) Deletion of 15q24-26 in prenatally detected diaphragmatic hernia: increasing evidence of a candidate region for diaphragmatic development. Prenat Diagn 21:289–292 10.1002/pd.50 [DOI] [PubMed] [Google Scholar]
- 164.Biggio JR Jr, Descartes MD, Carroll AJ, Holt RL (2004) Congential diaphragmatic hernia: is 15q26.1-26.2 a candidate locus: Am J Med Genet A 126:183–185 [DOI] [PubMed] [Google Scholar]
- 165.Hengstschlager M, Mittermayer C, Repa C, Drahonsky R, Deutinger J, Bernaschek G (2004) Association of deletions of the chromosomal region 15q24-ter and diaphragmatic hernia: a new case and discussion of the literature. Fetal Diagn Ther 19:510–512 10.1159/000080164 [DOI] [PubMed] [Google Scholar]
- 166.Tumer Z, Harboe TL, Blennow E, Kalscheuer VM, Tommerup N, Brondum-Nielsen K (2004) Molecular cytogenetic characterization of ring chormosome 15 in three unrelated patients. Am J Med Genet A 130:340–344 10.1002/ajmg.a.30035 [DOI] [PubMed] [Google Scholar]
- 167.Chen CP, Shih JC, Chern SR, Lee CC, Wang W (2004) Prenatal diagnosis of mosaic trisomy 16 associated with congenital diaphragmatic hernia and elevated maternal serum alpha-fetoprotein and human chorioni gonadotrophin. Prenat Diagn 24:63–66 10.1002/pd.796 [DOI] [PubMed] [Google Scholar]
- 168.Johnson P, Duncan K, Blunt S, Bell G, Ali Z, Cox P, Moore GE (2000) Apparent confined placental mosaicism of trisomy 16 and multiple fetal anomalies: case report. Prenat Diagn 20:417–421 [DOI] [PubMed] [Google Scholar]
- 169.Baldermann C, Taege C, Musil A, Rath F, Hansmann I (2000) Ring chromosome 17 with monosomy 17 associated with unusual severe malformations. Am J Hum Genet Suppl 2 67:160 [Google Scholar]
- 170.Hayashi S, Hyodo M, Kinutani M, Fujiwara H, Date K, Mizuone T, Kisaka Y, Urabe T, Miharu N, Ohama K (1997) Characterization of isochromosome 18q in prenatal diagnosis by G-banding method and fluorescence in situ hybridization. Jpn J Hum Genet 42:127 [Google Scholar]
- 171.Geneix A, Goburdhun J, Fallet C, Lacroute G, Satge D (2001) A fetus with pseudo Fryns syndrome and t(18;22) translocation. Genet Couns 12:169–171 [PubMed] [Google Scholar]
- 172.Smith NM, Fernandez H, Chambers HM, Callen DF (1992) Necropsy findings in a fetus with a 46,XY,dic t(X;21)(p11.1;p11.1). J Med Genet 29:503–506 [PMC free article] [PubMed] [Google Scholar]
- 173.Kim EH, Cohen RS, Ramachandran P, Mineta AK, Babu VR (1992) Trisomy 22 with congenital diaphragmatic hernia and absense of corpus callosum in a liveborn premature infant. Am J Med Genet 44:437–438 10.1002/ajmg.1320440410 [DOI] [PubMed] [Google Scholar]
- 174.Ladonne JM, Gaillard D, Carre-Pigeon F, Gabriel R (1996) Fryns syndrome phenotype and trisomy 22. Am J Med Genet 61:68–70 [DOI] [PubMed] [Google Scholar]
- 175.Phillipson J, Benirschke K, Bogart M (1990) Two live-born infants with trisomy 22. Pediatr Pathol 10:1001–1005 [DOI] [PubMed] [Google Scholar]
- 176.Golombek S, Shaw R (1994) Trisomy 22 in an Iowa newborn. Iowa Med 84:31–33 [PubMed] [Google Scholar]
- 177.Ramsing M, Gillessen-Kaesbach G, Holzgreve W, Fritz B, Rehder H (2000) Variability in the phenotypic expression of Fryns syndrome: a report of two sibships. Am J Med Genet 95:415–424 [DOI] [PubMed] [Google Scholar]
- 178.Van Voss VH, Foerster W, Arnold W, Knoll G, Somville T, Muntefering H, Kemperdick H (1982) Multiple Miβbildungen bei einem frühgeborenen mit kompletter Trisomie 22. Der Kinderarzt 13:693–695 [Google Scholar]
- 179.Plaja A, Vendrell T, Sarret E, Toran N, Mediano C (1994) Terminal deletion of Xp in a dysmorphic anencephalic fetus. Prenat Diagn 14:410–412 10.1002/pd.1970140512 [DOI] [PubMed] [Google Scholar]
- 180.Dawani NM, Al Madhoob AR, Ali FA, Shabib F (2004) Fryns syndrome: a case associated with karyotype XO. Ann Saudi Med 24:129–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ee X, Zhang L, Lu J, Tso P, Blaner WS, Levin MS, Li E (2002) Increased neotnatal mortality in mice lacking cellular retinal-binding protein II. J Biol Chem 277:36617–36623 10.1074/jbc.M205519200 [DOI] [PubMed] [Google Scholar]
- 182.Ghyselinck NB, Bavik C, Sapin V, Mark M, Bonnier D, Hindelang C, Dierich A, Nilsson CB, Hakansson H, Sauvant P, et al (1999) Cellular retinal-binding protein I is essential for vitamin A homeostasis. EMBO J 18:4903–4914 10.1093/emboj/18.18.4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Laziuk GI, Ostrovskaia TI, Lurie IV, Kirillova IA, Kravtsova GI (1979) Pathologic anatomy of the Wolf-Hirschorn syndrome (partial monosomy 4p). Arkh Patol 41:40–45 [PubMed] [Google Scholar]
- 184.Reddy KS (1999) A paternally inherited terminal deletion, del(8)(p23.1)pat, detected prenatally in an amniotic fluid sample: a review of deletion 8p23.1 cases. Prenat Diagn 19:868–872 [DOI] [PubMed] [Google Scholar]
- 185.Devriendt K, Van Schoubroeck D, Eyskens B, Gewillig M, Vandenberghe K, Fryns JP (1998) Prenatal diagnosis of a terminal short arm deletion of chromosome 8 in a fetus with an atrioventricular septal defect. Prenat Diagn 18:65–67 [DOI] [PubMed] [Google Scholar]
- 186.Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, et al (2003) GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424:443–447 10.1038/nature01827 [DOI] [PubMed] [Google Scholar]
- 187.Okubo A, Miyoshi O, Baba K, Takagi M, Tsukamoto K, Kinoshita A, Yoshiura K, Kishino T, Ohta T, Niikawa N, et al (2004) A novel GATA4 mutation completely segregated with atrial septal defect in a large Japanese family. J Med Genet 41:e97 10.1136/jmg.2004.018895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Moog U, Engelen JJM, Albrechts JCM, Baars LGM, De Die-Smulders CEM (2000) Familial dup(8)(p12p21.1): mild phenotypic effect and review of partial 8p duplications. Am J Med Genet 94:306–310 [DOI] [PubMed] [Google Scholar]
- 189.Wilson WG, Wyandt HE, Shah H (1983) Interstitial deletion of 8q: occurrence in a patient with multiple exostoses and unusual facies. Am J Dis Child 137:444–448 [PubMed] [Google Scholar]
- 190.Mowery-Rushton PA, Stadler MP, Kochmar SJ, McPherson E, Surti U, Hogge WA (1997) The use of interphase FISH for prenatal diagnosis of Pallister-Killian syndrome. Prenat Diagn 17:255–265 [DOI] [PubMed] [Google Scholar]
- 191.Peltomaki P, Knuutila S, Ritvanen A, Kaitila I, De La Chapelle A (1987) Pallister-Killian syndrome: cytogenetic and molecular studies. Clin Genet 31:399–405 [DOI] [PubMed] [Google Scholar]
- 192.Greer JJ, Babiuk RP, Thebaud B (2003) Etiology of congenital diaphragmatic hernia: the retinoid hypothesis. Pediatr Res 53:726–730 10.1203/01.PDR.0000062660.12769.E6 [DOI] [PubMed] [Google Scholar]
- 193.Wilson JG, Roth CB, Warkany J (1953) An analysis of the syndrome of malformations induced by maternal vitamin A deficiency: effects of restoration of vitamin A at various times during gestation. Am J Anat 92:189–217 10.1002/aja.1000920202 [DOI] [PubMed] [Google Scholar]
- 194.Anderson D (1941) Incidence of congenital diaphragmatic hernia in the young of rats bred on a diet deficient in vitamin A. Am J Dis Child 62:888–889 [Google Scholar]
- 195.Anderson D (1949) Effect of diet during pregnancy upon the incidence of congenital hereditary diaphragmatic hernia in the rat. Am J Pathol 25:163–185 [PMC free article] [PubMed] [Google Scholar]
- 196.Mey J, Babiuk RP, Clugston R, Zhang W, Greer JJ (2003) Retinal dehydrogenase-2 is inhibited by compounds that induce congenital diaphragmatic hernia in rodents. Am J Pathol 162:673–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Migliazza L, Otten C, Xia H, Rodriguez JI, Diez-Pardo JA, Tovar JA (1999) Cardiovascular malformation in congenital diaphragmatic hernia: human and experimental studies. J Pediatr Surg 34:1352–1358 10.1016/S0022-3468(99)90010-6 [DOI] [PubMed] [Google Scholar]
- 198.Mendelsohn C, Lohnes D, Decimo D, Lufkin T, LeMeur M, Chambon P, Mark M (1994) Function of the retinoic acid receptors (RARs) during development (II): multiple abnormalities at various stages of organogenesis in RAR double mutants. Development 120:2749–2771 [DOI] [PubMed] [Google Scholar]
- 199.You LR, Takamoto N, Yu CT, Tanaka T, Kodama T, Demajo FJ, Tsai SY, Tsai MJ (2005) Mouse lacking COUP-TFII as an animal model of Bochdalek-type congenital diaphragmatic hernia. Proc Natl Acad Sci USA 102:16351–16356 10.1073/pnas.0507832102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Major D, Cadenas M, Fournier L, Leclerc S, Lefebre M, Cloutier R (1998) Retinol status of newborn infants with congenital diaphragmatic hernia. Pediatr Surg Int 13:547–549 10.1007/s003830050399 [DOI] [PubMed] [Google Scholar]
- 201.Qui Y, Krishnan V, Pereira S, Tsai SY, Tsai MJ (1996) Chicken ovalbumin upstream promotor-transcription factors and their regulation. J Steroid Biochem Mol Biol 56:81–85 10.1016/0960-0760(95)00225-1 [DOI] [PubMed] [Google Scholar]
- 202.Tsai SY, Tsai MJ (1997) Chicken ovalbumin upstream promoter-transcription factors (COUP-TFs): coming of age. Endocr Rev 18:229–240 10.1210/er.18.2.229 [DOI] [PubMed] [Google Scholar]
- 203.Huggins GS, Bacani CJ, Boltax J, Aikawa R, Leiden JM (2001) Friend of GATA 2 physically interacts with chicken ovalbumin upstream promotor-TF2 (COUP-TFII) and COUP-TFIII and represses COUP-TFII-dependent activation of the atrial natriuretic factor promotor. J Biol Chem 276:28029–28036 10.1074/jbc.M103577200 [DOI] [PubMed] [Google Scholar]
- 204.Arceci RJ, King AA, Simon MC, Orkin SH, Wilson DB (1993) Mouse GATA-4: a retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol Cell Biol 13:2235–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Crispino JD, Lodish MB, Thurberg BL, Litovsky SH, Collins T, Molkentin JD, Orkin SH (2001) Proper coronary vascular development and heart morphogenesis depend on interaction of GATA-4 with FOG cofactors. Genes Dev 15:839–844 10.1101/gad.875201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Pritchard-Jones K, Fleming S, Davidson D, Bickmore W, Porteous D, Gosden C, Bard J, Buckler A, Pelletier J, Housman D, et al (1990) The candidate Wilm’s tumour gene is involved in genitourinary development. Nature 346:194–197 10.1038/346194a0 [DOI] [PubMed] [Google Scholar]
- 207.Devriendt K, Deloof E, Moerman P, Legius E, Vanhole C, De Zegher F, Proesmans W, Devlieger H (1995) Diaphragmatic hernia in Denys-Drash syndrome. Am J Med Genet 57:97–101 10.1002/ajmg.1320570120 [DOI] [PubMed] [Google Scholar]
- 208.Cho HY, Lee BS, Kang CH, Kim WH, Ha IS, Cheong HI, Choi Y (2006) Hydrothorax in a patient with Denys-Drash syndrome associated with a diaphragmatic defect. Pediatr Nephrol 21:1909–1912 10.1007/s00467-006-0273-5 [DOI] [PubMed] [Google Scholar]
- 209.Denamur E, Bacquet N, Baudouin V, Da Silva F, Veitia R, Peuchmaur M, Elion J, Gubler MC, Fellous M, Niaudet P, et al (2000) WT1 splice-site mutations are rarely associated with primary steroid-resistant focal and segemental glomerulosclerosis. Kidney Int 57:1868–1872 10.1046/j.1523-1755.2000.00036.x [DOI] [PubMed] [Google Scholar]
- 210.Reardon W, Smith S, Suri M, Grant J, O’Neill D, Kelehan P, Fitzpatrick D, Hastie N (2004) WT1 mutation is a cause of congenital diaphragmatic hernia associated with Meacham syndrome. Paper presented at the American Society of Human Genetics Annual Meeting, Los Angeles, October 26–30 [Google Scholar]
- 211.Kreidberg JA, Sariola H, Loring JM, Maeda M, Pelletier J, Housman D, Jaenisch R (1993) WT-1 is required for early kidney development. Cell 74:679–691 10.1016/0092-8674(93)90515-R [DOI] [PubMed] [Google Scholar]
- 212.Yuan W, Rao Y, Babiuk RP, Greer JJ, Wu JY, Ornitz DM (2003) A genetic model for a central (septum transversum) congenital diaphragmatic hernia in mice lacking Slit3. Proc Natl Acad Sci USA 100:5217–5222 10.1073/pnas.0730709100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Liu J, Zhang L, Wang D, Shen H, Jiang M, Mei P, Hayden PS, Sedor JR, Hu H (2003) Congenital diaphragmatic hernia, kidney agenesis and cardiac defects associated with Slit3-deficiency in mice. Mech Dev 120:1059–1070 10.1016/S0925-4773(03)00161-8 [DOI] [PubMed] [Google Scholar]
- 214.Pasutto F, Sticht H, Hammersen G, Gillessen-Kaesbach G, Fitzpatrick DR, Nurnberg G, Brasch F, Schirmer-Zimmermann H, Tolmie JL, Chitayat D, et al (2007) Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation. Am J Hum Genet 80:550–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kawaguchi R, Yu J, Honda J, Hu J, Whitelegge J, Ping P, Wiita P, Bok D, Sun H (2007) A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science 315:820–825 10.1126/science.1136244 [DOI] [PubMed] [Google Scholar]