Abstract
A proper understanding of population genetic stratification—differences in individual ancestry within a population—is crucial in attempts to find genes for complex traits through association mapping. We report on genomewide typing of ∼10,000 single-nucleotide polymorphisms in 297 individuals, to explore population structure in Europeans of known and unknown ancestry. The results reveal the presence of several significant axes of stratification, most prominently in a northern-southeastern trend, but also along an east-west axis. We also demonstrate the selection and application of EuroAIMs (European ancestry informative markers) for ancestry estimation and correction. The Coriell Caucasian and CEPH (Centre d'Étude du Polymorphisme Humain) Utah sample panels, often used as proxies for European populations, are found to reflect different subsets of the continent’s ancestry.
Genomewide association studies are becoming key tools in attempts to map genes underlying complex traits. However, the presence of population stratification or individual ancestry differences within samples may confound the promise of such approaches. In particular, discordant ancestry levels between cases and controls can lead to false-positive association with a trait and/or reduced power to detect such associations. The issue is most acute and widely recognized in individuals who differ in continental origin or who are admixed between such populations. Ancestry informative markers (AIMs), typically SNPs, that show large frequency differences among intercontinental groups can be used to detect and correct for such stratification. However, the study of intracontinental structure is less well explored or understood.
Among the continents, Europe is remarkable for its relatively small size, dearth of migration barriers, and abundance of historical population movements.1 These features, along with low levels of genetic differentiation, suggest a relatively homogenous continental population. Consequently, it has been argued that European population stratification does not represent a significant source of bias in epidemiological studies.2 However, recent autosomal SNP studies have highlighted significant patterns of structure within Europe along a north-south axis.3 The potentially confounding influence of this stratification on association-mapping studies in European-derived population samples was also recently demonstrated.4,5 Beyond these first insights, little is known about the geographic distribution and complexity of European genetic structure; the identification of additional significant patterns requires more-extensive population samples and a greater numbers of markers.
To address this issue further, we typed 297 individuals from 21 European and world populations for ∼10,000 autosomal SNPs, primarily using Affymetrix 10K mapping arrays. The European population samples represent a broad range of the geographic and linguistic diversity of the continent (full details can be found in table 1). In brief, they consisted of western Irish (n=6), eastern English (n=8), French (n=1), German (n=8), Valencian Spanish (n=20), Basque Spanish (n=8), Italian (n=9), Polish (n=8), Greek (n=8), Finnish (n=7), Armenian (n=8), and Ashkenazi Jewish (n=5) subjects. The Italian, Ashkenazi Jewish, and Greek samples include 2, 1, and 1 individuals, respectively, from the Coriell Cell Repository. For broader context, the European populations were examined together with two African population samples (Mende from Sierra Leone [n=22] and Burunge from Tanzania [n=20]) as well as several Asian populations (Brahmin [n=11] and Mala [n=11]) from India and Central Asian Altaian (n=20). One Middle Eastern and two North African individuals from the Coriell panel were also included. Some individuals were typed for 11,071 autosomal SNPs with use of the Affymetrix 10K Xba 131 array and others on the newer 10K 2.0 Xba array (table 1). A total of 9,724 SNPs overlapped between the two platforms, and these formed the core data for analysis. Two European-derived population samples (Coriell Caucasians and European Americans) are described further below. In an effort to minimize the effect of missing data, each analysis includes only SNPs that had genotypes for at least one individual in each population sample. This resulted in slightly different sets for each comparison, but the average missing-data rate per individual never exceeded 3.5%.
Table 1. .
Individual Samples Description[Note]
| Nation of Origin/Ethnicity | Language Family/Subfamily |
No. of Subjects |
Origin/Collected by | Affymetrix Mapping Array |
| “Caucasian” | Unknown | 42 | Coriell14 | 10K Xba 131 |
| Central France | Romance/Italic | 1 | M.B. | 10K Xba 131 |
| Valencia, Spain | Romance/Italic | 20 | E. Parra (University of Toronto)14 | 10K Xba 131 |
| Italy | Romance/Italic | 2 | Coriell | 10K Xba 131 |
| Southern Italy and Sicily | Romance/Italic | 5 | M.B., B.M., M.D.S. | 10K 2.0 Xba |
| Utah | Germanic | 74 | CEPH | 100K |
| Hanover, Germany | Germanic | 8 | R.D. | 10K 2.0 Xba |
| Eastern England | Germanic | 8 | B.M. | 10K 2.0 Xba |
| Ashkenazi Jewish (USA) | Germanic | 4 | M.B., E.E.Q., L.N.P., M.D.S. | 10K 2.0 Xba |
| Poland | Slavic | 8 | M.B., B.M., M.D.S. | 10K 2.0 Xba |
| Greece (set 1) | Hellenic | 7 | B.M. | 10K 2.0 Xba |
| Greece (set 2) | Hellenic | 1 | Coriell | 10K Xba 131 |
| Basque region (France and Spain) | Basque | 8 | S. Alonso (University of the Basque Country, Bilbao, Spain) | 10K 2.0 Xba |
| Connaught, Ireland | Celtic | 6 | M.D.S., B.M. | 10K 2.0 Xba |
| Armenia (one person per province) | Armenian | 8 | T.S. | 10K 2.0 Xba |
| Finland | Finno-Ugric | 7 | A. de la Chapelle (Ohio State University) | 10K 2.0 Xba |
| Ashkenazi Jewish | Semitic (probably) | 1 | Coriell | 10K Xba 131 |
| Middle East | Semitic (probably) | 2 | Coriell | 10K Xba 131 |
| North Africa | Semitic (probably) | 1 | Coriell | 10K Xba 131 |
| Mende (West Africa) | Niger-Congo | 22 | G. Argyropoulos (Pennington Biomedical Research Center)14 | 10K Xba 131 |
| Burunge (East Africa) | Cushitic | 20 | S. Tishkoff (University of Maryland)14 | 10K Xba 131 |
| Altai Republic (Central Asia) | Turkic/Altaic | 20 | T. Schurr (University of Pennsylvania, Philadelphia)14 | 10K Xba 131 |
| Andhra Pradesh (India): | ||||
| Brahmin (upper caste) | Indic | 11 | L. Jorde, M. Bamshad (University of Utah)14 | 10K Xba 131 |
| Mala (lower caste) | Indic | 11 | L. Jorde, M. Bamshad (University of Utah)14 | 10K Xba 131 |
Note.— All samples described here were collected with appropriate human subject approvals from the various institutions involved and under the principle of informed consent.
We first examined the European populations in the context of the other worldwide samples, using principal coordinate analysis6 (PCoA), which summarizes the variance in multivariate data sets into trends of maximum relevance known as principal components (PCs). PCoA was chosen over principal component analysis, since it was shown to have better power to identify clusters7 and is more robust to missing genotype data. We used R software’s ade4 package,8 to conduct PCoA on the matrix of allele-sharing distances (ASDs)9,10 between all pairs of individuals. The ASD between two individuals for a given SNP is 0 if they have identical genotypes, 0.5 if they share one allele, and 1 if they have no allele in common. Overall ASD between two individuals was calculated by averaging these distances over all nonmissing SNPs in common.
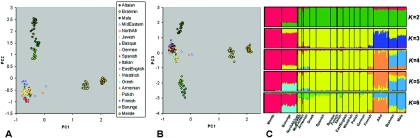
The PCoA clearly identifies four widely dispersed groupings corresponding to Europe, South Asia, Central Asia, and Africa (figs. 1A, 1B, and 2). In these figures, PC1 appears to separate the Africans from the other populations, whereas PC2 divides the Asians from the Europeans and Africans and PC3 splits the Central Asians apart from the South Asians. The wide gaps observed among the four clusters may reflect the absence of geographically intermediate groups in the analysis, an explanation hinted at by the intermediate position of the North African and Middle Eastern individuals included. However, it is unknown whether the inclusion of further geographic samples would produce a smooth continuum or series of clusters. In line with previous studies,11 there is low apparent diversity in Europe, with the entire continentwide sample only marginally more dispersed than single-population samples from elsewhere in the world. The Spanish and Basque groups are the farthest away from other continental groups, which is consistent with the suggestion that the Iberian Peninsula holds the most ancient European genetic ancestry.12,13 It is also clear that geography is not the sole marker of differentiation. The Mala and Brahmin are low and high caste groups, respectively, from the same region of India, yet they still show significant separation supporting some degree of socially maintained stratification (figs. 1 and 2). A complementary Bayesian approach that uses the program STRUCTURE14,15 supports the PCoA findings (fig. 1C). This method generates admixture components from individual genotype data without consideration of previous population labels, essentially with the use of departures from Hardy-Weinberg equilibrium. When the number of putative populations (K) is set at four, the groups largely correspond to the same four regional divides apparent from the PCoA.
Figure 1. .
Population structure in European, African, and Asian individuals. A and B, PCoA results based on average interindividual ASD, with use of 9,100 SNPs. PC1, 2, and 3 explain 11.6%, 3.4%, and 1.4%, respectively, of the variation. C, Bayesian clustering results with use of STRUCTURE14,15 and the same markers and individuals. Each individual is represented as a vertical line divided into, at most, K colored segments, where K is the prespecified number of populations into which the data are to be divided. STRUCTURE runs consisted of 80,000 iterations, with a previous burn-in of 40,000 steps, and were performed under the admixture model, which allows fractional assignment of the genome to different populations. Because of the historical and geographic proximity of European populations, the correlated-allele-frequencies model was also employed. The STRUCTURE plots shown above were generated using the companion program DiStruct.29
Figure 2. .
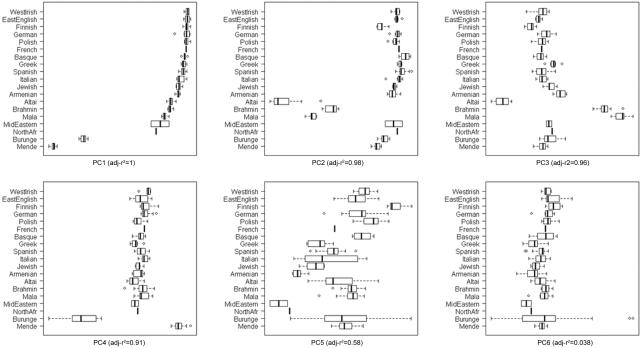
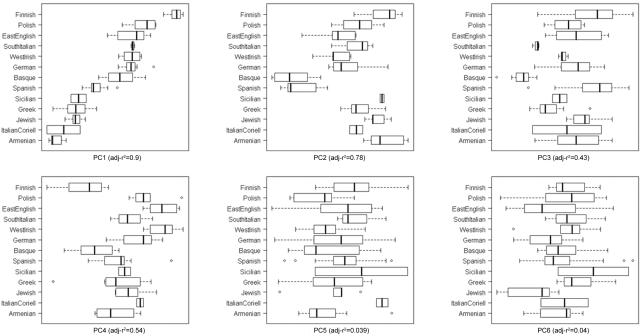
PCoA boxplots for the first six PCs in samples from Europe and neighboring continents. Bold vertical bars represent the median PC values of each group; the two hinges are the first and third quartile, and notches give an ∼95% CI for the difference in two medians. The overall correlation between group membership and PC value is reported by ANOVA’s adjusted r2 for each PC. The few subsequent PCs that are also significant pertain to Europe and are best observed in figures 4 and 5.
We next investigated European individuals in more detail, using a similar approach. An initial Mantel test16 between matrices of interindividual geographic and genetic distances was highly significant (P<.001), suggesting some degree of geographic substructure despite the relatively limited diversity. PCoA was then performed on the sample of European individuals alone, with the use of additional measures of significance to describe the more subtle patterns. Although the amount of variation explained by each PC is an indication of its importance, these proportions are not measures of statistical significance and are typically small for very large numbers of markers, as in the present data set. Therefore, we evaluated PC significance, using two independent methods. First, we tested linear correlation of PC axes with population or group membership through an analysis of variance (ANOVA) test, with each PC as dependent variable and population or group membership as predictor. The second method, which we refer to as the “split karyotype test” (SKT), does not rely on individual population assignment.17,18 The SKT is a form of split-half reliability test, where the SNP data are divided into two independent (nonsyntenic) marker sets (e.g., markers on odd vs. even chromosomes18), and PCoA is performed on each set separately. Under a null hypothesis of no population structure, there is not expected to be any correlation between the PCoA results obtained from each set of SNPs. We test this in the SKT, by calculating the Spearman correlation coefficient between individual PCs for each SNP set. If the structure or stratification represented by a PC is robust and significant, then the two independent marker sets should produce correlated individual PCs. However, the use of a single combination of two nonsyntenic SNP sets presents an increasing risk of type I or II errors when the number of markers and their overall informativeness diminish (data not shown). Such is the case in these analyses of European population samples, which are less differentiated than the worldwide samples to which the test was previously applied.18 Therefore, the test was extended to 100 permutations of nonsyntenic SNP sets. We used the program and formula from Dr. Zaykin’s Web site to calculate Fisher’s combined P value19 from the 100 Spearman P values. It is important to note that PC nonsignificance can reflect either absence of structure or inability of a particular marker set to detect structure. This extended SKT method was also evaluated using simulated groups of individuals whose four grandparents came from the same populations. Preliminary results indicated that, when the informativeness of markers is sufficiently high to see clear stratification, the number of significant PC axes is a good indication of the number of differentiable parental populations (M. Bauchet, unpublished data).
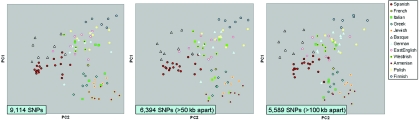
The SKT and ANOVA test were conducted on the PCoA results with use of the full SNP data set (9,111 SNP markers) and on two subsets of the total data including only SNPs at least 50 kb (6,349 SNPs) or 100 kb (5,555 SNPs) apart, to ensure that close marker spacing did not affect the results (table 2 and fig. 3). The first four PCs were found to be consistently significant across all tests and marker sets (table 2) and are likely to represent real structure. The stability of the findings across different marker-separation sets (50 kb and 100 kb) suggests that geographic structure is distributed throughout the data and that nearby markers in the 10K arrays are redundant in terms of ancestry informativeness.
Table 2. .
PCoA Significance Tests[Note]
| All SNPs(n=9,111) |
SNPs >50 kb Apart(n=6,349) |
SNPs >100 kb Apart(n=5,555) |
||||
| Test | Adjusteda r2 (P) | SKT Pb | Adjusteda r2 (P) | SKT Pb | Adjusteda r2 (P) | SKT Pb |
| PC1 | .90 (<.001) | <.0001 | .89 (<.001) | <.0001 | .90 (<.001) | <.0001 |
| PC2 | .78 (<.001) | <.0001 | .74 (<.001) | <.0001 | .72 (<.001) | <.0001 |
| PC3 | .43 (<.001) | <.0001 | .50 (<.001) | <.0001 | .35 (<.001) | <.0001 |
| PC4 | .54 (<.001) | <.0001 | .30 (<.001) | <.0001 | .19 (<.01) | <.01 |
| PC5 | <.1 (NS) | NS | <.1 (NS) | NS | .13 (<.05) | <.001 |
| PC6 | <.1 (NS) | NS | <.1 (NS) | NS | .18 (<.01) | <.001 |
| PC7 | <.1 (NS) | NS | .17 (<.01) | NS | .18 (<.01) | <.01 |
| PC8 | <.1 (NS) | NS | <.1 (NS) | NS | <.1 (NS) | NS |
| PC9 | <.1 (NS) | <.01 | <.1 (NS) | NS | <.1 (NS) | NS |
| PC10 | .13 (<.05) | NS | <.1 (NS) | NS | <.1 (NS) | NS |
Note.— Significance tests using the ANOVA and SKT. PCoA was conducted separately for each SNP set. The French singleton and the German outlier were excluded. Percentages of the variance explained by each PC are generally the same in all three cases (fig. 4A). NS = not significant at the .05 level.
ANOVA correlation coefficient (adjusted r2) in bold.
Combined P value calculated for SKT, as described in the text.
Figure 3. .
Stability of the PC1-PC2 distribution of European individuals across marker sets with different minimum intermarker separation (50 kb and 100 kb).
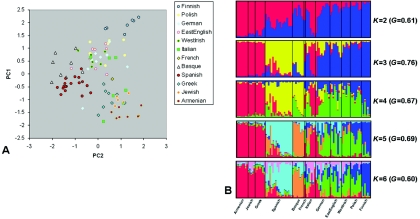
PC1 largely separates northern from southeastern individuals (fig. 4A) and is consistent with the clines observed in classic gene-frequency,13,20 Y-chromosome,21 mtDNA,22,23 and whole-genome3 studies of European diversity. PC2 reflects mainly east-west geographic separation and, particularly, identifies the two Iberian populations (Spanish and Basques) in our analysis as distinct (fig. 4A). Furthermore, PC3 and PC4 emphasize the separation of the Basques and Finns, respectively, from other Europeans (fig. 5). The Basques are known to have unusual allele frequencies for several marker systems24 and speak a unique non–Indo-European language. In line with their non–Indo-European Uralic language and previous study of their Y-chromosomes,25 the Finns show evidence of an increased affinity to the Central Asian populations when placed in an intercontinental context (fig. 1A and 1B). Overall, STRUCTURE analysis of the European populations is highly consistent with PCoA; for example, when the number of populations (K) is 3, the major divisions correspond to the northern, southeastern, and Iberian populations (fig. 4B). In cases of higher K values, first the Finns (K=4) and then the Basques (K=5) emerge as distinctive.
Figure 4. .
Population structure in European individuals. A, PCoA based on average interindividual ASD across 9,114 SNPs. PC1, 2, 3, and 4 (fig. 5) explain 2.05%, 1.7%, 1.6%, and 1.5%, respectively, of the variation and were highly significant by SKT and ANOVA testing (see table 2). B, Bayesian clustering analysis using STRUCTURE and the same markers and European individuals. Each individual is represented as a vertical line divided into K colored segments, where K is the prespecified number of populations into which the data are to be divided. The clusteredness (measured by G)—the extent to which individuals belong to a single cluster rather than a combination of clusters30—is also given alongside each K value. See the legend of figure 1 for further details of STRUCTURE runs and conditions.
Figure 5. .
PCoA boxplots for the first six PCs in European samples. Bold vertical bars represent the median PC values of each group; the two hinges are the first and third quartile, and notches give an ∼95% CI for the difference in two medians. The overall correlation between group membership and PC value is reported by ANOVA’s adjusted r2 for each PC. The subsequent PCs (up to 10) are not significant. The single French sample and the German outlier were excluded.
Within the two broad northern (Polish, Irish, English, Germans, and some Italians) and southeastern (Greeks, Armenians, Jews, and some Italians) clusters, further reliable structure is less obvious because individuals from different population samples are often interspersed with each other. Thus, in some cases, geographic distance or physical barriers are not well reflected. For instance, despite their insular origin, Irish and English individuals cluster with the continental Germans and Poles. Similarly, large geographical gaps, such as that between Greece and Armenia, are much less obvious at the genetic level. Conversely, Italy appears to be a zone of sharp differentiation over small distances. Some Italians cluster with the northern Europeans, whereas others fall into the southeastern grouping (fig. 4A). The SKT confirms significant stratification within those metaclusters, as suggested by the wide amount of PCoA space occupied by each (fig. 4A). Significant SKT stratification is also observed within the Spanish and the Italian samples. However, Mantel correlations between genetic and geographic distance were not significant within northern and southeastern metaclusters. It is likely that additional populations, additional individuals for some populations, and an increased number of markers will be required to investigate the nature and extent of these more subtle patterns.
Correction for stratification in association studies is dependent on the identification of and adjustment for relevant axes of ancestry that vary within the study population. Although a large number of arbitrary SNPs can be used for this purpose, a more efficient and informative approach is to identify subsets of ancestry-informative SNPs—for example, European AIMs, or “EuroAIMs.” One use of the AIMs that is not possible with arbitrary markers, such as sets from mapping arrays, is the separate investigation of particular axes of ancestry. We focus here on the main recognizable trend in our European data set—PC1, or the northern-southeastern ancestry axis—and measure it in two cohorts of European-derived population samples.
The first of these, the Coriell Caucasian panel (n=42), curated by the Coriell Cell Repositories, was typed using the Affymetrix 10K Xba 131 array. This panel has been used to portray European variation; for example, it was the core European representative sample in the SNP Consortium allele-frequency project. However, the genetically and socially ill-defined term “Caucasian” leaves doubt as to which population(s) this sample represents and how well it does so. The second European proxy sample we investigated is the CEPH panel, composed of European-American Utah residents, sampled in 1980, who declared ancestry from northern and western Europe; this panel forms one of the four populations used in the international HapMap project. Our CEPH Utah panel is made up of 74 unrelated individuals from family trios, 32 of which overlap with the HapMap European CEU individuals. Each individual was genotyped on the Affymetrix 100K Mapping array for ∼100,000 SNPs, but only the 6,207 markers overlapping with the 10K data set were considered here.
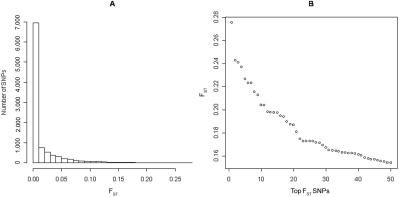
We identified EuroAIMs from the original panel of Europeans (fig. 4) by defining northern (n=36) and southeastern (n=31) cohorts of individuals on the basis of extreme polar values in PC1 (>0.5 or <−0.5). The northern cohort included all Finnish and Polish; most German, Irish, and English; as well as some Basque and Italian individuals. The southeastern cohort included all Armenians, Jews, Greeks, and the other Italians. Weir’s unbiased FST was then calculated for each SNP as a measure of genetic distance between the two groups (fig. 6).26 All SNPs were ranked by FST, with those showing the highest values likely to represent the best northern-southeastern EuroAIMs. The 20 SNPs presenting the highest FST levels are listed in table 3, along with allele-frequency differences between cohorts. Table 3 also shows that these top 20 EuroAIMs have levels of divergence comparable to a SNP (rs4988235) that was shown elsewhere to induce false-positive results due to stratification in European populations along the same geographical axis.4 The top 1,200 EuroAIMs can be downloaded from the Shriver Lab Web site.
Figure 6. .
Distribution of FST between northern (n=36) and southeastern (n=31) cohorts of individuals selected from PC1 values in figure 4 (>0.5 or <−0.5). A, Histogram with use of all 9,721 SNPs available. B, Plot of top 50 SNPs of highest FST (also see table 3).
Table 3. .
Top 20 Northern-Southeastern EuroAIMs[Note]
| SNP | Chromosome | Weir FST | Δa |
| rs988436 | 5 | .2755 | .295 |
| rs942793 | 10 | .2428 | .342 |
| rs1368136 | 8 | .2412 | .379 |
| rs2060983 | 8 | .2373 | .377 |
| rs4988235b | 2 | .2352 | .374 |
| rs1404402 | 1 | .2267 | .354 |
| rs1016120 | 2 | .2232 | .269 |
| rs1414411 | 1 | .2232 | .365 |
| rs2014303 | 4 | .2156 | .332 |
| rs1030626 | 8 | .2126 | .355 |
| rs1517661 | 12 | .2041 | .348 |
| rs764138 | 16 | .2039 | .349 |
| rs2218497 | 13 | .1981 | .345 |
| rs725379 | 2 | .1980 | .338 |
| rs1377724 | 15 | .1974 | .230 |
| rs1406121 | 2 | .1973 | .345 |
| rs869538 | 4 | .1945 | .309 |
| rs1905471 | 13 | .1940 | .236 |
| rs764681 | 16 | .1898 | .321 |
| rs1280100 | 4 | .1873 | .320 |
| rs723211 | 10 | .1867 | .333 |
Note.— The full set of 1,200 EuroAIMs between the northern and southeastern cohort is available at the Shriver Lab Web site.
Allele frequency difference between cohorts.
This SNP is not in our original set but was part of a study in which it showed significant stratification between northwestern and southeastern Europeans.4
To assess the potential impact of the observed European stratification in case-control association studies, we calculated the factor by which association statistics might be inflated (i.e., how much more likely false-positive results are to arise).27 A simple estimator for this inflation factor λ is the mean allele-frequency correlation between cases and controls (χ2) across null loci (i.e., loci thought to not influence the trait or condition).28 We examined the most extreme scenario supported by our data, where the case and control groups are composed of northern and southeastern individuals, respectively. We simulated 1,000 cases and 1,000 controls on the basis of these cohorts’ observed allele frequencies, and the mean χ2 across all loci at least 50 kb apart with an allele count of at least 5 (6,312 SNPs) was calculated. We multiplied the result by 1.03, which, in this case, is the maximum factor by which λ can exceed the mean χ2 at the 95% confidence level.28 By this method, we obtain a value λmax≈48, which substantially exceeds the null hypothesis (zero stratification) expectation of 1. In other words, a conservative P value for a candidate SNP can be obtained after dividing the χ2 value (or any association statistic) by 48.28 Similar calculations were made for the EuroAIMs panels yielding λmax values gradually increasing from 163 (1,200 EuroAIMS) to 407 (50 EuroAIMS). These observations confirm the practical importance of PC1 stratification in European populations and the utility of the selected EuroAIMs panels to control for it. Therefore, these marker sets were further tested in PCoA and STRUCTURE analysis of the Coriell and CEPH individuals.
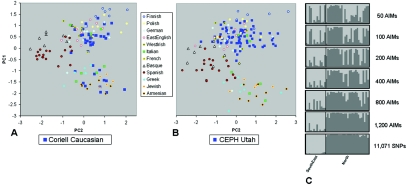
PCoA of the European samples together with the Coriell Caucasian panel (fig. 7A) reveals the latter to be divided by the same northern-southeastern structure evident in the general sample. It also contains some of the substructure observed within the southeastern cluster, as we verified with SKT significance testing and PCoA (not shown). Although it shows clear evidence of having derived from multiple European populations, the Coriell Caucasian sample lacks the full range of variation observed in Europeans, specifically that observed in the Spanish, Basque, and Finnish individuals. Northern-southeastern stratification is also evident in STRUCTURE analysis based on the full set of SNPs (fig. 7C, bottom) and was used to classify the Coriell Caucasian panel into a northern group and a southeast group, which correspond to the PCoA clusters (fig. 7A). This structure may largely be captured using ∼10-fold fewer SNPs, provided these markers are EuroAIMs selected as most informative on the northern-southeastern axis from the full 10K set. Using <1,200 EuroAIMs of the type available in this panel gradually leads to loss of consistent structure and a corresponding increase in misclassification of individual origins (fig. 7C). We could also have selected EuroAIMs specific for other PCs but elected not to do so because we lacked additional human samples that could be used to verify the utility of such AIM panels. Since European stratification is relatively modest, a larger number of SNPs will have to be screened to produce smaller but still efficient EuroAIM sets for PC1 and to generate useful sets for the investigation of PC2, PC3, and PC4.
Figure 7. .
Population structure in panels of European-derived ancestry within the context of European individuals (from fig. 4A). A, PCoA of the Coriell Caucasian panel (n=42), together with Europeans of known ancestry, based on all 9,114 SNPs in common. B, PCoA of the CEPH Utah individuals (n=74) and Europeans with use of all 6,207 SNPs in common. C, STRUCTURE runs using the Coriell Caucasian sample based on the full SNP data set (bottom) as well as sets of different numbers of north-southeast EuroAIMs (available from the Shriver Lab Web site).
Finally, the CEPH Utah individuals cluster with northwestern Europeans, in line with their more restricted described origins within Europe (north and west), and represent only a fraction of the northern-southeastern variation (PC1) observed in the Coriell Caucasian panel (fig. 7B). Furthermore, despite the dispersion of CEPH individuals along PC2, the SKT did not detect any significant stratification along this axis. STRUCTURE analysis also failed to detect meaningful structure with use of either EuroAIMs or the full set of available markers for this data set (6,207 SNPs).
Our genomewide investigation of European ancestry, in combination with other recent studies, demonstrates the importance of considering population stratification in studies using European and European-American individuals. Further examination of additional population samples, more individuals per population, and a larger number of markers will allow refinement of the important axes of variation. This will in turn enable the selection of efficient EuroAIM sets for measuring and correcting for stratification within European-derived population samples, as well as inform the debate on the population history of Europe.
Acknowledgments
We are grateful to the research subjects and to all who helped by collecting or providing human samples—in particular, S. Alonso, G. Argyropoulos, M. Bamshad, C. Batini, S. Beleza, M. Bower, C. Brady, A. de la Chapelle, V. Coia, G. Destro-Bisol, L. Jorde, R. Kaczanowski, R. Kittles, E. Parra, T. Schurr, and S. Tishkoff. We thank Dr. Zaykin for providing the program to calculate the combined P value. We thank our financial supporters: National Institutes of Health National Human Genome Research Institute grant HG002154, Science Foundation of Ireland Walton fellowship 04/W4/B643 (to M.D.S.), Health Research Board-Ireland grant RP/2004/155 (to B.M.), and the Hill and Weiss fellowships (to M.B.).
Web Resources
The URLs for data presented herein are as follows:
- Dr. Zaykin’s Web site, http://statgen.ncsu.edu/zaykin/tpm (for the program and formula to calculate Fisher’s combined P value for the SKT)
- Shriver Lab Web site, http://www.anthro.psu.edu/biolab/euroaims.pc1.xls (for dbSNP numbers of the top 1,200 EuroAIMs describing the northern-southeastern axis [PC1 in ], ordered by decreasing FST; other measures of informativeness were also used for comparison and provided nearly identical panels [not shown])
References
- 1.Davies N (1998) Europe: a history. Harper Perennial, New York [Google Scholar]
- 2.Wacholder S, Rothman N, Caporaso N (2002) Counterpoint: bias from population stratification is not a major threat to the validity of conclusions from epidemiological studies of common polymorphisms and cancer. Cancer Epidemiol Biomarkers Prev 11:513–520 [PubMed] [Google Scholar]
- 3.Seldin MF, Shigeta R, Villoslada P, Selmi C, Tuomilehto J, Silva G, Belmont JW, Klareskog L, Gregersen PK (2006) European population substructure: clustering of northern and southern populations. PLoS Genetics 2:e143 10.1371/journal.pgen.0020143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell CD, Ogburn EL, Lunetta KL, Lyon HN, Freedman ML, Groop LC, Altshuler D, Ardlie KG, Hirschhorn JN (2005) Demonstrating stratification in a European American population. Nat Genet 37:868–872 10.1038/ng1607 [DOI] [PubMed] [Google Scholar]
- 5.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- 6.Gower JC (1966) Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53:325–338 [Google Scholar]
- 7.Chae SS, Warde WD (2006) Effect of using principal coordinates and principal components on retrieval of clusters. Comput Stat Data Anal 50:1407–1417 10.1016/j.csda.2005.01.013 [DOI] [Google Scholar]
- 8.Chessel D, Dufour AB, Thioulouse J (2004) The ade4 package-I: one-table methods. R News 4:5–10 [Google Scholar]
- 9.Chakraborty R, Jin L (1993) A unified approach to study hypervariable polymorphisms: statistical considerations of determining relatedness and population distances. In: Pena SDJ, Chakraborty R, Epplen J, Jeffreys AJ (eds) DNA fingerprinting: current state of the science. Birkhauser, Basel, Switzerland, pp 153–175 [DOI] [PubMed] [Google Scholar]
- 10.Mountain JL, Cavalli-Sforza LL (1997) Multilocus genotypes, a tree of individuals, and human evolutionary history. Am J Hum Genet 61:705–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavalli-Sforza LL, Feldman MW (2003) The application of molecular genetic approaches to the study of human evolution. Nat Genet 33:266–275 10.1038/ng1113 [DOI] [PubMed] [Google Scholar]
- 12.Belle EMS, Landry PA, Barbujani G (2006) Origins and evolution of the Europeans’ genome: evidence from multiple microsatellite loci. Proc Biol Sci 273:1595–1602 10.1098/rspb.2006.3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavalli-Sforza L, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton, NJ [Google Scholar]
- 14.Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mantel N (1967) Detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220 [PubMed] [Google Scholar]
- 17.Shriver MD, Kennedy J, Parra EJ, Lawson HA, Sonpar V, Huang J, Akey JM, Jones KW (2004) The genomic distribution of population substructure in four populations using 8,525 autosomal SNPs. Hum Genomics 1:274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shriver MD, Mei R, Parra EJ, Sonpar V, Halder I, Tishkoff SA, Schurr TG, Zhadanov SI, Osipova LP, Brutsaert TD, et al (2005) Large-scale SNP analysis reveals clustered and continuous patterns of human genetic variation. Hum Genomics 2:81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaykin DV, Zhivotovsky LA, Westfall PH, Weir BS (2002) Truncated product method for combining P-values. Genet Epidemiol 22:170–185 10.1002/gepi.0042 [DOI] [PubMed] [Google Scholar]
- 20.Menozzi P, Piazza A, Cavalli-Sforza L (1978) Synthetic maps of human gene frequencies in Europeans. Science 201:786–792 10.1126/science.356262 [DOI] [PubMed] [Google Scholar]
- 21.Chikhi L, Nichols RA, Barbujani G, Beaumont MA (2002) Y genetic data support the Neolithic demic diffusion model. Proc Natl Acad Sci USA 99:11008–11013 10.1073/pnas.162158799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McEvoy B, Richards M, Forster P, Bradley DG (2004) The longue durée of genetic ancestry: multiple genetic marker systems and Celtic origins on the Atlantic facade of Europe. Am J Hum Genet 75:693–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards M, Macaulay V, Torroni A, Bandelt HJ (2002) In search of geographical patterns in European mitochondrial DNA. Am J Hum Genet 71:1168–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bauduer F, Feingold J, Lacombe D (2005) The Basques: review of population genetics and Mendelian disorders. Hum Biol 77:619–637 10.1353/hub.2006.0001 [DOI] [PubMed] [Google Scholar]
- 25.Kittles RA, Perola M, Peltonen L, Bergen AW, Aragon RA, Virkkunen M, Linnoila M, Goldman D, Long JC (1998) Dual origins of Finns revealed by Y chromosome haplotype variation. Am J Hum Genet 62:1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370 10.2307/2408641 [DOI] [PubMed] [Google Scholar]
- 27.Devlin B, Roeder K (1999) Genomic control for association studies. Biometrics 55:997–1004 10.1111/j.0006-341X.1999.00997.x [DOI] [PubMed] [Google Scholar]
- 28.Reich DE, Goldstein DB (2001) Detecting association in a case-control study while correcting for population stratification. Genet Epidemiol 20:4–16 [DOI] [PubMed] [Google Scholar]
- 29.Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138 10.1046/j.1471-8286.2003.00566.x [DOI] [Google Scholar]
- 30.Rosenberg NA, Mahajan S, Ramachandran S, Zhao CF, Pritchard JK, Feldman MW (2005) Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet 1:e70 10.1371/journal.pgen.0010070 [DOI] [PMC free article] [PubMed] [Google Scholar]