Abstract
FANCD2 is an evolutionarily conserved Fanconi anemia (FA) gene that plays a key role in DNA double-strand–type damage responses. Using complementation assays and immunoblotting, a consortium of American and European groups assigned 29 patients with FA from 23 families and 4 additional unrelated patients to complementation group FA-D2. This amounts to 3%–6% of FA-affected patients registered in various data sets. Malformations are frequent in FA-D2 patients, and hematological manifestations appear earlier and progress more rapidly when compared with all other patients combined (FA–non-D2) in the International Fanconi Anemia Registry. FANCD2 is flanked by two pseudogenes. Mutation analysis revealed the expected total of 66 mutated alleles, 34 of which result in aberrant splicing patterns. Many mutations are recurrent and have ethnic associations and shared allelic haplotypes. There were no biallelic null mutations; residual FANCD2 protein of both isotypes was observed in all available patient cell lines. These analyses suggest that, unlike the knockout mouse model, total absence of FANCD2 does not exist in FA-D2 patients, because of constraints on viable combinations of FANCD2 mutations. Although hypomorphic mutations arie involved, clinically, these patients have a relatively severe form of FA.
Fanconi anemia (FA) is a rare genome-instability disorder with the variable presence of congenital malformations, progressive bone-marrow failure (BMF), predisposition to malignancies, and cellular hypersensitivity to DNA-interstrand crosslinking agents.1 There are at least 13 complementation groups (FA-A [MIM 607139], -B [MIM 300515], -C [MIM 227645], -D1 [MIM 600185], -D2 [MIM 227646], -E [MIM 600901], -F [MIM 603467], -G [MIM 602956], -I [MIM 609053], -J [MIM 609054], -L [MIM 608111], -M [MIM 609644], and -N [MIM 610355]), each of which is associated with biallelic or hemizygous mutations in a distinct gene.2–4 To date, 12 of the underlying genes have been identified: FANCA, FANCB, FANCC, FANCD1/BRCA2, FANCD2, FANCE, FANCF, FANCG/XRCC9, FANCJ/BRIP1, FANCL/PHF9, FANCM/HEF, and FANCN/PALB2.3–7 Eight of the FA proteins (FANCA, -B, -C, -E, -F, -G, -L, and -M) and other components assemble in a nuclear complex, the FA “core complex,” that is required for the monoubiquitination of FANCD2 at amino acid residue K561.8,9 Monoubiquitination occurs in response to DNA damage and during the S phase of the cell cycle.9,10 The monoubiquitinated FANCD2 isoform (FANCD2-L, as opposed to FANCD2-S) is targeted to nuclear foci containing proteins such as BRCA1 [MIM 113705], BRCA2, and RAD51 [MIM 179617] that are involved in DNA-damage signaling and recombinational repair.11–14 The precise role of FANCD2 remains unknown, but FANCD2-deficient DT40 cells show defects in homologous recombination-mediated DNA double-strand break (DSB) repair, translesion synthesis, and gene conversion.11,15,16 Therefore, FANCD2 is thought to play a central role in the maintenance of genome stability.11,16,17
The human and murine Fancd2 genes show a higher degree of homology than do the corresponding Fanca, Fancc, Fance, Fancf, and Fancg genes.18 Fancd2-knockout mice suffer from perinatal lethality, microphthalmia, and early epithelial cancers,19 but it remains controversial whether the murine FA-D2 phenotype in general is more severe than the corresponding murine knockouts of the other FA genes.19,20 Fancd2 is required for survival after DNA damage in Caenorhabditis elegans.21 Fancd2-deficient zebrafish embryos display severe developmental defects due to increased apoptosis, which underscores the importance of Fancd2 function during vertebrate ontogenesis.22 Finally, knock-down of Drosophila Fancd2 causes pupal lethality.23 In humans, it has been estimated that complementation group FA-D2 accounts for between <1%24 and 3%25 of all patients with FA. The presence of FANCD2 pseudogenes complicates mutation analysis, which may explain why there has been just one other report of a single human FANCD2 mutation since the original description.26,27 As a concerted effort among nine laboratories, we present a comprehensive mutation profile of the FANCD2 gene (Ensembl Genome Browser [accession number ENSG00000144554]). We show that the FA phenotype resulting from FANCD2 deficiency is relatively severe. In contrast to all other FA genes, (1) the mutation spectrum of FANCD2 is dominated by splicing mutations, and (2) residual FANCD2 protein exists in all tested cell lines from FA-D2 patients, which suggests lethality of biallelic null mutations.
Patients, Material, and Methods
Diagnostic Procedures
Anticoagulated peripheral blood and skin-biopsy samples were referred to the participating laboratories for diagnostic testing. Confirmation of the diagnosis of FA, subtyping, and mutation analysis were performed with informed consent according to the Declaration of Helsinki. The study was approved by the institutional review boards of the participating centers. Clinical suspicion of FA was confirmed by the detection of cellular hypersensitivity to DNA-crosslinking agents following published procedures.28–32 In patients with increasing and/or long-term stable blood counts, the possibility of somatic reversion leading to mosaicism of hematopoietic cells was considered, and cultured fibroblasts were used for mitomycin-C (MMC) sensitivity testing and for complementation studies.
Patient Statistics
A total of 29 fully informative FA-D2 patients (patients 1–29) were included in the present genotype-phenotype study. A fetal case (number 19) and five patients with hematopoietic mosaicism (patients 3, 14, 15, 25, and 26) were excluded from clinical follow-up studies. Four additional FA-D2 patients (patients 30–33) with incomplete clinical data were not part of the phenotype analysis, but results concerning their mutations are indicated in the text, tables, and figures.
Three end points were evaluated to determine hematologic severity: time to hematological onset, defined as “cell count of at least one lineage constantly below normal range”; period from BMF to hematological stem-cell transplantation (HSCT); and time to HSCT. Kaplan-Meier estimates were computed for the length of overall survival. Birth was taken as the date of FA onset for all these calculations. Comparisons were made with patients in the International Fanconi Anemia Registry (IFAR), as reported elsewhere,33 by means of log-rank test statistics. Multivariate and competing-risk analyses were not possible because of the limited number of informative patients.
Cell Culture
Epstein-Barr virus (EBV)–transformed lymphoblastoid cell lines (LCLs) were established using cyclosporin A, as described elsewhere.34 All blood-derived cell cultures were maintained in RPMI 1640 medium with GlutaMAX (Gibco) supplemented with 15% fetal bovine serum (FBS) (Sigma). Fibroblast strains were established using standard cell-culture procedures and were propagated in Earle’s minimal essential medium with GlutaMAX (Gibco) and 15% FBS. All cultures were kept in high-humidity incubators in an atmosphere of 5% (v/v) CO2 and, in the case of fibroblasts, 5% (v/v) O2 by replacing ambient air with nitrogen.35 MMC treatment was for 48 h at 12 ng/ml (fibroblasts) or 15 ng/ml (LCLs) to cause cell-cycle arrest, or for 12 h at 100 ng/ml to induce monoubiquitination of the protein, FANCD2-L. In some cases, RNA stabilization was achieved by the addition of cycloheximide (CHX) to cell cultures at a final concentration of 250 μg/ml 4.5 h before RNA isolation.
Retroviral Complementation
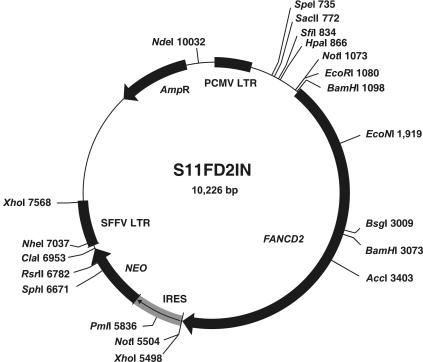
For construction of the D2-IRES (internal ribosomal entry site)–neo retroviral expression vector S11FD2IN, the D2-IRES-puro plasmid pMMP-FANCD226 was cut using SalI. The ends were blunted, and the fragment containing the FANCD2 ORF was cut out with EcoRI and was ligated into S11IN, which was cut with BamHI, was blunted, and was cut again with EcoRI (fig. 1). S11 vectors are based on the spleen focus–forming virus and are derived from the GR plasmid.36 Sequencing of the retroviral plasmid S11FD2IN revealed three reported polymorphisms in the FANCD2 ORF—c.1122A→G, c.1509C→T, and c.2141C→T26—and another silent base substitution, c.3978C→T. Stable retroviral packaging cells were generated by infection of PG13 cells and selection in G418 (Sigma), as described elsewhere.37 In addition, the cDNAs encoding enhanced green fluorescent protein (GFP) and FANCA cDNAs were separately cloned into the vector S11IN for transduction of the cells under study, with GFP serving to monitor complete selection and FANCA serving as negative complementation control.
Figure 1. .
Circular map of vector S11FD2IN. The retroviral-expression vector S11FD2IN contains a bicistronic construct of the full-length FANCD2 cDNA (“FANCD2”) and the neomycin resistance gene (NEO). Translation of the latter is ensured by an IRES. Also shown are the long terminal repeats (LTR), the restriction sites and their positions, and the bacterial resistance (AmpR). Used for cloning of FANCD2 into the target vector S11IN were the 5′ EcoRI and the 3′ SalI (insert) and BamHI (vector) sites; the latter two were destroyed by blunting.
Retroviral transduction of cultured cells followed published protocols.38,39 Selection of transduced cells was in G418 (Sigma) at a final concentration of 0.8–1.2 mg/ml for ∼10 d. Transduced cells were analyzed for their sensitivity to MMC, with use of flow cytometry, to assess survival rates and cell-cycle arrest.39,40
Immunoblotting
FANCD2 immunoblotting was performed as described elsewhere,9 with minor modifications. Detection was by the chemiluminescence technique with use of standard enhanced chemoluminescence reagent (Amersham) or SuperSignalWestFemto (Pierce).
Mutation and Haplotype Characterization
Primers used for cDNA amplification are shown in table 1, and those additionally used for cDNA sequencing are shown in table 2 (GenBank accession numbers NM_033084 and AF340183).
Table 1. .
FANCD2 cDNA Amplification Primers
| Forward |
Reverse |
||||||
| PCR Fragment |
Designation | Binding Position | Sequence (5′→3′) |
Designation | Binding Position | Sequence (5′→3′) |
PCR Product Size (bp) |
| 1 | FA-D2, Fr.1 F | −47 to −27 | GCGACGGCTTCTCGGAAGTAA | FA-D2, Fr.1 R | 998 to 976 | CTGTAACCGTGATGGCAAAACAC | 998 |
| 2 | FA-D2, Fr.2 F | 763 to 787 | GACCCAAACTTCCTATTGAAGGTTC | FA-D2, Fr.2 R | 1996 to 1975 | CTACGAAGGCATCCTGGAAATC | 1,234 |
| 3 | FA-D2, Fr.3 F | 1757 to 1777 | CGGCAGACAGAAGTGAATCAC | FA-D2, Fr.3 R | 2979 to 2958 | GTTCTTGAGAAAGGGGACTCTC | 1,223 |
| 4 | FA-D2, Fr.4 F | 2804 to 2829 | TTCTACATTGTGGACTTGTGACGAAG | FA-D2, Fr.4 R | 3942 to 3922 | GTCTAGGAGCGGCATACATTG | 1,139 |
| 5 | FA-D2, Fr.5(L) F | 3761 to 3781 | CAGCAGACTCGCAGCAGATTC | FA-D2, Fr.5(L) R | 4700 to 4679 | GACTCTGTGCTTTGGCTTTCAC | 940 |
Table 2. .
FANCD2 cDNA Sequencing Primers
| Designation | Binding Position |
Sequence (5′→3′) |
| sFA-D2, 244 F | 244 to 263 | ACCCTGAGGAGACACCCTTC |
| sFA-D2, 545 F | 545 to 566 | GGCTTGACAGAGTTGTGGATGG |
| sFA-D2, 1011 F | 1011 to 1033 | CAGCGGTCAGAGCTGTATTATTC |
| sFA-D2, 1308 F | 1308 to 1327 | GTCGCTGGCTCAGAGTTTGC |
| sFA-D2, 1574 F | 1574 to 1596 | CCCCTCAGCAAATACGAAAACTC |
| sFA-D2, 2142 F | 2142 to 2162 | GGTGACCTCACAGGAATCAGG |
| sFA-D2, 2381 F | 2381 to 2404 | GAGAGATTGTAAATGCCTTCTGCC |
| sFA-D2, 2679 F | 2679 to 2699 | TGACCCTACGCCATCTCATAG |
| sFA-D2, 3268 F | 3268 to 3288 | GCCCTCCATGTCCTTAGTAGC |
| sFA-D2, 3573 F | 3573 to 3594 | GCACACAGAGAGCATTCTGAAG |
| sFA-D2, 4049 F | 4049 to 4069 | ACACGAGACTCACCCAACATG |
| sFA-D2, 4303 F | 4303 to 4323 | GAGTCTGGCACTGATGGTTGC |
| sFA-D2, 367 R | 367 to 347 | CATCCTGCAGACGCTCACAAG |
| sFA-D2, 621 R | 621 to 600 | CAGGTTCTCTGGAGCAATACTG |
| sFA-D2, 951 R | 951 to 929 | CTGTAACCGTGATGGCAAAACAC |
| sFA-D2, 1158 R | 1183 to 1158 | TCTGAGTATTGGTGCTATAGATGATG |
| sFA-D2, 1414 R | 1414 to 1396 | CCTGCTGGCAGTACGTGTC |
| sFA-D2, 1704 R | 1704 to 1684 | GAATACGGTGCTAGAGAGCTG |
| sFA-D2, 2253 R | 2253 to 2232 | CTCCTCCAAGTTTCCGTTATGC |
| sFA-D2, 2526 R | 2526 to 2505 | GTTTCCAAGAGGAGGGACATAG |
| sFA-D2, 3346 R | 3346 to 3328 | GGACGCTCTGGCTGAGTAG |
| sFA-D2, 3674 R | 3674 to 3653 | GTAGGGAATGTGGAGGAAGATG |
| sFA-D2, 4159 R | 4159 to 4139 | CCAGCCAGAAAGCCTCTCTAC |
| sFA-D2, 4409 R | 4409 to 4387 | GGGAATGGAAATGGGCATAGAAG |
A total of seven large amplicons (superamplicons) were generated with primers that are unique to certain regions of the functional FANCD2 gene. These primers and the sizes of the superamplicons I–VII are shown in table 3. The superamplicons served as templates in place of genomic DNA. They were used to amplify the genomic sequence before sequencing; an exception to this was superamplicon V, which was used for direct sequencing. Genomic primers for the amplification of all FANCD2 exons and adjacent intron regions and their sizes are displayed in table 4. Additional genomic mutation-specific primers are shown in table 5.
Table 3. .
FANCD2 Superamplicon Primers
| Forward | Reverse | |||||
| Superamplicon | Containing Exon(s) | Designation | Sequence(5′→3′) | Designation | Sequence(5′→3′) | PCR Product Size(bp) |
| I | 1 and 2 | hFANCD2_exon1_F | TATGCCCGGCTAGCACAGAA | hFANCD2_super_1_2_R | GGCCCACAGTTTCCGTTTCT | 4,346 |
| II | 3 | hFANCD2_super_3_3_F | GTGTCACGTGTCTGTAATCTC | hFANCD2_super_3_3_R | CTGGGACTACAGACACGTTTT | 2,323 |
| III | 7, 8, and 9 | hFANCD2_super_7_14_F | TGGGTTTGGTAGGGTAATGTC | hFANCD2_exon9_R | TACTCATGAAGGGGGGTATCA | 4,595 |
| IV | 10, 11, 12, 13, and 14 | hFANCD2_exon10_F | GCCCAGCTCTGTTCAAACCA | hFANCD2_super_7_14_R | TTAAGACCCAGCGAGGTATTC | 5,635 |
| V | 13, 14, 15, 16, and 17 | FA-D2, sup13-I17 F | CATGGCAGGAACTCCGATCTTG | FA-D2, sup13-I17 F | CTCCCTTAAAAGCTCAAAGCTCAAGTTC | 8,858 |
| VI | 19 and 20 | hFANCD2_super_19_22_F | ACGTAATCACCCCTGTAATCC | hFANCD2_exon20_R | TGACAGAGCGAGACTCTCTAA | 2,749 |
| VII | 21, 22, and 23 | FA-D2, 21_23, F | GCTTCTAGTCACTGTCAGTTCACCAG | FA-D2, 21_23, R | ACGTTGGCCAGAAAGTAATCTCAG | 2,518 |
| VIII | 23, 24, 25, and 26 | hFANCD2_super_23_29_F | GGCCTTGTGCTAAGTGCTTTT | hFANCD2_exon26_R | TCAGGGATATTGGCCTGAGAT | 3,252 |
| IX | 27, 28, and 29 | hFANCD2_exon27_F | GCATTCAGCCATGCTTGGTAA | hFANCD2_super_23_29_R | CACTGCAAACTGCTCACTCAA | 3,371 |
| X | 30 | hFANCD2_super_30_32_F | CCAAAGTACTGGGAGTTTGAG | hFANCD2_exon30_R | TACCCAGTGACCCAAACACAA | 2,186 |
| XVI | 31 and 32 | hFANCD2_exon31_F | CCATTGCGAACCCTTAGTTTC | hFANCD2_super_30_32_R | ACCCTGGTGGACATACCTTTT | 299 |
| XII | 33 and 34 | hFANCD2_super_33_36_F | GAGCAATTTAGCCTGTGGTTTT | hFANCD2_exon34_R | TATAGCAAGAGGGCCTATCCA | 3,457 |
| XIII | 35 and 36 | hFANCD2_exon35_F | TTAGACCGGGAACGTCTTAGT | hFANCD2_super_33_36_R | TCTGGGCAACAGAACAAGCAA | 2,040 |
| XIV | 43a | hFANCD2_super_43_44_F | AGGGTCCTGAGACTATATACC | hFANCD2_exon43a_R | AGCATGATCTCGGCTCACCA | 2,040 |
| XV | 44 | hFANCD2_exon44_F | CACCCAGAGCAGTAACCTAAA | hFANCD2_super_43_44_R | ACCATCTGGCCGACATGGTA | 464 |
Table 4. .
FANCD2 Exon Primers
| Forward | Reverse | ||||
| Exon | Designation | Sequence(5′→3′) | Designation | Sequence(5′→3′) | PCR Product Size(bp) |
| 1 | hFANCD2_exon1_F | TATGCCCGGCTAGCACAGAA | hFANCD2_exon1_R | TCCCATCTCAGGGCAGATGA | 324 |
| 2 | hFANCD2_exon2_F | CCCCTCTGATTTTGGATAGAG | hFANCD2_exon2_R | TCTCTCACATGCCTCACACAT | 258 |
| 3 | hFANCD2_exon3_F | GACACATCAGTTTTCCTCTCAT | hFANCD2_exon3_R | AAGATGGATGGCCCTCTGATT | 354 |
| 4 | hFANCD2_exon4_F | TGGTTTCATCAGGCAAGAAACT | hFANCD2_exon4_R | AATCATTCTAGCCCACTCAACT | 253 |
| 4/5 | FA-D2, exon 4 II F | GAGAAGGAAAACTATGGTAGGAAAC | FA-D2, exon 5 II R | GTGTAAGCTCTGTTTTCCTCAGAG | 509 |
| 5 | hFANCD2_exon5_F | GCTTGTGCCAGCATAACTCTA | hFANCD2_exon5_R | AGCCCCATGAAGTTGGCAAAA | 298 |
| 6 | hFANCD2_exon6_F | GAGCCATCTGCTCATTTCTGT | hFANCD2_exon6_R | GCTGTGCTAAAGCTGCTACAA | 341 |
| 7 | hFANCD2_exon7_F | AATCTCGGCTCACTGCAATCT | hFANCD2_exon7_R | CAGAGAAACCAATAGTTTTCAG | 280 |
| 8 | hFANCD2_exon8_F | TAGTGCAGTGCCGAATGCATA | hFANCD2_exon8_R | AGCTAATGGATGGATGGAAAAG | 333 |
| 9 | hFANCD2_exon9_F | TTCACACGTAGGTAGTCTTTCT | hFANCD2_exon9_R | TACTCATGAAGGGGGGTATCA | 323 |
| 10 | hFANCD2_exon10_F | GCCCAGCTCTGTTCAAACCA | hFANCD2_exon10_R | CATTACTCCCAAGGCAATGAC | 229 |
| FA-D2, exon10, F | GTCTGCCCAGCTCTGTTCAAAC | FA-D2, exon10, R | ATTACTCCCAAGGCAATGACTGACTG | 232 | |
| 11 | hFANCD2_exon11_F | GTGGGAAGATGGAGTAAGAGA | hFANCD2_exon11_R | AGCTCCATTCTCTCCTCTGAA | 341 |
| FA-D2, exon11, F | CAGTTCAGTACAAAGTTGAGGTAGTG | FA-D2, exon11, R | CCGGATTAGTCAGTATTCTCAGTTAG | 267 | |
| 12 | hFANCD2_exon12_F | TGCCTACCCACTATGAATGAG | hFANCD2_exon12_R | TCTGACAGTGGGATGTCAGAA | 211 |
| 13 | hFANCD2_exon13_F | CAGGAACTCCGATCTTGTAAG | hFANCD2_exon13_R | ATGTGTCCATCTGGCAACCAT | 321 |
| FA-D2, exon 13 F P1+2 | CCGATCTTGTAAGTTCTTTTCTGGTACG | FA-D2, exon 13 R P1+2 | TGGCAACCATCAGCTATCATTTCCAC | 302 | |
| 14 | hFANCD2_exon14_F | CGTGTTTCGCTGATGTGTCAT | hFANCD2_exon14_R | TGGAGGGGGGAGAAAGAAAG | 186 |
| 15 | hFANCD2_exon15a_F | GTGTTTGACCTGGTGATGCTT | hFANCD2_exon15a_R | GGAAGGCCAGTTTGTCAAAGT | 325 |
| hFANCD2_exon15b_F | GTGGAACAAATGAGCATTATCC | hFANCD2_exon15b_R | CTTATTTCTTAGCACCCTGTCAA | 204 | |
| FA-D2, exon 15 F uniq | GGAACAAATGAGCATTATCCATTCTGTG | FA-D2, exon 15 R/ P1 | CTCAATGGGTTTGAACAATGGACTG | 363 | |
| 16 | hFANCD2_exon16_F | AGGGAGGAGAAGTCTGACATT | hFANCD2_exon16_R | TTCCCCTTCAGTGAGTTCCAA | 332 |
| FA-D2, exon 16 F P1 | GTCTGACATTCCAAAAGGATAAGCAAC | FA-D2, exon 16 R | CTTGAGACCCAGGTCAGAGTTC | 344 | |
| 17 | hFANCD2_exon17_F | GATGGGTTTGGGTTGATTGTG | hFANCD2_exon17_R | GATTAGCCTGTAGGTTAGGTAT | 422 |
| FA-D2, exon 17 F P1+2 | CTGGCATATTCCTAAATCTCCTGAAG | FA-D2, exon 17 R | GCCTGTAGGTTAGGTATAAAGAAGTG | 472 | |
| 18 | hFANCD2_exon18_F | GGCTATCTATGTGTGTCTCTTT | hFANCD2_exon18_R | CCAGTCTAGGAGACAGAGCT | 282 |
| 19 | hFANCD2_exon19_F | CGATATCCATACCTTCTTTTGC | hFANCD2_exon19_R | ACGATTAGAAGGGAACATGGAA | 328 |
| 20 | hFANCD2_exon20_F | CACACCAACATGGCACATGTA | hFANCD2_exon20_R | TGACAGAGCGAGACTCTCTAA | 239 |
| 21 | hFANCD2_exon21_F | AAAGGGGCGAGTGGAGTTTG | hFANCD2_exon21_R | GAGACAGGGTAGGGCAGAAA | 339 |
| 22 | hFANCD2_exon22_F | ATGCACTCTCTCTTTTCTACTT | hFANCD2_exon22_R | GTAACTTCACCAGTGCAACCAA | 279 |
| 23 | hFANCD2_exon23_F | TTCCCTGTAGCCTTGCGTATT | hFANCD2_exon23_R | ACAAGGAATCTGCCCCATTCT | 356 |
| 24 | hFANCD2_exon24_F | CTCCCTATGTACGTGGAGTAA | hFANCD2_exon24_R | CCCCACATACACCATGTATTG | 258 |
| 25 | hFANCD2_exon25_F | AGGGGAAAGTAAATAGCAAGGA | hFANCD2_exon25_R | GTGGGACATAACAGCTAGAGA | 350 |
| 26 | hFANCD2_exon26_F | GACATCTCTCAGCTCTGGATA | hFANCD2_exon26_R | TCAGGGATATTGGCCTGAGAT | 324 |
| 27 | hFANCD2_exon27_F | GCATTCAGCCATGCTTGGTAA | hFANCD2_exon27_R | CCAATTACTGATGCCATGATAC | 324 |
| 28 | hFANCD2_exon28_F | TCTACCTCTAGGCAGTTTCCA | hFANCD2_exon28_R | GATTACTCCAACGCCTAAGAG | 354 |
| FA-D2, exon 28 F | TCTACCTCTAGGCAGTTTCCA | FA-D2, exon 28 R | GATTACTCCAACGCCTAAGAG | 354 | |
| 29 | hFANCD2_exon29_F | CTTGGGCTAGAGGAAGTTGTT | hFANCD2_exon29_R | TCTCCTCAGTGTCACAGTGTT | 384 |
| 30 | hFANCD2_exon30_F | GAGTTCAAGGCTGGAATAGCT | hFANCD2_exon30_R | TACCCAGTGACCCAAACACAA | 348 |
| FA-D2, exon 30 F | CATGAAATGACTAGGACATTCCTG | FA-D2, exon 30 F | GCAAGATGAATATTGTCTGGCAATACG | 319 | |
| 31 | hFANCD2_exon31_F | CCATTGCGAACCCTTAGTTTC | hFANCD2_exon31_R | ACCGTGATTCTCAGCAGCTAA | 341 |
| 32 | hFANCD2_exon32_F | CCACCTGGAGAACATTCACAA | hFANCD2_exon32_R | AGTGCCTTGGTGACTGTCAAA | 336 |
| 33 | hFANCD2_exon33_F | CACGCCCGACCTCTCAATTC | hFANCD2_exon33_R | TACTGAAAGACACCCAGGTTAT | 340 |
| 34 | hFANCD2_exon34_F | TTGGGCACGTCATGTGGATTT | hFANCD2_exon34_R | TATAGCAAGAGGGCCTATCCA | 349 |
| FA-D2, exon 34 II F | GGCAATCTTCTTGGGCTTATTACTGAG | FA-D2, exon 34 II R | CAACTTCCAAGTAATCCAAAGTCCACTTC | 327 | |
| 35 | hFANCD2_exon35_F | TTAGACCGGGAACGTCTTAGT | hFANCD2_exon35_R | GTCCAGTCTCTGACAAACAAC | 300 |
| 36 | hFANCD2_exon36_F | CCTCTGGTTCTGTTTTATACTG | hFANCD2_exon36_R | GGCCAAGTGGGTCTCAAAAC | 398 |
| 37 | hFANCD2_exon37_F | CTTCCCAGGTAGTTCTAAGCA | hFANCD2_exon37_R | TCTGGGCAACAGAACAAGCAA | 277 |
| FA-D2, exon 37 II F | CATCCTCTTACTAAGGACCCTAGTGAAAG | FA-D2, exon 37 II R | CAGCAACTTCCAAGTAATCCAAAGTCCAC | 288 | |
| 38 | hFANCD2_exon38_F | GCACTGGTTGCTACATCTAAG | hFANCD2_exon38_R | AAGCCAGGACACTTGGTTTCT | 274 |
| 39 | hFANCD2_exon39_F | TGCTCAAAGGAGCAGATCTCA | hFANCD2_exon39_R | GCATCCATTGCCTTCCCTAAA | 236 |
| 40 | hFANCD2_exon40_F | CCTTGGGCTGGATGAGACTA | hFANCD2_exon40_R | CAGTCCAATTTGGGGATCTCT | 309 |
| 41 | hFANCD2_exon41_F | GATTGCAAGGGTATCTTGAATC | hFANCD2_exon41_R | CCCCAATAGCAACTGCAGATT | 214 |
| 42 | hFANCD2_exon42_F | AACATACCGTTGGCCCATACT | hFANCD2_exon42_R | GCTTAGGTGACCTTCCTTACA | 356 |
| 43 | hFANCD2_exon43a_F | GTGGCTCATGCTTGTAATCCT | hFANCD2_exon43a_R | AGCATGATCTCGGCTCACCA | 366 |
| hFANCD2_exon43b_F | CTGCCACCTTAGAGAACTGAA | hFANCD2_exon43b_R | TCAGTAGAGATGGGGTTTCAC | 358 | |
| hFANCD2_exon43c_F | TAGAATCACTCCTGAGTATCTC | hFANCD2_exon43c_R | CTCAAGCAATCCTCCTACCTT | 405 | |
| hFANCD2_exon43d_F | AGTTGGTGGAGCAGAACTTTG | hFANCD2_exon43d_R | CAGCTTCTGACTCTGTGCTTT | 367 | |
| hFANCD2_exon43e_F | TCAACCTTCTCCCCTATTACC | hFANCD2_exon43e_R | CTCGAGATACTCAGGAGTGAT | 381 | |
| hFANCD2_exon43f_F | GGTATCCATGTTTGCTGTGTTT | hFANCD2_exon43f_R | AGTTCTGCTCCACCAACTTAG | 306 | |
| 44 | hFANCD2_exon44_F | CACCCAGAGCAGTAACCTAAA | hFANCD2_exon44_R | GAAAGGCAAACAGCGGATTTC | 213 |
| FA-D2, exon 44 II F | CTAGGAGCTGTATTCCAGAGGTCAC | FA-D2, exon 44 II R | GGATCCTACCAGTAAGAAAGGCAAAC | 250 | |
Table 5. .
FANCD2 Mutation-Specific Primers
| Designation | Sequence(5′→3′) | PCR/Sequencing |
| FA-D2, exon4-6 F | GAAGGAAAACTATGGTAGGAAACTGGTG | PCR/Sequencing |
| FA-D2, exon 6 R | CAGATGTATTAGGCTAATAAGCACAG | PCR/Sequencing |
| FA-D2, exon4-i6 R | CCAGAAGCAGTTTGATGAGACTCTTAG | PCR/Sequencing |
| FA-D2, exon i4F | GCTTTCCAAAAGAAGCTCTTTCAGAC | PCR/Sequencing |
| FA-D2, exon4-IVS F | GGAGACACCCTTCCTATCCCAAAG | PCR/Sequencing |
| FA-D2, exon 5F | GAGTGGGCTAGAATGATTTTTAACAGC | PCR/Sequencing |
| FA-D2, exon 5 R | CTCTGAGGAAAACAGAGCTTACAC | PCR/Sequencing |
| D2_AluYb9 F | GCAATCTCGGCTCACTGCAAGCTC | PCR/Sequencing |
| D2_IVS4/AluYb9, R | GCTGTTAAAAATCATTCTACTTTGGGAGG | PCR/Sequencing |
| FA-D2, ex 10 F | GACTTGACCCAAACTTCCTATTGAA* | PCR/Sequencing |
| FA-D2, ex 14 F | TCGTGTTTCGCTGATGTGT | PCR/Sequencing |
| FA-D2, ex 11 R | CCGGATTAGTCAGTATTCTCAGTTAG | PCR/Sequencing |
| IVS14+2411 R | CGAGACCATCCTGACTAACACG | PCR/Sequencing |
| IVS14+2512 R | GATACCCCTTAAGAATACAGAGC | PCR/Sequencing |
| FANCD2_16S | AGAGCTAGGGAGGAGAAGTCTGA | PCR |
| FANCD2_18A | GAGCTGAGATCGTGCCAACT | PCR |
| FANCD2_17S | TGGTCAAGTTACACTGGCATATT | Sequencing |
| FANCD2_17A | CCATCCTTCAGCAATCACTC | Sequencing |
| D2_P2_21_23 F | GTTTTCTGATACTTGGAAACTACTGGCTTG | PCR/Sequencing |
| D2_P1_21_23 R | GACACAGAGGTAGCAAAGGATGTTC | PCR/Sequencing |
| FA-D2, ex21_23, int1 | CTATGATGAATTTGCCAACCTGATCC | Sequencing |
| FA-D2, ex21_23, int2 | GAGGGCTCCTTCACTTAATAACAATC | Sequencing |
| FA-D2, ex21_23, int3 | GTATTGTTTACCTGCTGGCTGGTTG | Sequencing |
| FA-D2_sup_exon26 II F uniq | TAGGGTCACAAGCCTAATCTCCTTT | PCR |
| FA-D2_sup_exon26 II R uniq | GGCCATGATGAATAATCTTTCTTTTGTTTG | PCR |
For haplotyping, four microsatellite markers flanking FANCD2 on chromosome 3 were studied: D3S1597, a dinucleotide repeat at 9.340 Mb; D3S1038, a dinucleotide repeat at 10.488 Mb; D3S3611, another dinucleotide repeat at 10.529 Mb; and D3S1675, a tetranucleotide repeat at 10.643 Mb. Primers used for microsatellite amplifications are specified in table 6. Two of the forward primers were 5′-Cy5-; the other two 5′-Cy5.5- primers were labeled for multiplex fragment analysis with DNA Sizing standards on a CEQ 8000 Genetic Analysis System (both Beckman Coulter).
Table 6. .
Microsatellite Primers
| Primer Sequence(5′→3′) |
|||
| STR | Genomic Position (Mb) |
Sense | Antisense |
| D3S1597 | 9.34 | AGTACAAATACACACAAATGTCTC | CAATTCGCAAATCGTTCATTGCT |
| D3S1038 | 10.49 | AAAGGGGTTCAGGAAACCTG | CCCTCCAGTAAGAGGCTTCCTAG |
| D3S3611 | 10.53 | GCTACCTCTGCTGAGCATATTC | CACATAGCAAGACTGTTGGGGGC |
| D3S1675 | 10.64 | GGATAGATGGATGAATGGATGGC | CCTCTCTAACTACCAATTCATCCA |
Results
Assignment to and Frequency of Group FA-D2
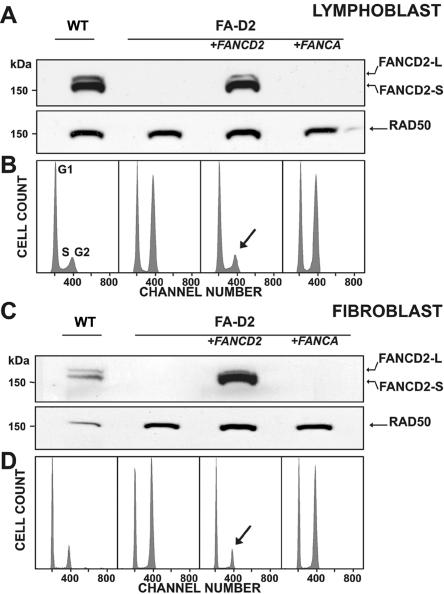
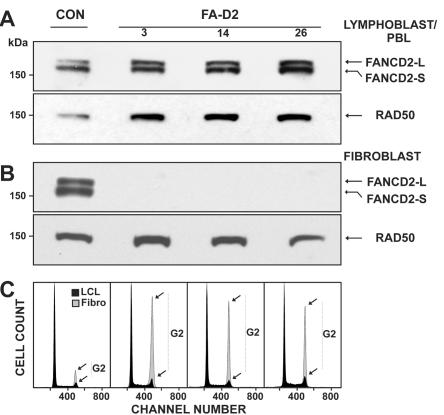
Figure 2 demonstrates our strategy for the assignment of cultured FA cells to group FA-D2. MMC sensitivity was demonstrated by cell-cycle analysis that showed G2-phase arrest of the tested LCLs (fig. 2B, lane 2).30–32 The apparent absence of FANCD2 bands on standard-exposure immunoblots suggested their belonging to complementation group FA-D2 (fig. 2A, lane 2).41 Transduction of putative FA-D2 LCLs with FANCD2 cDNA with use of the retroviral vector S11FD2IN restored FANCD2 expression and function, as reflected by the emergence of both FANCD2 isoforms (FANCD2-S and -L) (fig. 2A, lane 3); simultaneously, the MMC sensitivity of transduced cells returned to normal control-cell levels, as evidenced by the reduction of G2 phase cell–cycle fractions (fig. 2A, lane 1, and fig. 2B, lanes 1 and 3). Transduction of D2 LCLs with GFP or FANCA resulted in neither the restoration of either FANCD2 isoform nor a normalization of G2 phase arrest, as exemplified for FANCA with use of S11FAIN, as shown in figure 2A and 2B, lanes 4. In cases with suspected hematopoietic mosaicism, cultured fibroblasts were assayed using a corresponding strategy (fig. 2C and 2D). As shown in table 7, only a minority of patients were assigned to group FA-D2 by primary mutation analysis. This group includes four affected siblings of four different index patients and an unrelated deceased patient with only DNA available.
Figure 2. .
Delineation of FA-D2. A, Assignment to group FA-D2 on the basis of the absence of either FANCD2 band on immunoblots after exposure of the patients’ cells to MMC, here shown for an LCL from patient 6 (lane 2). Transduction with FANCD2 cDNA with use of S11FD2IN restores both isoforms of FANCD2—S and L (lane 3)—similar to a nontransduced normal control (lane 1). Transduction with FANCA cDNA in the same vector fails to show such restoration (lane 4). B, Assignment to group FA-D2 on the basis of cell-cycle analysis. After exposure to MMC, the LCL of the same patient shows pronounced G2-phase arrest (56.6%) (lane 2; Hoechst 33342 staining). Transduction with FANCD2 cDNA by use of S11FD2IN reduces the G2 phase to normal (14.9%) (lane 3, arrow), similar to the nontransduced normal control (16.6%) (lane 1). Transduction with FANCA cDNA in the same vector fails to reverse the G2-phase arrest (53.1%) (lane 4). Panels C and D are analogous to panels A and B and show complementation with cultured fibroblasts from patient 10; staining in panel D was with 4',6-diamidino-2-phenylindole (DAPI). G2-phase proportions in panel D are 20.3% (lane 1, control), 61.3% (lane 2, nontransduced FA), 19.9% (lane 3, FANCD2-transduced FA), and 58.5% (lane 4, FANCA-transduced FA). RAD50 [MIM 604040] was used as the loading control in panels A and C. WT=wild type.
Table 7. .
Laboratory Diagnostic Data for the Cohort of 29 FA-D2 Patients[Note]
| G2-Phase Arrest (%), G2/GF | Breaks/Cell | FANCD2 Mutation | ||||||||
| Patient Number | Kindred Sibling | Cell Type of Laboratory Diagnosis | Spontaneous | MMC/DEB | Spontaneous | MMC/DEB | Technique of Complementation Group Assignment | Allele 1 | Allele 2 | Somatic Mosaicism |
| 1 | 1/I | Lymphocyte | 65.7 | ND | .07 | 4.5 (MMC)/6.6 (MMC) | IB of LCL | c.696-121C→G (exonization) p.S232insQNNFX | c.696-121C→G (exonization) p.S232insQNNFX | None (residual protein) |
| 2 | 2/I | Lymphocyte | 54.3 | 70.1 (MMC) | .09 | 1.4 (MMC)/1.5 (DEB) | IB and RC of LCL | c.1948-6C→A (exon 22 skipping) p.E650X | c.3599delT p.I1200KfsX12 | None (residual protein) |
| 3 | 3/I | Lymphocyte (before mosaicism) | 38.6 (before mosaicism) | 46.6 (MMC) (before mosaicism) | .04 | .06 (MMC) | RC of fibroblasts | c.1948-16T→G (exon 22 skipping) p.E650X | c.1948-16T→G (exon 22 skipping) p.E650X | 1954G→A (exon 22), V652I, reconstitutes exon 22 recognition (blood, BM, LCL) |
| 4 | 4/I | Lymphocyte | 45.7 | 63.6 (MMC) | ND | ND | RC of fibroblasts | c.1948-16T→G (exon 22 skipping) p.E650X | c.1948-16T→G (exon 22 skipping) p.E650X | None (no LCL) |
| 5 | 4/II | Lymphocyte | 44.5 | 58.9 (MMC) | ND | ND | RC of fibroblasts | c.1948-16T→G (exon 22 skipping) p.E650X | c.1948-16T→G (exon 22 skipping) p.E650X | None (residual protein) |
| 6 | 5/I | Lymphocyte | 34.5 | 64.7 (MMC) | .05 | 4.7 (MMC)/ 5.6 (DEB) | IB and RC of LCL | c.274-57_−56insinvAluYb8nt36_319 +dup c.274–69_–57 (exon 5 skipping) p.I92YfsX7 | c.3803G→A p.W1268X | None (residual protein) |
| 7 | 6/I | Lymphocyte | 34.8 | 51.5 (MMC) | ND | ND | IB of LCL | c.904C→T p.R302W | c.1092G→A p.W364X | None (residual protein) |
| 8 | 7/I | Lymphocyte | 45.6 | 58.4 (MMC) | .06 | 1.3 (MMC) | IB of LCL | c.990-1G→A (aberrant splicing) p.S330RfsX16 | c.1948-6C→A (exon 22 skipping) p.E650X | None (residual protein) |
| 9 | 8/I | Lymphocyte | 65.3 | 70.9 (MMC) | .12 | ND | IB of LCL | c.810_812delGTC p.S271del | c.1948-16T→G (exon 22 skipping) p.E650X | None (residual protein) |
| 10 | 9/I | Lymphocyte | 38.4 | 58.4 (MMC) | ND | ND | RC of fibroblasts | c.1948-16T→G (exon 22 skipping) p.E650X | c.2715+1G→A (aberrant splicing) p.E906LfsX4 | None (residual protein) |
| 11 | 10/I | Lymphocyte | 55.4 | 65.8 (MMC) | ? (Wien) | ? (Wien) | IB of LCL | c.3707G→A (aberrant splicing) p.H1229EfsX7 | c.2835dupC p.D947RfsX3 | None (residual protein) |
| 12 | 11/I | Lymphocyte | 40.2 | 61.1 (MMC) | ND | ND | IB of LCL | c.274-57_–56insinvAlu Yb8nt36_319 +dup c.274–69_–57 p.I92YfsX7 | c.3453_3456delCAAA p.N1151KfsX46 | None (residual protein) |
| 13 | 12/I | Lymphocyte | 27.6 | 57.8 | .11 | 1.08 (MMC)/2.9 (DEB) | IB of LCL | c.1948-16T→G (exon 22 skipping) p.E650X | c.1948-16T→G (exon 22 skipping) p.E650X | None (residual protein in T cells and LCL) |
| 14 | 13/I | Lymphocyte fibroblast | 20.9 | 32.1 (MMC) | ND | ND | Mutation analysis (by sibling) | c.1948-6C→A (exon 22 skipping) p.E650X | 2775_2776CC→TT p.R926X | 2775_2776 CC (blood, LCL) |
| 15 | 13/II | Lymphocyte fibroblast | 25.0 | 35.4 (MMC) 69.2 (MMC) | 0 | .11 (MMC)/.02 (DEB) | RC of fibroblasts | c.1948-6C→A (exon 22 skipping) p.E650X | 2775_2776CC→TT p.R926X | Recombination |
| 16 | 14/I | Lymphocyte | ND | ND | .04 | 1.78 (DEB) | IB of LCL | c.2444G→A p.R815Q | c.2444G→A p.R815Q | None (residual protein) |
| 17 | 14/II | Lymphocyte | ND | ND | .12 | 3.1 (DEB) | Mutation analysis (by sibling) | c.2444G→A p.R815Q | c.2444G→A p.R815Q | None (no LCL) |
| 18 | 15/I | Lymphocyte | ND | ND | .12 | 1.5 (DEB) | IB of LCL | c.696-2A→T (exon 10 skipping) p.S232RfsX6 | c.1321_1322delAG (aberrant splicing) p.V379_K515del | None (residual protein) |
| 19 | 16/I | Fetal blood | ND | ND | ND | 3.7 (DEB) | RC of fetal fibroblasts | c.692T→Gpat p.L231R | c.2444G→Amat p.R815Q | Not done |
| 20 | 17/I | Lymphocyte | ND | ND | .02 | 8.4 (DEB) | IB and RC of LCL | c.1948–6C→Amat, (exon 22 skipping) p.E650X | 2660delApat p.E888RfsX16 | None (residual protein) |
| 21 | 18/I | Lymphocyte | ND | ND | .02 | 5.4 (DEB)10.3 (DEB) | IB of LCL | c.2404C→T p.Q802X | c.2444G→A p.R815Q | None (residual protein) |
| 22 | 19/I | Lymphocyte | ND | ND | .04 | 3.7 (DEB) | RC of fetal fibroblasts from 880/2 (early spontaneous abortion) | c.2444G→Apat p.R815Q | c.2715+1G→Amat (aberrant splicing) p.E906LfsX4 | None (residual protein) |
| 23 | 20/I | Lymphocyte | ND | ND | .08 | 7.4 (DEB) | IB of LCL | c.757C→T p.R253X | c.1367T→G p.L456R | None (residual protein) |
| 24 | 20/II | Lymphocyte | ND | ND | .20 | 8.9 (DEB) | IB of LCL | c.757C→T p.R253X | c.1367T→G p.L456R | None (residual protein) |
| 25 | 21 | Lymphocyte | ND | ND | Data missing | Data missing | Mutation analysis | c.1948–16T→G (exon 22 skipping) p.E650X | c.1948–16T→G (exon 22 skipping) p.E650X | 1953G→T (W651C) (blood, LCL) |
| 26 | 22/I | Fibroblast | 22.2 (fibroblast) | 54 (fibroblast, 300 nM ∼ 100 ng/ml MMC) | .04 | .16 (300 nM ∼ 100 ng/ml MMC) | Mutation analysis in fibroblasts (by sibling) | c.376A→G (aberrant splicing) p.S126RfsX12 | c.3803G→A p.W1268X | 376A (blood) |
| 27 | 22/II | Lymphocyte | ND | ND | .12 | >10 (MMC) | IB and IP of LCL | c.376A→G (aberrant splicing) p.S126RfsX12 | c.3803G→A p.W1268X | None (residual protein) |
| 28 | 23/I | Lymphocyte | ND | ND | .10 | 6.0 (MMC) | IB, IP and RC of LCL | c.206–2A→T (exon 4 skipping) p.A69DfsX7 | g.22875_23333del459 (c.1414–71_c.1545+256del459) p.E472_K515del | None (residual protein) |
| 29 | 23/II | Lymphocyte | ND | ND | .12 | 8.1 (MMC) | Mutation analysis (by sibling) | c.206-2A→T (exon 4 skipping) p.A69DfsX7 | g.22875_23333del459 (c.1414–71_c.1545+256del459) p.E472_K515del | None (residual protein) |
Note.— DEB=diepoxybutane; RC=retroviral complementation; IB=immunoblotting; IP=immunoprecipitation; ND=not determined; G2=G2 phase fraction of the cell cycle; GF=growth fraction; G2/GF=ratio G2 phase fraction over GF.
In the North American IFAR collection, of 630 classified patients with FA, 7 (patients 19–25) were fully informative clinically and were included in the present cohort; another (patient 32) is among the four additional patients. Results of immunoblotting data alone suggested that 10 more IFAR patients belonged to the FA-D2 group, for a maximum estimate of 18 FA-D2 subjects in the IFAR group of 630. Among the patients referred to the two German laboratories, 15 of 243 patients with FA were assigned to complementation group FA-D2. The latter figures suggest that the proportion of patients who meet criteria for complementation group D2 may be higher than reported elsewhere.2,24,25
Clinical Data for FA-D2 Patients
Including, where appropriate, information from a prenatal case (number 19), phenotypic details in the present cohort of 29 FA-D2 patients with adequate clinical information are shown in tables 8 and 9. With the exception of two families (those of patients 19 and 22), there was no general tendency for increased rates of spontaneous abortions in this study. Among the 28 fully informative FA-D2 patients, there was only a single malignancy observed (acute myelogenous leukemia [AML] in patient 1), and there was no apparent overrepresentation of malignancies in the parents or grandparents of the FA-D2 patients in our cohort.
Table 8. .
Clinical Phenotype of the Cohort of FA-D2 Patients
| Malformation | No. of Affected/Totala |
Frequency (%) |
| Symptom: | ||
| Microcephaly | 25/28 | 89 |
| (Intrauterine) growth retardation | 25/29 | 86 |
| Anomalies of skin pigmentation | 21/28 | 75 |
| Radial-ray defects | 21/29 | 72 |
| Microphthalmia | 17/28 | 61 |
| Renal anomalies | 10/28 | 36 |
| Malformations of the external ear | 9/28 | 32 |
| Brain anomalies | 9/29 | 31 |
| Including hydrocephalus | 5/29 | 17 |
| Psychomotor retardationb | 8/28 | 29 |
| Hypogonadism/genital anomalies | 7/28 | 25 |
| Hip dysplasia/dislocation | 6/28 | 21 |
| Heart anomalies | 4/28 | 14 |
| Malformations of the gastrointestinal tract | 4/28 | 14 |
| Distinct syndromic association: | ||
| Michelin-tire baby syndrome (MIM 156610) | 2/28 | 7 |
| VACTERL-like association (MIM 192350) | 1/28 | 4 |
| Holoprosencephaly (MIM 236100) | 1/28 | 4 |
| Kartagener syndrome (MIM 244400) | 1/28 | 4 |
A prenatal case (number 19) was partially informative. Pertinent information was used where applicable.
Including attention-deficit/hyperactivity disorder.
Table 9. .
Clinical Diagnostic Data for the Cohort of 29 FA-D2 Patients[Note]
| Patient Number | Kindred/Sibling | Consanguinity | Sex | Ethnicity/Nationality | Age at Diagnosis | Clinical Presentation | Hematologic Manifestation(s) | Survival at Last Follow-Up | Family History |
| 1 | 1/I | Unknown | F | Asian Indian | 6 mo | IUGR, patent ductus arteriosus, pigmentation anomalies, microcephaly, low-set ears, hypoplastic thumb with duplicate nail (R), radial-ray aplasia with cutaneous thumb (L), pelvic kidney (R), congenital hip dislocation (L), and aplasia of the corpus callosum | BMF as of age 2 years and 4 mo, transfusions from age 3 years and 2 mo, AML at age 7.0 years | Deceased at age 7 years and 6 mo (AML, pneumonia) | No SABs; no known cancer |
| 2 | 2/I | Absent | F | While German | 5 Years and 7 mo | GR, pigmentation anomalies, microcephaly, microphtalmia, low-set thumbs, duplicate kidney (R), dysplastic hips | BMF as of age 5 years and 7 mo, cortisol from age 8 years, transfusions from age 8 years and 4 mo, androgen from age 9 years and 2 mo | Deceased at age 11 years and 4 mo (subarachnoidic hemorrhage) | 1 SAB; MGM: cervical cancer at age 40 years |
| 3 | 3/I | Cousins of 1st° | M | White Turk | 1 Years and 11 mo | IUGR, pigmentation anomalies, microcephaly, hypoplastic thumbs (L>R), syndactyly II/III toes, hypogenitalism, and glomerulosclerosis | Stable partial mosaicism, BMF as of age 11 years, cortisol and androgen from age 12 years, transfusions from age 18 years and 9 mo, and BMT at age 19 years and 7 mo | Deceased at age 20 years and 7 mo (viral encephalitis after BMT) | No SABs, no known cancer |
| 4 | 4/I | Cousins of 2nd° | F | White Turk | 5 years and 10 mo | IUGR, pigmentation anomalies, microcephaly, microphtalmia, hypoplastic thumb (R), hydocephalus internus, hypoplastic corpus callosum, mental retardation, and hyperactivity/attention-deficit disorder | BMF as of age 2 years and 6 mo, transfusions from age 2 years and 6 mo, subdural hemorrhage at age 6 years, and BMT at 7 years | 8 years and 3 mo | No SABs; no known cancer |
| 5 | 4/II | Cousins of 2nd° | M | White Turk | 4 Years and 5 mo | IUGR, microcephaly, microphtalmia, strabismus, mental retardation, and hyperactivity/attention-deficit disorder | BMF as of age 3 years and 3 mo, transfusions from age 3 years and 3 mo, and oxymetholon from age 5 years and 9 mo | 6 Years and 11 mo | No SABs; no known cancer |
| 6 | 5/I | Absent | M | White German | 2 Years and 6 mo | GR, microcephaly, microphtalmia, absent antihelix (R), radial-ray hypoplasia, preaxial hexadactyly (R), duplicate pelvic kidney (R), maldescensus of the testes, micropenis, dysplastic hips, hypoplastic corpus callosum, misshaped brain ventricles, and psychomotor retardation | BMF as of age 2 years and 9 mo, and BMT at age 3 years and 3 mo. | 4 Years and 4 mo | No SABs; PGM cancer at age 70 years; otherwise, no cancer history |
| 7 | 6/I | Absent | F | White Italian | 2 Years | IUGR, pigmentation anomalies, microcephaly, microphtalmia, absent thumbs, short radii, absent antihelix (R), and closed auditory canals | BMF as of age 4.5 Years | 12 Years | No SABs; no cancer history |
| 8 | 7/I | Absent | M | White German | 3 Years and 9 mo | Pigmentation anomalies, microcephaly, “flat” auricles-absent antihelix?, ptosis, short thumbs, and hyperactivity/attention-deficit disorder | BMF as of age 4 years | 4 Years and 4 mo | No SABs; no cancer history |
| 9 | 8/I | Absent | F | White Czech | 2 Years and 11 mo | IUGR, microcephaly, brain atrophy, patent ductus arteriosus, esophagus atresia, tracheoesophageal fistula (IIIb), hypoplastic kidneys, polycystic ovary (L), triphangeal digitalized thumbs, pedes equinovari, and rib anomaly (VACTERL-like association) | BMF as of age 2 years and 10 mo and transfusions from age 2 years and 11 mo | Deceased at age 5 years and 10 mo (hemorrhage) | 1 SAB (first trimester); no cancer in the family |
| 10 | 9/I | Absent | F | White Turk | 7 mo | IUGR, pigmentation anomalies, microcephaly, hydrocephalus internus, absent corpus callosum, microphtalmia, small mouth, low-set ears, hypoplastic thumbs, unilateral triphalangeal (R), pelvic kidney (L), hip luxation, and psychomotor retardation | BMF as of age 2 years | 2 years and 3 mo | 1 SAB (first trimester); 1 pregnancy terminated because of hydrocephalus and renal agenesis; PGF bronchus cancer |
| 11 | 10/I | Absent | F | White Austrian | 10 Years and 10 mo | IUGR, pigmentation anomalies, microcephaly, hypoplastic thumbs, and ectopic kidney (R) | BMF as of age 10 years and 10 mo, transfusions from age 10 years and 10 mo, MDS (RAEB-t) with del(7)(q32) at age 10 years and 10 mo, BMT at age 11 years and 1 mo | 11 Years and 11 mo | No SABs; no cancer history |
| 12 | 11/I | Absent | M | White Dane | 3 Mo | IUGR, atresia of the duodenum, microcephaly, dilated lateral ventricles and stenosis of the aquaeduct (hydrocephalus), hypoplasia of the corpus callosum, microphtalmia, closed auditory channels, hypoplastic thumbs, and micropenis | BMF as of age 2 wk | 5 Mo | No SABs; PGM and MPGM breast cancer, MGGF prostate cancer |
| 13 | 12/I | Cousins of 1st° | M | White Turk | 5 Years and 5 mo | IUGR, pigmentation anomalies, microcephaly, microphalmia, psychomotor retardation, and Michelin-tire baby syndrome | BMF as of age 1 years and 5 mo | 5 Years and 8 mo | No SABs; no cancer history |
| 14 | 13/I | Absent | F | White German | 34 Years and 2 mo | IUGR, microcephaly, and mild radial-ray hypoplasia | None | 34 Years | No SABs; no cancer history |
| 15 | 13/II | Absent | F | White German | 21 Years and 11 mo | IUGR, microcephaly, radial -ray hypoplasia, dysplasia of mandibula, anomalies of the teeth, dysplasia of hip (R), and mental retardation | Transfusions from age 17 years and 6 mo, MDS (RARS-RAEB) at age 17 years and 6 mo | Deceased at age 23 years and 5 mo (pneumonia, invasive aspergillosis, and hemorrhage) | No SABs; no cancer history |
| 16 | 14/I | Cousins of 3° | M | White Spanish | 6 Years | Patent ductus arteriosus, pigmentation anomalies, bifid thumb (R), and hypogonadism | BMF as of age 7 years (very mild hypoplasia of the myeloid series) | 25 Years | No SABs; MGF lung, and PGF stomach cancer |
| 17 | 14/II | Cousins of 3° | M | White Spanish | 8 Mo | Pigmentation anomalies, microphtalmia, hypoplastic thumb (R), absent os metacarpale I (L), and glandular hypospadia | Blood cell counts at low-range normal levels | 20 Years | No SABs; MGF lung, and PGF stomach cancer |
| 18 | 15/I | Absent | F | White Spanish | 5 Years and 3 mo | IUGR, pigmentation anomalies, microcephaly, microphtalmia, hypotelorism, and annular pancreas | BMF as of age 5 years and 3 mo, androgen, G-SCF, EPO and transfusions from age 5 years and 3 mo, BMT at age 10 years and 11 mo | Deceased at age 11 years and 1 mo (graft failure/did not take) | No SABs; MMGM colon cancer |
| 19 | 16/I | Absent | M | Caucasian, maternal Irish and English, paternal Irish and Italian | 22 Wk of gestation | IUGR, absent thumb and radial aplasia (R), and lateral cerebral ventricular dilation (hydrocephalus) | NA | NA, terminated with diagnosis of FA | 3 First-trimester SABs, 4th fetus with IUGR, radial aplasia, cystic hygromas, encephalocele, probably heart defects, terminated; MGM pancreas, MMGM breast, MGF melanoma and basal cell cancer |
| 20 | 17/I | Absent | M | White maternal German, paternal Dutch | 4 Years and 5 mo | IUGR, pigmentation anomalies, microcephaly, and microphtalmia | BMF as of age 2 years, BMT at age 5 years | 9 Years (4 years post BMT) | 1 SAB, M 3× basal cell, MMMGM melanoma, MMGF breast, PGM bowel cancer |
| 21 | 18/I | Absent | M | White Hispanic (Mexican) | 7 Mo | IUGR, café-au-lait spots, microcephaly, microphtalmia, hearing loss (auditory canals? sensory hearing impairment?), absent thumbs and radii, intestinal atresia, renal defects, genital abnormalities (undescended testes), and learning disabilities | None yet | 10 Years and 3 mo | No SAB; one cancer |
| 22 | 19/I | Absent | M | White maternal Irish, Dutch Yugoslavian French and Native American, paternal Irish and Sicilian | Newborn | Patent ductus arteriosus, pigmentation anomalies, low-set ears, malformed auricle (R), constriction bands of mid forearms (Michelin-tire baby syndrome?), preaxial hexadactyly (R), and hypoplastic thumb with ponce flottant (L) | BMF as of age 1 year and 4 mo | 2 Years and 6 mo | 5 Miscarriages with one positive for FA; PGM breast, MPGM cervix and lung cancer, MMGM brain tumor |
| 23 | 20/I | Absent | M | Maternal African American/Caucasian, paternal African American | (1 Mo) | IUGR, microcephaly, microphtalmia, hypoplastic thumb (L), hypoplastic metacarpal I (R), and horseshoe kidney | None | 1 Years and 4 mo | GM: 2 miscarriages; cancer only in great-GP generation |
| 24 | 20/II | Absent | F | Maternal African American/Caucasian, paternal African American | 4 Years and 6 mo | IUGR, café-au-lait spots, microcephaly, and microphtalmia | BMF starting from age 4 years and 6 mo | 5 Years and 9 mo | GM: 2 miscarriages; cancer only in great-GP generation |
| 25 | 21/I | Cousins of 1st° | M | White Turk | 5 Years and 5 mo | IUGR, pigmentation anomalies, microcephaly, microphtalmia, and pelvic kidney | BMF starting from age 4 years and oxymetholone and prednisone from age 5 years and 5 mo | Deceased at age 9 years (intracranial hemorrhage) | No SAB; no cancer history |
| 26 | 22/I | Absent | F | White Dutch | 5 Years | IUGR (asymmetrical); pigmentation anomalies; microcephaly; ventriculomegaly (hydrocephalus) and multiple developmental anomalies of the brain, possibly holoprosencephaly; hypotelorism; microphtalmia; narrow auditory canals; hypoplastic os metacarpale I; renal aplasia (R); dysplasia of the hip (L); and growth-hormone deficiency | BMF as of age 5 years, transfusions from age 6 years and 9 mo, and GCSF and BMT at age 7 years and 10 mo | 9 Years | No SAB; no cancer history |
| 27 | 22/II | Absent | M | White Dutch | 3 Years | IUGR, pigmentation anomalies, microcephaly, hypoplastic corpus callosum; hypertelorism, blepharophimosis, and preaxial hexadactyly (L) | BMF as of age 2 years and 1 mo, transfusions from age 5 years and 8 mo; BMT at age 7 years and 7 mo | 7 Years and 9 mo | No SAB; no cancer history |
| 28 | 23/I | Absent | M | White Dutch | 8 Years and 6 mo | IUGR, pigmentation anomalies, microcephaly, Kartagener syndrome with situs inversus, and mild mental retardation | BMF as of age 8 years and 5 mo, no transfusions, BMT at age 9 years and 5 mo | Deceased at age 10 years and 1 mo (gastrointestinal hemorrhage due to necrotizing enterocolitis post BMT) | 1 SAB; no cancer history |
| 29 | 23/II | Absent | M | White Dutch | 5 Years and 8 mo | IUGR, pigmentation anomalies, microcephaly, and microphtalmia | BMF as of age 5 years and 8 mo and BMT at age 6 years and 7 mo | 7 Years and 10 mo | 1 SAB; no cancer history |
Note.— L=left; R=right; GR=growth retardation; IUGR=intrauterine growth retardation; BMT=bone-marrow transplantation; MDS=myelodysplastic syndrome; SAB=spontaneous abortion; MGM=maternal grandmother; PGF=paternal grandfather; MPGM=maternal and paternal grandmother; MGGF=maternal greatgrandfather; MGF=maternal grandfather; MMGM=mother of the maternal grandmother; MMGF=mother of the maternal grandfather; MMMGM=mother of the maternal greatgrandmother; GP=grandparent.
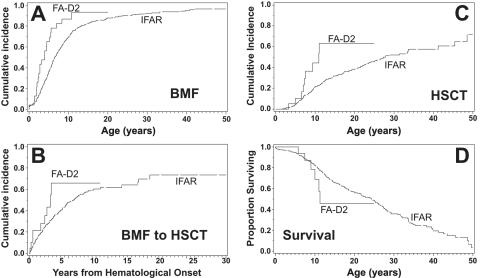
The median age at diagnosis of these FA-D2 patients was 4 years and 5 mo (n=29). When the fetal case (number 19) and five mosaic patients (patients 3, 14, 15, 25, and 26) were excluded, the median age at transfusion dependency was 10 years and 10 mo (n=23). Figure 3 compares the progressive hematological course and the outcome of our group of FA-D2 patients with that reported elsewhere for 754 North American IFAR patients.42 BMF in our D2 group (n=23) occurred at an earlier age (median for FA-D2 patients was 2.4 years vs. 6 years and 7 mo for IFAR; P=.001) (fig. 3A), and the period from BMF to HSCT was shorter (median age at HSCT for FA-D2 patients [n=9] was 5 years and 6 mo vs. 11 years and 4 mo for IFAR [n=218]; P<.08) (fig. 3B). Age at HSCT for our FA-D2 patients was earlier than in the IFAR patients of combined groups (median age for FA-D2 patients was 10 years and 11 mo vs. 27 years and 11 mo for IFAR; P<.01) (fig. 3C). Of 23 FA-D2 patients, 9 patients of our cohort received HSCT. Kaplan-Meier estimates (fig. 3D) suggest that our FA-D2 patients (n=23) may have a shorter life span than the IFAR patients overall, since their survival curve falls below that of the IFAR patients after age 9 years; however, the difference of median survival (11 years and 4 mo for FA-D2 patients vs. 24 years and 3 mo for IFAR) was not significant, because the number of nonmosaic FA-D2 patients (two) of our cohort who reached adulthood was too low for statistical support.
Figure 3. .
Clinical course of 23 fully informative, nonmosaic FA-D2 patients in this study. A, The cumulative incidence of BMF of the FA-D2 patients in the present study (FA-D2) precedes that of all patients with FA in the IFAR42 (P=.001). B, The period from BMF to HSCT, which was shorter in the patients of the present study than in those of the IFAR42 (trending, P<.08). C, Cumulative incidence of HSCT of the FA-D2 patients in our study, which likewise antedates that of all patients in the IFAR42 (P<.01). D, Kaplan-Meier curves of survival, which suggest higher death rates of the FA-D2 patients than of all patients in the IFAR aged >10 years.
FANCD2 and the FANCD2 Pseudogenes
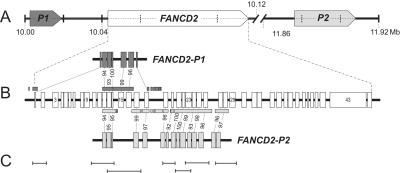
BLAT searches (Human BLAT Search) identified two pseudogene regions: FANCD2-P1 spanning 16 kb, located ∼24 kb upstream of FANCD2, and FANCD2-P2 spanning 31.9 kb, located ∼1.76 Mb downstream of FANCD2 (fig. 4A). P1 and P2 are in the same orientation as the functional gene. They are characterized by high sequence homology with certain FANCD2 exons and have retained ordered succession of their exon/intron equivalents, compared with the functional gene. On the other hand, the exon replicas of FANCD2-P1 and FANCD2-P2 have acquired numerous deletions and insertions. Striking sequence similarity of the D2 pseudogenes extends into some FANCD2 introns, particularly in the regions of IVS21-IVS26. Thus, P1 and P2 reveal recognizable patterns of conserved gene structure (fig. 4B). FANCD2-P1 is roughly a copy of the front portion of FANCD2, including, with intermittent gaps, the region of exons 1–18 (homology with FANCD2 exons 1 and 12–16 and the 3′ portion of exon 18). The region upstream of FANCD2-P1 shares homology with the putative FANCD2 promoter predicted within the CpG-rich region of ∼800 bp upstream of the start codon of the functional gene.60 The corresponding region upstream of P1 is interrupted by an AluY element (RepeatMasker). FANCD2-P2 is an approximate match of the middle portion of FANCD2. Including gaps, it spans the region of exons 12–28 (homology with FANCD2 exons 12–14 and 17–28).
Figure 4. .
Topography of FANCD2, its pseudogenes, and the superamplicons. A, The two pseudogenes—FANCD2-P1 and FANCD2-P2—located upstream and downstream, respectively, of the functional FANCD2 gene. All three have the same orientation. The scale denotes Mb on chromosome 3. B, FANCD2 exons and their pseudogene equivalents, connected by dashed lines, indicating percentages of nucleotide identity. Homology also extends into many introns nearby, as indicated by the boxes beyond and below the active gene. C, Graphic presentation of the positions and sizes of 7 superamplicons relative to the active gene shown in panel B. These amplicons represent FANCD2 exon-exon or exon-intron regions. Unique primer-binding sites ensure specific amplification.
Mutations in FANCD2
Unique amplification of the functional FANCD2 gene was achieved using primers that exclude pseudogene sequences. In FANCD2 regions sharing extensive homology with FANCD2-P1 and -P2, seven superamplicons (fig. 4C) were used for genomic mutation screens. Studies at the RNA level were implemented to guide the genomic analyses. All identified mutations and their predicted consequences at the protein level are compiled in table 10. The distribution of the mutations among the individual patients is shown in table 7.
Table 10. .
Identified FANCD2 Mutations and Their Effects
| Mutationa |
Consequencea |
||
| Location and Patient No(s). | Genomic DNA | RNA | Protein |
| Exon 2: | |||
| 32 | c.2T→C | r.2T→C | Failure of normal translation initiation |
| Intron 3: | |||
| 28 and 29 | c.206-2A→T (IVS3-2 A→T) | r.206_273del68 (exon 4 skipping) | p.A69DfsX7 |
| Intron 4: | |||
| 6, 12, and 30 | c.274-57_-56insinvAluYb8nt36_319 +dup c.274-69_−57b | r.274_377del104 (exon 5 skipping) | p.I92YfsX7 |
| Exon 5: | |||
| 26 and 27 | c.376A→G | r.376A→G+r.377_378ins13 (aberrant splicing) | p.S126RfsX12 |
| Exon 9: | |||
| 19 | c.692T→G | r.692T→G | p.L231R |
| Intron 9: | |||
| 1 | c.696-121C→G (IVS9-12 1C→G) | r.695+1619_696-126ins34 (exonization) | p.S232insQNNFX |
| 18 | c.696-2A→T (IVS9-2 A→T) | r.696_783del88 (exon 10 skipping) | p.S232RfsX6 |
| Exon 10: | |||
| 23 and 24 | c.757C→T | r.757C→T | p.R253X |
| 31 and 33 | c.782A→T | r.696_783del88 (exon 10 skipping) | p.S232RfsX6 |
| Exon 11: | |||
| 9 | c.810_812delGTC | r.810_812delGTC | p.S271del |
| Exon 12: | |||
| 7 | c.904C→T | r.904C→T | p.R302W |
| Intron 12: | |||
| 8 | c.990-1G→A (IVS12-1 G→A) | r.990del8 (aberrant splicing) | p.S330RfsX16 |
| Exon 13: | |||
| 7 | c.1092G→A | r.1092G→A | p.W364X |
| Intron 14: | |||
| 33 | g.13377_17458dup4082 (duplication, including exons 11–14) | r.784_1134dup (duplication of 351 nt in-frame) | p.262_378dup (duplication of 117 aa) |
| Exon 16: | |||
| 18 | c.1321_1322delAG | r.1135_1545del411 (exon 15–17 skipping) | p.V379_K515del |
| 23 and 24 | c.1367T→G | r.1367T→G | p.L456R |
| 31 | c.1370T→C | r.1370T→C | p.L457P |
| Exon 17: | |||
| 28, 29 | g.22875_23333del459 (c.1414-71_c.1545+256del459) | r.1414_1545del132 | p.E472_K515del |
| Intron 21: | |||
| 3, 4, 5, 9, 10, 13, and 25 | c.1948-16T→G (IVS21-16 T→G) | r.1948_2021del74 (exon 22 skipping) | p.E650X |
| 2, 8, 14, 15, and 20 | c.1948-6C→A (IVS21-6 C→A) | r.1948_2021del74 (exon 22 skipping) | p.E650X |
| Exon 26: | |||
| 21 | c.2404C→T | r.2404C→T | p.Q802X |
| 16, 17, 19, 21, 22, and 30 | c.2444G→A | r.2444G→A | p.R815Q |
| Exon 28: | |||
| 20 | c.2660delA | r.2660delA | p.E888RfsX16 |
| Intron 28: | |||
| 10 and 22 | c.2715+1G→A (IVS28+1G→A) | r.2715_2716ins27 (aberrant splicing) | p.E906LfsX4 |
| Exon 29: | |||
| 14 and 15 | c. 2775_2776CC→TT | r. 2775_2776CC→TT | p.R926X |
| 11 | c.2835dupC | r.2835dupC | p.D947RfsX3 |
| Exon 34: | |||
| 12 | c.3453_3456delCAAA | r.3453_3456delCAAA | p.N1151KfsX46 |
| Exon 36: | |||
| 2 | c.3599delT | r.3599delT | p.I1200KfsX12 |
| Exon 37: | |||
| 32 | c.3706C→A | r.3684_3707del24 (aberrant splicing) | p.R1228S_F1235del |
| 11 | c.3707G→A | r.3684_3727del44 (aberrant splicing) | p.H1229EfsX7 |
| Exon 38: | |||
| 6, 26, and 27 | c.3803G→A | r.3803G→A | p.W1268X |
Nomenclature is according to the Human Genome Variation Society.
This Alu was identical to the evolutionary young subfamily Yb8.43,44 It was lacking its annotated nucleotides 1–35, had integrated in reverse orientation (with its poly-A tail toward the 5′ end of FANCD2), and had duplicated the 13-nt sequence c.274–69 to c.274–57 of FANCD2 IVS4, such that this duplicated sequence flanked the Alu repeat on both sides. Altogether, the insertion length was 298 bp.
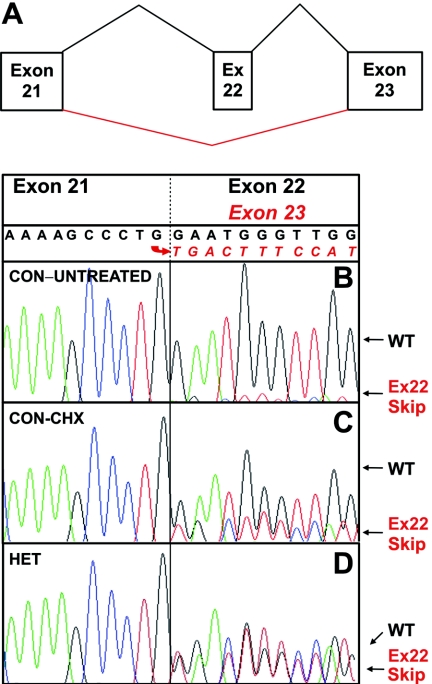
Mutations Affecting Pre-mRNA Splicing
In peripheral-blood lymphocytes (PBLs), LCLs, and cultured fibroblasts from unaffected controls, two species of FANCD2 cDNAs were consistently detected by sequence analysis of the regions corresponding to exon 22 (fig. 5A and 5B) and exons 15–17 (data not shown). This observation suggests low-level skipping of these exons, consistent with FANCD2 RNA being subject to alternative splicing. mRNA stabilization via CHX treatments of cultured cells resulted in a relative increase of the alternatively spliced mRNA species (fig. 5A and 5C), implying instability of the alternatively spliced FANCD2 mRNAs. On genomic sequencing, alternative splicing of exon 22 was not associated with any mutation or variant in exon 22 or the adjacent intronic regions in 25 controls.
Figure 5. .
Exon 22 splicing. A, Schematic depiction of the splicing patterns resulting from exon 22 retention or skipping. B, cDNA sequencing in an LCL from a normal control (CON), showing predominance of exon 22 sequence following that of exon 21 but also low levels of underlying sequence readable as exon 23. C, Treatment of the same LCL from a normal control with CHX for 4 h before cDNA synthesis, which increases the relative level of sequence with exon 22 skipping. D, cDNA sequencing in an LCL from a compound heterozygote (HET) for splice-acceptor mutation in intron 21, c.1948-16T→G (patient 9), which shows comparable levels of inclusion and exclusion of exon 22 sequence following that of exon 21. Ex=exon; WT=wild type.
Without CHX treatments, cell lines from patients 2, 8, 9, 10, 14, 15, and 20 in our cohort displayed almost equal levels of exon 22 skipping and retention (fig. 5A and 5D). Patients 9 and 10, with balanced levels of exon 22 skipping and retention, were compound heterozygous carriers of the mutation c.1948-16T→G. A different base substitution preceding exon 22, c.1948-6C→A, was present on one allele of the compound-heterozygous patients 2, 8, 14, 15, and 20, likewise resulting in similar levels of exon 22 skipping and retention. Both mutations—c.1948-16T→G and 1948-6C→A—are predicted to disrupt the splice-acceptor recognition in intron 21 suggested by impaired scores of the 3′ splice site relative to the wild type (see, e.g., table 11). Both result in skipping of exon 22.
Table 11. .
FANCD2 3′ Splice-Acceptor Calculations
| Exon/Intron and Designation |
Sequence | MaxEntScan Scorea |
| 4: | ||
| Consensus | ctcttcttttttctgcatagCTG | 9.12 |
| c.206-2A→T | ctcttcttttttctgcattgCTG | .76 |
| 10: | ||
| Consensus | tctttttctaccattcacagTGA | 7.39 |
| c.696-2A→T | tctttttctaccattcactgTGA | −.97 |
| 13: | ||
| Consensus | ttcctctctgctacttgtagTTC | 6.19 |
| c.990-1G→A | ttcctctctgctacttgtatTTC | −2.56 |
| 22: | ||
| Consensus | tgtttgtttgcttcctgaagGAA | 6.43 |
| c.1948-16T→G | tgttggtttgcttcctgaagGAA | 5.58 |
| c.1948-6C→A | tgtttgtttgcttcatgaagGAA | 4.51 |
| 37: | ||
| Consensus | ACTTTTGTTGTTTTCTTCCGTGT | 2.10 |
| c.3706C→A | ACTTTTGTTGTTTTCTTCAGTGT | 10.14 |
MaxEntScan::score3ss for human splice sites.
Three apparently unrelated patients (patients 6, 12, and 30) showed balanced levels of skipping and retention of exon 5 due to heterozygous insertional mutagenesis by an Alu element between positions c.274–57 and c.274–56 into an AT-rich target sequence in IVS4. Integration site, type, length, and orientation of the Alu and the duplicated FANCD2 intron sequence were identical in all three patients. We did not detect any such Alu insertions in 300 unaffected control alleles.
Aberrant splicing of exons 4, 5, 10, 13, 15–17, 28, and 37 was observed in other patients also (analysis of the regulatory splec sequences by ESEfinder and Rescue-ESE). Patients 28 and 29 showed skipping of exon 4 caused by a base substitution in the preceding canonical splice-acceptor site (c.206-2A→T). Patients 26 and 27 had a base substitution in exon 5 (c.376A→G) abrogating the downstream splice donor. This change led to the inclusion of 13 bp of IVS5 into the transcript by activating a cryptic 5′ splice site in intron 5 (r.377_378ins13) also reported elsewhere.26 Patient 18 showed skipping of exon 10 due a base substitution in the upstream splice acceptor (c.696-2A→T). Exon 10 skipping was observed in patient 31, who had a substitution of the second-to-the-last base of exon 10 (c.782A→T). In patient 8, we detected a splice-acceptor mutation upstream of exon 13 (c.990-1G→A). This change results in the activation of a cryptic splice acceptor 8 bp downstream and the exclusion of the corresponding sequence from the mature mRNA. A 2-bp deletion in exon 16 (c.1321_1322delAG) in patient 18 causes skipping of exons 15–17. In that case, aberrant splicing occurs in the same position as low-grade alternative splicing in normal controls, but at heterozygous levels. Patients 10 and 22 showed inclusion of a 27-bp sequence of intron 28 into mRNA because of a splice-donor mutation (c.2715+1G→A) and the use of a cryptic splice-donor downstream. Patient 11 had a base substitution in exon 37 (c.3707G→A), reported elsewhere,26 that abrogates the normal splice acceptor 25 bp upstream and activates a cryptic site 19 bp downstream of the mutation, resulting in skipping of 44 bp. Interestingly, an adjacent base substitution (c.3706C→A) in patient 32 generates a new splice acceptor that is used instead of the normal one 23 bp upstream, leading to skipping of the 24 nt in between. With the exception of those of patient 1, all of these splicing aberrations were due to heterozygous mutations. Patient 1 showed homozygous exonization of an IVS9 fragment because of a mutation in intron 9 (c.696-121C→G), which activates cryptic splice sites. Predicted scores and consequences of some of these splice mutations are computed in tables 11 and 12. Apart from 1321_1322delAG, which causes skipping of exons 15–17, all mutations affecting splicing in the patients in this study result in frameshifts and subsequent premature termination of translation. More than half—that is, 30 of 58 mutated alleles of the 29 clinically informative FA-D2 patients, or 34 of all 66 alleles—were splicing mutations, which makes that the most prevalent type of mutation.
Table 12. .
FANCD2 Splice-Donor Calculations
| Exon/Intron and Designation |
Sequence | Score (Splicefinder) | Difference | Result |
| 5: | ||||
| Consensus | CAGgtgtggag | LC4 12||3|2| | ||
| c.376A→G | CGGgtgtggag | LC2 2|8||3|2 | Large | Malfunction |
| Cryptic splice donor | GAGgcatggaaa | HC1 |12|3|2| | Gain of function | |
| 9: | ||||
| Consensus | acggtaactta | LC4 ||12|2|| | ||
| c.696-121C→G | ACGgtaagtta | HC3 ||17|| | Large | Gain of function |
| 10: | ||||
| Consensus | AAGgtagaaaa | LC4 |12|||2| | ||
| c.782A→T | ATGgtagaaaa | LC3 ||10|||2| | Small | Malfunction |
| 28: | ||||
| Consensus | AAGgtattgga | LC4 |12||||| | ||
| c.2715+1G→A | AAGatattgga | No score | Large | Malfunction |
| Cryptic splice donor | AAGgtttgtgaa | LC4 |10||5|| | Gain of function |
Mutation-activated cryptic splice donor; calculation available only for the consensus dinucleotide gt.
Other Mutations
There were five different heterozygous nonsense mutations in nine patients from six families (c.757C→T in siblings 23 and 24, c.1092G→A in patient 7, c.2404C→T in patient 21, c.2775_2776CC→TT in siblings 14 and 15, and c.3803G→A in patient 6 and siblings 26 and 27) (tables 7 and 10). In addition, we detected five different missense mutations in 11 patients from nine families (c.692T→G in patient 19; c.904C→T in patient 7, identical to a mutation reported elsewhere26; c.1367T→G in siblings 23 and 24; c.1370T→C in patient 31; and c.2444G→A in siblings 16 and 17 and patients 19, 21, 22, and 30). These amino acid substitutions were classified as missense mutations because of their absence from unaffected controls, their absence from FA-D2 patients of our cohort with other biallelic mutations, and their occurrence at evolutionarily conserved residues (ClustalW; ExPASy). Missense mutations were either compound heterozygous in combination with other types of FANCD2 mutations or were homozygous in consanguineous families. Three unrelated patients had small deletions (c.2660delA in patient 20, c.3453_3456delCAAA in patient 12, and c.3599delT in patient 2) resulting in frameshifts. Another small deletion was in frame and affected a single codon (c.810_812delGTC in patient 9). There was only a single small frameshift duplication (c.2835dupC in patient 11). A large genomic deletion (g.22875_23333del459) spanning the entire exon 17 (similar to a mutation reported elsewhere without defined breakpoints26) and the adjacent 71 bp of intron 16 and 256 bp of intron 17 was found in sibling pair 28 and 29. This deletion resulted in a net loss of 41 aa. A large genomic duplication in patient 33 included exons 11–14 and resulted in the insertion of 132 aa. Both gross gene rearrangements retained the reading frame. In all of our patients, nonsense mutations, deletions, and insertions affected exclusively single alleles in combination with splice or missense mutations. A unique case was a compound heterozygous start codon mutation (c.2T→C) in patient 32.
Figure 6 illustrates the distribution of FANCD2 mutations that were identified in this study, including those of four FA-D2 patients who were reported elsewhere.26,27
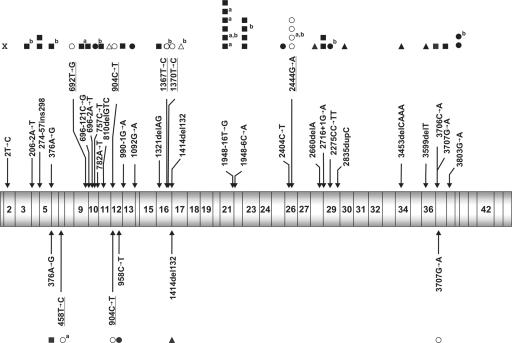
Figure 6. .
Positions and identity of mutations detected in FANCD2. Mutations identified in the present study are shown above, mutations reported elsewhere24,45 are indicated below the schematic display of FANCD2 cDNA. Blackened squares (▪) represent mutations resulting in aberrant splicing patterns, blackened circles (•) represent nonsense mutations, unblackened circles (○) represent missense mutations, blackened triangles (▴) represent frameshift deletions or duplications, and unblackened triangles (Δ) represent in-frame deletions or duplications. Missense mutations are depicted above or below the other mutations and are underlined. Superscript a at the right upper corner of a symbol denotes homozygous occurrence (2 alleles); superscript b denotes an affected sibling (relationship bias). Mutation 3707G→A was originally reported as a missense mutation,26 whereas we characterized it as a splicing mutation.
Ethnic Associations and Shared Alleles
Relatively severe birth defects and early hematological onset were observed in three patients (4, 5, and 13) who were homozygous for the splice mutation c.1948-16T→G with exon 22 skipping. These three patients and two other homozygotes with reverse mosaicism in the hematopoietic system (patients 3 and 25) were all from four consanguineous Turkish families. Of two other FA-D2 patients who were compound heterozygotes for this mutation, one was also of Turkish origin; the other came from the eastern Czech Republic. The splice mutation c.1948-6C→A, likewise leading to exon 22 skipping, was detected in five patients (2, 8, 14, 15, and 20), including two sisters (patients 14 and 15). These patients came from three families in northern Germany and an American family of German ancestry (patient 20). They presented with intermediate phenotypic and hematological severity. Relatively mild birth defects and a protracted hematological course into adulthood was observed in two siblings, from a consanguineous Spanish family (patients 16 and 17), with the homozygous missense substitution c.2444G→A. Of four compound heterozygotes for this mutation with mild disease manifestations, one had mixed ethnicity (patient 19), one was Hispanic American (patient 21), one had Sicilian ancestry (patient 22), and another had Spanish and Portuguese ancestry (patient 30). An allele with the insertion of an AluYb8 element was found in compound heterozygous patients of German (patient 6), Danish (patient 12), and Spanish/Portuguese (patient 30) descent. No other mutations occurred in more than two families.
On haplotype analysis using markers flanking FANCD2, all patients homozygous for the intron 21 mutation detected in the Turkish population were homozygous for markers D3S1597, D3S1938, D3S3611, and D3S1675. The resulting haplotype was shared, in a heterozygous state, with the nonconsanguineous compound heterozygous Turkish patient (patient 10). The Czech patient (patient 9) with the same mutation had a different haplotype. Lack of homozygotes for the intron 21 mutation prevalent in the German population (c.1948-6C→A; patients 2, 8, 14, 15, and 20) and unavailability of patients’ parents precluded construction of a mutation-associated haplotype. However, all patients with this mutation had one or two identical marker(s) on at least one side of their mutated FANCD2 gene. This finding suggests that c.1948-6C→A is an old mutation, with erosion of an ancient haplotype. The consanguineous siblings (patients 16 and 17) homozygous for the mutation prevalent in Spanish or southern European populations (c.2444G→A) were also homozygous for the set of markers used. Of their common haplotype, the microsatellite markers adjacent to FANCD2 were shared with a Hispanic patient (patient 21), a patient with Sicilian ancestry (patient 22), and a patient of Spanish/Portuguese descent (patient 30), all compound heterozygotes for this mutation. Additional support for a conserved haplotype came from linkage disequilibrium. All of the patients homo- or heterozygous for the mutation c.2444G→A were also homo- or heterozygous for the polymorphism c.2702G→T (p.G901V). Sequence analysis of the parents indicated that both substitutions were on the same allele. A single patient (patient 19) with the mutation c.2444G→A shared neither the haplotype nor the polymorphism c.2702G→T. Apart from c.2702G→T, which was also observed without association with the mutation c.2444G→A, the only new FANCD2 polymorphisms detected in our study were c.3978C→T (synonymous base substitution, exon 41) and c.4478A→G (3′ UTR, according to GenBank accession number NM_033084, or intron 43, according to accession number AF340183). All others have been reported elsewhere.26 Despite clear ethnic association of the patients with the insertion of an AluYb8 element in intron 4, two of these patients (6 and 12) shared all of the four markers studied. Patient 30, with the same mutation, had retained a single identical marker adjacent to FANCD2. A base substitution in the Alu sequence, 260G→A, present in all three cases but in <10% of complete AluYb8 elements in the human genome (Human BLAT Search) further suggests that the Alu insertion goes back to a single event and is an ancient rather than a recurrent mutation.
Reverse Mosaicism
Among the 28 fully informative FA-D2 patients in this study (excluding fetal case number 19), five (patients 3, 14, 15, 25, and 26) developed reverse mosaicism in the hematopoietic system. Mosaic patients were recognized by the fact that they had levels of both FANCD2-S and -L in protein from LCLs that were comparable to those of unaffected controls (fig. 7A). They also had low chromosome breakage rates in blood and blood-derived LCLs (table 7), and they had lost the typical G2-phase arrest of their lymphocytes after exposure to MMC (fig. 7C). Nonetheless, these patients displayed the characteristic clinical FA phenotype. Their cultured fibroblasts failed to show either FANCD2 isoform on standard immunoblots (fig. 7B). They were sensitive to MMC, as indicated by elevated chromosome breakage and G2-phase arrest (fig. 7C). Molecular studies confirmed these findings. Two patients with heterozygous base substitutions in the coding sequence, resulting in a nonsense (patient 14) and a splice (patient 26) mutation, showed reversion to the respective wild-type bases in primary blood cells and LCLs. The mechanism of these reversions is not clear and could involve back mutation, recombination with loss of heterozygosity, or recombination with gene conversion. Intragenic mitotic crossover is the likely but not proven mechanism of mosaicism in the sibling of patient 14 (patient 15) who had retained her dinucleotide substitution in her peripheral-blood cells. Two patients (3 and 25) with the c.1948-16T→G splice mutation had different second-site mutations nearby. The compensatory mutation of patient 3 was c.1954G→A (p.V652I), detected in blood, bone marrow, and an LCL. The compensatory mutation of patient 25 was c.1953G→T (p.W651C), detected in blood. c.1954G→A restored exon 22 retention correctly. c.1953G→T cDNA was not available. Clinically, three of five mosaic patients (3, 14, and 15) in the present cohort experienced a mild or protracted hematological course. The other two of five patients (patients 25 and 26) had no apparent benefit from their mosaicism; one of them required relatively early HSCT, and the other died of intracranial hemorrhage (table 9). The rate of 17% mosaic FA-D2 patients in our study is within the 15%46 to 20%47 or 25%48 range reported for other complementation groups. With a rate comparable to FANCA, FANCD2 appears to be another FA gene particularly prone to reverse mosaicism.
Figure 7. .
Reverse mosaicism. Blood-derived cells from FA-D2 patients with reverse mosaicism of the hematopoietic system (patients 3 and 26, LCLs; patient 14, stimulated PBL; panel A, lanes 2, 3, and 4) reveal both FANCD2 bands at levels similar to a random normal control (lane 1) after exposure to MMC. In contrast, neither FANCD2 band was present in fibroblasts from the same patients, but only in the control (B). RAD50 was used as loading control in panels A and B. The LCLs and PBL used in panel A fail to show G2-phase arrest on flow cytometric cell cycle distributions in response to MMC (panel C) (black histograms indicate DAPI stain; control (CON), 8.0% G2; patient 3, 8.8% G2; patient 14, 8.8% G2; patient 26, 10.4% G2), whereas the corresponding cultured FA-D2 fibroblasts retain high G2-phase accumulations, which is again in contrast to the non-FA control (superimposed gray histograms; CON, 22.6% G2; patient 3, 53.2% G2; patient 14, 56.0% G2; patient 26, 54.8% G2). PBLs in panel A were stimulated with anti-CD3, anti-CD28, and Il-2 and, in panel B, with PHA.
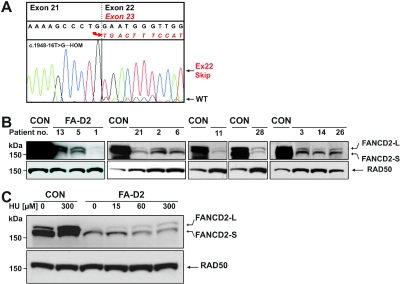
Residual FANCD2 Protein
cDNA sequencing of LCLs of patients 3, 4, 5, and 13 showed nearly complete exon 22 skipping. However, we consistently observed a small amount of correctly spliced mRNA retaining exon 22 (fig. 8A; compare with fig. 5A). Genomic sequencing identified homozygosity of these patients for the common underlying mutation, the base substitution c.1948-16T→G in IVS21. Homozygosity for this mutation was also observed in the deceased patient 25 with no cDNA available. All of these patients were products of consanguineous matings. In all LCLs that were homozygous for the above splicing mutation, we were able to demonstrate minute amounts of FANCD2 protein. A more surprising finding, however, was the presence of residual FANCD2 protein in PBLs and LCLs of every tested FA-D2 patient. Detection of residual protein required overexposure of FANCD2 immunoblots (fig. 8B). Unlike standard exposures that revealed no FANCD2 bands in most of the FA-D2 cell lines (see, e.g., fig. 2), both FANCD2-S and FANCD2-L bands were detected when films were exposed overnight. As the study progressed, it became evident that the cell lines initially detected with residual protein were those with the highest levels. When we systematically re-examined all of our FA-D2 lines, all 21 LCLs available from our 29 fully informative FA-D2 patients had minute but unequivocal amounts of residual protein (table 7). This was also true for CD3/CD28/IL-2–stimulated PBL cultures from patient 13. Patients 4, 17, and 19 with no available LCLs remained untested. In contrast, our mosaic patients displayed levels of FANCD2 protein in the control range. They had normal chromosome-breakage rates and lacked G2-phase arrest, whereas the nonmosaic patients had high chromosome breakage and G2 arrest. We therefore consider it unlikely that undetected mosaicism accounts for the presence of residual protein in the remainder of our patients. In support of this conclusion, we were able to detect residual protein, including both -S and -L bands, in fibroblasts from patients 3, 14, and 26 when the blot of figure 7B was overexposed (data not shown). Densitometry suggested reductions of residual FANCD2 protein on the order of 1 of 100 to 1 of 1,000 relative to wild type, with the degree of expression differing greatly among individual LCLs (fig. 8B). FA-D2 LCLs with the highest levels of residual FANCD2 were used to examine its characteristics on overexposed blots. The intensity of the FANCD2-L band increased as a function of the concentration of the DNA crosslinking agent (fig. 8C) and the period of treatment (not shown). This time and concentration dependency suggests genuine biochemical activity of the residual FANCD2 protein, implying that most, if not all, cases of FA-D2 result from functionally hypomorphic mutations.
Figure 8. .
Residual FANCD2 protein. A, Exon 23 sequence following that of exon 21 (exon 22 exclusion, aberrant splicing), which prevails in cDNA from homozygotes for the splice-acceptor mutation in intron 21, c.1948-16T→G, but, at low level, underlying sequence is readable as exon 22 (exon inclusion). Depicted are results from patient 5. B, Blood-derived cells from nonmosaic FA-D2 patients (exemplified 13, 5, 1, 21, 2, 6, 11, and 28) show faint but conspicuous FANCD2 bands of both species in response to MMC exposure exclusively on overexposed immunoblots, as indicated by the very intense FANCD2 signals of the normal controls (CON) (patient 13, stimulated PBL; patients 5, 1, 21, 2, 6, 11, and 28, LCLs; loading control RAD50). The individual abundance of residual protein varies considerably at low levels. C, LCLs were subjected to the indicated concentrations of hydroxyurea (HU) for 16 h. On an overexposed blot, the FANCD2-L band of the residual protein in the LCL from patient 21 increases with the HU concentration in a dose-dependent response. Normal control LCLs are distinctive by their prominent FANCD2 signals.
Discussion
Our results suggest that FA-D2 is a more frequent FA complementation group than previously reported, apparently influenced by the high proportion of patients from specific ethnic groups in the present study. 2,24,25 The relatively large proportion of FA-D2 patients with Turkish ancestry in the present study appears to be due to a founder effect for the FANCD2 mutation c.1948-16T→G among individuals of Turkish origin. This is similar to the disparity in the frequency of FA-C patients in the IFAR database compared with the European FA population. The proportion of FA-C patients in the IFAR is 15%,49 compared with only 10% in the European data set.2 This is due to the relatively high frequency of Ashkenazi Jewish patients with FA in the IFAR who have the prevalent FANCC mutation c.456+4A→G (formerly referred to as “IVS4+4A→G”),33 comprising 7.5% of all IFAR patients and 50% of these FA-C patients. On the basis of our present data, we estimate that ∼6% of patients with FA belong to complementation group D2. This estimate is supported by recent studies with figures of 4 of 53 (∼7.5%),46 3 of 73 (∼4.1%),50 and 5%.49
The FA-D2 patients in our cohort displayed anomalies and malformations typical of FA such that there were no exceptional clinical features that had not been observed elsewhere.51 However, it is remarkable that not a single FA-D2 patient lacked phenotypical manifestations, whereas the proportion of patients with FA without anomalies and malformations is generally estimated to be as high as 30%.24 Growth retardation was present in 86% of the present cohort, substantially higher than the 58%52 and 63%24 reported. Microcephaly was present in 89% of the FA-D2 cases; in contrast, Faivre et al.52 found anomalies of the head in only 56%. Anomalies of skin pigmentation were present in 75% of our FA-D2 cohort, compared with 71% and 64% reported elsewhere.24,52 Of our FA-D2 patients, 72% had radial-ray defects, in contrast to only 47%52 or 49.1%51 of all patients with FA. Of the patients in the present study, 61% had microphthalmia, whereas 38% has been the reported percentage in other patients with FA.24 As with these rather common phenotypic alterations, FA-D2 patients also showed higher rates of rare FA features, such as psychomotor retardation and hyperactivity/attention-deficit disorder. Psychomotor retardation was present in 29% of our FA-D2 cohort versus 12% or 10% such individuals in other studies.24,52 A third of our FA-D2 patients had anomalies of the brain, whereas other studies report such alterations on the order of 4.5%,52 7.7%,51 and 8%24 of their patients with FA. Of our FA-D2 patients with brain anomalies, 17% had hydrocephalus, in contrast to 4.6% reported elsewhere.51 Since several laboratories contributed to the present study and since all of our FA-D2 patients came from groups of patients with previously unassigned FA, it is unlikely that our rates reflect major biases. However, a more severe D2 phenotype has also been observed in Drosophila, comparing Fancd2 and Fancl knock-down.23 Given the high frequency of phenotypic alterations, it is not surprising that, in 30% of our FA-D2 patients, the diagnosis of FA was made by the time the patients were aged 2 years. The median age at diagnosis in our cohort was 4.5 years, which is considerably younger than for other patients with FA, for whom the diagnosis is made in only 30% of patients before onset of hematological manifestations at the median age of 7.6 years.53 In addition to an earlier median age of hematological onset (i.e., BMF) in our patients with FA, there was a shorter median period between BMF and HSCT, earlier HSCT, and a tendency toward shorter median survival than of all FA-affected patients listed in the IFAR.42 Because of relatively small numbers and the relative deficit of older patients in our cohort, statistical significance was not reached for all of these end points. HSCT appears to be a rather frequent therapeutic option in FA-D2 patients. In theory, however, deficient ATM [MIM 607585]/ATR [MIM 601215]–dependent phosphorylation of FANCD245,54,55 could involve additional toxicity of conditioning regimens. Collectively, our data suggest that FA-D2 patients represent a group with frequent but typical congenital anomalies and malformations and with relatively early hematological manifestations, compared with most other FA complementation groups.
Among the FA proteins, FANCD2 is unique, since the presence of residual protein and the demonstration of its activation can be accomplished in a single assay. In our cohort, LCLs and PBLs from 21 fully informative, nonmosaic study FA-D2 patients showed traces of residual FANCD2 protein. Importantly, the residual protein always consisted of both FANCD2 isoforms, and the typical time- and dose-dependent induction of FANCD2-L was maintained, suggesting a preserved function. Differences in expression levels of residual FANCD2 between individual LCLs might result from variations of conserved splice-site recognition, in mRNA and protein stability, and, very clearly, from differences in cell growth. FANCD2 is highly expressed and monoubiquitinated in the S phase of the cell cycle.10,56 The proportion of S-phase cells is a function of cell growth, such that differences in cell proliferation between individual cell lines account for the wide variation of FANCD2 protein levels. These differences render any quantitative mutation-specific comparisons of residual FANCD2 protein levels close to impossible. The existence of residual protein has been described elsewhere for other FA-D2 patients,9,26,27,46 but our study confirms residual protein as a consistent and, in all likelihood, essential feature of cells of FA-D2 patients. Somatic reversion as a cause of residual protein levels could be excluded in many of the patients in this study, because the diagnosis of FA in these patients was based on hypersensitivity to crosslinking agents of their cells in vitro. It could be argued that these LCLs might include very small, undetected amounts of reverted cells and thus might actually represent low-proportion mosaics. However, reverse mosaicism has been widely observed only at certain rates (<25%) and not at the frequency at which we detected residual FANCD2 protein. Moreover, reverted cells, once present, tend to outgrow nonreverted FA cells rather rapidly, at least in LCLs, which never occurred in our long-term studies. Finally, detection of residual protein with both FANCD2-S and -L bands in some of our FA-D2 fibroblast lines without the possibility of mosaicism strongly argues in favor of residual protein and against somatic reversion. Of note, the vast majority of our FA-D2 patients are homozygous or compound heterozygous for at least a missense mutation or a nucleotide substitution affecting splicing without altering the canonical AG and GT dinucleotides of splice sites. Missense mutations might lead only to a partial degradation of FANCD2, whereas the other type of mutations causing exon skipping or aberrant splicing might compete against but not abrogate the original splice site, guaranteeing a residual amount of FANCD2.
FANCD2 is targeted to chromatin after DNA damage-dependent monoubiquitination. In this process, FANCE binds to FANCD2, providing the critical bridge linking FANCD2 to FANCC and the rest of the FA core complex.57 The binding site of FANCD2 for FANCE has not been determined. Our data suggest that this binding site, in addition to K561 and apart from an intact FA nuclear core complex, must be retained by mutated residual FANCD2 protein to become monoubiquitinated.
Despite a rather severe phenotype in most of the FA-D2 patients, the vast majority of our FA-D2 patients were found to carry leaky mutations, affecting merely splicing, and displayed residual FANCD2 protein of both isotypes in their cell lines. Splicing mutations have become an increasingly successful target for experimental therapeutic approaches. Modified and antisense oligonucleotides have been used to inhibit cryptic exons or to activate regular exons weakened by mutations via targeting of the oligonucleotides to the desired transcript. This approach could eventually lead to effective therapies for the correction of erroneous splicing (reviewed in the work of Garcia-Blanco et al.58). The tight regulation of FANCD2 expression and activation and the presence of low-abundant wild-type gene products associated with FANCD2 mutations should render FANCD2 an ideal candidate for RNA-reprogramming strategies such as spliceosome-mediated RNA trans-splicing (SMaRT) (reviewed in the work of Mansfield et al.59).
Acknowledgments
We thank Richard Friedl (Wurzburg), for expert flow cytometry; Birgit Gottwald (Wurzburg), for dedicated cell culture work; Daniela Endt (Wurzburg), for sequencing; Dr. Sabine Herterich (Wurzburg), for microsatellite analyses; and Kerstin Goettsche and Silke Furlan (Dusseldorf), for assistance with retroviral vectors. We are grateful to Dr. Heidemarie Neitzel (Berlin), for providing patient DNAs, and to Ralf Dietrich (Unna), for facilitating personal contacts with FA-affected families and for arranging for insight into their medical histories. The GR plasmid for construction of diagnostic retroviral vectors was kindly provided by Dr. Christopher Baum (Hamburg). We thank Dr. Birgit Pils (Oxford, United Kingdom), for help with database searches; Dr. Heiner Schaal (Dusseldorf), Dr. Adrian Krainer (Cold Spring Harbor, NY), and Dr. Chris Smith (Cambridge, United Kingdom), for advice with the characterization of some of the splice-site mutations. Dr. Markus Schmugge (Zurich) and Dr. Eva Seemanova (Prague) are gratefully acknowledged for providing clinical information, as are Drs. John Wagner (Minneapolis), David Williams (Cincinnati), Farid Boulad (New York), and many other physicians who provided clinical data for the IFAR. We are deeply obliged to all of the participating patients and families and to the many clinicians who contributed to the present work through their patient care. This work was supported in part by grants from the Deutsche Fanconi-Anämie-Hilfe (to H. Hoehn and D.S.), the Schroeder-Kurth Fund (to R.K. and D.S.), and the Senator Kurt und Inge Schuster Foundation (to R.K.) and by National Institutes of Health grants R37 HL32987 (to A.D.A.) and R01 CA82678 (to M.B. and A.D.A.). The work of J.S. was funded by European Union grant FI6R-CT-2003-508842; Spanish Ministries of Science grant SAF2006-03340; Spanish Ministries of Health grants PI051205, G03/073, and PI061099; and the La Caixa Foundation Oncology Programme. The Centro de Investigaciones Energéticas, Medioambientales y Tecnológicas has been supported by Spanish Ministry of Health grant G03/073; VI Framework Program of the E.U. grant CONSERT, Ref. 005242; and Spanish Interministerial Commission for Science and Technology grant SAF 2005-00058). J.P.d.W., H.J., and H. Hanenberg were supported by the Fanconi Anemia Research Fund; J.P.d.W. and H.J. were supported by the Dutch Cancer Society.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- ClustalW, ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX (for the polypeptide sequences compared using the Windows interface ClustalX [v. 1.81, for the multiple sequence-alignment program])
- CpG Island Explorer, http://bioinfo.hku.hk/cpgieintro.html (for promoter analyses [v. 1.9, at the settings GC 60%, CpG O/E ratio 0.7, and minimum length 500 nt)
- Ensembl Genome Browser, http://www.ensembl.org/ (for FANCD2 genomic sequences [accession number ENSG00000144554] and Fancd2 sequence information for other species)
- ESEfinder, http://rulai.cshl.edu/tools/ESE/ (for analysis of regulatory splice sequences)
- ExPASy, http://www.expasy.org/sprot/ (for the Swiss-Prot Fancd2 protein sequences of different species, including Homo sapiens)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for FA-D2 [accession number NM_033084, 43 exons]) and FANCD2 [accession number AF340183, 44 exons], used as the human FANCD2 cDNA reference)
- Human BLAT Search, http://genome.ucsc.edu/cgi-bin/hgBlat (for genomic FANCD2 sequences)
- Human Genome Variation Society, http://hgvs.org/ (for the mutation nomenclature)
- MaxEntScan, http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq_acc.html (for estimation of deduced splice-acceptor function, with use of a maximum entropy model)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for FA-A, -B, -C, -D1, -D2, -E, -F, -G, -I, -J, -L, -M, -N, BRCA1, RAD51, RAD50, ATM/ATR, RAD50, Michelin-tire baby syndrome, VACTERL-like association, holoprosencephaly, and Kartagener syndrome)
- RepeatMasker, http://www.repeatmasker.org/ (for analysis of repetitive elements)
- Rescue-ESE (http://genes.mit.edu/burgelab/rescue-ese (for analysis of regulatory splice sequences)
- Splicefinder, http://www.uni-duesseldorf.de/rna/ (for predicted splice-donor performance)
References
- 1.Joenje H, Patel KJ (2001) The emerging genetic and molecular basis of Fanconi anaemia. Nat Rev Genet 2:446–457 10.1038/35076590 [DOI] [PubMed] [Google Scholar]
- 2.Levitus M, Rooimans MA, Steltenpool J, Cool NF, Oostra AB, Mathew CG, Hoatlin ME, Waisfisz Q, Arwert F, de Winter JP, et al (2004) Heterogeneity in Fanconi anemia: evidence for 2 new genetic subtypes. Blood 103:2498–2503 10.1182/blood-2003-08-2915 [DOI] [PubMed] [Google Scholar]
- 3.Reid S, Schindler D, Hanenberg H, Barker K, Hanks S, Kalb R, Neveling K, Kelly P, Seal S, Freund M, et al (2007) Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet 39:162–164 10.1038/ng1947 [DOI] [PubMed] [Google Scholar]
- 4.Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J, et al (2007) Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet 39:159–161 10.1038/ng1942 [DOI] [PubMed] [Google Scholar]
- 5.Levran O, Attwooll C, Henry RT, Milton KL, Neveling K, Rio P, Batish SD, Kalb R, Velleuer E, Barral S, et al (2005) The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat Genet 37:931–933 10.1038/ng1624 [DOI] [PubMed] [Google Scholar]
- 6.Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, Steltenpool J, Stone S, Dokal I, Mathew CG, et al (2005) A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet 37:958–963 10.1038/ng1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levitus M, Waisfisz Q, Godthelp BC, de Vries Y, Hussain S, Wiegant WW, Elghalbzouri-Maghrani E, Steltenpool J, Rooimans MA, Pals G, et al (2005) The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat Genet 37:934–935 10.1038/ng1625 [DOI] [PubMed] [Google Scholar]
- 8.Meetei AR, Sechi S, Wallisch M, Yang D, Young MK, Joenje H, Hoatlin ME, Wang W (2003) A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol Cell Biol 23:3417–3426 10.1128/MCB.23.10.3417-3426.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, Grompe M, D’Andrea AD (2001) Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell 7:249–262 10.1016/S1097-2765(01)00173-3 [DOI] [PubMed] [Google Scholar]
- 10.Taniguchi T, Garcia-Higuera I, Andreassen PR, Gregory RC, Grompe M, D’Andrea AD (2002) S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood 100:2414–2420 10.1182/blood-2002-01-0278 [DOI] [PubMed] [Google Scholar]
- 11.Thompson LH, Hinz JM, Yamada NA, Jones NJ (2005) How Fanconi anemia proteins promote the four Rs: replication, recombination, repair, and recovery. Environ Mol Mutagen 45:128–142 10.1002/em.20109 [DOI] [PubMed] [Google Scholar]
- 12.Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D’Andrea AD, Wang ZQ, Jasin M (2005) Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci USA 102:1110–1115 10.1073/pnas.0407796102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ (2004) The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell 15:607–620 10.1016/j.molcel.2004.08.009 [DOI] [PubMed] [Google Scholar]
- 14.Digweed M, Rothe S, Demuth I, Scholz R, Schindler D, Stumm M, Grompe M, Jordan A, Sperling K (2002) Attenuation of the formation of DNA-repair foci containing RAD51 in Fanconi anaemia. Carcinogenesis 23:1121–1126 10.1093/carcin/23.7.1121 [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto K, Hirano S, Ishiai M, Morishima K, Kitao H, Namikoshi K, Kimura M, Matsushita N, Arakawa H, Buerstedde JM, et al (2005) Fanconi anemia protein FANCD2 promotes immunoglobulin gene conversion and DNA repair through a mechanism related to homologous recombination. Mol Cell Biol 25:34–43 10.1128/MCB.25.1.34-43.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mirchandani KD, D’Andrea AD (2006) The Fanconi anemia/BRCA pathway: a coordinator of cross-link repair. Exp Cell Res 312:2647–2653 10.1016/j.yexcr.2006.06.014 [DOI] [PubMed] [Google Scholar]
- 17.Levitus M, Joenje H, de Winter JP (2006) The Fanconi anemia pathway of genomic maintenance. Cell Oncol 28:3–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blom E, van de Vrugt HJ, de Winter JP, Arwert F, Joenje H (2002) Evolutionary clues to the molecular function of Fanconi anemia genes. Acta Haematol 108:231–236 10.1159/000065659 [DOI] [PubMed] [Google Scholar]
- 19.Houghtaling S, Timmers C, Noll M, Finegold MJ, Jones SN, Meyn MS, Grompe M (2003) Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev 17:2021–2035 10.1101/gad.1103403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carreau M (2004) Not-so-novel phenotypes in the Fanconi anemia group D2 mouse model. Blood 103:2430 10.1182/blood-2003-11-3946 [DOI] [PubMed] [Google Scholar]
- 21.Dequen F, St-Laurent JF, Gagnon SN, Carreau M, Desnoyers S (2005) The Caenorhabditis elegans FancD2 ortholog is required for survival following DNA damage. Comp Biochem Physiol B Biochem Mol Biol 141:453–460 10.1016/j.cbpc.2005.05.010 [DOI] [PubMed] [Google Scholar]
- 22.Liu TX, Howlett NG, Deng M, Langenau DM, Hsu K, Rhodes J, Kanki JP, D’Andrea AD, Look AT (2003) Knockdown of zebrafish Fancd2 causes developmental abnormalities via p53-dependent apoptosis. Dev Cell 5:903–914 10.1016/S1534-5807(03)00339-3 [DOI] [PubMed] [Google Scholar]
- 23.Marek LR, Bale AE (2006) Drosophila homologs of FANCD2 and FANCL function in DNA repair. DNA Repair (Amst) 5:1317–1326 [DOI] [PubMed] [Google Scholar]
- 24.Tischkowitz M, Dokal I (2004) Fanconi anaemia and leukaemia—clinical and molecular aspects. Br J Haematol 126:176–191 10.1111/j.1365-2141.2004.05023.x [DOI] [PubMed] [Google Scholar]
- 25.Taniguchi T, D’Andrea AD (2006) The molecular pathogenesis of Fanconi anemia: recent progress. Blood 107:4223–4233 10.1182/blood-2005-10-4240 [DOI] [PubMed] [Google Scholar]
- 26.Timmers C, Taniguchi T, Hejna J, Reifsteck C, Lucas L, Bruun D, Thayer M, Cox B, Olson S, D’Andrea AD, et al (2001) Positional cloning of a novel Fanconi anemia gene, FANCD2. Mol Cell 7:241–248 10.1016/S1097-2765(01)00172-1 [DOI] [PubMed] [Google Scholar]
- 27.Borriello A, Locasciulli A, Bianco AM, Criscuolo M, Conti V, Grammatico P, Cappellacci S, Zatterale A, Morgese F, Cucciolla V, et al (2007) A novel Leu153Ser mutation of the Fanconi anemia FANCD2 gene is associated with severe chemotherapy toxicity in a pediatric T-cell acute lymphoblastic leukemia. Leukemia 21:72–78 10.1038/sj.leu.2404468 [DOI] [PubMed] [Google Scholar]
- 28.Joenje H (1997) Fanconi anemia: cytogenetic diagnosis. Free University of Amsterdam, Amsterdam [Google Scholar]
- 29.Auerbach AD (2003) Diagnosis of Fanconi anemia by diepoxybutane analysis. In: Current protocols in human genetics, supplement 37. John Wiley, New York and London, pp 8.7.1–8.7.15 [Google Scholar]
- 30.Berger R, Le Coniat M, Gendron MC (1993) Fanconi anemia: chromosome breakage and cell cycle studies. Cancer Genet Cytogenet 69:13–16 10.1016/0165-4608(93)90104-T [DOI] [PubMed] [Google Scholar]
- 31.Seyschab H, Friedl R, Sun Y, Schindler D, Hoehn H, Hentze S, Schroeder-Kurth T (1995) Comparative evaluation of diepoxybutane sensitivity and cell cycle blockage in the diagnosis of Fanconi anemia. Blood 85:2233–2237 [PubMed] [Google Scholar]
- 32.Heinrich MC, Hoatlin ME, Zigler AJ, Silvey KV, Bakke AC, Keeble WW, Zhi Y, Reifsteck CA, Grompe M, Brown MG, et al (1998) DNA cross-linker-induced G2/M arrest in group C Fanconi anemia lymphoblasts reflects normal checkpoint function. Blood 91:275–287 [PubMed] [Google Scholar]
- 33.Kutler DI, Auerbach AD (2004) Fanconi anemia in Ashkenazi Jews. Fam Cancer 3:241–248 10.1007/s10689-004-9565-8 [DOI] [PubMed] [Google Scholar]
- 34.Neitzel H (1986) A routine method for the establishment of permanent growing lymphoblastoid cell lines. Hum Genet 73:320–326 10.1007/BF00279094 [DOI] [PubMed] [Google Scholar]
- 35.Schindler D, Hoehn H (1988) Fanconi anemia mutation causes cellular susceptibility to ambient oxygen. Am J Hum Genet 43:429–435 [PMC free article] [PubMed] [Google Scholar]
- 36.Hildinger M, Abel KL, Ostertag W, Baum C (1999) Design of 5′ untranslated sequences in retroviral vectors developed for medical use. J Virol 73:4083–4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanenberg H, Xiao XL, Dilloo D, Hashino K, Kato I, Williams DA (1996) Colocalization of retrovirus and target cells on specific fibronectin fragments increases genetic transduction of mammalian cells. Nat Med 2:876–882 10.1038/nm0896-876 [DOI] [PubMed] [Google Scholar]
- 38.Hanenberg H, Hashino K, Konishi H, Hock RA, Kato I, Williams DA (1997) Optimization of fibronectin-assisted retroviral gene transfer into human CD34+ hematopoietic cells. Hum Gene Ther 8:2193–2206 [DOI] [PubMed] [Google Scholar]
- 39.Hanenberg H, Batish SD, Pollok KE, Vieten L, Verlander PC, Leurs C, Cooper RJ, Gottsche K, Haneline L, Clapp DW, et al (2002) Phenotypic correction of primary Fanconi anemia T cells with retroviral vectors as a diagnostic tool. Exp Hematol 30:410–420 10.1016/S0301-472X(02)00782-8 [DOI] [PubMed] [Google Scholar]
- 40.Chandra S, Levran O, Jurickova I, Maas C, Kapur R, Schindler D, Henry R, Milton K, Batish SD, Cancelas JA, et al (2005) A rapid method for retrovirus-mediated identification of complementation groups in Fanconi anemia patients. Mol Ther 12:976–984 10.1016/j.ymthe.2005.04.021 [DOI] [PubMed] [Google Scholar]
- 41.Shimamura A, de Oca RM, Svenson JL, Haining N, Moreau LA, Nathan DG, D’Andrea AD (2002) A novel diagnostic screen for defects in the Fanconi anemia pathway. Blood 100:4649–4654 10.1182/blood-2002-05-1399 [DOI] [PubMed] [Google Scholar]
- 42.Kutler DI, Singh B, Satagopan J, Batish SD, Berwick M, Giampietro PF, Hanenberg H, Auerbach AD (2003) A 20-year perspective on the International Fanconi Anemia Registry (IFAR). Blood 101:1249–1256 10.1182/blood-2002-07-2170 [DOI] [PubMed] [Google Scholar]
- 43.Carter AB, Salem AH, Hedges DJ, Keegan CN, Kimball B, Walker JA, Watkins WS, Jorde LB, Batzer MA (2004) Genome-wide analysis of the human Alu Yb-lineage. Hum Genomics 1:167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roy-Engel AM, Carroll ML, Vogel E, Garber RK, Nguyen SV, Salem AH, Batzer MA, Deininger PL (2001) Alu insertion polymorphisms for the study of human genomic diversity. Genetics 159:279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ho GP, Margossian S, Taniguchi T, D’Andrea AD (2006) Phosphorylation of FANCD2 on two novel sites is required for mitomycin C resistance. Mol Cell Biol 26:7005–7015 10.1128/MCB.02018-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soulier J, Leblanc T, Larghero J, Dastot H, Shimamura A, Guardiola P, Esperou H, Ferry C, Jubert C, Feugeas JP, et al (2005) Detection of somatic mosaicism and classification of Fanconi anemia patients by analysis of the FA/BRCA pathway. Blood 105:1329–1336 10.1182/blood-2004-05-1852 [DOI] [PubMed] [Google Scholar]
- 47.Callen E, Casado JA, Tischkowitz MD, Bueren JA, Creus A, Marcos R, Dasi A, Estella JM, Munoz A, Ortega JJ, et al (2005) A common founder mutation in FANCA underlies the world’s highest prevalence of Fanconi anemia in Gypsy families from Spain. Blood 105:1946–1949 10.1182/blood-2004-07-2588 [DOI] [PubMed] [Google Scholar]
- 48.Lo Ten Foe JR, Kwee ML, Rooimans MA, Oostra AB, Veerman AJ, van Weel M, Pauli RM, Shahidi NT, Dokal I, Roberts I, et al (1997) Somatic mosaicism in Fanconi anemia: molecular basis and clinical significance. Eur J Hum Genet 5:137–148 [PubMed] [Google Scholar]
- 49.Kennedy RD, D’Andrea AD (2005) The Fanconi anemia/BRCA pathway: new faces in the crowd. Genes Dev 19:2925–2940 10.1101/gad.1370505 [DOI] [PubMed] [Google Scholar]
- 50.Casado JA, Callen E, Jacome A, Rio P, Castella M, Lobitz S, Ferro T, Munoz A, Sevilla J, Cantalejo A, et al (2006) A comprehensive strategy for the subtyping of Fanconi anemia patients: conclusions from the Spanish Fanconi Anemia research network. J Med Genet (electronically published November 14, 2006; accessed March 27, 2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Auerbach AD, Buchwald M, Joenje H (2001) Fanconi anemia, 8th ed. Vol 1. McGraw-Hill, New York, pp 753–768 [Google Scholar]
- 52.Faivre L, Guardiola P, Lewis C, Dokal I, Ebell W, Zatterale A, Altay C, Poole J, Stones D, Kwee ML, et al (2000) Association of complementation group and mutation type with clinical outcome in fanconi anemia. Blood 96:4064–4070 [PubMed] [Google Scholar]
- 53.Huret JL (2002) Fanconi anaemia. In: Atlas Genet Cytogentic Oncol Haematol (http://atlasgeneticsoncology.org/kprones/FA10001.html) (accessed March 27, 2007)
- 54.Andreassen PR, D’Andrea AD, Taniguchi T (2004) ATR couples FANCD2 monoubiquitination to the DNA-damage response. Genes Dev 18:1958–1963 10.1101/gad.1196104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pichierri P, Rosselli F (2004) The DNA crosslink-induced S-phase checkpoint depends on ATR-CHK1 and ATR-NBS1-FANCD2 pathways. EMBO J 23:1178–1187 10.1038/sj.emboj.7600113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holzel M, van Diest PJ, Bier P, Wallisch M, Hoatlin ME, Joenje H, de Winter JP (2003) FANCD2 protein is expressed in proliferating cells of human tissues that are cancer-prone in Fanconi anaemia. J Pathol 201:198–203 10.1002/path.1450 [DOI] [PubMed] [Google Scholar]
- 57.Pace P, Johnson M, Tan WM, Mosedale G, Sng C, Hoatlin M, de Winter J, Joenje H, Gergely F, Patel KJ (2002) FANCE: the link between Fanconi anaemia complex assembly and activity. EMBO J 21:3414–3423 10.1093/emboj/cdf355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcia-Blanco MA, Baraniak AP, Lasda EL (2004) Alternative splicing in disease and therapy. Nat Biotechnol 22:535–546 10.1038/nbt964 [DOI] [PubMed] [Google Scholar]
- 59.Mansfield SG, Chao H, Walsh CE (2004) RNA repair using spliceosome-mediated RNA trans-splicing. Trends Mol Med 10:263–268 10.1016/j.molmed.2004.04.007 [DOI] [PubMed] [Google Scholar]
- 60.Wang Y, Leung FC (2004) An evaluation of new criteria for CpG islands in the human genome as gene markers. Bioinformatics 20:1170–1177 10.1093/bioinformatics/bth059 [DOI] [PubMed] [Google Scholar]