Abstract
In response to a number of genotoxic stimuli that induce DNA damage in cells, the tumour suppressor p53 is activated resulting in cell cycle arrest or apoptosis. In this study, we have identified stimulated with retinoic acid 13 (Stra13), a basic helix–loop–helix transcription factor, as a regulator of ionizing-radiation-induced apoptosis. We show that Stra13 is induced in response to several DNA-damaging agents in a p53-independent manner. Stra13−/− thymocytes show impaired apoptosis in response to ionizing radiation, and consistently, p53 levels and also expression of its key transcriptional targets Puma and Noxa are reduced in the mutant thymocytes. In vitro, Stra13 regulates p53 levels in a mouse double mutant 2 (Mdm2)-dependent manner by physically interacting with p53 and preventing Mdm2-mediated ubiquitination and nuclear export. Together, our studies provide evidence that Stra13 is involved in DNA-damage-induced apoptosis and indicate its role in tumorigenesis.
Keywords: DNA damage, p53, thymocytes, apoptosis, Stra13, radiation
Introduction
Transcription factors belonging to the basic helix–loop–helix (bHLH) family are important regulators of cellular differentiation, cell-cycle arrest and apoptosis. Stimulated with retinoic acid 13 (Stra13; also known as differentially expressed in chondrocytes/enhancer of split and hairy related protein 2 (Dec1/Sharp2)), a bHLH factor, is widely expressed during mouse embryogenesis (Boudjelal et al, 1997). Studies of overexpression have implicated Stra13 in many biological responses including cell-cycle arrest, differentiation and apoptosis (Sun & Taneja, 2000; Seimiya et al, 2002; Ivanova et al, 2004). Conversely, Stra13−/− mice show a defect in Fas-mediated apoptosis of activated T and B cells, leading to the development of a lupus-like autoimmune disorder (Sun et al, 2001) and impaired skeletal muscle regeneration in response to injury (Sun et al, 2007). Although Stra13 transcriptionally regulates its target genes, it also interacts with several proteins including Usf (upstream stimulatory factor), Stat3 (signal transducer and activator of transcription 3) and Clock/Bmal (clock/brain and muscle arnt-like protein), and modulates their activity by transcription-independent mechanisms (Dhar & Taneja, 2001; Ivanova et al, 2004; Sato et al, 2004). Recent studies have shown that Stra13 is upregulated in response to stress stimuli such as serum deprivation, hypoxia and various cytokines (Sun & Taneja, 2000; Ivanova et al, 2001, 2004; Yun et al, 2002), indicating that it might mediate cellular responses to stress. However, the signalling pathways and molecular mechanisms by which it participates in stress responses are not completely understood.
A pivotal target that mediates DNA-damage-induced cellular responses of growth arrest and apoptosis is the tumour suppressor p53. p53 is a short-lived protein that is normally expressed at low levels owing to its interaction with mouse double mutant 2 (Mdm2), which signals its degradation. Mdm2, an E3 ubiquitin ligase, interacts with the amino-terminus of p53 and promotes its degradation by ubiquitination (Haupt et al, 1997). In response to DNA damage, p53 becomes phosphorylated and this prevents its interaction with Mdm2, leading to its stabilization and activation (Ko & Prives, 1996; Giaccia & Kastan, 1998; Oren, 1999). p53, in its active form, transcriptionally upregulates the expression of target genes that are important for cell-cycle arrest, such as p21Waf1 (Brugarolas et al, 1995), and apoptosis, such as Puma, Noxa and Bax (McCurrach et al, 1997; Villunger et al, 2003).
In this study, we examined the role of Stra13 in DNA-damage-induced cellular responses. We show that Stra13 is upregulated by ionizing radiation and genotoxic agents both in vitro and in vivo in a p53-independent manner. Stra13-deficient thymocytes show resistance to γ-irradiation-induced apoptosis and express reduced levels of p53 and its target genes Puma and Noxa after DNA damage. Conversely, overexpression of Stra13 results in the accumulation of p53 in an Mdm2-dependent manner by protecting it from Mdm2-mediated ubiquitination and degradation. Our studies provide evidence that Stra13 is involved in p53-dependent apoptosis in response to DNA damage. Thus, these results provide new observations that link Stra13 to DNA-damage-induced cellular responses.
Results And Discussion
Stra13 is upregulated by genotoxic stimuli
Our previous studies have shown that Stra13−/− mice are defective in Fas-mediated apoptosis of activated T and B cells (Sun et al, 2001). As Stra13 is upregulated in response to stress stimuli, such as serum deprivation and hypoxia (Sun & Taneja, 2000; Ivanova et al, 2001; Yun et al, 2002), we investigated whether Stra13 is involved in other stress response pathways such as those induced by DNA damage. We first analysed whether Stra13 expression is altered in response to DNA-damaging agents. NIH3T3 cells were treated with etoposide, which elicits double-stranded DNA breaks, or with cisplatin, a drug that causes interstrand crosslinks. Stra13 expression was induced in response to both etoposide and cisplatin treatment (Fig 1A). Moreover, treatment of NIH3T3 cells (Fig 1B) and p53-null 10.1 T cells (Fig 1C) with ionizing radiation also led to an increase in its expression, indicating that Stra13 is upregulated in response to ionizing radiation in a p53-independent manner. To verify that Stra13 is induced in vivo in response to DNA damage, we examined Stra13 expression in radiation-sensitive tissues such as thymus and spleen from wild-type mice after irradiation. Western blot analysis of both tissues showed an apparent increase in Stra13 expression in response to ionizing radiation (Fig 1D,E). Together, these data indicate that Stra13 expression is induced in response to genotoxic stress in vitro and in vivo and suggest that it might have a role in DNA-damage-induced responses.
Figure 1.
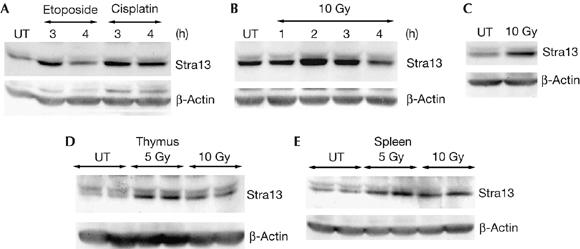
Stra13 is induced in response to DNA damage. (A) NIH3T3 cells were treated with etoposide or cisplatin and stimulated with retinoic acid 13 (Stra13) expression was analysed by western blot at the indicated time points. Untreated (UT) cells were used as controls. Membranes were reprobed with β-actin antibody. (B,C) NIH3T3 cells (B) and p53-null 10.1 T cells (C) were irradiated (10 Gy), and Stra13 expression was analysed by western blot at the indicated times. 10.1 T cells were analysed 2 h after irradiation. (D,E) Wild-type mice were exposed to ionizing radiation and killed after 2.5 or 6 h. Stra13 expression was analysed in the thymus (6 h) and spleen (2.5 h). Untreated mice were used as controls. Two mice were used for each dose.
Impaired radiation-induced apoptosis in Stra13−/− mice
In response to ionizing radiation, cells undergo cell-cycle arrest, which allows for repair of damaged DNA, or undergo apoptosis, which results in the elimination of cells with a damaged genome (Kastan et al, 1991). As Stra13 is induced by ionizing-radiation, we examined ionizing-radiation-induced apoptosis and cell-cycle arrest in Stra13−/− thymocytes. At the same time point after irradiation, 40–50% of wild-type thymocytes were apoptotic, whereas only 20–30% of Stra13−/− thymocytes underwent apoptosis (Fig 2A). Consistently, 14 h after irradiation, the percentage of viable cells was about twofold higher in Stra13−/− thymocytes (supplementary Fig 1A online). These results indicate that thymocyte apoptosis after DNA damage is defective in the absence of Stra13. However, no significant difference in ionizing-radiation-induced cell-cycle arrest was seen in Stra13−/− thymocytes (supplementary Fig 1B online).
Figure 2.
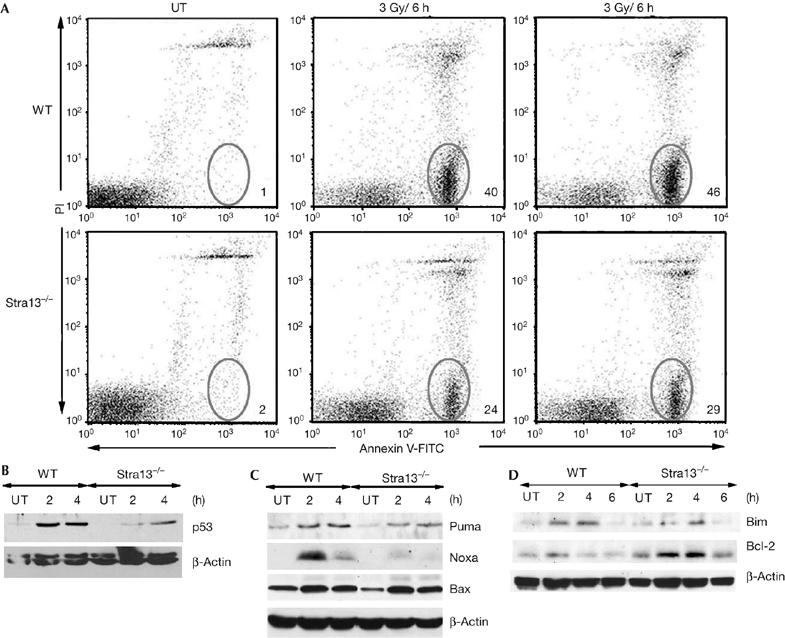
Defective γ-radiation-induced apoptosis and p53 activity in Stra13−/− thymocytes. (A) Thymocytes from wild-type (WT) and Stra13−/− mice were left untreated (UT) or γ-irradiated. After 6 h, cells were stained with Annexin V–FITC and propidium iodide (PI). Numbers represent the percentage of apoptotic cells. The results are representative of eight experiments. (B) Thymocytes were treated with ionizing radiation (5 Gy) or left untreated (UT), and p53 expression was analysed by western blotting. (C,D) Untreated (UT) or irradiated (5 Gy) thymocytes were analysed for expression levels of Puma, Noxa and Bax (C), Bim and Bcl-2 (D) by immunoblotting. Stra13, stimulated with retinoic acid 13.
As thymocytes undergo apoptosis in response to ionizing radiation in a p53-dependent manner (Lowe et al, 1993), we analysed whether the p53 response to DNA damage is changed in Stra13−/− thymocytes. As expected, p53 was highly induced in wild-type thymocytes, whereas p53 levels were reduced in Stra13−/− cells consistent with defective apoptosis (Fig 2B). In addition, Stra13−/− thymocytes expressed reduced levels of p53 target genes Puma and Noxa, whereas Bax expression was not significantly altered (Fig 2C). Consistent with reduced apoptosis and increased viability, Stra13−/− thymocytes also expressed reduced levels of Bim and increased levels of Bcl-2 compared with wild-type controls (Fig 2D). These results are in agreement with previous findings indicating: a role for Noxa, Puma and Bim in ionizing-radiation-induced apoptosis (Bouillet et al, 1999; Villunger et al, 2003); a normal radiation response of Bax−/− thymocytes (Knudson et al, 1995); and protection of cells from radiation-induced apoptosis by Bcl-2 (Sentman et al, 1991).
Stra13 regulates p53 in an Mdm2-dependent manner
To identify the mechanism underlying reduced p53 levels in Stra13−/− thymocytes, we examined the effect of Stra13 on p53 levels by western blotting. Stra13 expression enhanced the amount of p53 protein (two-, three- and fourfold) in a dose-dependent manner in NIH3T3 cells (Fig 3A) but not in p53−/−/Mdm2−/− mouse embryonic fibroblasts (MEFs; Fig 3B), indicating that Stra13 influences p53 in an Mdm2-dependent manner. To determine whether Stra13 regulates Mdm2 at the transcriptional level, we examined the effect of Stra13 expression on Mdm2 mRNA levels in human colorectal carcinoma (HCT)116 cells, which express wild-type p53, and also in HCT116 p53−/− cells. Mdm2 messenger RNA levels were not changed by Stra13 expression (Fig 3C). Stra13 expression also had no effect on p53 mRNA levels in HCT116 cells (Fig 3C) although it elevated endogenous p53 protein (Fig 3D). These results indicated that Stra13 does not regulate p53 levels by modulating Mdm2 or p53 expression.
Figure 3.
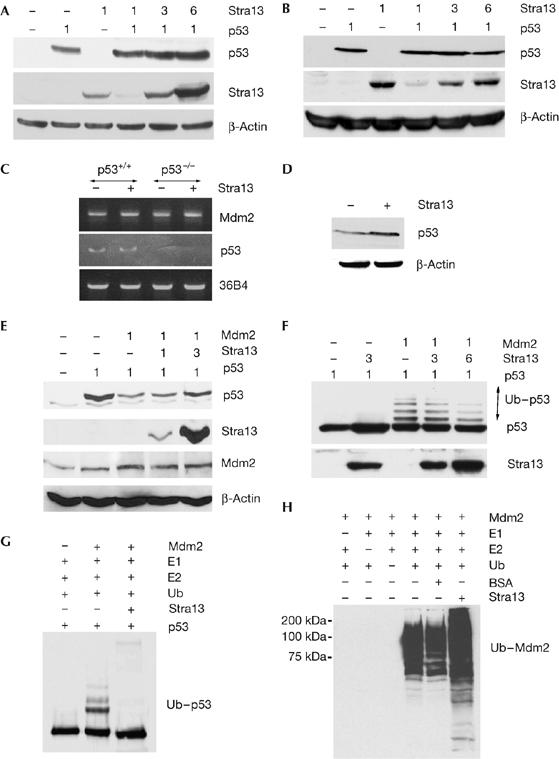
Stra13 modulates p53 in an Mdm2-dependent manner. (A,B) Western blot analysis in NIH3T3 cells (A) and p53−/−/Mdm2−/−MEFs (B), transfected with p53 and Flag–Stra13 at the indicated ratios. p53 and Stra13 expression were determined by western blot using p53 and Flag antibodies. (C) HCT116 cells (p53+/+) and HCT116 p53−/− (p53−/−) were transfected with vector (−) or Flag–Stra13 (+). Mdm2 and p53 mRNA levels were analysed by RT–PCR. 36B4 transcripts were analysed as an internal control. (D) HCT116 cells were transfected with vector (−) or Flag–Stra13 (+) and analysed for endogenous p53 protein levels by western blot. (E) NIH3T3 cells were transfected with p53, Mdm2 or Flag-Stra13 as indicated and immunoblotted with p53, Flag and Mdm2 antibodies. (F) NIH3T3 cells were transfected with p53, Mdm2 and Flag–Stra13 as indicated. Cells were treated with ALLN for 4 h before they were collected. Lysates were immunoblotted using p53 and Flag antibodies. The ubiquitinated form of p53 (Ub–p53) is indicated. (G) Purified p53 was incubated with Mdm2, E1, E2 and ubiquitin (Ub) in the absence or presence of in vitro-translated Stra13. p53 ubiquitination was detected with p53 antibody. (H) GST–Mdm2 was tested for auto-ubiquitination in the absence or presence of Stra13. Ubiquitinated Mdm2 was detected by ubiquitin antibody. ALLN, N-acetyl-leucyl-leucyl-norleucinal; GST, glutathione-S-transferase; Mdm, mouse double mutant; MEF, mouse embryonic fibroblast; RT–PCR, reverse transcriptase–PCR; Stra13, stimulated with retinoic acid 13.
As Mdm2 ubiquitinates and degrades p53 (Haupt et al, 1997; Kubbutat et al, 1997), we tested whether Stra13 protects p53 from Mdm2-mediated degradation. NIH3T3 cells were transfected with p53, Mdm2 and increasing amounts of Stra13. Mdm2 led to p53 degradation as expected, whereas expression of Stra13 partly protected p53 from degradation (Fig 3E), indicating that Stra13 might interfere with Mdm2-mediated ubiquitination of p53. To investigate this possibility, we expressed p53 with Mdm2 in the presence of increasing amounts of Stra13. Transfected cells were treated with the proteasome inhibitor N-acetyl-leucyl-leucyl-norleucinal (ALLN) to prevent p53 degradation, and then analysed by western blotting with p53 antibody (Fig 3F). Coexpression of Stra13 with p53 resulted in increased p53 levels, consistent with our previous observations. As expected, expression of Mdm2 together with p53 resulted in the accumulation of ubiquitinated p53. Interestingly, the addition of increasing amounts of Stra13 in the presence of Mdm2 reduced p53 ubiquitination. Consistently, Stra13 expression blocked Mdm2-mediated p53 ubiquitination in vitro (Fig 3G). However, Mdm2 self-ubiquitination was not altered, indicating that the reduction of p53 ubiquitination by Stra13 is not due to a general inhibition of the E3 ubiquitin-ligase activity of Mdm2 (Fig 3H).
Stra13 interacts with p53 and prevents its nuclear export
An alternative mechanism by which Stra13 might influence p53 levels is through interaction with p53 and/or Mdm2. We first examined whether Stra13 colocalizes with p53 in untreated and etoposide-treated cells. After etoposide treatment, Stra13 was found in nuclear foci and colocalized with p53 both in the absence and presence of DNA damage (Fig 4A). We then tested whether Stra13 interacts with p53 and Mdm2 by co-immunoprecipitation assays. Co-immunoprecipitation analysis of p53−/−/Mdm2−/− cells transfected with Stra13 and p53 showed clearly an interaction in the absence of Mdm2 (Fig 4B). Stra13 also interacted with Mdm2 in the absence of p53 (Fig 4C). Nevertheless, the association of p53 with Mdm2 was not altered by expression of Stra13 (Fig 4D). To determine whether the interaction with p53 is direct and to map the domains of the interaction, full-length glutathione-S-transferase (GST)–Stra13 (1–411) and deletion mutants containing only the N-terminal bHLH domain (1–127) or the carboxy-terminal repression region (111–411) were incubated with in vitro-translated p53 (Fig 4E). Full-length Stra13 and the 1–127 mutant interacted with p53, whereas no interaction was seen with the 111–411 mutant. Interestingly, in correlation with the interaction data, expression of full-length Stra13 and the 1–127 mutant elevated endogenous p53 levels (Fig 4F), whereas the C-terminal mutant that failed to interact with p53 did not increase endogenous p53 levels. These results indicate that bHLH domain in Stra13 interacts with p53 and is sufficient to modulate its levels. To map the domains in p53 that interact with Stra13, various GST–p53 deletion mutants were tested for interaction with Stra13 (Fig 4G). Stra13 interacted with full-length p53 (1–393) and a mutant containing only the C-terminal residues 319–393, indicating that the predominant interaction occurs with this region. In addition, a weak interaction was observed with the 1–160 mutant. The C terminus of p53 contains lysine residues that are ubiquitinated by Mdm2, which then signals its nuclear export (Lohrum et al, 2001). As Stra13 interacts predominantly with the C terminus of p53 and inhibits Mdm2-mediated ubiquitination and degradation, we reasoned that Stra13 might regulate Mdm2-mediated nuclear export of p53. We therefore determined cellular localization of p53 in the presence of Mdm2 and Stra13. In the absence of Mdm2, p53 was mostly nuclear and 15% of transfected cells showed cytoplasmic p53. Consistent with previous reports, expression of Mdm2 increased cytoplasmic localization of p53 in 35% of transfected cells (Fig 4H), whereas coexpression of Stra13 with Mdm2 relocalized p53 in the nucleus, and only 18% of cells had cytoplasmic p53. Thus, Stra13 protects p53 by preventing its ubiquitination and subsequent nuclear export.
Figure 4.
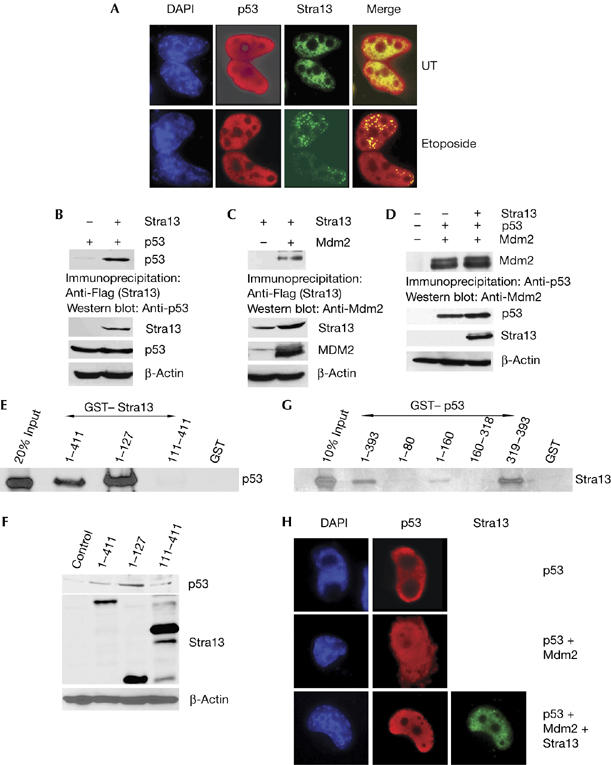
Stra13 interacts with p53 and prevents its Mdm2-mediated nuclear export (A) H1299 cells were transfected with p53 and Flag–Stra13, and stained with p53 and Flag antibodies. Nuclei were detected with DAPI. (B) p53/Mdm2−/− cells were transfected with p53 and Flag–Stra13. Lysates were immunoprecipitated with Flag antibody and immunoblotted with p53 antibody. The lower panels show western blot analysis of Stra13 and p53 expression. (C) H1299 cells were transfected with Mdm2 and Flag–Stra13. Lysates were immunoprecipitated with Flag antibody and immunoblotted with Mdm2 antibody. The lower panels show western blot analysis of Stra13 and Mdm2 expression. (D) H1299 cells transfected with Mdm2 and p53 in the absence or presence of Flag–Stra13. Lysates were immunoprecipitated with p53 antibody and immunoblotted with Mdm2 antibody. The lower panels show western blot analysis of p53, Mdm2 and Stra13 expression. (E) GST–Stra13 mutants were tested for interaction with in vitro-translated p53. GST was used as a control. 20% input was run on the gel. (F) NIH3T3 cells were transfected with p53 and His–Stra13 mutants, as indicated. Control cells were transfected with vector alone. Endogenous p53 was analysed by western blot analysis. The middle panel shows Stra13 expression by western blot. (G) Stra13 interacts with the carboxyl terminus of p53. GST–p53 mutants were tested for interaction with in vitro-translated Stra13. GST was used as a control. 10% input was run on the gel. (H) Stra13 prevents Mdm2-mediated nuclear export of p53. H1299 cells were transfected with p53, Mdm2 and Flag–Stra13 and analysed for subcellular localization of p53 by immunofluorescence. Nuclei were stained with DAPI. DAPI, 4,6-diamidino-2-phenylindole; GST, glutathione-S-transferase; Mdm, mouse double mutant; Stra13, stimulated with retinoic acid 13.
Taken together, our results show a role for Stra13 in ionizing-radiation-induced apoptosis by regulation of p53. Similar to Stra13, many factors, including Hif1α (hypoxia inducible factor 1α), Atf3 (activating transcription factor 3), Stat1, Yy1 (yin yang 1) and Pml (promyelocytic leukaemia protein) (Chen et al, 2003; Townsend et al, 2004; Yan et al, 2005), modulate p53 levels by direct interaction with p53 and/or Mdm2. Stra13 interacts with both p53 and Mdm2, and its expression results in reduced p53 ubiquitination, nuclear export and degradation. As Stra13 interacts directly with the C-terminal end of p53 that is subject to post-translational modifications, it is likely that this association results in masking the lysine residues that are subject to Mdm2-mediated ubiquitination. However, we cannot exclude that its association with Mdm2 also contributes to stabilization of p53. Additionally, the weak interaction with the 1–160 region of p53 indicates that Stra13 regulates p53 by mechanisms that are as yet unknown.
DNA damage responses have an important role in maintaining genomic stability, and genetic defects in this response can result in premature ageing, immune defects and development of cancers. The induction of Stra13 in response to several genotoxic stimuli, combined with its ability to regulate radiation-induced apoptosis and p53 levels, indicates that its altered expression or activity might be important in radiation sensitivity and oncogenesis.
Methods
Mice. Stra13−/− mice have been described previously (Sun et al, 2001). All animal protocols followed institutional guidelines.
DNA damage. Four- to six-week-old wild-type and Stra13−/− mice, or cultured cells, were γ-irradiated using a 137Cs source as indicated. NIH3T3 cells were treated with 20 μg/ml etoposide (Sigma, St Louis, MO, USA) or 20 μg/ml cisplatin (Sigma) for various durations as indicated.
Apoptosis of thymocytes. Thymocytes from wild-type and Stra13−/− mice were irradiated, stained with AnnexinV–fluorescein isothiocyanate (BD PharMingen, San Diego, CA, USA) and PI (Sigma) and analysed by flow cytometry (Becton Dickinson FACScan, San Jose, CA, USA).
GST pull-down assays, co-immunoprecipitation and antibodies. GST pull-down assays and co-immunoprecipitations were carried out as described previously (Sun & Taneja, 2000). GST pull-down assays were carried out with equivalent amounts of various GST–p53 mutants or GST alone, and 35S-labelled in vitro-translated Stra13. Conversely, GST–Stra13 mutants (Sun & Taneja, 2000) were used for the interaction with 35S-labelled in vitro-translated p53. For co-immunoprecipitation assays, H1299 or p53/Mdm2−/− MEFs were transfected with p53, Mdm2 and Flag–Stra13, as indicated. Lysates were immunoprecipitated and detected by western blotting with appropriate antibodies. β-Actin was used to confirm equivalent loading of proteins. The primary antibodies were Stra13 (Dec1; kindly provided by Dr B. Yan, University of Rhode Island), p53 (FL393 and DO-1; Santa Cruz, Santa Cruz, CA, USA), Flag (M2; Sigma); Mdm2 (SMP14 and N-20; Santa Cruz), ubiquitin (P4D1; Santa Cruz) and β-actin (Sigma).
Reverse transcriptase–PCR. Semiquantitative reverse transcriptase–PCR was carried out as described (Boudjelal et al, 1997). HCT116 and HCT116 p53−/− cells were transfected with vector (pCS2) alone or pCS2–Stra13. Total RNA was isolated using Trizol. The following primers were used: p53: 5′-GCTGCTCAGATAGCGATG-3′ and 5′-GGCATCCTTGAGTTCCAAG-3′ for 30 cycles; Mdm2: 5′-GGAAATGCACTTCATG-3′ and 5′-CAGGGCTTATTCCTTTTC-3′ for 30 cycles; and 36B4: 5′-CAGCTCTGGAGAAACTGCTG-3′ and 5′-GTGTACTCAGTCTCCACAGA-3′ for 19 cycles. PCR products were run on a 1.5% agarose gel and visualized by ethidium bromide staining.
Ubiquitination assays. For in vitro ubiquitination assays, 10 ng of purified p53 (ProteinOne, Bethesda, MD, USA) was incubated with 25 ng E1 (Boston Biochemicals, Cambridge, MA, USA), 100 ng E2 (GST–UbcH5c; Boston Biochemicals), 150 ng GST–Mdm2, 10 μg His–ubiquitin, 5 μM ubiquitin aldehyde and 2 mM ATPγS in the absence or presence of 6 μl (∼60 ng) of in vitro-translated Stra13 for 2 h at 37°C in a buffer containing 40 mM Tris–HCl, 5 mM MgCl2 and 2 mM dithiothreitol. Reactions were analysed by western blotting using p53 antibody. For in vivo experiments, p53, Mdm2 and Flag–Stra13 were transfected in NIH3T3 cells. At 24 h after transfection, cells were treated with the proteasome inhibitor (ALLN; Calbiochem, San Diego, CA, USA) for 4 h and analysed by western blot using p53 antibody. For Mdm2 ubiquitination assays in vitro, 250 ng of GST–Mdm2 was incubated with E1 (25 ng), E2 (100 ng) and His–ubiquitin (5 μg) for 1 h at 37°C in a buffer containing 40 mM Tris–HCl, 5 mM MgCl2, 2 mM dithiothreitol and 2 mM ATP. BSA or in vitro-translated Stra13 (750 ng) was added as indicated. Reactions were analysed by immunoblotting with ubiquitin antibody.
Immunofluorescence microscopy. H1299 cells were transfected with Flag-Stra13 and p53. After 24 h, cells were treated with 20 μM etoposide and immunostained with Flag and p53 antibodies. p53 subcellular localization in the presence or absence of Mdm2 and Stra13 was similarly analysed. Nuclei were detected with 4,6-diamidino-2-phenylindole (DAPI) staining. A total of 1,000 nuclei were counted for cellular distribution of p53.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank S. Jones, J. Manfredi, M. Oren, T. Shenk, B. Volgestein and B. Yan for the generous gifts of reagents, and Z.-Q. Pan for advice. This work was supported by funds from the National Institutes of Health and a Scholar Award from the leukaemia and Lymphoma Society (R.T). T.H.T was supported by an National Cancer Institute postdoctoral training grant (CA88796).
References
- Boudjelal M, Taneja R, Matsubara S, Bouillet P, Dollé P, Chambon P (1997) Overexpression of Stra13, a novel retinoic acid-inducible gene of the basic helix–loop–helix family, inhibits mesodermal and promotes neuronal differentiation of P19 cells. Genes Dev 11: 2052–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, Adams JM, Strasser A (1999) Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 286: 1735–1738 [DOI] [PubMed] [Google Scholar]
- Brugarolas J, Chandrasekaran C, Gordon JI, Beach D, Jacks T, Hannon GJ (1995) Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature 377: 552–557 [DOI] [PubMed] [Google Scholar]
- Chen D, Li M, Luo J, Gu W (2003) Direct interactions between HIF-1α and Mdm2 modulate p53 function. J Biol Chem 278: 13595–13598 [DOI] [PubMed] [Google Scholar]
- Dhar M, Taneja R (2001) Cross-regulatory interaction between Stra13 and USF results in functional antagonism. Oncogene 20: 4750–4756 [DOI] [PubMed] [Google Scholar]
- Giaccia AJ, Kastan MB (1998) The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev 12: 2973–2983 [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M (1997) MDM2 promotes the rapid degradation of p53. Nature 387: 296–299 [DOI] [PubMed] [Google Scholar]
- Ivanova AV, Ivanov SV, Danilkovitch-Miagkova A, Lerman MI (2001) Regulation of STRA13 by the von Hippel–Lindau tumor suppressor protein, hypoxia, and the UBC9/ubiquitin proteasome degradation pathway. J Biol Chem 276: 15306–15315 [DOI] [PubMed] [Google Scholar]
- Ivanova AV, Ivanov SV, Zhang X, Ivanov VN, Timofeeva OA, Lerman MI (2004) STRA13 interacts with STAT3 and modulates transcription of STAT3-dependent targets. J Mol Biol 340: 641–653 [DOI] [PubMed] [Google Scholar]
- Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, Craig RW (1991) Participation of p53 protein in the cellular response to DNA damage. Cancer Res 51: 6304–6311 [PubMed] [Google Scholar]
- Knudson CM, Tung KS, Tourtellotte WG, Brown GA, Korsmeyer SJ (1995) Bax-deficient mice with lymphoid hyperplasia and male germ cell death. Science 270: 96–99 [DOI] [PubMed] [Google Scholar]
- Ko LJ, Prives C (1996) p53: puzzle and paradigm. Genes Dev 10: 1054–1072 [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH (1997) Regulation of p53 stability by Mdm2. Nature 387: 299–303 [DOI] [PubMed] [Google Scholar]
- Lohrum MA, Woods DB, Ludwig RL, Balint E, Vousden KH (2001) C-terminal ubiquitination of p53 contributes to nuclear export. Mol Cell Biol 21: 8521–8532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T (1993) p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 362: 847–849 [DOI] [PubMed] [Google Scholar]
- McCurrach ME, Connor TM, Knudson CM, Korsmeyer SJ, Lowe SW (1997) Bax-deficiency promotes drug resistance and oncogenic transformation by attenuating p53-dependent apoptosis. Proc Natl Acad Sci USA 94: 2345–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren M (1999) Regulation of the p53 tumor suppressor protein. J Biol Chem 274: 36031–36034 [DOI] [PubMed] [Google Scholar]
- Sato F, Kawamoto T, Fujimoto K, Noshiro M, Honda KK, Honma S, Honma K, Kato Y (2004) Functional analysis of the basic helix–loop–helix transcription factor DEC1 in circadian regulation. Interaction with BMAL1. Eur J Biochem 271: 4409–4419 [DOI] [PubMed] [Google Scholar]
- Seimiya M et al. (2002) Clast5/Stra13 is a negative regulator of B lymphocyte activation. Biochem Biophys Res Commun 292: 121–127 [DOI] [PubMed] [Google Scholar]
- Sentman CL, Shutter JR, Hockenberry D, Kanagawa O, Korsmeyer SJ (1991) Bcl-2 inhibits multiple forms of apoptosis but not negative selection in thymocytes. Cell 67: 879–888 [DOI] [PubMed] [Google Scholar]
- Sun H, Taneja R (2000) Stra13 expression is associated with growth arrest and represses transcription through histone deacetylase-dependent and -independent mechanisms. Proc Natl Acad Sci USA 97: 4058–4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Lu B, Li RQ, Flavell RA, Taneja R (2001) Defective T cell activation and autoimmune disorder in Stra13-deficient mice. Nat Immunol 2: 1040–1047 [DOI] [PubMed] [Google Scholar]
- Sun H, Li L, Vercherat C, Gulbagci N, Acharjee S, Li J, Chung T-K, Thin TH, Taneja R (2007) Stra13 regulates satellite cell activation by antagonizing Notch signaling. J Cell Biol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend PA, Scarabelli TM, Davidson SM, Knight RA, Latchman DS, Stephanou A (2004) STAT-1 interacts with p53 to enhance DNA damage-induced apoptosis. J Biol Chem 279: 5811–5820 [DOI] [PubMed] [Google Scholar]
- Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ, Adams JM, Strasser A (2003) p53- and drug-induced apoptotic responses mediated by BH3-only proteins Puma and Noxa. Science 302: 1036–1038 [DOI] [PubMed] [Google Scholar]
- Yan C, Lu D, Hai T, Boyd DD (2005) Activating transcription factor 3, a stress sensor, activates p53 by blocking its ubiquitination. EMBO J 24: 2425–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun Z, Maecker HL, Johnson RS, Giaccia AJ (2002) Inhibition of PPAR γ2 gene expression by the HIF-1-regulated gene DEC1/Stra13: a mechanism for regulation of adipogenesis by hypoxia. Dev Cell 2: 331–341 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


