Despite their central role in maintaining genetic integrity during cell division, there is still considerable uncertainty about how DNA sequences are selected to be replication origins in eukaryotes (DePamphilis, 1999; Gilbert, 2001; Machida et al, 2005). The budding yeast Saccharomyces cerevisiae uses a limited set of efficient origins, each of which contains a conserved sequence motif and are used fairly reproducibly in different cell cycles. By contrast, the replication origins of Xenopus and Drosophila early cleavage embryos are located randomly with respect to the DNA sequence, although mechanisms might exist to ensure that they are spaced apart (Blow et al, 2001; Hyrien et al, 2003). The somatic cells of metazoans organize their replication origins in yet another way, which resembles aspects of those of both budding yeast and cleavage embryos. Many—perhaps most (Mesner et al, 2006)—initiation events occurs in ‘initiation zones' that are up to several kilobases in size; these zones contain numerous inefficient initiation sites that are used in different cell cycles. Origins differ in the efficiency with which they are used; therefore, some sites are utilized more frequently than others. As yet, no consensus DNA sequence has been found at metazoan initiation origins.
The complement of proteins responsible for the assembly of functional replication forks is well conserved among eukaryotes (Diffley, 2004; Nishitani & Lygerou, 2004; Blow & Dutta, 2005). Binding of the mini chromosome maintenance 2–7 (Mcm2–7) complex to origin DNA during late mitosis and G1 phase ‘licenses' the origin for use in the subsequent S phase. The loading of Mcm2–7 onto DNA requires the origin recognition complex (ORC), and together they form part of the ‘pre-replicative complex' (pre-RC). With the exception of the S. cerevisiae ORC, pre-RC proteins bind DNA with little or no sequence preference; therefore, it is unclear how the pattern of origins seen in other organisms is established. One possibility is that some aspect of chromatin or chromosome structure directs pre-RC assembly. Another possibility is that licensed origins are widely distributed over chromosomal DNA, but that only a few are selected for use during S phase. Consistent with this idea, the number of Mcm2–7 complexes assembled onto DNA is approximately 10 times the number of origins typically used in a normal S phase (Hyrien et al, 2003; Blow & Dutta, 2005).
Several recent papers have addressed how replication origins are distributed in the fission yeast Schizosaccharomyces pombe, which is a unicellular eukaryote that is only distantly related to the budding yeasts. In addition to work by Patel and colleagues, and Feng and colleagues (Patel et al, 2006; Feng et al, 2006), two new fission yeast microarray-based genome-wide studies, one by the Paul Nurse group (Heichinger et al, 2006) and the other by the Hisao Masukata group (Hayashi et al, 2007), have been published in the 1 November 2006 and 7 March 2007 issues of The EMBO Journal, respectively. Furthermore, in another paper published in the same March issue, the cell biology of DNA replication in S. pombe is examined by Susan Gasser and colleagues, who show how replication sites are spatially and temporally organized in the nucleus (Meister et al, 2007).
Heichinger and colleagues exploited the increase in DNA content that occurs during replication to map replication origins (Fig 1C). They observed 401 strong origins used during a normal S phase, although they estimated that, on average, each origin is used in only approximately 30% of cell cycles. Treatment of cells with the DNA synthesis inhibitor hydroxyurea (HU) revealed an additional 503 inefficient origins. By slowing fork progression, HU might allow inefficient origins—which are usually inactivated by passive replication from a nearby origin—to fire.
Figure 1.
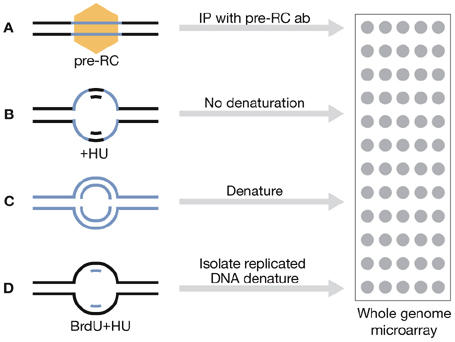
Strategies for mapping replication origins. A small section of chromosomal DNA (blue and black lines) containing a replication origin is shown. Blue lines show single-stranded DNA that is ultimately used to detect sequences in a whole-genome microarray. (A) Origin DNA is immunoprecipitated (IP) using antibodies (ab) against pre-replication complex (pre-RC) proteins. (B) Forks are slowed with hydroxyurea (HU), exposing single-stranded DNA ahead of the fork that is used to probe the microarray without the denaturation that is normally needed. (C) Total DNA from cells synchronized at different stages of S phase is used to probe a whole-genome microarray. Immediately after initiation, origin DNA is present at twice the level of flanking DNA. (D) Newly synthesized DNA is labelled with bromodeoxyuridine (BrdU), which is then separated on density gradients.
Hayashi and colleagues identified replication origins by incorporation of the thymidine analogue bromodeoxyuridine (BrdU) in the presence of HU (Fig 1D), and correlated this with the chromatin binding of the pre-RC proteins Orc1 and Mcm6 (Fig 1A). They identified 307 early replication origins at which pre-RC proteins and BrdU incorporation co-localized. Another 153 late or inefficient origins that bound pre-RC proteins, but did not incorporate BrdU, were also found. Hayashi and colleagues identified 78% of the early/efficient origins published by Heichinger and colleagues, which included 80–90% of those characterized in other studies (Segurado et al, 2003; Dai et al, 2005; Feng et al, 2006).
As predicted by previous analyses (Segurado et al, 2003; Dai et al, 2005), the origins identified by Heichinger and colleagues, and Hayashi and colleagues, do not display a consensus sequence but have a high average A+T content (greater than 70%), similar to that of intergenes in fission yeast. (A+T)-rich sequences are the only common element found in the few origins of replication that have been characterized in metazoans (Aladjem & Fanning, 2004).
The average inter-origin distance found in the different genome-wide studies mentioned here is 30–40 kb; however, if the 503 inefficient origins described by Heichinger and colleagues are included, the average origin spacing is reduced to 14 kb. These authors suggest that the inefficient origins might be used only in a small percentage of cell cycles to bridge large gaps between more efficient origins, to guarantee complete genome replication. However, given that the weak origins were not identified in the other genome-wide studies, they might have no physiological relevance. Another possibility is that they represent some of the numerous additional sites where Mcm2–7 complexes are loaded at low efficiency, which do not clearly show up in the chromatin immunoprecipitation experiments of Hayashi and colleagues. The large zones containing weak origins alone have some resemblance to the initiation zones of somatic mammalian cells. There are approximately 10 times more Mcm2–7 complexes on DNA than ORC molecules or active origins (Blow & Dutta, 2005). All of the chromatin-bound Mcm2–7 complexes are likely to be able to initiate replication (Woodward et al, 2006); therefore, they might mark additional sites of replication initiation for use in various contingencies.
S phase checkpoints suppress the firing of late origins when replication forks are inhibited (Santocanale & Diffley, 1998; Shirahige et al, 1998; Kumar & Huberman, 2004). Hayashi and colleagues observed that approximately 50% (153 out of 307) of the sites containing bound Orc1 and Mcm6 did not incorporate BrdU in the presence of HU, indicating that these sites are either inefficiently used or are late origins that are suppressed by HU. In S phase checkpoint kinase cds1Δ mutant cells, BrdU was incorporated into some, but not all, of these late/inefficient origins as well as large sections of subtelomeric DNA. A similar increase in origin firing in cds1Δ cells has been reported (Feng et al, 2006); these authors mapped origins by looking for regions of DNA that become single-stranded in the presence of HU (Fig 1B). By contrast, Heichinger and colleagues found that only 7% of origins replicated late in S phase, many in subtelomeric regions. They also detected only a small increase in origin firing when checkpoint-defective rad3 mutant cells were treated with HU. One possible reason for this discrepancy is that the majority of late-firing origins in fission yeast do so only by virtue of being inefficiently—and stochastically—used throughout S phase. It is also possible that although rad3 is thought to act upstream of cds1, the different mutants affected late-origin firing to different extents.
Meister and colleagues provided new data on the dynamics of DNA replication by examining S. pombe cells carrying proliferating cell nuclear antigen (PCNA) tagged with the green fluorescent protein to mark sites of replication (Meister et al, 2007). This revealed that, as in other eukaryotes, DNA replication in S. pombe occurs in discrete structures called replication foci or factories, each containing an average of 14 replication forks. As in metazoan cells, the fission yeast factories showed reproducible changes in number and subnuclear distribution during S phase.
The DNA replicated in a single replication factory is thought to represent a contiguous block of chromosomal DNA (Berezney et al, 2000; Kitamura et al, 2006). Interestingly, the origin mapping studies of Hayashi and colleagues indicate that early/efficient origins and late/inefficient origins tend to distribute separately into large chromosome regions, potentially providing a basis for their replication in different factories throughout S phase (Fig 2). Hayashi and colleagues swapped the chromosomal location of an early/efficient origin with a late/inefficient origin and showed that the origin efficiency changed according to the location. This implies that the efficiency or timing of origin firing is dependent on local chromatin or chromosome structure. Molecular combing and observation of individual fission yeast DNA molecules have revealed considerable variability between the location of origins in different cell cycles, confirming the idea that many origins are inefficient (Patel et al, 2006; Rhind, 2006). Although the inefficiency of many fission yeast origins implies a significant degree of randomness in the location of replication origins, the specific patterns observed by whole-genome studies (Feng et al, 2006; Hayashi et al, 2007; Heichinger et al, 2006) indicate that non-random factors are also at work.
Figure 2.
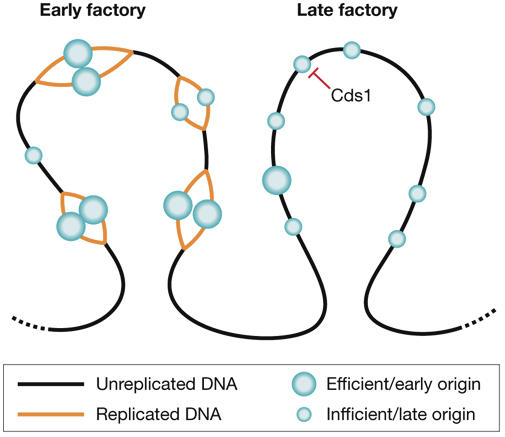
Organization of replication factories in fission yeast. Origins are found in clusters that replicate at different times in S phase and can be visualized as bright replication foci (factories). Origins can fire with different efficiencies. Efficient origins (large circles) fire early in S phase, whereas inefficient origins (small circles) can fire at different times throughout S phase, and so tend to be found in late factories. The S phase checkpoint kinase Cds1 might have a role in the replication timing programme by inhibiting the firing of defined clusters in early S phase.
The grouping of replication forks into factories might provide an environment in which newly replicated DNA can be assembled into specific chromatin states, thereby maintaining the epigenetic information that is otherwise disrupted by replication (Gilbert, 2001; Taddei et al, 2004). Meister and colleagues show that replication foci are highly dynamic, and can fuse, split and travel across the nucleus. These rapid movements might be driven by interaction with other nuclear components, which could be involved in assembling specific chromatin structures. cds1Δ mutant cells showed unusual patterns in which foci characteristic of both early and late S phase were seen together. This might be a manifestation of the firing of a subset of late/inefficient origins that are normally repressed by the checkpoint system during early S phase (Hayashi et al, 2007). When replication was slowed by HU, a relatively normal, although slower, progression of replication patterns was maintained. Treatment of cds1Δ mutant cells with HU, however, led to the complete dismantling of replication foci, although HU only slowed the normal progression of patterns in wild-type cells. The loss of foci seen in cds1Δ cells treated with HU might be attributable to the collapse of replication forks expected to occur under these conditions (Lopes et al, 2001; Tercero & Diffley, 2001; Cobb et al, 2005).
The papers of Heichinger and colleagues, Hayashi and colleagues, and Meister and colleagues provide a glimpse of the unexplored intricacies of how eukaryotic genomes are organized for replication. They show that the efficiency of origin usage is likely to be an important factor in explaining overall patterns of genome replication. They also imply that higher-order chromatin organization has a role in the spatial and temporal organization of replication. In metazoans, such as Xenopus, nuclear reprogramming is accompanied by changes in chromatin loop size and replication rates, indicating a functional link between the two (Lemaitre et al, 2005). A challenge now is to understand how this interplay between chromatin organization and replication comes about.


References
- Aladjem MI, Fanning E (2004) The replicon revisited: an old model learns new tricks in metazoan chromosomes. EMBO Rep 5: 686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney R, Dubey DD, Huberman JA (2000) Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108: 471–484 [DOI] [PubMed] [Google Scholar]
- Blow JJ, Dutta A (2005) Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol 6: 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Gillespie PJ, Francis D, Jackson DA (2001) Replication origins in Xenopus egg extract are 5–15 kilobases apart and are activated in clusters that fire at different times. J Cell Biol 152: 15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JA, Schleker T, Rojas V, Bjergbaek L, Tercero JA, Gasser SM (2005) Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev 19: 3055–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Chuang RY, Kelly TJ (2005) DNA replication origins in the Schizosaccharomyces pombe genome. Proc Natl Acad Sci USA 102: 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis ML (1999) Replication origins in metazoan chromosomes: fact or fiction? Bioessays 21: 5–16 [DOI] [PubMed] [Google Scholar]
- Diffley JF (2004) Regulation of early events in chromosome replication. Curr Biol 14: R778–R786 [DOI] [PubMed] [Google Scholar]
- Feng W, Collingwood D, Boeck ME, Fox LA, Alvino GM, Fangman WL, Raghuraman MK, Brewer BJ (2006) Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat Cell Biol 8: 148–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DM (2001) Making sense of eukaryotic DNA replication origins. Science 294: 96–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Katou Y, Itoh T, Tazumi M, Yamada Y, Takahashi T, Nakagawa T, Shirahige K, Masukata H (2007) Genome-wide localization of pre-RC sites and identification of replication origins in fission yeast. EMBO J 26: 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heichinger C, Penkett CJ, Bahler J, Nurse P (2006) Genome-wide characterization of fission yeast DNA replication origins. EMBO J 25: 5171–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O, Marheineke K, Goldar A (2003) Paradoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. Bioessays 25: 116–125 [DOI] [PubMed] [Google Scholar]
- Kitamura E, Blow JJ, Tanaka TU (2006) Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell 125: 1297–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Huberman JA (2004) On the slowing of S phase in response to DNA damage in fission yeast. J Biol Chem 279: 43574–43580 [DOI] [PubMed] [Google Scholar]
- Lemaitre JM, Danis E, Pasero P, Vassetzky Y, Mechali M (2005) Mitotic remodeling of the replicon and chromosome structure. Cell 123: 787–801 [DOI] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M (2001) The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412: 557–561 [DOI] [PubMed] [Google Scholar]
- Machida YJ, Hamlin JL, Dutta A (2005) Right place, right time, and only once: replication initiation in metazoans. Cell 123: 13–24 [DOI] [PubMed] [Google Scholar]
- Meister P, Taddei A, Ponti A, Baldacci G, Gasser S (2007) Replication foci dynamics: replication patterns are modulated by S phase checkpoint kinases in fission yeast. EMBO J 26: 1315–1326- [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesner LD, Crawford EL, Hamlin JL (2006) Isolating apparently pure libraries of replication origins from complex genomes. Mol Cell 21: 719–726 [DOI] [PubMed] [Google Scholar]
- Nishitani H, Lygerou Z (2004) DNA replication licensing. Front Biosci 9: 2115–2132 [DOI] [PubMed] [Google Scholar]
- Patel PK, Arcangioli B, Baker SP, Bensimon A, Rhind N (2006) DNA replication origins fire stochastically in fission yeast. Mol Biol Cell 17: 308–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N (2006) DNA replication timing: random thoughts about origin firing. Nat Cell Biol 8: 1313–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Diffley JF (1998) A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395: 615–618 [DOI] [PubMed] [Google Scholar]
- Segurado M, de Luis A, Antequera F (2003) Genome-wide distribution of DNA replication origins at A+T-rich islands in Schizosaccharomyces pombe. EMBO Rep 4: 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H (1998) Regulation of DNA-replication origins during cell-cycle progression. Nature 395: 618–621 [DOI] [PubMed] [Google Scholar]
- Taddei A, Hediger F, Neumann FR, Gasser SM (2004) The function of nuclear architecture: a genetic approach. Annu Rev Genet 38: 305–345 [DOI] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF (2001) Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553–557 [DOI] [PubMed] [Google Scholar]
- Woodward AM, Gohler T, Luciani MG, Oehlmann M, Ge X, Gartner A, Jackson DA, Blow JJ (2006) Excess Mcm2–7 license dormant origins of replication that can be used under conditions of replicative stress. J Cell Biol 173: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]


