Abstract
The signal-recognition particle (SRP) mediates the translocation of membrane and secretory proteins across the endoplasmic reticulum upon interaction with the SRP receptor. In trypanosomes, the main RNA molecule is the spliced-leader (SL) RNA, which donates the SL sequence to all messenger RNA through trans-splicing. Here, we show that RNA interference silencing of the SRP receptor (SRα) in Trypanosoma brucei caused the accumulation of SRP on ribosomes and triggered silencing of SL RNA (SLS). SLS was elicited due to the failure of the SL RNA-specific transcription factor tSNAP42 to bind to its promoter. SL RNA reduction, in turn, eliminated mRNA processing and resulted in a significant reduction of all mRNA tested. SLS was also induced under pH stress and might function as a master regulator in trypanosomes. SLS is reminiscent of, but distinct from, the unfolded protein response and can potentially act as a new target for parasite eradication.
Keywords: Trypanosoma brucei , signal recognition receptor;, SL RNA, trans-splicing
Introduction
In eukaryotes, translocation of secretory and membrane proteins across the endoplasmic reticulum is mediated primarily by the signal-recognition particle (SRP; Keenan et al, 2001). During protein translocation, the signal-peptide-binding protein of the SRP, SRP54, samples the signal peptide emerging from the ribosome nascent chain (RNC). The resulting SRP–RNC complex interacts with the SRP receptor SRα. Guanosine triphosphate (GTP) binding to SRα enhances the affinity between SRP and SR (Rapiejko & Gilmore, 1997). After signal sequence transfer to the translocon, both SRP54 and SRα act as reciprocal GTPase-activating proteins, resolving their association (Powers & Walter, 1995). The RNC binds to the membrane by interactions with the channel or Sec61 complex, and the protein is co-translationally translocated.
Trypanosomes are ancient protozoan parasites, which have a unique SRP that contains two RNA molecules—the 7SL RNA and a transfer RNA-like molecule—and lacks the Alu-domain-binding proteins (Liu et al, 2003; Lustig et al, 2005). The SRP pathway is essential for parasite survival (Liu et al, 2002; Lustig et al, 2005). Trypanosomes process their RNA by a unique mechanism, as all nuclear messenger RNA undergo trans-splicing, which involves the addition of a common spliced-leader (SL) sequence to the 5′ end of the mRNA. The source of the SL is a small RNA, the SL RNA. The SL RNA promoter is the only RNA polymerase II (pol II) promoter characterized in trypanosomes (Gilinger & Bellofatto, 2001; Das et al, 2005). It recruits the trypanosome snRNA-activating protein (tSNAP) complex, which contains three subunits: tSNAP26 and tSNAP50, which are related to factors that mediate small nuclear RNA (snRNA) transcription in metazoa, and tSNAP42, which is trypanosome specific (Das et al, 2005).
In this study, we show that the depletion of cells from the SRP receptor SRα elicits a new signalling pathway, leading to complete elimination of mRNA production through inhibition of SL RNA transcription. This novel pathway can also be induced under pH stress and might therefore be analogous to the unfolded protein response (UPR) that acts to protect cells from ambient stress (Schroder & Kaufman, 2005).
Results
SL RNA and mRNA production
To examine further the role of SRP in protein sorting, we knocked down the expression of the SRP receptor SRα by RNA interference (RNAi). The SRα gene was identified by homology searches of the Trypanosoma brucei genome. The gene (Tb11.01.1650) is composed of 582 amino acids, and shares 39% identity and 48% similarity to the human protein. Silencing was carried out by using expression of a stem–loop structure from an inducible promoter (Wang et al, 2000). Induction of SRα silencing was lethal and cells started dying 3 days after induction (Fig 1A). As a result of silencing, cells became ‘fat', as visualized by both confocal and scanning electron microscopy (SEM; Fig 1B). The level of SRα mRNA was reduced on silencing (Fig 1C). Interestingly, significant reduction of tubulin mRNA was also observed, which might explain the ‘fat' phenotype (Fig 1B).
Figure 1.

Deleterious effects of signal-recognition particle receptor α silencing. (A) SRα is essential for cell growth. The growth of uninduced cells was compared with induced cells. The number of uninduced cells (−Tet) is shown by open diamonds and that of induced cells (+Tet) by filled squares. Standard deviation from three different experiments is indicated by error bars. (B) Morphology of SRα-depleted cells. Cells uninduced (−Tet) and after 3 days of induction (+Tet) were visualized by confocal microscopy and by scanning electron microscope (insets). Scale bars, 5 μm. (C) Northern analysis of SRα messenger RNA upon silencing. RNA was prepared from induced (+Tet) and uninduced (−Tet) cells. Total RNA (20 μg) was subjected to northern analysis with random-labelled probes. Double stranded RNA production was inspected using the stuffer sequence of the stem–loop RNA (Pex). The transcripts are indicated by arrows. rRNA, ribosomal RNA; SRα, SRP receptor α; SRP, signal-recognition particle.
To investigate further the effect of SRα silencing on mRNA production, the level of several mRNAs was examined from cells 2–4 days after silencing (Fig 2A). In each case, the level of mRNA was reduced. The overall reduction in the level of mRNA is not due to total inhibition of RNA pol II-dependent transcription, as the level of EP procyclin, which is transcribed by RNA pol I, was similarly reduced (Fig 2A). The effect on mRNA was specific to SRα depletion, as no such effect was observed after SRP54 RNAi silencing. However, SRP54 depletion was efficient and its mRNA was completely eliminated (Fig 2A). Next, we examined whether the reduction of mRNA in SRα-depleted cells was due to changes in mRNA stability. Cells were treated with actinomycin D, before and after depletion, and the half-life of three mRNAs was determined. The results (supplementary Fig S1 online) show no effect on mRNA stability.
Figure 2.
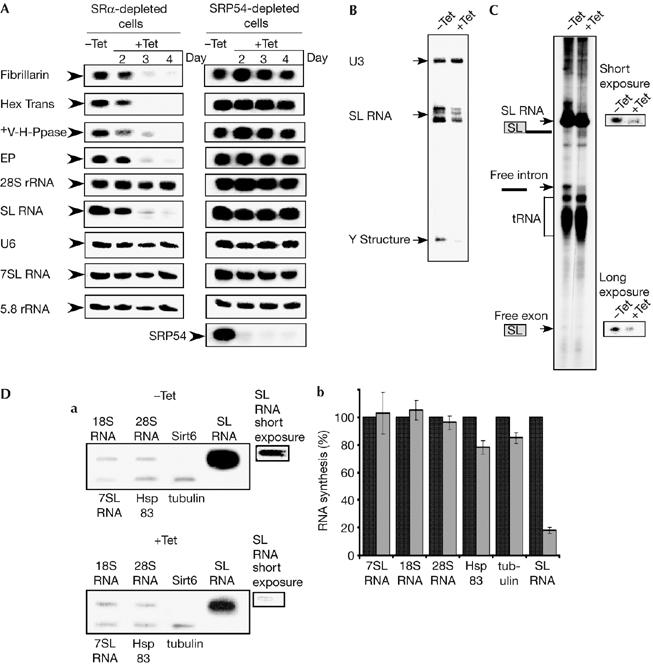
Signal-recognition particle receptor α silencing specifically inhibits spliced-leader RNA transcription and messenger RNA production. (A) The effect of SRα or SRP54 depletion on RNA level. RNA (20 μg) was prepared from uninduced (−Tet) and silenced cells 2–4 days after induction (+Tet) and subjected to northern analysis with random-labelled probes, as specified in the supplementary information online. (B) SL RNA level in SRα-depleted cells. RNA was prepared from uninduced cells (−Tet) and silenced cells 3 days after induction (+Tet) and subjected to primer extension with radiolabelled oligonucleotides complementary to the SL RNA and U3 snoRNA. Complementary DNA was separated on 6% (w/v) denaturing gel. U3, SL RNA and Y structure are indicated. (C) RNA synthesis in SRα-depleted cells. Permeable cells were prepared from the same number of cells carrying the SRα silencing construct, without induction (−Tet) or after tetracycline induction for 3 days (+Tet), as described in the Methods. The RNA was fractioned on 6% (w/v) denaturing gel. The identity of the RNAs are as indicated. (D) Slot-blot analysis of transcripts synthesized in permeable cells. (a) RNA was prepared from permeable cells, as described in (C), and was used for hybridizing with a blot carrying DNA encoding for the genes, as indicated (18S and 28S rRNA, 7SL RNA, Sirt6, tubulin, Hsp83 and SL RNA). Short and long exposures of the SL RNA signals are shown. (b) Quantitative analysis shows the percentage reduction in the level of RNA synthesis, as determined by densitometry of three independent experiments. s.d. is indicated by error bars. Black and grey bars represent uninduced and induced cells, respectively. SL RNA, spliced-leader RNA; snoRNA, small nucleolar RNA; SRα, SRP receptor α; SRP, signal-recognition particle.
Inhibition of SL RNA synthesis, which is transcribed by pol II, should eliminate mRNA production by blocking trans-splicing. We examined next the level of SL RNA and compared it to other small RNAs transcribed by pol I and pol III, such as 5.8S ribosomal RNA, U6 snRNA and 7SL RNA. Significant RNA reduction was observed only for SL RNA (Fig 2A).
To examine trans-splicing during SRα depletion directly, the level of the Y structure intermediate was determined in steady-state mRNA. This resulted in two principal extension products: the Y structure and the mature capped SL RNA. The results (Fig 2B) indicate that the level of Y structure was reduced by 78%±5 compared with uninduced cells, and this corresponds to the decrease in the level of SL RNA. This suggests that during SRα depletion, the reduction in the Y structure is not the result of splicing defects, but is due to a reduction in the level of SL RNA.
Next we examined, using the permeable cell system, whether nascent SL RNA transcription is affected in the silenced cells (Tschudi & Ullu, 1990; Ullu & Tschudi, 1990). SL RNA is the major radiolabelled transcript in this system, but nascent tRNA, rRNA and mRNA can also be detected. The results (Fig 2C) indicated an 80%±4 reduction in the level of newly transcribed SL RNA. The decrease in the level of SL RNA was also reflected by a decrease in the level of the free intron and SL exon. This suggests that the mechanism/machinery of trans-splicing was not abrogated, and that the reduction reflects only a reduction in SL RNA production.
We wanted to explore further whether SRα depletion elicited a general shut-off of RNA transcription or whether transcription shut-off was specific to SL RNA. RNA extracted from permeable cells—before and after depletion—was analysed by slot-blot hybridization. We examined the level of rRNA, 7SL RNA and mRNA—tubulin and Hsp83—transcription, and compared it with that of the SL RNA (Fig 2D). The human gene Sirt6 was used as a control for nonspecific hybridization. Densitometric-based quantification of these data—based on long exposure—shows that mRNA levels were reduced by only 15–20% (as illustrated in Fig 2D), whereas SL RNA transcription—based on short exposure—was reduced by 82%±2 as a result of SRα depletion. Thus, there was a specific shut-off of SL RNA transcription as a consequence of the loss of SRα.
Changes in tSNAP complex during SRα depletion
To investigate how SL RNA transcription was specifically shut down, we examined some specific effects, such as amount and localization, on SL RNA transcription factors. Proteins present in nuclear extracts, made from cells before or after SRα knockdown, were studied by using western analysis. Figure 3A shows a significant reduction in the level of two of the tSNAPc subunits, tSNAP50 and tSNAP26 which most probably reflects mRNA elimination. Indeed, a significant reduction in protein synthesis examined by in vivo labelling was observed after SRα depletion (see supplementary Fig S2 online). By contrast, a significant increase in the third tSNAPc subunit, tSNAP42, was observed (Fig 3A). The localization of tSNAP42 showed a marked change in the subnuclear localization on silencing (Fig 3B). In uninduced cells, tSNAP42 localized to a distinct ‘dot', which marks the unique site of SL RNA transcription (Dossin & Schenkman, 2005). Following SRα silencing, tSNAP42 was not concentrated as this dot, but was spread throughout the nucleus. tSNAP42 might be the target to receive the signal to shut off SL RNA transcription, by losing its ability to bind to the SL RNA promoter. Indeed, a chromatin immunoprecipitation (ChIP) assay carried out on DNA from cells before and after silencing, indicated that the SL RNA transcription complex was not formed in SRα-depleted cells, as no binding of tSNAP42 to the SL RNA promoter was observed, as in uninduced cells. The specificity of binding to the SL RNA promoter was controlled by a lack of binding to the rRNA (Fig 3C). To examine whether the mRNA reduction observed in the SRα-silenced cells was entirely correlated with SL RNA shut-off, the tSNAP42 expression was silenced by RNAi. The results (supplementary Fig S3 online) show that in tSNAP42-silenced cells, SL RNA and mRNA levels were proportionally reduced.
Figure 3.

Inhibition of spliced-leader RNA synthesis is linked to changes in the tSNAP complex. (A) The level of tSNAP proteins during SRP receptor α silencing. A 50 μg portion of proteins from induced (+Tet) and uninduced (−Tet) nuclear extracts (as described in the Methods), and affinity purified tSNAPc complex, prepared as previously described (Das et al, 2005), were fractioned on a 10% (w/v) SDS–polyacrylamide gel and subjected to western analysis with tSNAP26 (1:3,000), tSNAP42 (1:7,500) and tSNAP50 (1:5,000) antibodies. (B) The effect of SRα silencing on tSNAP42 localization. Uninduced cells (−Tet) and silenced cells (+Tet) on day three after induction were fixed with 4% (v/v) formaldehyde for 25 min, incubated with tSNAP42 antibodies and detected by an FITC-conjugated second antibody (indicated by arrows). The nucleus was stained with DAPI. Scale bars, 5 μm. Enlargement of the nuclear area is framed. (C) Binding of tSNAP42 to the SL RNA gene. ChIP was carried out on cells depleted of SRα (+Tet) or uninduced cells (−Tet), using tSNAP42 antibody (Ab) or without antibody (no Ab) as a control. PCR on total input (input) and the immunoprecipitates is shown. The input and pellet were diluted 1:4,000 and 1:20, respectively. PCR was carried out by using primers specific (a) to the SL RNA promoter or (b) to the ribosomal DNA locus. The positions of the PCR products are indicated (not in scale) above a scheme depicting the transcription loci. Nucleotides are numbered with respect to the transcription start site. DAPI, 4,6-diamidino-2-phenylindole; DIC, differential interference contrast; FITC, fluorescein isothiocyanate; SL RNA, spliced-leader RNA; SRα, SRP receptor α; SRP, signal-recognition particle.
To control for off-target silencing of SRα, we constructed a second stem–loop construct to a different domain of the gene. The results (supplementary Fig S4 online) show that this silencing has the same phenotype such as SL RNA reduction, tSNAP42 accumulation and aberrant localization. As the regulation observed in this study affected mostly SL RNA transcription, we termed this novel process spliced-leader RNA silencing (SLS).
Inducers of SLS
To investigate the source of the signal that elicited SLS, we examined the status of SRP on ribosomes. Release of the SRP–RNC complex requires GTPase activity, which in turn requires the interaction of SRP54–SRα; therefore, it is possible that the absence of the SRα receptor might cause the SRP–RNC complex to become immobilized. Accordingly, SRP might become fixed onto ribosomes. Extracts were prepared to separate free SRP present in the post-ribosomal supernatant (PRS) from ribosomal-bound SRP. Figure 4A shows that during silencing, the level of ribosomal-bound SRP, as detected by northern analysis of 7SL RNA, increased from 22%±4 in the uninduced cells to 65%±3 in the silenced cells. The quantity and quality of ribosomes in each fraction was determined using the 5.8S rRNA probe. These results indicate that, in the absence of the SRα receptor, SRP bound to ribosomes is not transient and that the interaction of SRP–RNC is stable and can withstand high-salt extraction. We examined next the localization of these SRP–RNC complexes in the cell by in situ hybridization with 7SL RNA. The results indicate that during silencing, all the SRP that are normally spread in the cytoplasm became concentrated in a defined area (Fig 4B). These data indicate that in SLS the signal is transmitted from the SRP stalled on the ribosomes to the nucleus to affect SL RNA transcription.
Figure 4.
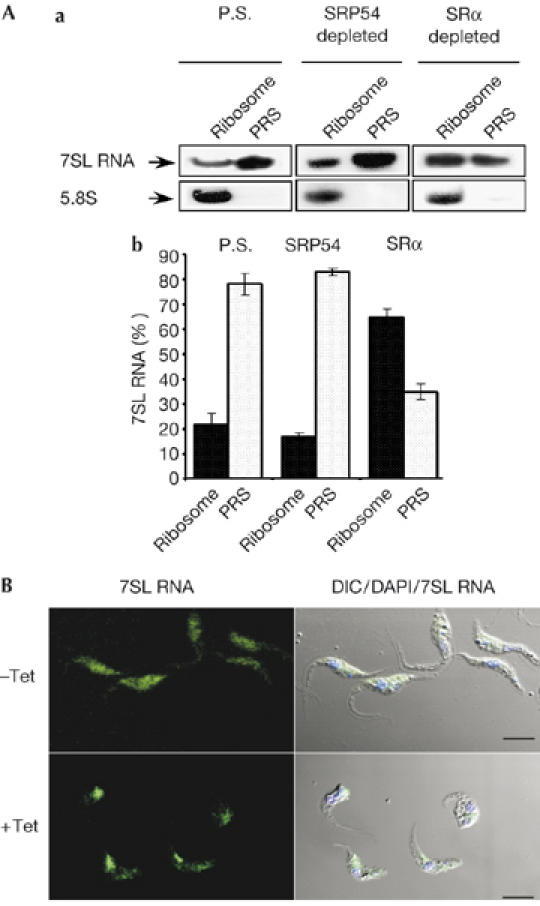
Changes in signal-recognition particle localization and ribosome association during signal-recognition parcticle receptor α depletion. (A) Binding of SRP to the RNC. (a) Extracts, PRS and ribosome fractions were prepared as described in the Methods from parental strain cells (P.S), and from cells depleted of SRP54 or SRα, 3 days after induction. RNA prepared from PRS and ribosomal fractions was subjected to northern analysis with 7SL RNA and 5.8S rRNA probes (indicated with arrows). (b) Quantitative analysis shows the percentage of 7SL RNA in PRS and ribosomal fractions, as determined by densitometry of three independent experiments. Standard deviation is indicated by error bars. (B) Localization of 7SL RNA during SRα silencing. Cells uninduced (−Tet) and after 3 days of induction (+Tet) were fixed and hybridized with DIG-labelled PCR probe to 7SL RNA, which was detected by FITC-conjugated DIG antibodies. The nucleus was stained with DAPI. Scale bars, 5 μm. DIC, differential interference contrast; DIG, digoxigenin 11; FITC, flourescein isothiocyanate; PRS, post-ribosomal supernatant; RNC, ribosome nascent chain; SRα, SRP receptor α; SRP, signal-recognition particle.
To investigate whether SLS can be induced under physiological conditions, parasites were exposed to low pH and the level of SL RNA was examined. The results show a significant reduction in SL RNA (Fig 5Aa) and mRNAs (Fig 5Ab). The reduction is attributed to SLS, as elevation in tSNAP42 was observed (Fig 5B), indicating that SLS is induced by the parasite in response to a relevant physiological stress.
Figure 5.

Spliced-leader RNA silencing is activated under pH stress. (A) SL RNA and mRNA reduction under low pH. (a) RNA was prepared from parental strain cells grown for 2–3 days in pH 7.0 or 5.5 and subjected to northern analysis with SL RNA probe and 5.8S rRNA probe (used as a control for equal loading). (b) The same as in (a), but using mRNA probes, as indicated. (B) The level of tSNAP42 under low pH conditions. Proteins extracted from the same number of cells (1 × 107 cells), as described in (A), were subjected to western analysis with tSNAP42 antibody. SL RNA, spliced-leader RNA.
Discussion
In this study, we describe a novel regulatory circuit that responds to stress by the elimination of mRNA production, elicited by shut-off of SL RNA transcription. This is an immediate and extreme response, which leads to parasite death.
SLS was induced by a major stress to the protein translocation machinery; this is unlikely to occur in nature but might mimic other stresses that the parasite encounters. This indicates that the signalling pathway leading to SLS might be induced by different cues. At present, the signalling pathway involved in SLS is unknown. It is also unknown whether the inability of tSNAP42 to bind to its promoter is because it is modified directly by SLS or if SLS affects yet another factor that does not allow tSNAP42 binding.
SLS is activated under low pH, and at present we are investigating the effect of various stresses on SLS induction. The ability to transmit defects in sorting and folding of proteins to shut off a cellular process, such as transcription observed in this study, resembles UPR found in other eukaryotic cells. The UPR elicited by various stresses induces upregulation of the folding capacity of the endoplasmic reticulum and downregulation of the biosynthetic load, by shutting off protein synthesis at the transcriptional and translational levels (Schroder & Kaufman, 2005). Trypanosomes might carry two lines of defence against stress: a UPR-like mechanism and SLS. These pathways might be activated, depending on the severity of stress. Severe stress might activate SLS to quickly eliminate these ‘sick' parasites from the population.
It is interesting to examine whether the phenomenon described here is specific to trypanosomes, or whether similar transcription shut-off can be induced in metazoa on depletion of the SRP receptor. This is the first demonstration that SL RNA expression by SLS is a master regulator in the trypanosome cell. This discovery opens a new path towards a therapy based on SLS pathway activation. This could lead to stymied parasitic growth, and to relief from the devastating diseases caused by these parasites.
Methods
Cell growth, transfection and extract preparation. Procyclic T. brucei strain 29-13 (Wirtz et al, 1999) were grown in SDM-79 medium and transfected as described by Liu et al (2002). Extracts and preparation of ribosome and post-ribosomal supernatant have been described by Ben Shlomo et al (1999). Nuclear extracts were prepared as described previously (Das et al, 2005).
RNA analysis. Northern analysis for mRNA has been described by Liu et al (2002). Small RNAs were fractionated on a 10% (w/v) polyacrylamide gel containing 7 M urea. The RNA blots were hybridized to oligonucleotides or random primer DNA probes as specified in the supplementary Table I online. Primer extension was as described by Liu et al (2003).
Microscopy. Immunofluorescence was as described by Liu et al (2002). Cell nuclei were stained with 4′-6-diamidino-2-phenylindole (DAPI) for 5 min. For in situ hybridization, digoxigenin-labelled DNA probes were used as described previously (Lustig et al, 2005). Cells were visualized using a Zeiss LSM 510 META inverted microscope (Carl Zeiss AG 73446, Oberkochen, Germany). SEM was carried out as described by Rothmann et al (2000).
Cell permeabilization. The procedure was similar to that described by Tschudi & Ullu (1990). The only deviation from the published protocol is that transcription buffer TB × 1 was used (150 mM sucrose, 20 mM potassium, L-glutamate (Sigma, St Louis, MO, USA), 10 mM HEPES-KOH (pH 7.9), 2.5 mM MgCl2, 1 mM dithiotheritol, 10 μg/ml leupeptin).
For slot-blot analysis of the RNA synthesized in permeable cells, plasmid DNA or PCR products were used. Hybridization was carried out at 55–60°C in 60% (v/v) formamide, 2 × SSC (0.3 M sodium chloride, 0.03 M sodium citrate), 100 μg/ml salmon sperm DNA, 0.1% (w/v) Sarcosyl, with the entire RNA fraction extracted from permeable cells. After hybridization, filters were washed twice in 2 × SSC and 0.1% (w/v) SDS at 65°C for 30 min.
Chromatin immunoprecipitation. ChIP was carried out as described by Lowell & Cross (2004). Immunoprecipitated material after cross link reversal was deproteinized and subjected to PCR analysis using primers (see the supplementary information online). Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Information
Acknowledgments
We thank M. Mandelboim for her help in the permeable cell expression. This study was supported by an International Research Scholar grant from the Howard Hughes Research Institute to S.M.
References
- Ben Shlomo H, Levitan A, Shay NE, Goncharov I, Michaeli S (1999) RNA editing associated with the generation of two distinct conformations of the trypanosomatid Leptomonas collosoma 7SL RNA. J Biol Chem 274: 25642–25650 [DOI] [PubMed] [Google Scholar]
- Das A, Zhang Q, Palenchar JB, Chatterjee B, Cross GA, Bellofatto V (2005) Trypanosomal TBP functions with the multisubunit transcription factor tSNAP to direct spliced-leader RNA gene expression. Mol Cell Biol 25: 7314–7322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dossin FM, Schenkman S (2005) Actively transcribing RNA polymerase II concentrates on spliced leader genes in the nucleus of Trypanosoma cruzi. Eukaryot Cell 4: 960–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilinger G, Bellofatto V (2001) Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms. Nucleic Acids Res 29: 1556–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Stroud RM, Walter P (2001) The signal recognition particle. Annu Rev Biochem 70: 755–775 [DOI] [PubMed] [Google Scholar]
- Liu L, Liang XH, Uliel S, Unger R, Ullu E, Michaeli S (2002) RNA interference of signal peptide-binding protein SRP54 elicits deleterious effects and protein sorting defects in trypanosomes. J Biol Chem 277: 47348–47357 [DOI] [PubMed] [Google Scholar]
- Liu L, Ben Shlomo H, Xu YX, Stern MZ, Goncharov I, Zhang Y, Michaeli S (2003) The trypanosomatid signal recognition particle consists of two RNA molecules, a 7SL RNA homologue and a novel tRNA-like molecule. J Biol Chem 278: 18271–18280 [DOI] [PubMed] [Google Scholar]
- Lowell JE, Cross GA (2004) A variant histone H3 is enriched at telomeres in Trypanosoma brucei. J Cell Sci 117: 5937–5947 [DOI] [PubMed] [Google Scholar]
- Lustig Y, Goldshmidt H, Uliel S, Michaeli S (2005) The Trypanosoma brucei signal recognition particle lacks the Alu-domain-binding proteins: purification and functional analysis of its binding proteins by RNAi. J Cell Sci 118: 4551–4562 [DOI] [PubMed] [Google Scholar]
- Powers T, Walter P (1995) Reciprocal stimulation of GTP hydrolysis by two directly interacting GTPases. Science 269: 1422–1424 [DOI] [PubMed] [Google Scholar]
- Rapiejko PJ, Gilmore R (1997) Empty site forms of the SRP54 and SRα GTPases mediate targeting of ribosome-nascent chain complexes to the endoplasmic reticulum. Cell 89: 703–713 [DOI] [PubMed] [Google Scholar]
- Rothmann C, Levinshal T, Timan B, Avtalion RR, Malik Z (2000) Spectral imaging of red blood cells in experimental anemia of Cyprinus carpio. Comp Biochem Physiol A Mol Integr Physiol 125: 75–83 [DOI] [PubMed] [Google Scholar]
- Schroder M, Kaufman RJ (2005) ER stress and the unfolded protein response. Mutat Res 569: 29–63 [DOI] [PubMed] [Google Scholar]
- Tschudi C, Ullu E (1990) Destruction of U2, U4, or U6 small nuclear RNA blocks trans splicing in trypanosome cells. Cell 61: 459–466 [DOI] [PubMed] [Google Scholar]
- Ullu E, Tschudi C (1990) Permeable trypanosome cells as a model system for transcription and trans-splicing. Nucleic Acids Res 18: 3319–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Morris JC, Drew ME, Englund PT (2000) Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J Biol Chem 275: 40174–40179 [DOI] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol 99: 89–101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


