Abstract
Defects in chromosome replication can lead to translocations that are thought to result from recombination events at stalled DNA replication forks. The progression of forks is controlled by an essential DNA helicase, which unwinds the parental duplex and can stall on encountering tight protein–DNA complexes. Such pause sites are hotspots for recombination and it has been proposed that stalled replisomes disassemble, leading to fork collapse. However, in both prokaryotes and eukaryotes it now seems that paused forks are surprisingly stable, so that DNA synthesis can resume without recombination if the barrier protein is removed. Recombination at stalled forks might require other events that occur after pausing, or might be dependent on features of the surrounding DNA sequence. These findings have important implications for our understanding of the regulation of genome stability in eukaryotic cells, in which pausing of forks is mediated by specific proteins that are associated with the replicative helicase.
Keywords: checkpoint, DNA replication fork barriers, mini-chromosome maintenance helicase, recombination, replisome
Introduction
DNA replication forks are established from a single origin in the circular chromosome of Escherichia coli and at multiple origins on each linear chromosome in eukaryotic cells. An assembly of proteins termed the replisome is associated with each fork, and comprises the replicative helicase together with DNA polymerases, a primase to initiate each new DNA fragment and other accessory factors. Progression of a fork normally continues until it encounters another fork coming in the opposite direction, at which point termination occurs and the replisome is disassembled.
In the presence of certain kinds of alkylation on the DNA template, or if the production of nucleotides is inhibited, the replicative helicase progresses much more slowly, so that the fork is said to have ‘stalled' (Katou et al, 2003; Tercero & Diffley, 2001). Forks can also stall in response to agents that inhibit polymerases but that allow the helicase to continue unwinding the parental duplex (Pacek et al, 2006; Walter & Newport, 2000). Eukaryotic cells activate checkpoint kinases under such conditions, in response to altered features of the fork, including the exposure of more single-stranded DNA (ssDNA; Branzei & Foiani, 2005; Li & Zou, 2005; McGowan & Russell, 2004). Checkpoint kinases have an essential role in such situations because they prevent an irreversible collapse of the stalled forks, which would otherwise be fatal for the cell (Lopes et al, 2001; Tercero & Diffley, 2001).
Forks also pause during the normal process of chromosome replication at ‘replication fork barriers' (RFBs), where particular proteins bind tightly to DNA. In the E. coli genome, a range of pause sites have been identified. Terminator (Ter) sequences are located on the opposite side of the circular chromosome to the origin and these represent programmed pause sites that are bound by a protein called Tus. The Tus–Ter complex forms a polar barrier that blocks progression of the replicative helicase when a fork arrives in the non-permissive direction (Hill & Marians, 1990). The stalled fork is resolved when a fork arrives from the opposite direction and triggers termination (Fig 1A). Other RFBs in E. coli represent accidental pause sites, which are usually less efficient. For example, forks might pause on encountering the Lac or Tet repressors bound to their cognate operators, but efficient stalling requires tandem arrays of many such complexes (Payne et al, 2006; Possoz et al, 2006). Finally, a bacterial fork moves more slowly when it encounters the ribosomal RNA (rRNA) transcriptional machinery moving in the opposite direction (French, 1992) and pauses for longer periods at sites where the transcription machinery has stalled in DNA sequences that are rich in GC repeats (Krasilnikova et al, 1998).
Figure 1.
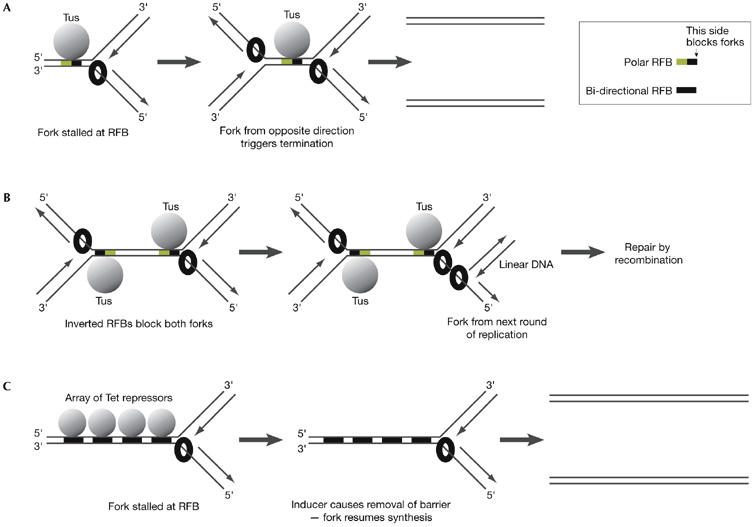
Pausing of forks at replication fork barriers in Escherichia coli. (A) A fork normally pauses at a Tus protein–terminator sequence (Tus–Ter) barrier until another fork arrives from the opposite direction and induces termination. (B) Artificial inversion of Tus–Ter barriers blocks forks in both directions. Arrival of a new fork from the next round of replication produces linear DNA that might drive repair by recombination. (C) Forks stall at large arrays of Tet operators bound by the repressor protein, but removal of the barrier by an inducer (anhydrotetracycline) allows the rapid resumption of DNA synthesis in the absence of recombination. RFB, replication fork barrier.
RFBs also exist in eukaryotic cells and can be programmed or accidental, polar or bi-directional. The best examples of programmed RFBs are the polar barriers that ensure that the rDNA is largely replicated in the same direction as transcription (Brewer & Fangman, 1988; Krings & Bastia, 2005; Linskens & Huberman, 1988; Sanchez-Gorostiaga et al, 2004) and the replication termination sequence 1 (RTS1) barrier, which causes unidirectional replication of the fission yeast mating-type locus (Dalgaard & Klar, 2001). Eukaryotic forks also encounter a range of weaker accidental RFBs, such as the polar barriers that form at transfer RNA (tRNA) genes (Deshpande & Newlon, 1996), and the bi-directional barriers that are present at centromeres (Greenfeder & Newlon, 1992). Stalling is not normally observed when eukaryotic forks encounter the RNA polymerase II (Pol II) machinery moving in the opposing direction through linear chromosomes (Brewer & Fangman, 1988; Ivessa et al, 2003), but can be seen when such clashes occur in circular plasmids, suggesting that an additional torsional contribution is required (Prado & Aguilera, 2005). As in E. coli, eukaryotic forks also pause on encountering sites where Pol II stalls in GC-rich DNA, perhaps owing to the encounter with defective messenger ribonucleoprotein complexes (Wellinger et al, 2006). Finally, forks also pause transiently at other protein–DNA complexes, such as pre-replicative complexes at origins of replication and protein complexes at, or near, telomeres (Makovets et al, 2004; Miller et al, 2006; Wang et al, 2001).
In both prokaryotes and eukaryotes, RFB sites are often associated with an increased frequency of recombination, leading to the theory that paused forks stimulate recombination directly through the rapid disassembly of the stalled replisome and the collapse of the fork (see, for example, Defossez et al, 1999; Admire et al, 2006; Kobayashi et al, 1998; Lambert et al, 2005). When forks pause at RFBs they do not seem to elicit or require a checkpoint response (Calzada et al, 2005; Lambert et al, 2005), probably owing to the lack of substantial amounts of ssDNA (Gruber et al, 2000; Lucchini & Sogo, 1994). This lack of stabilization by checkpoint kinases has been taken as a potential explanation for the collapse of paused forks that can lead to recombination (Admire et al, 2006; Lambert et al, 2005).
Nevertheless, there is a growing body of evidence from both prokaryotes and eukaryotes to support an alternative view. It seems that paused forks and their associated replisomes are surprisingly stable, so that the act of pausing per se is unlikely to induce collapse of the fork. Furthermore, there is evidence, in several cases, to indicate that hotspots of recombination are dependent on other factors, in addition to the pausing of a DNA replication fork. The literature concerning these issues is both fascinating and highly complex, and forms the topic of this review.
Recombination at paused forks in E. coli
Several studies have shown that the pausing of forks at Tus–Ter complexes can stimulate recombination at these sites. A Ter sequence acts as a deletion hotspot when placed in a plasmid (Bierne et al, 1991), and the λ prophage excises at a higher frequency at the terminus than at other sites in the E. coli chromosome (Louarn et al, 1991). Furthermore, a screen for ‘hot' DNAs that stimulate recombination identified Ter sequences (Nishitani et al, 1993) and such hot activity requires the Tus protein, suggesting a link to paused forks (Horiuchi et al, 1994).
Insertion of additional Ter sequences into the E. coli chromosome—so as to create a region that forks cannot enter from either direction—produces strains that grow slowly and are dependent on the RecA protein, which mediates strand invasion during recombination (Horiuchi & Fujimura, 1995; Sharma & Hill, 1995). Such strains accumulate linear DNA in the presence of Tus and have increased rates of recombination adjacent to the pause sites, indicating that the stalled forks lead to the formation of double-strand breaks in DNA (DSBs; Bierne et al, 1997; Michel et al, 1997). However, DSBs are not produced by the collapse of the fork that initially pauses at the Tus–Ter complex; instead, they result from the arrival of a new fork in the subsequent round of chromosome replication (Fig 1B). Replication of the daughter strand from the first round then produces a linear molecule that stimulates recombination (Bidnenko et al, 2002; Sharma & Hill, 1995).
The presence of the Lac promoter in an E. coli plasmid also stimulates recombination in a manner that is apparently linked to the replication of the plasmid (Vilette et al, 1992). Insertion of a tandem array of 34 Lac operators into the E. coli chromosome makes the resultant strain dependent on RecA for viability when the Lac repressor is expressed (Payne et al, 2006). Furthermore, a large array of 240 Tet operator sites blocks growth altogether in the presence of the Tet repressor protein (Possoz et al, 2006). Importantly, however, two-dimensional DNA gels show that a paused DNA replication fork can resume DNA synthesis within 5 min of the addition of inducer to remove the Tet repressor protein from the array (Possoz et al, 2006). This is true even in the absence of RecA, indicating that restart in this case might simply involve the direct resumption of DNA synthesis by the stalled replisome (Fig 1C).
Clashes between forks and the transcriptional machinery can also promote the formation of deletions in E. coli plasmids (Vilette et al, 1992), and many genes in the chromosome are orientated so as to avoid such clashes with DNA replication forks (Brewer, 1988). Nevertheless, this seems to correlate more with the essential nature of the genes involved than with the level of expression, indicating that such preferential orientation represents a kind of ‘insurance policy' to protect essential genes, whereas cells are still able, in most cases, to deal with clashes between replication forks and active RNA polymerase (Rocha & Danchin, 2003).
Together, these data indicate that the stalling of forks at protein–DNA barriers is, indeed, associated with increased recombination in E. coli. Paused forks do not necessarily collapse, however, and DSBs are only produced in an indirect manner by the subsequent round of DNA replication. Paused bacterial replisomes have not been examined directly; however, it seems likely that they persist at RFBs, as DNA synthesis resumes rapidly after removal of a barrier protein.
Pausing of forks and recombination in budding yeast
The link between replication and recombination in eukaryotic cells has been studied most intensively with regard to the rDNA locus in budding yeast (Fig 2). The rDNA array comprises several hundred repeats of a 9-kb unit that contains the genes encoding the rRNAs. Recombination in the rDNA is regulated in a highly complex manner, as recombination between repeats is required to maintain the copy number of the array, but must also be restrained to prevent the excision of extra-chromosomal circles (ERCs). These comprise one or more rDNA repeats and are thought to contribute to ageing (Defossez et al, 1999).
Figure 2.
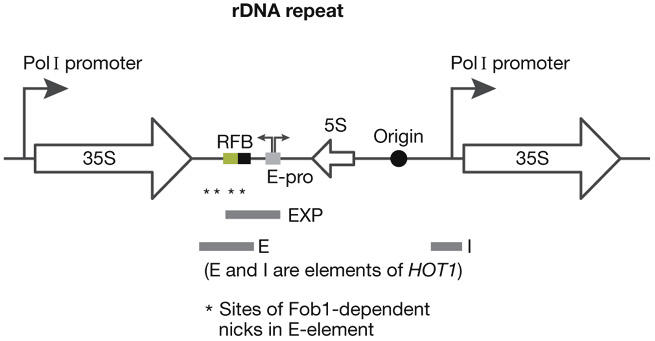
The ribosomal DNA locus in budding yeast. The replication fork barrier (RFB) ensures that the 35S gene is replicated in the same direction as RNA polymerase I (Pol I) transcription. Maintenance of the copy number of the rDNA repeats is dependent on the EXP sequence (expansion of rDNA repeats) that comprises the RFB and the bi-directional Pol II promoter E-pro (expressing two non-coding mRNAs). The enhancer of rDNA transcription (E) element (containing the RFB and associated 5′ sequences) and the initiation of rDNA transcription (I) element (containing the Pol I promoter) of HOT1 stimulate recombination when placed at other chromosomal loci. Fob1, fork blocking 1; rDNA, ribosomal DNA.
The first insight into the mechanisms of rDNA recombination came from a screen for hot DNAs that can stimulate recombination when placed in a plasmid. This led to the identification of HOT1, which is a fragment derived from the rDNA repeats (Keil & Roeder, 1984). The HOT1 sequence was subsequently found to comprise two key elements. The first component is the promoter for RNA Pol I (I element), which transcribes the rRNA genes; HOT1 only stimulates recombination when Pol I transcribes into the recombining sequences (Huang & Keil, 1995; Voelkel-Meiman et al, 1987). The second component functions as an enhancer of Pol I transcription in the HOT1 fragment (E element; Voelkel-Meiman et al, 1987), although it is not required for Pol I transcription in the rDNA (Burkhalter & Sogo, 2004; Wai et al, 2001). This enhancer element was found to contain a polar RFB that ensures unidirectional replication throughout the rDNA repeats (Brewer & Fangman, 1988; Linskens & Huberman, 1988). A screen for factors required for HOT1 activity identified the fork blocking 1 (Fob1) protein, which is required for forks to pause at the RFB (Kobayashi & Horiuchi, 1996), and binds directly to the RFB sequence and surrounding sites (Huang & Moazed, 2003; Kobayashi, 2003; Mohanty & Bastia, 2004).
These findings initially suggested that pausing of forks at the RFB might drive recombination in the rDNA in collaboration with Pol I transcription. Consistently, subsequent studies showed that Pol I is required to maintain the high copy number of the rDNA repeats (Brewer et al, 1992; Kobayashi et al, 1998), similar to the Rad52 (Radiation sensitive 52) protein, which is required for almost all recombination events in budding yeast (Gangloff et al, 1996). Fob1 also has a crucial role in the regulation of rDNA copy number; however, the situation is complex, as Fob1 is required for both the decrease of copy number that occurs after the loss of Pol I and for the increase after the subsequent re-introduction of the enzyme (Kobayashi et al, 1998). The formation of monomeric ERCs is reduced in the absence of Fob1 and the lifespan is increased, which is consistent with Fob1 having a crucial role in regulating recombination within the rDNA (Defossez et al, 1999).
The link between the RFB and recombination is enigmatic, however, as the insertion of HOT1 outside the rDNA promotes recombination in a manner that is dependent on Fob1 and Pol I, even when the direction of replication is such that forks do not pause at the RFB (Ward et al, 2000). This indicates that Fob1 contributes to HOT1 recombination independently of its role in pausing forks at the RFB. In addition, although HOT1 recombination normally requires Fob1, this is not the case after hyperactivation of Pol I transcription in a strain with a reduced number of rDNA repeats (Serizawa et al, 2004).
Within the endogenous rDNA, the maintenance of copy number requires an element called expansion of rDNA repeats (EXP), which comprises not only the RFB sequence, but also the adjacent 400 bp (Fig 2). The 400 bp region is not required for RFB function, but instead contains a non-coding bidirectional promoter, E-Pro, which can stimulate the dissociation of cohesin from the surrounding region (Kobayashi & Ganley, 2005).
The Sir2 deacetylase represses transcription by E-Pro within the EXP element (Kobayashi & Ganley, 2005) and limits unequal sister-chromatid recombination within rDNA (Kobayashi et al, 2004). In the absence of Sir2, a plasmid containing the Fob1–RFB integrates into the rDNA with high frequency, but only if the RFB is oriented such that forks pause at the RFB (Benguria et al, 2003). However, recombination under such conditions requires the full EXP element, and so pausing of forks at the RFB is only part of the mechanism.
In addition to mediating fork pausing at the RFB, it is now clear that Fob1 has other functions that could explain its role in HOT1 recombination and the maintenance of rDNA copy number. An elegant study showed that Fob1 is required for the formation of nicks at specific locations within the rDNA enhancer (Burkhalter & Sogo, 2004). These nicks are present even during G1 phase and occur at sites that are distinct from the RFB but are important for HOT1 activity. The formation of such nicks was subsequently found to require topoisomerase I, in addition to Fob1, and an in vivo footprint at the corresponding sites was found to be dependent on Fob1 (Di Felice et al, 2005).
Together, these studies show that recombination at both HOT1 and the endogenous rDNA is stimulated by a multitude of factors and activities. Stalling of forks at the RFB might be one important element; however, it is clear that a paused fork is not sufficient to stimulate recombination between the rDNA repeats and that other events are also required. These are dependent on both surrounding DNA sequence elements and the activity of other proteins.
Sites where forks pause on encountering stalled Pol II complexes are also hotspots for recombination (Prado & Aguilera, 2005; Wellinger et al, 2006). Recombination at such sites requires transcription to occur during S phase, and both pausing and recombination are stimulated in the absence of the Rrm3 (ribosomal DNA recombination mutation 3) helicase (Prado & Aguilera, 2005; Wellinger et al, 2006), which aids the passage of forks past protein–DNA barriers (Ivessa et al, 2003). This is consistent with a mechanistic link between the stalling of forks and the stimulation of recombination. However, plasmids with active pause sites of this kind can be maintained even in cells lacking either the RecA homologue Rad51 or the Rad52 protein, and the presence of replication intermediates in two-dimensional DNA gels is not altered by the absence of these proteins, indicating that the paused forks often recover without recombination (Prado & Aguilera, 2005).
Recombination after fork pausing in fission yeast
The pausing of DNA replication forks has a crucial role in a specific recombination reaction that mediates the switching of mating type in fission yeast (Dalgaard & Klar, 1999, 2000). A genomic imprint is formed at the recipient mat1 locus on chromosome 2 and promotes recombination with the donor loci. Formation of the imprint requires that mat1 is replicated by a fork moving towards the centromere; this is achieved by the RTS1 replication fork barrier that is located before mat1 on the centromere proximal side, and that blocks forks from this direction (Dalgaard & Klar, 2001). RTS1 therefore contributes indirectly to the subsequent recombination reaction of mating-type switching, by determining the direction of replication of the mating-type region.
Two studies have shown, however, that transposition of RTS1 to other loci on chromosome 3 stimulates recombination at these sites (Fig 3A). In the first study, insertion of either one or two copies of RTS1 at the ura4 locus produced a notable phenotype, in which optimal viability of cells became dependent on Rad22—the fission yeast homologue of Rad52—and the frequency of chromosomal translocations was greatly increased (Lambert et al, 2005). A similar phenotype was observed in cells in which endogenous RTS1 was deleted, so that RTS1–ura4 on chromosome 3 was the only copy of the RTS1 sequence (Lambert et al, 2005). The Rad22 protein could be detected by chromatin immunoprecipitation (ChIP) at the RTS1–ura4 locus, in contrast to the endogenous RTS1 locus on chromosome 2 (Lambert et al, 2005).
Figure 3.
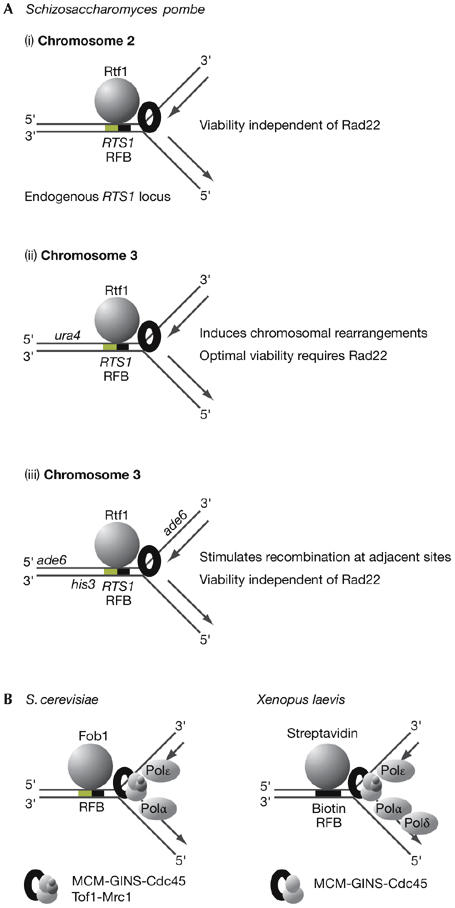
Pausing of forks at eukaryotic replication fork barriers. (A) Pausing of forks at the replication termination sequence 1 (RTS1) barrier in Schizosaccharomyces pombe can have different outcomes depending on the chromosomal context (for (ii) see Lambert et al, 2005; for (iii) see Ahn et al, 2005). (B) Chromatin immunoprecipitation studies have shown that replisome components persist at paused forks in Saccharomyces cerevisiae (see Calzada et al, 2005) and Xenopus laevis (see Pacek et al, 2006). Fob1, fork blocking 1; Pol, DNA polymerase; Rtf1, replication termination factor 1. The MCM (mini-chromosome maintenance) helicase is required for the progression of eukaryotic DNA replication forks, together with the Cdc45 protein and the four-protein GINS complex (Sld5–Psf1–Psf2–Psf3). In budding yeast, MCM–GINS–Cdc45 also interact with the regulatory proteins Tof1 and Mrc1.
In a second study, transposition of RTS1 to the ade6 locus on chromosome 3 stimulated recombination at adjacent sequences, but did not make cells dependent on Rad22 for optimal viability (Ahn et al, 2005).
These studies confirm the link between paused replication forks and increased recombination at adjacent loci; however, they also indicate that the consequences of a DNA replication fork stalling at a particular protein–DNA barrier might be influenced by the chromosomal context surrounding the RFB.
Preservation of the paused replisome at RFBs
To understand the molecular events that result from pausing of replication forks at RFBs, it is necessary to study the kinetics of pausing at specific loci during a single round of chromosome replication (Fig 3B).
One such approach in eukaryotic cells was the creation of a budding yeast strain with two opposed Fob1–RFBs between two highly active and early origins of replication on chromosome 3 (Calzada et al, 2005). Synchronous cell cultures were used to show that forks arrive at each barrier with similar kinetics, pause for an extended period and then pass the barrier before completing replication of the intervening region. ChIP experiments showed that replisome components can be detected at each RFB throughout the period during which pausing occurs. Pausing and recovery are independent of checkpoint kinases and Rad52, and might simply involve the resumption of synthesis by the stalled fork after removal of the barrier. Consistently, the Rrm3 helicase specifically accumulates at both barrier sites during the period of pausing (Calzada et al, 2005)—note that a recent study also detected Rrm3 at normal replication forks (Azvolinsky et al, 2006).
In another study using extracts of Xenopus eggs, an ‘accidental' RFB was created at a specific site on a plasmid, in the form of a biotin–streptavidin complex (Pacek et al, 2006). ChIP experiments showed that replisome components accumulated at the RFB during the period of pausing, as seen at the budding yeast Fob1–RFB.
Together, these experiments indicate that paused eukaryotic replisomes do not simply disassemble on pausing, but instead can remain stable for extended periods. Therefore, although collapse and recombination represent one possible outcome of stalling, it seems likely that eukaryotic forks can often simply resume synthesis once the barrier has been removed, just as stalled forks in the E. coli genome are able to recover after barrier removal at large Tet arrays (Possoz et al, 2006).
Stalling of DNA replication forks at fragile sites
Specific sites in eukaryotic chromosomes are known to be particularly sensitive to breakage when chromosome replication is perturbed, resulting in the formation of chromosomal translocations (Richards, 2001).
In mammalian cells, common fragile sites can be visualized by treating cells with the polymerase inhibitor aphidicolin and then subsequently examining metaphase spreads. Reduced expression of the checkpoint kinase ATM and Rad3-related (ATR) increases the frequency of rearrangements at fragile sites, even in the absence of aphidicolin (Casper et al, 2002). A related phenomenon might exist in budding yeast, in which absence of the Mec1 (Mitosis entry checkpoint 1) kinase—which is functionally similar to ATR—causes increased breakage of chromosomes at particular loci that correspond to ‘replication slow zones' (Cha & Kleckner, 2002).
Two screens for fragile sites in budding yeast showed that they are usually associated with sequences that contain both Ty transposons and tRNAs (Admire et al, 2006; Dunham et al, 2002). Fragile sites have also been detected in budding yeast by reducing the expression of DNA polymerase-α. Such fragile sites are associated with the inverted repeat sequences of Ty elements (Lemoine et al, 2005). A model has been proposed in which the breakage of fragile sites requires two factors: slowing of replication and the presence of sequence elements, such as inverted repeats, which can form potentially recombinogenic secondary structures when unwound (Lemoine et al, 2005). Impairing polymerase-α function should increase the amount of ssDNA on the lagging-strand template, thereby favouring secondary-structure formation when the fragile site is unwound.
tRNAs act as polar RFBs; therefore, the association of yeast fragile sites with Ty elements and tRNAs raises the possibility that the stalling of forks at tRNA RFBs might initiate the formation of fragile sites (Admire et al, 2006). It seems likely that nearby sequence elements, such as the long terminal repeats of the Ty elements, will also contribute to fragile-site formation. Perhaps the role of checkpoint kinases in protecting cells against fragile sites might relate to the stabilization of DNA secondary structures that are formed when such elements are unwound, particularly as checkpoint kinases are not required for pausing or recovery of forks per se at the budding yeast Fob1–RFB (Calzada et al, 2005), or at the fission yeast RTS1–ura4 RFB (Lambert et al, 2005).
Mechanisms of fork pausing at RFBs
Pausing of replication forks at accidental barriers, such as Tet repressor/operator arrays in E. coli, might simply result from the physical clash between the replisome and the protein–DNA complex at the RFB. At the Tus–Ter barrier, however, pausing occurs by a more sophisticated mechanism. One possibility is that specific interactions between Tus and the replicative helicase dnaB might contribute to pausing (Mulugu et al, 2001). However, a recent study has identified a different mechanism in which unwinding of the RFB from the non-permissive side springs a ‘molecular mousetrap', which allows a conserved base in the displaced strand to ‘flip-out' so that it can be bound tightly by Tus (Mulcair et al, 2006).
Eukaryotic cells seem to have evolved a different strategy to deal with the pausing of forks at protein–DNA barriers. Pausing at the fission yeast mating-type locus, at the rDNA of both budding and fission yeast, and also at a budding yeast tRNA has been found to require two specific proteins: Swi1/3 (identified through mutations that inhibit switching of mating type) in fission yeast and Tof1/Csm3 (Topoisomerase I interacting factor 1/Chromosome segregation in meiosis 3) in budding yeast (Calzada et al, 2005; Dalgaard & Klar, 2000; Krings & Bastia, 2004; Mohanty et al, 2006; Tourriere et al, 2005). These two factors bind each other and associate with the replicative mini-chromosome maintenance (MCM) helicase at DNA replication forks (Gambus et al, 2006; Katou et al, 2003; Lee et al, 2004; Mayer et al, 2004; Noguchi et al, 2004).
It therefore seems that pausing of eukaryotic forks is—at least in these particular cases—an active process mediated by the replisome itself. This could allow eukaryotic cells to do other important things at paused replication forks, such as establishing the imprint at the fission yeast mating-type locus. It is interesting to note that two studies have shown that recombination rates are increased in fission yeast cells lacking Swi1/3 (Ahn et al, 2005; Sommariva et al, 2005). Furthermore, both fission yeast and budding yeast cells lacking Swi1/Tof1 often have subnuclear foci of recombination proteins (Noguchi et al, 2004; Tourriere et al, 2005). This indicates that the ability of eukaryotic DNA replication forks to pause their progression at RFBs might help to preserve genome stability. Perhaps proteins such as Swi1/Tof1 assist the fork in pausing stably at tight protein–DNA barriers until helicases, such as Rrm3, unwind the RFB and displace the barrier protein, so that the replicative helicase can proceed and allow the fork to resume synthesis.
Conclusions
It is clear that much remains to be learnt about the mechanisms by which eukaryotic forks pause at protein–DNA barriers, and about the molecular events that follow the stalling of forks at such RFBs. It will be fascinating to discover whether ‘active' pausing by Tof1/Swi1 is a general feature of all eukaryotic RFBs, and if this mechanism helps to preserve genome stability or has evolved for other reasons. It is clear that pause sites can be hotspots for recombination, although this seems to be linked to other factors besides the act of pausing itself. Perhaps there is competition at protein–DNA barriers between the inherent stability of a replisome that arrests its own progression through Tof1/Swi1, and the possibility that collapse and recombination might occur stochastically or be promoted by other features at fragile sites. If this is the case, although eukaryotic cells often hold forks, they do not necessarily have a knife in the other hand.
Acknowledgments
We gratefully acknowledge the financial support of Cancer Research UK and the EMBO Young Investigator Programme.
References
- Admire A, Shanks L, Danzl N, Wang M, Weier U, Stevens W, Hunt E, Weinert T (2006) Cycles of chromosome instability are associated with a fragile site and are increased by defects in DNA replication and checkpoint controls in yeast. Genes Dev 20: 159–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn JS, Osman F, Whitby MC (2005) Replication fork blockage by RTS1 at an ectopic site promotes recombination in fission yeast. EMBO J 24: 2011–2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A, Dunaway S, Torres JZ, Bessler JB, Zakian VA (2006) The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev 20: 3104–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benguria A, Hernandez P, Krimer DB, Schvartzman JB (2003) Sir2p suppresses recombination of replication forks stalled at the replication fork barrier of ribosomal DNA in Saccharomyces cerevisiae. Nucleic Acids Res 31: 893–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidnenko V, Ehrlich SD, Michel B (2002) Replication fork collapse at replication terminator sequences. EMBO J 21: 3898–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H, Ehrlich SD, Michel B (1991) The replication termination signal terB of the Escherichia coli chromosome is a deletion hot spot. EMBO J 10: 2699–2705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H, Ehrlich SD, Michel B (1997) Deletions at stalled replication forks occur by two different pathways. EMBO J 16: 3332–3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei D, Foiani M (2005) The DNA damage response during DNA replication. Curr Opin Cell Biol 17: 568–575 [DOI] [PubMed] [Google Scholar]
- Brewer BJ (1988) When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell 53: 679–686 [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL (1988) A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55: 637–643 [DOI] [PubMed] [Google Scholar]
- Brewer BJ, Lockshon D, Fangman WL (1992) The arrest of replication forks in the rDNA of yeast occurs independently of transcription. Cell 71: 267–276 [DOI] [PubMed] [Google Scholar]
- Burkhalter MD, Sogo JM (2004) rDNA enhancer affects replication initiation and mitotic recombination: Fob1 mediates nucleolytic processing independently of replication. Mol Cell 15: 409–421 [DOI] [PubMed] [Google Scholar]
- Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K (2005) Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev 19: 1905–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper AM, Nghiem P, Arlt MF, Glover TW (2002) ATR regulates fragile site stability. Cell 111: 779–789 [DOI] [PubMed] [Google Scholar]
- Cha RS, Kleckner N (2002) ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297: 602–606 [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ (1999) Orientation of DNA replication establishes mating-type switching pattern in S. pombe. Nature 400: 181–184 [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ (2000) swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell 102: 745–751 [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ (2001) A DNA replication-arrest site RTS1 regulates imprinting by determining the direction of replication at mat1 in S. pombe. Genes Dev 15: 2060–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, Silver PA, Keil RL, Guarente L (1999) Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell 3: 447–455 [DOI] [PubMed] [Google Scholar]
- Deshpande AM, Newlon CS (1996) DNA replication fork pause sites dependent on transcription. Science 272: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Di Felice F, Cioci F, Camilloni G (2005) FOB1 affects DNA topoisomerase I in vivo cleavages in the enhancer region of the Saccharomyces cerevisiae ribosomal DNA locus. Nucleic Acids Res 33: 6327–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig F, Botstein D (2002) Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 99: 16144–16149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French S (1992) Consequences of replication fork movement through transcription units in vivo. Science 258: 1362–1365 [DOI] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, Edmondson RD, Labib K (2006) GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8: 358–366 [DOI] [PubMed] [Google Scholar]
- Gangloff S, Zou H, Rothstein R (1996) Gene conversion plays the major role in controlling the stability of large tandem repeats in yeast. EMBO J 15: 1715–1725 [PMC free article] [PubMed] [Google Scholar]
- Greenfeder SA, Newlon CS (1992) Replication forks pause at yeast centromeres. Mol Cell Biol 12: 4056–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber M, Wellinger RE, Sogo JM (2000) Architecture of the replication fork stalled at the 3′ end of yeast ribosomal genes. Mol Cell Biol 20: 5777–5787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TM, Marians KJ (1990) Escherichia coli Tus protein acts to arrest the progression of DNA replication forks in vitro. Proc Natl Acad Sci USA 87: 2481–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T, Fujimura Y (1995) Recombinational rescue of the stalled DNA replication fork: a model based on analysis of an Escherichia coli strain with a chromosome region difficult to replicate. J Bacteriol 177: 783–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi T, Fujimura Y, Nishitani H, Kobayashi T, Hidaka M (1994) The DNA replication fork blocked at the Ter site may be an entrance for the RecBCD enzyme into duplex DNA. J Bacteriol 176: 4656–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GS, Keil RL (1995) Requirements for activity of the yeast mitotic recombination hotspot HOT1: RNA polymerase I and multiple cis-acting sequences. Genetics 141: 845–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Moazed D (2003) Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev 17: 2162–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA (2003) The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein–DNA complexes. Mol Cell 12: 1525–1536 [DOI] [PubMed] [Google Scholar]
- Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K (2003) S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083 [DOI] [PubMed] [Google Scholar]
- Keil RL, Roeder GS (1984) Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell 39: 377–386 [DOI] [PubMed] [Google Scholar]
- Kobayashi T (2003) The replication fork barrier site forms a unique structure with Fob1p and inhibits the replication fork. Mol Cell Biol 23: 9178–9188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Ganley AR (2005) Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science 309: 1581–1584 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Horiuchi T (1996) A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells 1: 465–474 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Heck DJ, Nomura M, Horiuchi T (1998) Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev 12: 3821–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Nomura M, Horiuchi T (2001) Identification of DNA cis elements essential for expansion of ribosomal DNA repeats in Saccharomyces cerevisiae. Mol Cell Biol 21: 136–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Horiuchi T, Tongaonkar P, Vu L, Nomura M (2004) SIR2 regulates recombination between different rDNA repeats, but not recombination within individual rRNA genes in yeast. Cell 117: 441–453 [DOI] [PubMed] [Google Scholar]
- Krasilnikova MM, Samadashwily GM, Krasilnikov AS, Mirkin SM (1998) Transcription through a simple DNA repeat blocks replication elongation. EMBO J 17: 5095–5102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings G, Bastia D (2004) swi1- and swi3-dependent and independent replication fork arrest at the ribosomal DNA of Schizosaccharomyces pombe. Proc Natl Acad Sci USA 101: 14085–14090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings G, Bastia D (2005) Sap1p binds to Ter1 at the ribosomal DNA of Schizosaccharomyces pombe and causes polar replication fork arrest. J Biol Chem 280: 39135–39142 [DOI] [PubMed] [Google Scholar]
- Lambert S, Watson A, Sheedy DM, Martin B, Carr AM (2005) Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell 121: 689–702 [DOI] [PubMed] [Google Scholar]
- Lee BS, Grewal SI, Klar AJ (2004) Biochemical interactions between proteins and mat1 cis-acting sequences required for imprinting in fission yeast. Mol Cell Biol 24: 9813–9822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD (2005) Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell 120: 587–598 [DOI] [PubMed] [Google Scholar]
- Li L, Zou L (2005) Sensing, signaling, and responding to DNA damage: organization of the checkpoint pathways in mammalian cells. J Cell Biochem 94: 298–306 [DOI] [PubMed] [Google Scholar]
- Linskens MH, Huberman JA (1988) Organization of replication in the ribosomal DNA in Saccharomyces cerevisiae. Mol Cell Biol 8: 4927–4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Pellicioli A, Cotta-Ramusino C, Liberi G, Plevani P, Muzi-Falconi M, Newlon C, Foiani M (2001) The checkpoint response stabilizes stalled DNA replication forks. Nature 412: 599–602 [DOI] [PubMed] [Google Scholar]
- Louarn JM, Louarn J, Francois V, Patte J (1991) Analysis and possible role of hyperrecombination in the termination region of the Escherichia coli chromosome. J Bacteriol 173: 5097–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini R, Sogo JM (1994) Chromatin structure and transcriptional activity around the replication forks arrested at the 3′ end of the yeast ribosomal-RNA genes. Mol Cell Biol 14: 318–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makovets S, Herskowitz I, Blackburn EH (2004) Anatomy and dynamics of DNA replication fork movement in yeast telomeric regions. Mol Cell Biol 24: 4019–4031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML et al. (2004) Identification of protein complexes required for efficient sister chromatid cohesion. Mol Biol Cell 15: 1736–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan CH, Russell P (2004) The DNA damage response: sensing and signaling. Curr Opin Cell Biol 16: 629–633 [DOI] [PubMed] [Google Scholar]
- Michel B, Ehrlich SD, Uzest M (1997) DNA double-strand breaks caused by replication arrest. EMBO J 16: 430–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Rog O, Cooper JP (2006) Semi-conservative DNA replication through telomeres requires Taz1. Nature 440: 824–828 [DOI] [PubMed] [Google Scholar]
- Mohanty BK, Bastia D (2004) Binding of the replication terminator protein Fob1p to the Ter sites of yeast causes polar fork arrest. J Biol Chem 279: 1932–1941 [DOI] [PubMed] [Google Scholar]
- Mohanty BK, Bairwa NK, Bastia D (2006) The Tof1p–Csm3p protein complex counteracts the Rrm3p helicase to control replication termination of Saccharomyces cerevisiae. Proc Natl Acad Sci USA 103: 897–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcair MD, Schaeffer PM, Oakley AJ, Cross HF, Neylon C, Hill TM, Dixon NE (2006) A molecular mousetrap determines polarity of termination of DNA replication in E. coli. Cell 125: 1309–1319 [DOI] [PubMed] [Google Scholar]
- Mulugu S, Potnis A, Shamsuzzaman Taylor J, Alexander K, Bastia D (2001) Mechanism of termination of DNA replication of Escherichia coli involves helicase–contrahelicase interaction. Proc Natl Acad Sci USA 98: 9569–9574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitani H, Hidaka M, Horiuchi T (1993) Specific chromosomal sites enhancing homologous recombination in Escherichia coli mutants defective in RNase H. Mol Gen Genet 240: 307–314 [DOI] [PubMed] [Google Scholar]
- Noguchi E, Noguchi C, McDonald WH, Yates JR, Russell P (2004) Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol Cell Biol 24: 8342–8355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC (2006) Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell 21: 581–587 [DOI] [PubMed] [Google Scholar]
- Payne BT, van Knippenberg IC, Bell H, Filipe SR, Sherratt DJ, McGlynn P (2006) Replication fork blockage by transcription factor–DNA complexes in Escherichia coli. Nucleic Acids Res 34: 5194–5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possoz C, Filipe SR, Grainge I, Sherratt DJ (2006) Tracking of controlled Escherichia coli replication fork stalling and restart at repressor-bound DNA in vivo. EMBO J 25: 2596–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Aguilera A (2005) Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J 24: 1267–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards RI (2001) Fragile and unstable chromosomes in cancer: causes and consequences. Trends Genet 17: 339–345 [DOI] [PubMed] [Google Scholar]
- Rocha EP, Danchin A (2003) Essentiality, not expressiveness, drives gene-strand bias in bacteria. Nat Genet 34: 377–378 [DOI] [PubMed] [Google Scholar]
- Sanchez-Gorostiaga A, Lopez-Estrano C, Krimer DB, Schvartzman JB, Hernandez P (2004) Transcription termination factor reb1p causes two replication fork barriers at its cognate sites in fission yeast ribosomal DNA in vivo. Mol Cell Biol 24: 398–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa N, Horiuchi T, Kobayashi T (2004) Transcription-mediated hyper-recombination in HOT1. Genes Cells 9: 305–315 [DOI] [PubMed] [Google Scholar]
- Sharma B, Hill TM (1995) Insertion of inverted Ter sites into the terminus region of the Escherichia coli chromosome delays completion of DNA replication and disrupts the cell cycle. Mol Microbiol 18: 45–61 [DOI] [PubMed] [Google Scholar]
- Sommariva E, Pellny TK, Karahan N, Kumar S, Huberman JA, Dalgaard JZ (2005) Schizosaccharomyces pombe Swi1, Swi3, and Hsk1 are components of a novel S-phase response pathway to alkylation damage. Mol Cell Biol 25: 2770–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF (2001) Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553–557 [DOI] [PubMed] [Google Scholar]
- Tourriere H, Versini G, Cordon-Preciado V, Alabert C, Pasero P (2005) Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell 19: 699–706 [DOI] [PubMed] [Google Scholar]
- Vilette D, Uzest M, Ehrlich SD, Michel B (1992) DNA transcription and repressor binding affect deletion formation in Escherichia coli plasmids. EMBO J 11: 3629–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K, Keil RL, Roeder GS (1987) Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell 48: 1071–1079 [DOI] [PubMed] [Google Scholar]
- Wai H, Johzuka K, Vu L, Eliason K, Kobayashi T, Horiuchi T, Nomura M (2001) Yeast RNA polymerase I enhancer is dispensable for transcription of the chromosomal rRNA gene and cell growth, and its apparent transcription enhancement from ectopic promoters requires Fob1 protein. Mol Cell Biol 21: 5541–5553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Newport J (2000) Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase α. Mol Cell 5: 617–627 [DOI] [PubMed] [Google Scholar]
- Wang Y, Vujcic M, Kowalski D (2001) DNA replication forks pause at silent origins near the HML locus in budding yeast. Mol Cell Biol 21: 4938–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward TR, Hoang ML, Prusty R, Lau CK, Keil RL, Fangman WL, Brewer BJ (2000) Ribosomal DNA replication fork barrier and HOT1 recombination hot spot: shared sequences but independent activities. Mol Cell Biol 20: 4948–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RE, Prado F, Aguilera A (2006) Replication fork progression is impaired by transcription in hyperrecombinant yeast cells lacking a functional THO complex. Mol Cell Biol 26: 3327–3334 [DOI] [PMC free article] [PubMed] [Google Scholar]


