Abstract
Stat4 is required for Th1 development, although how a transiently activated factor generates heritable patterns of gene expression is still unclear. We examined the regulation of IL-18Rα expression to define a mechanism for Stat4-dependent genetic programming of a Th1-associated gene. Although Stat4 binds the Il18r1 promoter following IL-12 stimulation and transiently increases acetylated histones H3 and H4, patterns of histone acetylation alone in Th1 cells may not be sufficient to explain cell-type-specific patterns of gene expression. The level of DNA methylation and recruitment of Dnmt3a to Il18r1 inversely correlate with IL-18Rα expression, and blocking DNA methylation increases IL-18Rα expression. Moreover, there was decreased Il18r1–Dnmt3a association and DNA methylation following transient trichostatin A-induced histone hyperacetylation in Stat4−/−Th1 cultures. Increased association of Dnmt3a and the Dnmt3a cofactor Dnmt3L with the promoters of several Stat4-dependent genes was found in Stat4−/− Th1 cultures, providing a general mechanism for Stat4-dependent gene programming. These data support a mechanism wherein the transient hyperacetylation induced by Stat4 prevents the recruitment of DNA methyltransferases and the subsequent repression of the Il18r1 locus.
Keywords: chromatin, differentiation, DNA methylation, STAT proteins, T helper cell
Introduction
IL-4 promotes development of Th2 cells and atopic responses, whereas IL-12 induces Th1 differentiation and cell-mediated immunity (Glimcher and Murphy, 2000). The signal transducer and activator of transcription, Stat4, is required for most IL-12-stimulated functions and for development of IFN-γ-secreting Th1 cells from naïve CD4+ T cells (Kaplan et al, 1996; Thierfelder et al, 1996). As a result, Stat4-deficient mice mount greatly attenuated inflammatory responses in vivo, rendering them susceptible to a variety of infections but resistant to T-cell-mediated autoimmune disease (Kaplan, 2005). This phenotype arises from a requirement for transient Stat4 activation in programming the expression of multiple Th1 effector genes including the IL-18Rα chain (Hoey et al, 2003).
A central question in the biology of Th1 development is how a transiently activated factor mediates a heritable genetic program. One hypothesis suggests that Stat4 promotes chromatin accessibility, allowing other transcription factors to mediate tissue-specific expression (Avni et al, 2002; Fields et al, 2002). In this model, T-cell receptor signaling initiates expression of Th1-restricted genes and Stat4 maintains expression through the differentiation process by targeting increased histone acetylation. This model, based on examination of the Ifng gene, is supported by the presence of DNase hypersensitivity sites and Stat4-dependent histone acetylation and chromatin remodeling factor recruitment in Th1 cultures but lacking in Th2 cells (Agarwal and Rao, 1998; Fields et al, 2002; Zhang and Boothby, 2006). However, examination of additional Th1-restricted genes suggests that histone acetylation patterns alone cannot solely define T helper (Th)cell-restricted gene expression (Morinobu et al, 2004). Other epigenetic modifications, such as DNA methylation-mediated gene repression, also play a role in Th1- and Th2-restricted cytokine expression (Bird et al, 1998; Lee et al, 2001, 2002; Hutchins et al, 2002; Mullen et al, 2002; Makar et al, 2003; Jones and Chen, 2006; Murayama et al, 2006; Northrop et al, 2006), but it is not clear whether these are STAT-regulated processes.
IL-18Rα (IL-1Rrp) is expressed in Th1 but not Th2 cells (Xu et al, 1998b). Coupled with the IL-18Rβ chain (AcPL), IL-18Rα forms an IL-18 signaling complex that activates IFN-γ expression (Okamura et al, 1995; Born et al, 1998). IL-18 strongly synergizes with IL-12 in the induction of IFN-γ, providing additional Th1 effector functions and contributing to polarized immune responses (Okamura et al, 1995; Smeltz et al, 2001). IL-18Rα may have additional IL-18-independent functions that are important in inflammatory immunity (Gutcher et al, 2006). IL-18Rα expression is induced by IL-12 in a Stat4-dependent manner and, compared with wild-type (WT) Th1 cells, is considerably reduced in Stat4-deficient Th1 cultures (Lawless et al, 2000; Nakahira et al, 2001; Hoey et al, 2003).
To further explore Stat4-dependent mechanisms of gene expression, we have examined the Il18r1 locus, encoding IL-18Rα, for Stat4-dependent epigenetic modifications. We observed that Stat4 induced a transient histone hyperacetylation and limited DNA methylation of the Il18r1 promoter by decreasing the level of DNA methyltransferases associated with the locus. This supports a model wherein Stat4 regulates chromatin modifications and association of multiple modifying factors in programming a gene for cell-type-specific expression.
Results
Stat4-dependent regulation of the Il18r1 gene
The Il18r1 (IL-18Rα) gene is closely linked to Il1rl1 (T1/ST2/IL-33R) and Il18rap (IL-18Rβ) on chromosome 1 in the mouse genome within a cluster of IL-1R-related genes, although each gene has a distinct pattern of expression in Th subsets (Figure 1A) (Dale and Nicklin, 1999; Schmitz et al, 2005). Both IL-18R genes are restricted to Th1 cells, whereas T1/ST2 expression is restricted to Th2 cells (Xu et al, 1998a). Normal IL-18Rα expression is dependent on Stat4 during the differentiation process and differentiated Th1 cultures (Figure 1A–C). Il18r1 mRNA expression is less than 15% of WT and level of expression is not altered even after culture for an additional week of differentiation (Figure 1C and data not shown). Thus, the Il18r1 gene provides a model for understanding how Stat4 contributes to Th1-specific gene expression.
Figure 1.
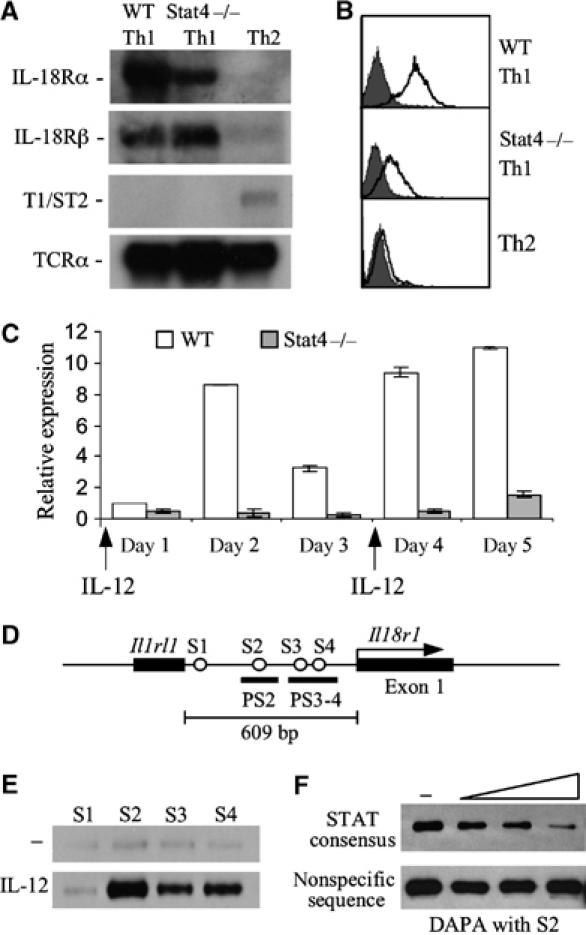
Regulation of the Il18r1 gene in T helper subsets. (A) Northern blot analysis of T1/ST2 (Il1rl1), IL-18Rα (Il18r1) and IL-18Rβ (Il18rap) mRNA in 1-week cultured WT Th1, Stat4−/− Th1 and Th2 cells. Northern blots were reprobed with TCRα as a loading control. (B) Analysis of IL-18Rα surface expression on 5-day C57BL/6 WT Th1, Stat4−/− Th1 and Th2 cells by flow cytometry. Shaded areas, control Ab staining; bold line, IL-18Rα Ab staining. (C) qPCR of Il18r1 mRNA levels during differentiation of WT and Stat4−/− Th1 cultures. RNA was isolated from cells of the indicated genotypes at each day of differentiation and used for real-time PCR. Results are relative to Il18r1 mRNA levels on day 1 in WT Th1 cells and were normalized to levels of β2-microglobulin. (D) Schematic of four putative STAT binding sites designated as S1, S2, S3 and S4 within the Il1rl1–Il18r1 intergenic region. (E) Stat4–DNA interactions were analyzed by DAPA with a biotinylated double-stranded DNA oligo encompassing each of the four putative STAT binding sites incubated with whole-cell extract isolated from anti-CD3 activated CD4 T cells cultured with or without 2 ng/ml IL-12 for 4 h. The precipitated Stat4 protein was detected using Western blot. (F) Binding specificity was determined by incubating the S2 oligo with cell extract in the presence of increasing concentrations of a STAT consensus competitor or a nonspecific competitor.
The ability of IL-12 to induce IL-18Rα expression in a Stat4-dependent manner suggested that Stat4 might bind directly to the Il18r1 locus. Sequence analysis identified four putative STAT consensus sites within the Il1rl1–Il18r1 intergenic region, designated S1–S4 (Figure 1D). DNA affinity precipitation assay (DAPA) results showed that all sites except S1 bound Stat4 protein following precipitation of extracts from IL-12-stimulated activated T cells (Figure 1E). The amount of S2 oligo precipitated Stat4 protein was decreased in the presence of a competitor containing a STAT consensus site but not an oligonucleotide with an irrelevant binding site, indicating the binding specificity of Stat4 for this DNA sequence (Figure 1F).
Il18r1 histone modifications in Th subsets
We have previously shown that transient activation of Stat4 (Figure 2A) mediates histone acetylation of the CD25 locus (O'Sullivan et al, 2004). To test whether Stat4 binding to the Il18r1 promoter resulted in histone modification in the locus, we performed chromatin immunoprecipitation (ChIP) assays over a time course of 48 h, with primers for the S3–S4 Stat4 binding sites and for a site in intron 1 with activated T cells stimulated with IL-12 for the indicated times (Figure 2B). Stat4 bound the Il18r1 promoter by 1 h after stimulation and association continued up to at least 24 h, correlating with the pattern of Stat4 activation (Figure 2A and C). Acetylation of histones H3 and H4 at the Il18r1 promoter increased over time at the intergenic and intron regions, the latter despite the lack of binding of Stat4 to the intron 1 region (Figure 2C). Acetylation of these regions was Stat4 dependent, as minimal acetylation was observed in Stat4-deficient T cells (Figure 2C).
Figure 2.
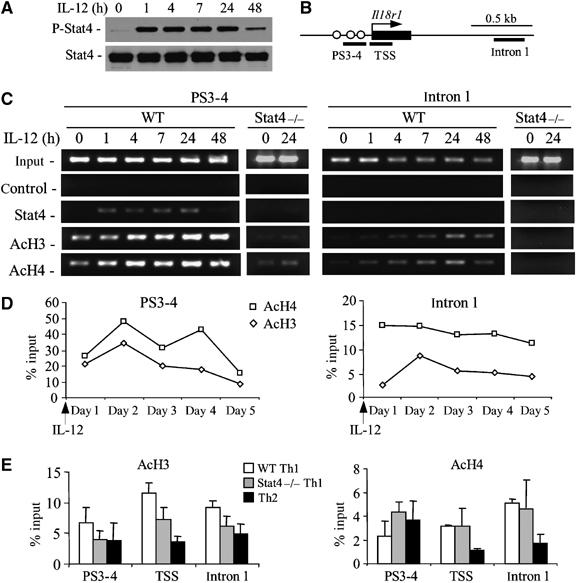
Stat4 induces transient histone hyperacetylation in the Il18r1 locus. (A) Phosphorylated Stat4 protein in anti-CD3 activated CD4 T cells following stimulation with 2 ng/ml IL-12 was assessed at the indicated time points by Western blot using antibodies against phosphorylated or total Stat4. (B) Schematic of the 5′ end of the Il18r1 gene indicating regions amplified for ChIP analysis. Rectangle, exon 1. (C) Dynamic changes of histone H3/H4 acetylation using ChIPs were assessed for PS3-4 and intron 1 regions with activated WT or Stat4−/− CD4 T cells treated or not with 2 ng/ml IL-12 for the indicated time points. Chromatin was precipitated with antibodies against Stat4, acetylated histones H3 and H4. (D) Levels of histone H3/H4 acetylation at the Il18r1 promoter and intron 1 regions were assessed in WT cultures over the 5-day differentiation period. Data are presented as percent of input following densitometric analysis. (E) Analysis of histone H3/H4 acetylation at regions of Stat4 binding sites (PS3–4), transcriptional start site (TSS) and intron 1 in 1-week cultured WT Th1, Stat4−/− Th1 and WT Th2 cells. Chromatin was precipitated using antibodies against acetylated histones H3 and H4, followed by semiquantitative PCR to amplify three genomic fragments indicated in (B). Data are presented as percent of input following densitometric analysis±s.e. of determination at several dilution factors.
Although there was clear Stat4-dependent histone acetylation at the Il18r1 locus, these effects seemed transient, particularly in the intron region (Figure 2C). During the 5 days of differentiation, histone acetylation levels peaked within 48 h and gradually fell over the 5 days of culture in both the promoter and intron 1 regions (Figure 2D). Thus, Stat4-dependent histone acetylation is a transient effect and the induced level of histone acetylation is not maintained throughout the differentiation culture.
Transient Stat4-dependent histone acetylation during differentiation (Figure 2C and D) suggested that acetylation patterns in differentiated Th1 or Th2 cultures might not be dramatically different. To test this, we performed ChIP assays for acetylated H3 and H4 in WT Th1, WT Th2 and Stat4−/− Th1 cultures using primers spanning the S3–S4 sites, transcriptional start site (TSS) and within intron 1 (Figure 2B). Compared with Th1 cells, there was decreased acetylation (2–3-fold) of histones H3 and H4 at the TSS and intron 1 sites in Th2 cells, although differences were modest (Figure 2E). Stat4−/− Th1 cultures had a level of histone H3 acetylation intermediate between WT Th1 and Th2 cultures in the TSS and intron 1 regions (Figure 2E). In contrast, histone H4 acetylation was essentially indistinguishable between WT and Stat4−/− Th1 cultures in the TSS and intron 1 regions. Thus, although Stat4 mediates a transient increase in acetylation during differentiation, the ability of Stat4 to program a Th1 cell for high-level IL-18Rα expression is not simply due to generation of stable histone acetylation patterns at the Il18r1 locus, as histone acetylation decreases over the period of differentiation.
DNA methylation near the Il18r1 promoter
To further explore Stat4-dependent IL-18Rα expression, we investigated other epigenetic changes of the Il18r1 locus. To examine the methylation state of CpG sites surrounding the TSS, we performed bisulfite sequence analysis using genomic DNA isolated from WT Th1, Stat4−/− Th1 and WT Th2 cells cultured for 1 week. After bisulfite conversion, genomic DNA was cloned from two PCR reactions amplifying regions between −495 to −228 and −228 to +71, spanning a total of 18 CpG dinucleotides. Little DNA methylation was detected in WT or Stat4−/− Th1 cells between −495 and −228, although there was increased methylation of this region in Th2 cells (Figure 3A). WT Th1 cells had limited cytosine methylation in the −228 to +71 region, whereas Stat4−/− Th1 cells had increased cytosine methylation in this region (Figure 3A and B). Th2 cells had even higher levels of DNA methylation in this region. The inverse correlation between level of methylation and expression of the IL-18Rα chain was particularly apparent when total methylation or methylation at cytosine +26 was considered (Figure 3A and B). Differential methylation at +26 and lack of methylation at −335 were confirmed using Southern analysis of MspI and HpaII restriction digests (Supplementary Figure 1).
Figure 3.
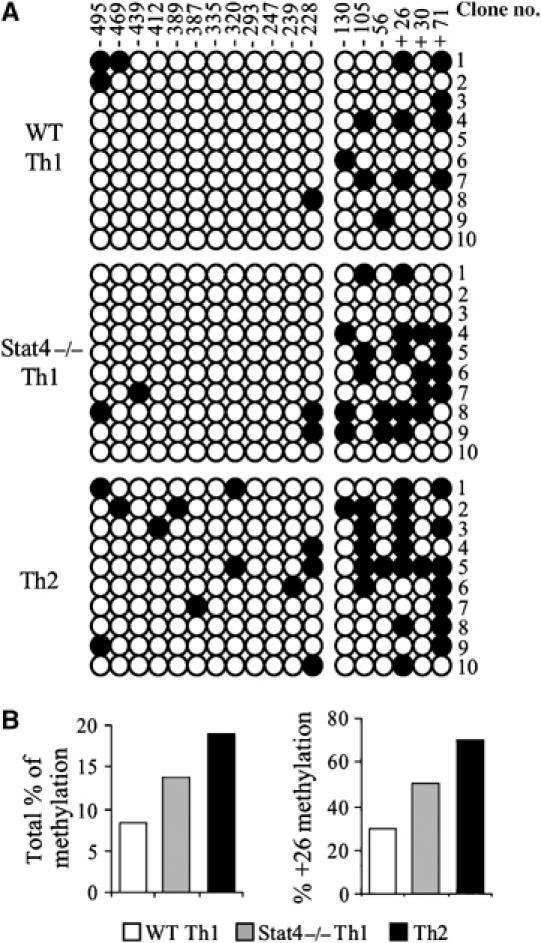
Site-specific cytosine methylation near the transcription start site accompanies helper T-cell differentiation. (A) Characterization of the cytosine methylation state of 18 CpG dinucleotides in the vicinity of the Il18r1 transcription start site by bisulfite sequencing analysis. The region was analyzed as two segments (12 CpG sites in –495 to –228 and six CpG sites in –130 to +71). Ten clones for both fragments were sequenced. Each row of circles represents an individual clone and the overall cytosine methylation at a given CpG site is indicated in a column. (B) Summary of the percentage of total methylcytosines across the region and methylation at CpG site +26, from analysis in (A).
Transient histone acetylation increases IL-18Rα expression in the absence of Stat4
As DNA methylation and transient induction of histone acetylation at the Il18r1 locus correlate with Stat4-dependent gene expression, we cultured Stat4-deficient T cells under Th1 conditions in the absence or presence of a histone deacetylase inhibitor, a DNA methylation inhibitor or both, to assess the ability of these drugs to alter IL-18Rα expression. Inhibitors were added to cultures only at the beginning of the differentiation period to generate a transient effect mimicking the condition when the Il18r1 locus would be exposed to the effects of Stat4. Treatment with the histone deacetylase inhibitor trichostatin A (TSA) increased expression of IL-18Rα following a transient increase in global histone acetylation (Figure 4A). The Il18r1 locus also showed transient increased acetylation, although after expansion of cells for an additional 48 h, the locus showed no difference in histone acetylation between vehicle and TSA-treated cells (data not shown). Incubation of cells with the cytosine methylation inhibitor 5-azacytidine (azaC) increased expression of IL-18Rα on Stat4−/− Th1 cells, confirming that DNA methylation had a negative effect on expression of IL-18Rα (Figure 4A). Incubation of cells with both inhibitors had an additive effect, suggesting a cooperative effect of histone modification and DNA methylation in regulating Il18r1 (Figure 4A).
Figure 4.
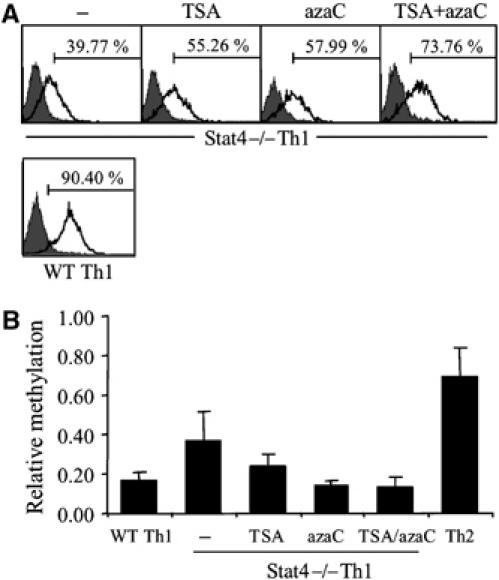
Inhibition of DNA methylation results in increased IL-18Rα expression in Stat4−/− Th1 cells. (A) FACS analysis of IL-18Rα expression on Stat4−/− Th1 cells cultured with the indicated inhibitors. Expression on WT Th1 cells is shown for comparison. (B) Quantitation of PCR analysis of HpaII-digested genomic DNA (indicated as relative methylation) from WT Th1, Th2 or Stat4−/− Th1 cultures treated as in (A).
The ability of TSA to mediate increased expression of IL-18Rα in the absence of sustained hyperacetylation was reminiscent of the ability of Stat4 to program this locus following transient hyperacetylation. As there are known links between DNA methylation and histone acetylation, we tested whether transient TSA-induced hyperacetylation had an effect on DNA methylation within the Il18r1 promoter. HpaII digestion of unmethylated genomic DNA limits PCR amplification of a fragment around the +26 HpaII site and was used as an assay to assess relative methylation in this region. There was low-level methylation of Th1 DNA and higher levels were detected from amplification of WT Th2 DNA, confirming results from bisulfite sequencing analysis (Figure 4B). Stat4−/− Th1 DNA had intermediate levels of methylation, and treatment of these cultures with either TSA or azaC reduced methylation in Stat4−/− Th1 cells to levels similar to those in WT Th1 cells (Figure 4B). These data support a model wherein transient histone acetylation at the Il18r1 locus results in decreased DNA methylation.
Stat4 limits recruitment of Dnmt3a to Th1 genes
Several chromatin modifications mediate the recruitment of DNA methyltransferases, including methylation of histone 3 lysines 9 and 27, whereas recruitment is inhibited by histone 3 lysine 9 acetylation (Jackson et al, 2002; Lehnertz et al, 2003; Lindroth et al, 2004; Abbosh et al, 2006). To determine if Stat4 regulates these chromatin modifications, we performed ChIP assays to test the presence of H3 modifications in WT and Stat4−/− T cells. IL-12 induced H3K9 acetylation in a Stat4-dependent manner (Figure 5A). H3K27 methylation was lower in WT cells than in Stat4−/− cells, although IL-12 stimulation did not have significant effects on this chromatin modification (Figure 5A). The physical associations of DNA methyltransferase (Dnmt) complexes with tri-methyl H3K27 have been shown in plants but not mammalian cells (Lindroth et al, 2004). We therefore used a peptide affinity precipitation assay to test this interaction in Th1 cell extract. A peptide containing the tri-methyl H3K27 modification was able to affinity purify two-fold more Dnmt3a, one of the enzymes that mediates de novo DNA methylation, compared with a peptide containing the acetylated H3K9 residue (Figure 5B). Although these interactions could be indirect, they support a physical interaction between modified histones and DNA methyltransferase complexes. We predicted that, based on differences in chromatin modifications and differences in the physical association of modified histone peptides with Dnmts, we would be able to see differential association of Dnmts with the Il18r1 locus in WT and Stat4−/− Th1 cells. To test this, we performed ChIP analysis with antibodies to Dnmt3a and Dnmt3b, the enzymes responsible for de novo cytosine methylation, using quantitative PCR (qPCR) to define levels present near the TSS. The level of Dnmt3a association with the Il18r1 promoter was reciprocally related to the level of IL-18Rα expression; low levels were observed in WT Th1 cultures, higher levels in Stat4−/− Th1 cells and the highest levels in Th2 cells (Figure 5C). Although Dnmt3b association with the promoter showed a similar pattern, there were overall much lower levels of Dnmt3b found, as assessed by qPCR, suggesting that Dnmt3a is the primary factor involved in methylation of this locus (Figure 5C).
Figure 5.
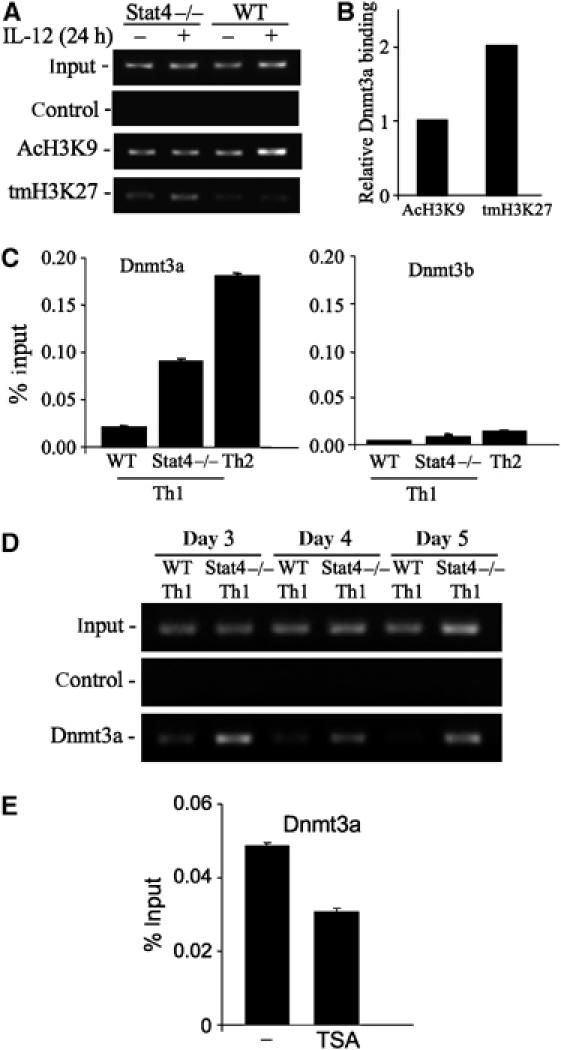
Increased repressive chromatin modification and Dnmt association at the Il18r1 locus in the absence of Stat4. (A) ChIP analysis of acetylated histone H3 lysine 9 (AcH3K9) and tri-methylated histone H3 lysine 27 (tmH3K27) at the Stat4 binding site PS3-4. Chromatin was prepared from activated WT or Stat4−/− CD4 cells, incubated with or without IL-12 for 24 h. (B) Affinity purification of Dnmt3a by peptides containing acetylated H3K9 or tri-methylated H3K27. Biotinylated peptides were bound to streptavidin–agarose and incubated with Th1 cell extract. Precipitated Dnmt3a was detected by immunoblot and densitometry was used for quantification. Representative of three experiments is shown. (C) Detection of DNA methyltransferases (Dnmt)3a and 3b near the TSS site in 5-day cultured WT Th1, Stat4−/− Th1 and Th2 cells. Chromatin was precipitated with antibodies against Dnmt3a or Dnmt3b followed by qPCR for DNA near the TSS site. (D) ChIP analysis of Dnmt3a binding to the Il18r1 locus during Th1 differentiation was performed with WT and Stat4−/− cells after 3, 4 and 5 days of culture. (E) Stat4−/− cells were cultured under Th1 conditions in the presence or absence of TSA as in Figure 4. ChIP for Dnmt3a and qPCR was performed as explained above. Representative of three experiments is shown.
As Dnmt3a demonstrated the largest differences in association with the Il18r1 among the cultures examined, we further defined the association of Dnmt3a with the Il18r1 gene during the process of Th1 differentiation. Although some Dnmt3a is present at the Il18r1 gene in WT Th1 cells at 3 days of culture, association is decreased over the period of differentiation, and by 5 days there is little Dnmt3a detected (Figure 5D). In contrast, higher levels of Dnmt3a association are observed with the Il18r1 gene in Stat4−/− Th1 cultures and levels are not diminished over the period of differentiation (Figure 5D). These results further suggested that TSA treatment of Stat4−/− Th1 cells, which was previously shown to increase IL-18Rα expression and decrease DNA methylation (Figure 4), should also decrease association of Dnmt3a with the Il18r1 promoter. We tested this using ChIP assay and observed a TSA-induced decrease in Dnmt3a association that was comparable in magnitude to TSA-induced decrease in DNA methylation (Figure 5E). Together, these results support our model that transient histone hyperacetylation, either targeted by Stat4 or globally by TSA, decreases the association of DNA methyltransferases and is sufficient to limit Il18r1 DNA methylation.
We next wanted to determine whether the effects of Stat4 observed at the Il18r1 promoter were also observed at other Stat4-dependent Th1-restricted gene promoters. To test this, we used ChIP assay to detect Dnmt3a and the Dnmt3-associated cofactor Dnmt3L at the promoters of multiple Th1 genes, including Ifng, Hlx, Ccr5, Furin, Etv5 (the gene encoding ERM) and the Stat4 binding site of the Il18r1 promoter. We observed increased association of Dnmt3a with each of these genes in Stat4−/− Th1 cells compared with WT Th1 cells, except for Etv5 (Figure 6). However, Dnmt3L, which greatly enhances the function of Dnmt3a (Gowher et al, 2005), was present at each of the gene promoters in Stat4−/− Th1 cells and was undetectable in WT Th1 cells (Figure 6), suggesting that in addition to the increased presence of Dnmt3a at these genes in the absence of Stat4, there may be even greater changes in DNA methyltransferase activity mediated by the presence of Dnmt3L. These data support a general role for Stat4 in programming Th1-specific gene expression through the prevention of Dnmt recruitment and DNA methylation.
Figure 6.

Stat4 prevents Dnmt3a and Dnmt3L association with multiple Th1 loci. WT and Stat4−/− Th1 cultures were subjected to ChIP analysis for Dnmt3a and Dnmt3L. Levels of association were determined by qPCR using primers for the Il18r1 Stat4 binding region or promoters for the Stat4-dependent Th1 genes Ifng, Hlx, Ccr5, Furin or Etv5. ND, not detected.
Discussion
The establishment of cell-type-specific patterns of gene expression has both genetic and epigenetic components. Trans-acting factors bind to DNA–chromatin complexes to initiate transcription of target genes and initiate epigenetic modifications of DNA and associated histones. Whereas many trans-acting factors mediate the activation of multiple target genes, relatively few transcription factors function as master regulators of cellular phenotype differentiation. Stat4 is one such critical factor for the development of Th1 cells. However, the mechanism through which Stat4 mediates this differentiation is still unclear. It likely involves activated Stat4 binding to target genes, which results in programming for restricted expression. We have shown that when Stat4 binds to a locus, it mediates hyperacetylation of histones H3 and H4 (Figure 2) (O'Sullivan et al, 2004). Importantly, this increase in acetylation is transient, coinciding with the pattern of Stat4 binding to the locus. Transient hyperacetylation of a locus could provide increased access for other DNA binding factors that subsequently maintain an activated gene state. However, this may not be the only mechanism of Stat4-mediated programming. We show here that a transient hyperacetylation, mediated either specifically by Stat4 or globally by TSA, results in decreased DNA methylation of the Il18r1 locus. Stat4 activity also limits the presence of DNA methyltransferases at the promoter of Il18r1 and other Th1-associated genes, thus protecting multiple loci from the repression associated with hypermethylated DNA.
DNA methylation in Th cells
DNA methylation also contributes to Th subset-specific expression of cytokine genes. The Ifng gene is differentially methylated between Th1 and Th2 cells (Yano et al, 2003; Winders et al, 2004; Jones and Chen, 2006). Similarly, IL-4 and other Th2 cytokines are regulated by methylation that requires the function of Dnmt1 (Lee et al, 2002; Makar et al, 2003). The Il4 locus in undifferentiated cells is in a methylated state and becomes progressively demethylated as cells become committed to the Th2 phenotype (Lee et al, 2002). DNA methylation results in the recruitment of repressive methyl DNA binding proteins such as MBD2, which may also compete for binding of trans-acting factors, such as GATA-3, to the DNA (Hutchins et al, 2002). DNA methylation could also prevent binding of transcription factors sensitive to CpG methylation (Yano et al, 2003; Winders et al, 2004; Jones and Chen, 2006). In this manner, even modest changes in DNA methylation, at specific sites, can have dramatic changes in gene expression (Jones and Chen, 2006; Murayama et al, 2006; Northrop et al, 2006). Thus, DNA methylation may have both direct effects, by decreasing the binding of trans-acting factors, and indirect effects, by recruiting methyl binding domain proteins that can recruit additional factors to potentiate a repressive chromatin structure. However, although DNA methylation is clearly required for appropriate expression of cytokines in Th subsets, the regulation of this process by transcription factors involved in Th cell differentiation has not been determined.
A model for Stat4-dependent gene programming
The data in this report provide a model of Stat4-mediated programming of Th1-specific gene expression (Figure 7). IL-18Rα is expressed at a basal level in unactivated CD4+ T cells. IL-12 induces Stat4 activation and binding to the Il18r1 promoter. Stat4 binding results in histone hyperacetylation, recruitment of chromatin modifying factors and transcription of the locus (Avni et al, 2002; Fields et al, 2002; O'Sullivan et al, 2004; Chang and Aune, 2005; Zhang and Boothby, 2006), not only allowing greater access to the locus, but also changing the chromatin landscape such that repressive factors are not recruited or maintained at the locus. This could be mediated by direct recruitment of a factor to target loci by Stat4, or indirectly, by Stat4 modifying chromatin in a manner that changes the association of repressive enzymes. Moreover, the effects of histone modification on DNA methylation could be either direct or indirect. In the absence of Stat4-mediated hyperacetylation, there is increased recruitment of Dnmt3a and increased DNA methylation, each contributing to the propagation of decreased locus transcription. The linking of temporal changes in histone acetylation to DNA methylation in our observations is similar to data demonstrating that TSA treatment of Neurospora results in a selective loss of DNA methylation (Selker, 1998). These processes are linked by histone methylation on H3 lysine 9 and 27, which target recruitment of Dnmts (Jackson et al, 2002; Lehnertz et al, 2003; Lindroth et al, 2004; Abbosh et al, 2006). Indeed, the tri-methyl H3K27 modification is found at Th2 cytokine loci in Th1 cells, indicating an association with gene repression (Koyanagi et al, 2005). These data also provide additional evidence for the function of Dnmt3a. Although Dnmts have been associated with DNA methylation in embryonic development, there is growing evidence that their recruitment to specific DNA sites is associated with differentiation of mature cells (Li, 2002; Jones and Chen, 2006). The gradient of Dnmt3a association to Il18r1 among Th1, Stat4−/− Th1 and Th2 cells suggests that the level of Dnmt recruitment to a gene and the subsequent DNA methylation can be critical determinants of Th subset gene expression.
Figure 7.

Model of Stat4 gene programming. Stat4 binds to the Il18r1 locus, promoting gene transient induction and hyperacetylation. Transient alterations of the locus limit association of Dnmts and subsequent DNA methylation allowing increased levels of expression. Th2 cells have higher levels of associated Dnmts and DNA methylation.
This model predicts that during Th1 differentiation, IL-12-induced Stat4 binds to multiple genes within the genome to pattern them for activation. Distinct factors promote the repression of Th1-associated genes, including Il18r1, during Th2 development (Smeltz et al, 2001), likely resulting in a higher level of DNA methylation and Dnmt3a association in Th2 cells compared with Stat4−/− Th1 cells. These data also fit with previous expression analysis of multiple Th1-restricted genes showing highest expression in WT Th1 cells, decreased but not absent expression in Stat4−/− cultures and undetectable expression in Th2 cells (Hoey et al, 2003). Thus, although Stat4 is not required for a basal level of Th1-restricted gene expression, it is required for mediating increased efficiency of transcription by patterning an induced signature on the target loci. This efficiency is reflected in vivo, where Stat4 deficiency results in a lack of cell-mediated immunity (Kaplan, 2005). Even in Stat4/Stat6 double deficient mice that lack the inhibition of Th1 gene expression by Th2-skewing signals, the absence of Stat4 prevents Th1-like cells that have only basal expression of Th1 genes from developing complete Th1 function (Kaplan et al, 1998).
The IL-33R–IL-18R locus
Our characterization of the regulation of the Il18r1 gene generates a starting point for investigating the regulation of the locus containing a Th2-restricted gene (Il1rl1/T1/ST2/IL-33R) and both Stat4-dependent and -independent Th1-restricted genes (Figure 1). Further analysis may identify cell type-specific regulatory elements and possible boundary elements that prevent transcription of the specific genes in Th1 and Th2 cells. It may also be intriguing to define the requirements for Stat4-independent cell-type-specific expression of Il18rap. Whereas Il18rap can be induced by IL-12 in a Stat4-dependent manner (Nakahira et al, 2001), programming for the Th1 state does not require Stat4 (Figure 1). Whether other genes also fall into this category and whether distinct transcription factors are required to regulate their expression should be examined in the future.
Although many of the transcription factors that mediate development of the Th1 phenotype are now recognized, how they confer programming for Th1-restricted expression is still unclear. In this report, we have investigated a mechanism for Stat4-dependent expression of Th1 genes. We have found that Stat4 binds directly to the Il18r1 locus, transiently increases acetylation of the locus and decreases Dnmt association and DNA methylation, resulting in higher expression of IL-18Rα in Th1 cells. This mechanism applies to a number of Th1-specific genes and represents a novel Stat4-dependent mechanism to generate a heritable Th1 phenotype.
Materials and methods
Mice
The generation of Stat4−/− mice was previously described (Kaplan et al, 1996). WT C57BL/6 and BALB/c mice were purchased from Harlan Bioproducts (Indianapolis, IN). Mice were maintained in pathogen-free conditions in barrier facilities in the Laboratory Animal Resource Center (Indiana University School of Medicine). All experiments were performed following approval of the IU Animal Care and Use Committee.
In vitro T-cell differentiation
CD4 cells were isolated from spleen and lymph nodes of mice using magnetic beads (Miltenyi Biotec). For Th differentiation, CD4 cells (1 × 106 cells/ml) were cultured with plate-bound anti-CD3 (4 μg/ml), 0.5 μg/ml soluble anti-CD28, under Th1 (2 ng/ml IL-12 and 10 μg/ml anti-IL-4) or Th2 (10 ng/ml IL-4 and 10 μg/ml anti-IFN-γ) skewing conditions and expanded after 3 days. After 5–7 days of culture, cells were harvested for FACS analysis with anti-IL-18Rα (R&D Systems, Minneapolis, MN) or other assays. In some experiments, Stat4−/− CD4 cells were cultured under Th1 conditions, without or with 20 nM trichostatin A treatment (Sigma, St Louis, MO), 10 μM azaC (Sigma), or both, were expanded with fresh media after 3 days of culture and collected for FACS, Western blot or bisulfite sequencing analysis after 5 days of culture. Quantitative RT–PCR was performed as described (Mathur et al, 2006).
DNA affinity precipitation assay
CD4+ T cells were activated with anti-CD3 (4 μg/ml) for 3 days, transferred to fresh plates and left unstimulated or stimulated with mouse IL-12 (2 ng/ml) for 4 h. Cells were lysed in cell lysis buffer (50 mM Tris–HCl pH 8, 0.5% Igepal, 150 mM NaCl, 1 mM EDTA, and 10% glycerol, supplemented with protease and phosphatase inhibitors) to generate total cell extracts. Biotinylated DNA oligonucleotides were incubated with streptavidin–agarose beads (Chemicon) in IP buffer (cell lysis buffer containing 15 mM NaCl) at 4°C for 2 h. The oligo–bead complex was washed and incubated with cell extract in IP buffer at 4°C overnight. The beads were washed two times, resuspended in SDS–PAGE loading buffer, boiled and run on SDS–PAGE gels before immunoblot.
Peptide affinity precipitation assay
Biotinylated acetylated H3K9 and tri-methylated H3K27 peptides (Chemicon) were immobilized to avidin beads. Total cell extracts from Th1 cells were precleared with avidin beads at 4°C for 2 h before incubating with 50 μl of the prepared avidin-conjugated peptides in peptide binding buffer (20 mM Hepes pH 7.5, 15 mM NaCl, 0.5% Ipegal and 0.5 mM dithiothreitol supplemented with proteinase and phosphatase inhibitors) overnight at 4°C. The complexes were washed three times with peptide binding buffer followed by immunoblotting for Dnmt3a.
Chromatin immunoprecipitation
Crosslinking of protein–chromatin complexes was achieved by adding formaldehyde into cell cultures to a final concentration of 1% and shaking at room temperature for 10 min. Glycine was added to a final 0.125 M to quench the crosslinking. Cells were washed once with ice-cold PBS, resuspended in cell lysis buffer (5 mM Pipes pH 8.0, 85 mM KCl and 0.5% NP-40) and incubated for 10 min on ice. Protease and phosphatase inhibitors were added to all buffers. Nuclei were harvested after centrifugation at 5000 r.p.m. for 5 min, resuspended in nuclear lysis buffer (50 mM Tris–HCl pH 8.0, 10 mM EDTA and 1% SDS) and incubated 10 min on ice. An ultrasonic processor (Vibra-Cell) was used to shear genomic DNA (150–300-bp fragments), with eight sets of 10-s 50 W bursts. Cell extracts were diluted five-fold in ChIP buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris–HCl pH 8.0 and 167 mM NaCl), precleared with salmon sperm DNA, BSA and protein A agarose bead slurry (50%) at 4°C for 1 h. The supernatant was incubated in the presence or absence of 5 μg antibody (anti-Stat4, Santa Cruz Biotechnology; anti-Dnmt3a and anti-Dnmt3b, Imgenex; anti-Dnmt3L, Abgent; all others from Chemicon) at 4°C overnight. The immunocomplex was precipitated with protein A agarose beads at 4°C for 1 h followed by centrifugation. The supernatant from the control precipitation was used as input material. The beads were washed consecutively with low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8 and 150 mM NaCl), high-salt wash buffer (as above with 500 mM NaCl), LiCl wash buffer (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA and 10 mM Tris–HCl pH 8), and twice in 1 mM EDTA, 10 mM Tris–HCl pH 8. Bound DNA was eluted from the beads with elution buffer (2% SDS, 2 mM EDTA and 20 mM Tris–HCl), vortexing and incubating at 37°C before centrifugation. The supernatant was collected, supplemented with 2 mM EDTA, 20 mM Tris–HCl and 10 mg/ml proteinase K and incubated at 37°C. DNA crosslinks were reversed by incubating precipitates at 65°C for 16 h. DNA was purified by phenol/chloroform extraction, ethanol precipitated and resuspended in H2O. DNA was then analyzed using either conventional or qPCR. qPCR was performed with site-specific primer sets using ABI PRISM7500. To quantify chromatin immunoprecipitates, a standard curve was generated from serial dilutions of a known amount of DNA generated from sonicated Th1 cells. To calculate ChIP results as a percentage of input, the amount of immunoprecipitated DNA from the isotype control antibody was subtracted from the amount of immunoprecipitated DNA from the specific antibody ChIP followed by normalizing against the amount of input DNA. The sequences of primer pairs and probes are listed as Supplementary data.
DNA methylation analysis
Genomic DNA was first cleaved by BamHI (Promega) to completion. The cleaved DNA was purified by phenol/chloroform extraction and ethanol precipitation. For HpaII PCR, BamHI-digested DNA was resuspended in water and further digested with HpaII. The BamHI/HpaII-digested genomic DNA was purified as described above followed by semiquantitative PCR. Bisulfite treatment of BamHI-digested DNA was performed as previously described (Frommer et al, 1992; Grunau et al, 2001). PCR fragments were cloned and sequenced.
Supplementary Material
Supplementary Information
Acknowledgments
We thank AO'Sullivan and ZY Wang for technical guidance and G Kansas, CH Chang, D Skalnik, N Laribee and members of the Kaplan laboratory for reviewing the manuscript. This work was supported by Public Health Service Award AI45515.
References
- Abbosh PH, Montgomery JS, Starkey JA, Novotny M, Zuhowski EG, Egorin MJ, Moseman AP, Golas A, Brannon KM, Balch C, Huang TH, Nephew KP (2006) Dominant-negative histone H3 lysine 27 mutant derepresses silenced tumor suppressor genes and reverses the drug-resistant phenotype in cancer cells. Cancer Res 66: 5582–5591 [DOI] [PubMed] [Google Scholar]
- Agarwal S, Rao A (1998) Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity 9: 765–775 [DOI] [PubMed] [Google Scholar]
- Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A (2002) T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol 3: 643–651 [DOI] [PubMed] [Google Scholar]
- Bird JJ, Brown DR, Mullen AC, Moskowitz NH, Mahowald MA, Sider JR, Gajewski TF, Wang CR, Reiner SL (1998) Helper T Cell differentiation is controlled by the cell cycle. Immunity 9: 229–237 [DOI] [PubMed] [Google Scholar]
- Born TL, Thomassen E, Bird TA, Sims JE (1998) Cloning of a novel receptor subunit, AcPL, required for interleukin-18 signaling. J Biol Chem 273: 29445–29450 [DOI] [PubMed] [Google Scholar]
- Chang S, Aune TM (2005) Histone hyperacetylated domains across the Ifng gene region in natural killer cells and T cells. Proc Natl Acad Sci USA 102: 17095–17100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale M, Nicklin MJ (1999) Interleukin-1 receptor cluster: gene organization of IL1R2, IL1R1, IL1RL2 (IL-1Rrp2), IL1RL1 (T1/ST2), and IL18R1 (IL-1Rrp) on human chromosome 2q. Genomics 57: 177–179 [DOI] [PubMed] [Google Scholar]
- Fields PE, Kim ST, Flavell RA (2002) Cutting edge: changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J Immunol 169: 647–650 [DOI] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA 89: 1827–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glimcher LH, Murphy KM (2000) Lineage commitment in the immune system: the T helper cell grows up. Genes Dev 14: 1693–1711 [PubMed] [Google Scholar]
- Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A (2005) Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J Biol Chem 280: 13341–13348 [DOI] [PubMed] [Google Scholar]
- Grunau C, Clark SJ, Rosenthal A (2001) Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res 29: e65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutcher I, Urich E, Wolter K, Prinz M, Becher B (2006) Interleukin 18-independent engagement of interleukin 18 receptor-alpha is required for autoimmune inflammation. Nat Immunol 7: 946–953 [DOI] [PubMed] [Google Scholar]
- Hoey T, Zhang S, Schmidt N, Yu Q, Ramchandani S, Xu X, Naeger LK, Sun YL, Kaplan MH (2003) Distinct requirements for the naturally occurring splice forms Stat4α and Stat4β in IL-12 responses. EMBO J 22: 4237–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins AS, Mullen AC, Lee HW, Sykes KJ, High FA, Hendrich BD, Bird AP, Reiner SL (2002) Gene silencing quantitatively controls the function of a developmental trans-activator. Mol Cell 10: 81–91 [DOI] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE (2002) Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416: 556–560 [DOI] [PubMed] [Google Scholar]
- Jones B, Chen J (2006) Inhibition of IFN-gamma transcription by site-specific methylation during T helper cell development. EMBO J 25: 2443–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan MH (2005) STAT4: a critical regulator of inflammation in vivo. Immunol Res 32: 231–241 [DOI] [PubMed] [Google Scholar]
- Kaplan MH, Sun Y-L, Hoey T, Grusby MJ (1996) Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature 382: 174–177 [DOI] [PubMed] [Google Scholar]
- Kaplan MH, Wurster AL, Grusby MJ (1998) A Stat4-independent pathway for the development of Th1 cells. J Exp Med 188: 1191–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyanagi M, Baguet A, Martens J, Margueron R, Jenuwein T, Bix M (2005) EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells. J Biol Chem 280: 31470–31477 [DOI] [PubMed] [Google Scholar]
- Lawless VA, Zhang S, Ozes ON, Bruns HA, Oldham I, Hoey T, Grusby MJ, Kaplan MH (2000) Stat4 regulates multiple components of IFN-γ-inducing signaling pathways. J Immunol 165: 6803–6808 [DOI] [PubMed] [Google Scholar]
- Lee DU, Agarwal S, Rao A (2002) Th2 lineage commitment and efficient IL-4 production involves extended demethylation of the IL-4 gene. Immunity 16: 649–660 [DOI] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Perez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB (2001) A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15: 763–774 [DOI] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH (2003) Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 13: 1192–1200 [DOI] [PubMed] [Google Scholar]
- Li E (2002) Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet 3: 662–673 [DOI] [PubMed] [Google Scholar]
- Lindroth AM, Shultis D, Jasencakova Z, Fuchs J, Johnson L, Schubert D, Patnaik D, Pradhan S, Goodrich J, Schubert I, Jenuwein T, Khorasanizadeh S, Jacobsen SE (2004) Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J 23: 4286–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makar KW, Perez-Melgosa M, Shnyreva M, Weaver WM, Fitzpatrick DR, Wilson CB (2003) Active recruitment of DNA methyltransferases regulates interleukin 4 in thymocytes and T cells. Nat Immunol 4: 1183–1190 [DOI] [PubMed] [Google Scholar]
- Mathur AN, Chang HC, Zisoulis DG, Kapur R, Belladonna ML, Kansas GS, Kaplan MH (2006) T-bet is a critical determinant in the instability of the IL-17-secreting T-helper phenotype. Blood 108: 1595–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinobu A, Kanno Y, O'Shea JJ (2004) Discrete roles for histone acetylation in human T helper 1 cell-specific gene expression. J Biol Chem 279: 40640–40646 [DOI] [PubMed] [Google Scholar]
- Mullen AC, Hutchins AS, High FA, Lee HW, Sykes KJ, Chodosh LA, Reiner SL (2002) Hlx is induced by and genetically interacts with T-bet to promote heritable T(H)1 gene induction. Nat Immunol 3: 652–658 [DOI] [PubMed] [Google Scholar]
- Murayama A, Sakura K, Nakama M, Yasuzawa-Tanaka K, Fujita E, Tateishi Y, Wang Y, Ushijima T, Baba T, Shibuya K, Shibuya A, Kawabe Y, Yanagisawa J (2006) A specific CpG site demethylation in the human interleukin 2 gene promoter is an epigenetic memory. EMBO J 25: 1081–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira M, Tomura M, Iwasaki M, Ahn HJ, Bian Y, Hamaoka T, Ohta T, Kurimoto M, Fujiwara H (2001) An absolute requirement for STAT4 and a role for IFN-gamma as an amplifying factor in IL-12 induction of the functional IL-18 receptor complex. J Immunol 167: 1306–1312 [DOI] [PubMed] [Google Scholar]
- Northrop JK, Thomas RM, Wells AD, Shen H (2006) Epigenetic remodeling of the IL-2 and IFN-gamma loci in memory CD8 T cells is influenced by CD4 T cells. J Immunol 177: 1062–1069 [DOI] [PubMed] [Google Scholar]
- O'Sullivan A, Chang HC, Yu Q, Kaplan MH (2004) STAT4 is required for interleukin-12-induced chromatin remodeling of the CD25 locus. J Biol Chem 279: 7339–7345 [DOI] [PubMed] [Google Scholar]
- Okamura H, Tsutsui H, Komatsu T, Yutsudo M, Hakura A, Tanimoto T, Torigoe K, Okura T, Nukada Y, Hattori K, Akita K, Namba M, Tanabe F, Konishi K, Fukuda S, Kurimoto M (1995) Cloning of a new cytokine that induces IFN-γ production by T cells. Nature 378: 88–91 [DOI] [PubMed] [Google Scholar]
- Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, Zurawski G, Moshrefi M, Qin J, Li X, Gorman DM, Bazan JF, Kastelein RA (2005) IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity 23: 479–490 [DOI] [PubMed] [Google Scholar]
- Selker EU (1998) Trichostatin A causes selective loss of DNA methylation in Neurospora. Proc Natl Acad Sci USA 95: 9430–9435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeltz RB, Chen J, Hu-Li J, Shevach EM (2001) Regulation of interleukin (IL)-18 receptor alpha chain expression on CD4(+) T cells during T helper (Th)1/Th2 differentiation. Critical downregulatory role of IL-4. J Exp Med 194: 143–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT, Sangster MY, Vignali DAA, Doherty PC, Grosveld GC, Ihle JN (1996) Requirement for Stat4 in interleukin-12 mediated responses of natural killer and T cells. Nature 382: 171–174 [DOI] [PubMed] [Google Scholar]
- Winders BR, Schwartz RH, Bruniquel D (2004) A distinct region of the murine IFN-gamma promoter is hypomethylated from early T cell development through mature naive and Th1 cell differentiation, but is hypermethylated in Th2 cells. J Immunol 173: 7377–7384 [DOI] [PubMed] [Google Scholar]
- Xu D, Chan WL, Leung BP, Huang F, Wheeler R, Piedrafita D, Robinson JH, Liew FY (1998a) Selective expression of a stable cell surface molecule on type 2 but not type 1 helper T cells. J Exp Med 187: 787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Chan WL, Leung BP, Hunter D, Schulz K, Carter RW, McInnes IB, Robinson JH, Liew FY (1998b) Selective expression and functions of interleukin-18 receptor on T helper (Th) type 1 but not Th2 cells. J Exp Med 188: 1485–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano S, Ghosh P, Kusaba H, Buchholz M, Longo DL (2003) Effect of promoter methylation on the regulation of IFN-gamma gene during in vitro differentiation of human peripheral blood T cells into a Th2 population. J Immunol 171: 2510–2516 [DOI] [PubMed] [Google Scholar]
- Zhang F, Boothby M (2006) T helper type 1-specific Brg1 recruitment and remodeling of nucleosomes positioned at the IFN-gamma promoter are Stat4 dependent. J Exp Med 203: 1493–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


