Abstract
To understand how mammalian genes are regulated from their natural chromosomal environment, we have analysed the molecular events occurring throughout a 150 kb chromatin segment containing the α globin gene locus as it changes from a poised, silent state in erythroid progenitors, to the fully activated state in late, erythroid cells. Active transcription requires the late recruitment of general transcription factors, mediator and Pol II not only to the promoter but also to its remote regulatory elements. Natural mutants of the α cluster show that whereas recruitment of the pre-initiation complex to the upstream elements occurs independently, recruitment to the promoter is largely dependent on the regulatory elements. An improved, quantitative chromosome conformation capture analysis demonstrates that this recruitment is associated with a conformational change, in vivo, apposing the promoter with its remote regulators, consistent with a chromosome looping mechanism. These findings point to a general mechanism by which a gene can be held in a poised state until the appropriate stage for expression, coordinating the level and timing of gene expression during terminal differentiation.
Keywords: erythropoiesis, globin, pre-initiation complex, RNA Pol II, transcription regulation
Introduction
An important aim of current biology is to understand how remote Cis-acting elements and the promoters they control switch genes on and off at the appropriate times in differentiation and development. Cis-acting regulatory sequences may be located tens or even hundreds of kilobases from the genes they control (Kleinjan and van Heyningen, 2005), but how they act over such long distances and the mechanisms by which they influence gene expression are poorly understood; understanding this represents a major challenge in the post-genome era. Perhaps the simplest way to address this is to focus specifically on how a remote enhancer interacts with its cognate promoter to influence transcription. To date, very few long-range promoter/enhancer interactions have been characterised in the natural environment of a mammalian chromosome.
The mammalian α globin gene locus provides a very well-characterised model system for studying this issue during erythropoiesis (Figure 1A). The α locus of all mammalian species analysed lies within a region of 135–155 kb of conserved synteny, with the α-like genes arranged along the chromosome in the order 5′-ζ-α-α-3′ (Figure 1B). The evolutionarily conserved, remote elements, which regulate their expression correspond to erythroid-specific DNase1 hypersensitive sites (HSs) located 40–60 kb upstream of the α genes (Hughes et al, 2005). Interestingly, these tissue-specific regulatory elements lie within the introns of a gene (c16orf35), which itself is transcribed in all cell types (Figure 1B; Vyas et al, 1995). When activated by these remote elements, the globin genes are exclusively expressed in erythroid cells and, therefore, the mechanism underlying the enhancer/promoter interaction can be studied in both erythroid cell lines and primary cells. Cells representing sequential stages of erythropoiesis (Figure 1A) can be used to analyse the order of events leading from the silenced locus, in early, non-committed progenitors, to the fully activated state in mature erythroblasts (Anguita et al, 2004).
Figure 1.
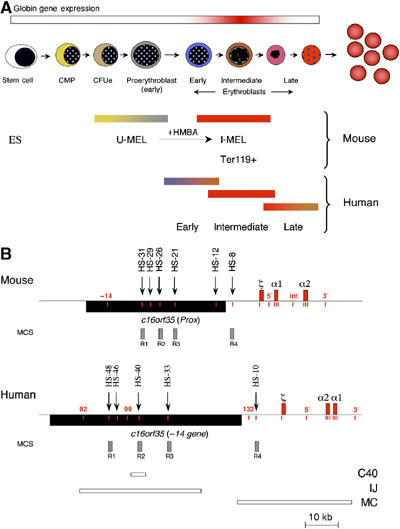
An outline of erythropoiesis, the cell lines used and schematics of the human and mouse α globin gene clusters. (A) The stages of differentiation as primary haematopoietic stem cells differentiate to mature erythrocytes and the corresponding cell lines used in this study. As a source of pluripotent mouse cells, we used embryonic stem cells (ES). Mouse erythroleukaemia (MEL) cells are well characterised, transformed erythroid cells that are blocked at the CFUe or early proerythroblast stage of differentiation (U-MEL). Induction of MEL cells by HMBA (hexa-methylene bis-acetamide) gives rise to terminally differentiated but still nucleated erythroid cells that express α and β globin mRNA at high levels (I-MEL). Primary mouse erythroblasts (Ter119+) were isolated from the spleens of phenylhydrazine-treated mice. Primary human erythroblasts at different stages of differentiation (early, intermediate and late) were isolated as previously described (Brown et al, 2006). Abreviations used: ES: embryonic stem cells; CMP: common myeloid progenitors; CFUe: colony forming units-erythroid; U-MEL: uninduced mouse erythroleukaemia cells ; I-MEL : induced MEL cells. (B) Chromosomal organisation of the mouse (top) and human (below) α globin clusters. The globin genes are shown as labelled red boxes. The positions of previously described DNase1 hypersensitive sites, discussed in the text, are shown as arrows. The widely expressed gene, c16orf35 (also known as −14 gene in human or the Prox gene in mouse), transcribed from the opposite strand to that of α globin is shown as a black box. Short vertical lines (red) indicate amplicons analysed in ChIP experiments. Grey boxes refer to previously defined multi-species conserved elements (MCS). Previously described deletions from the human α globin cluster (Bernet et al, 1995; Craddock et al, 1995) are shown as annotated (C40, IJ and MC) boxes.
We have recently shown that, during mouse erythropoiesis, transcription factor (TF) complexes initially bind the remote upstream elements progressing along the chromosome until, eventually, all elements, including the α globin promoters, are bound and the associated chromatin is modified (Anguita et al, 2004). Similar studies have confirmed these observations in human erythropoiesis (De Gobbi et al, in preparation). Interestingly, this priming process was shown to start in uncommitted, multipotent haemopoietic cells and, by the time these cells become fully committed proerythroblasts (Figure 1A), the α globin promoter and its upstream regulatory element(s) are ‘poised' for expression even though there is little or no α globin mRNA synthesis at this stage.
Here, we have studied what happens at the next stage of erythropoiesis as proerythroblasts undergo terminal differentiation to form intermediate and late erythroblasts and as transcription of the α globin genes becomes fully activated (Figure 1A). Activation of the α globin genes is accompanied by the recruitment of Sp/X-Kruppel-like transcription factors (Sp/X-KLFs) to the α globin promoters. The general transcription factors (GTFs), together with Pol II (collectively referred to as the pre-initiation complex (PIC)), are recruited not only to the α globin promoters but also to the upstream, cis-acting regulatory elements but only during the late stage of terminal, erythroid differentiation. Analysis of natural mutants that downregulate α globin expression (by removing either the α globin genes or their upstream regulatory elements) shows that the formation of these complexes at the upstream elements is essential to effectively recruit Pol II to the α globin promoters. Using a quantitative form of chromosome conformation capture (3C), we showed that gene activation is associated with a change in chromosomal structure (de Laat and Grosveld, 2003), bringing together the remote cis-elements, the promoters and the PIC. These findings suggest that a key role of the upstream elements in the α globin locus is to recruit the PIC to the α globin promoters at the appropriate stage of erythroid differentiation by bringing about a physical interaction between these elements. The α globin cluster thus provides a comprehensive model of how distant regulatory elements may communicate and activate gene expression at a specific time during differentiation.
Results
We have previously described the systems we use to study the process of erythropoiesis in mouse (Anguita et al, 2001) and human (Brown et al, 2006) and set out the importance of studying both cell lines and primary cells. Here, to study terminal erythroid differentiation, we used a mouse erythroleukaemia (MEL) cell line, which represents the proerythroblast stage of differentiation when there is little, if any, α globin mRNA synthesis. Upon induction, these cells differentiate to intermediate/late erythroblasts producing large amounts of α globin. Observations in induced MEL cells were confirmed by extensive analysis in primary (Ter119+) erythroblasts isolated from phenylhydrazine-treated mice (Anguita et al, 2004) and, when relevant, in human early, intermediate and late primary erythroblasts (Figure 1A) isolated from cultures of peripheral blood erythroid progenitors (Brown et al, 2006).
Sp/X-Kruppel-like factors are recruited to the α globin promoter late in differentiation
We previously defined the role of TF binding and chromatin modifications in generating the poised state of mouse and human α globin gene expression in uncommitted haematopoietic progenitors and erythroid progenitors (Anguita et al, 2001, 2004; De Gobbi et al, in preparation). This suggested that globin gene activation is driven by multiprotein complexes, nucleated by GATA1, which are progressively bound to regulatory elements (including promoters) throughout the α globin cluster during erythropoiesis. However, to date, no TFs have been found that are specifically recruited to the α globin regulatory elements at the time of transcriptional activation.
We therefore examined the expression and binding of Sp/X-KLFs that have been previously implicated in erythropoiesis and shown to interact with GATA1 (Merika and Orkin, 1995; Gregory et al, 1996). Using chromatin immunoprecipitation (ChIP), we found that widely expressed TFs, including CCAAT-box binding factor NFY (which is required for recruitment of the PIC; Huang et al, 2004; Kabe et al, 2005) and the KLF ZBP-89 (Woo et al, 2005), could be detected at the α globin promoter both before and after activation (Figure 2A).
Figure 2.
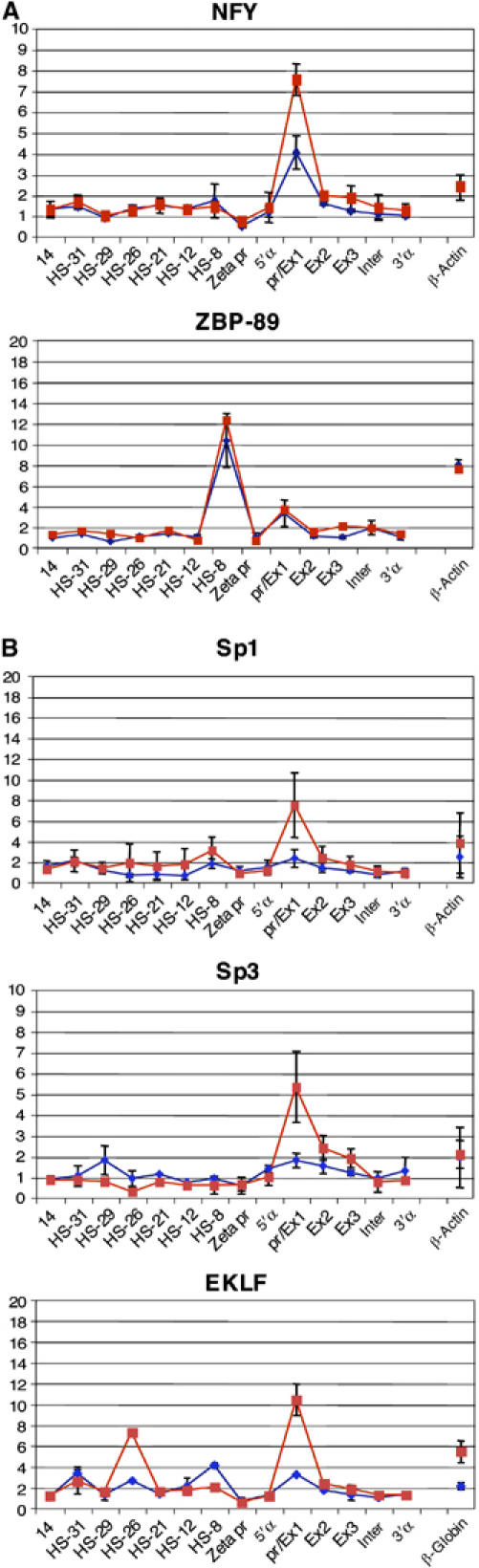
Recruitment of NFY and ZBP-89 (A) and Sp/X-Kruppel-like transcription factors (B) at the mouse α globin core promoter. Real-time PCR analysis of immunoprecipitated chromatin using the antibodies indicated in uninduced (blue) and induced (red) MEL cells. The y-axis represents enrichment over the input DNA, normalised to a control sequence in the GAPDH gene. The x-axis represents the positions of Taqman probes used. The coding sequence is represented by the three exons (promoter/Ex1, Ex2 and Ex3) of the α globin genes. Inter refers to the intergenic region (between α1 and α2). Negative controls 5′α and 3′α flank the α globin gene. β-Actin and β globin denote control sequences at the mouse β-actin gene and β globin promoter, respectively. Error bars correspond to ±1 s.d. from at least two independent ChIPs. Similar data were obtained from primary cells (Supplementary Figure S2).
On the other hand, other member(s) of the Sp/X-KLF family are recruited to cis-elements only in induced MEL cells, as transcription begins (Figure 2B). Sp1, Sp3 and EKLF are recruited to the promoter, whereas EKLF also appears to be recruited to the major enhancer HS-26 (Figure 2B). As these proteins are present in the cells throughout erythropoiesis (Supplementary Figure S1), they must be specifically recruited to cis-elements late in erythroid maturation and are, therefore, good candidates to be involved in triggering the transition from a poised to an active transcriptional state.
Of all Sp/X-KLF factors, EKLF has been most intensively studied in erythropoiesis where it plays a major role in β globin activation. Interestingly, a recent report has shown that EKLF is recruited to the mouse α globin promoter in induced MEL cells (Shyu et al, 2006).
PIC recruitment to the α globin gene promoters occurs only late in terminal erythroid differentiation
We analysed the recruitment of the PIC (GTFs and Pol II) at the promoter and throughout the body of the α globin gene using uninduced (proerythroblast) and induced (intermediate erythroblast) MEL cells (Figure 3). Although expressed throughout erythropoiesis, components of the PIC (e.g. Pol II, TFIIB and TFIIH; Supplementary Figure S1) were not detected at the α globin genes in uninduced cells; their recruitment only occurred late in differentiation. Some were predominantly recruited to the promoter. These included factors involved in positioning the PIC on the TATA box (TBP (Figure 3i), TFIIA (Figure 3ii) and TFIIB (Figure 3iii)), melting the core promoter DNA (TFIIE (Figure 3iv)) and the multifunctional TFIIH (Figure 3v and vi) component (also involved in creating an open promoter complex and phosphorylating the C-terminal domain [CTD] of Pol II). Similarly, we also detected components of the mediator complex (e.g. cdk8 (Figure 3xi)), which also phosphorylates the CTD) at the promoter only in induced cells.
Figure 3.
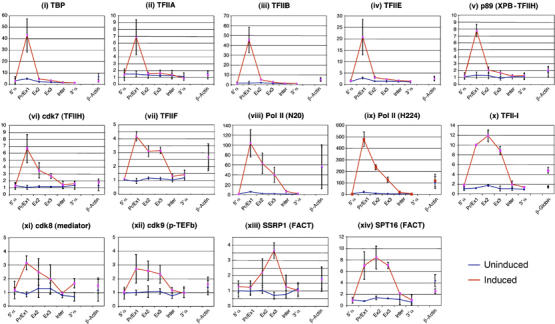
Recruitment of the pre-initiation complex at the mouse α globin core promoter during erythroid maturation. Real-time PCR analysis of immunoprecipitated chromatin using the antibodies indicated in uninduced (blue) and induced (red) MEL cells as in Figure 2. Error bars correspond to ±1 s.d. from at least two independent ChIPs. GTFs involved in recruitment of Pol II and initiation of transcription (TBP, TFIIA, TFIIB, TFIIE and TFIIH) were found at the promoter of the α globin gene only at the late stages (I-MEL) of terminal erythroid differentiation. We and others have found that the degree of enrichment observed for different transcription factors is not directly comparable. For example TFIIA (enriched 6 × ) and Pol II (enriched 100 × ) are both components of the same multiprotein complex. These differences may be caused by different epitope affinities or by differences in the degree of crosslinking for different components of such complexes.
Other factors were recruited throughout the entire α gene. The TFIIF–Pol II complex is not present at the α globin gene in uninduced MEL cells, but is present at the promoter and throughout the transcribed segment of the gene after induction (Figure 3vii, viii and ix). The recruitment and extension of Pol II binding in intermediate erythroblasts when globin is being transcribed is accompanied by a similar distribution of all tested components of the elongation machinery (e.g. cdk9, a component of p-TEFb (Figure 3xii), which switches CTD phosphorylation from ser 5 to ser 2), including two components (SSRP1 (Figure 3xiii) and SPT16 (Figure 3xiv)) of the protein complex, which facilitates chromatin transcription (FACT). Unexpectedly, the initiator binding protein TFII I (Figure 3x) was present throughout the gene, suggesting that it might also play an unpredicted role in elongation in vivo.
All relevant observations in MEL cells were confirmed by high-resolution ChIP analyses of the entire mouse α globin regulatory domain in Ter119+ cells, and the human α globin domain human primary erythroblasts and interspecific hybrids (MEL × human 16) containing a single copy of human chromosome 16, which can be induced to express both the mouse and human α globin genes. Mouse ES cells and human T lymphocytes served as negative controls for the primary cells used in these experiments (e.g Supplementary Figure S2; all data are available at http://www.imm.ox.ac.uk/mhu/home_pages/Higgs/vernimmen/).
Together, these results show that although many TFs are present and chromatin remodelling (as judged by the presence of erythroid-specific DHSs and activating chromatin marks) is established at the α globin promoters in proerythroblasts (Anguita et al, 2004), recruitment of the PIC/mediator and elongation machinery, required for maximal transcription, occurs only during the very last stages of differentiation when intermediate erythroblasts form.
Recruitment of the PIC to remote, conserved elements is necessary for recruitment to the α globin promoter
Enrichment of Pol II at the α globin promoter, assessed by two different antibodies (N20 and H224), was respectively ⩾80- and ⩾200-fold higher than at most other ‘negative' regions tested (Figure 3viii and ix). However, we also noted a small but consistent enrichment of Pol II and other components of the PIC (e.g. TFIIB) at the remote conserved elements (HS-31, -26, -21, -12 and -8) in induced MEL cells (data not shown), primary mouse Ter119+ cells (Supplementary Figure S2), induced mouse–human hybrids (HS-48, -40 and -33; Figure 4) and primary human erythroblasts (Supplementary Figure S2). These proteins were detectable at the upstream elements only using antibodies that performed very efficiently in ChIP experiments (e.g. anti-Pol II and -TFIIB). These results suggested that either the multiprotein TF complexes assembled at these sites (e.g. GATA-SCL complex and NF-E2; Anguita et al, 2004) recruit the PIC independently and/or that the upstream regions may be in close proximity and physically interact with the α globin promoter in erythroid cells. In the latter case, ChIP experiments using antibodies to components of the PIC might immunoprecipitate sequences at the upstream elements simply because they are in close proximity to the active promoters.
Figure 4.
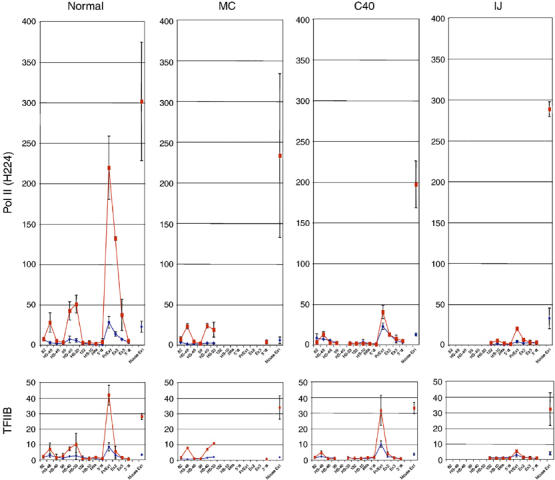
Binding of the pre-initiation complex at the upstream elements and the human α globin promoter. ChIP analyses using uninduced (blue) and induced (red) interspecific MEL hybrids containing a normal copy of chromosome 16 or copies of chromosome 16 from which HS-40 alone (C40; Figure 1B) or a larger segment (MC and IJ; Figure 1B) is deleted. Mouse Ex1 is an internal control corresponding to the mouse α globin promoter. The y-axis represents levels of enrichment of Pol II (top) and TFIIB (bottom) in mouse × human 16 hybrids. The x-axis represents the position of Taqman probes used across the human α globin locus, with the positions of erythroid-specific DNaseI hypersensitive sites. Error bars correspond to ±1 s.d. from two independent ChIPs.
To determine if the PIC is recruited independently at the promoter and the regulatory elements, we analysed interspecific hybrids derived from normal individuals or those with previously characterised, natural mutations of the α globin cluster. We analysed two types of mutation: those in which the remote upstream elements had been fully (IJ) or partially (C40) deleted but the α promoters remained intact (Figure 1B; Bernet et al, 1995; Craddock et al, 1995), and another in which all α-like genes were deleted (MC) but the upstream elements were still present (Figure 1B; Craddock et al, 1995).
Using these hybrids, we have previously shown that the formation of DHSs occurs independently at the α globin promoters and the upstream elements (Bernet et al, 1995; Craddock et al, 1995). Here, we have shown that key, tissue-restricted TFs (NF-E2 and SCL) still bind the upstream elements when the α globin promoters are deleted (MC hybrid; Supplementary Figure S3) and when the major α-regulatory element is missing (HS-40, C40 hybrid; Supplementary Figure S3). Similarly, the α globin promoter still binds the CCAAT-box binding TF (NFY) and recruits KLFs in the absence of HS-40 (C40 hybrid; Supplementary Figure S4A), and even in the absence of all upstream elements (IJ hybrid; Supplementary Figure S4A). These findings suggest that recruitment of TFs and the associated chromatin modifications, required to create the poised state, occur independently at the promoter and upstream elements and require no interaction between these two sets of elements.
We next analysed the recruitment of Pol II and GTFs in the same situations. Importantly, in the absence of the α globin promoters, Pol II and other components of the PIC (e.g. TFIIB) were still recruited to the upstream elements (HS-48, -40 and -33) in activated MEL cells (MC hybrid; Figure 4) at what appear to be similar, or slightly reduced, levels to those seen in the wild-type chromosomes. The significance of this slight reduction is not clear. However, the levels of TFIIB recruited to the upstream elements appear to be the same in the absence of the α globin promoters (Figure 4). This unequivocally shows that the PIC is specifically recruited to these elements during erythropoiesis and that binding observed using ChIP analysis is not the result of an indirect interaction with abundant Pol II and other components of the PIC binding at the promoter.
However, when all of the upstream elements are deleted (IJ hybrid; Figure 4), binding of Pol II and other components of the PIC (TFIIA, B, D, E and I) to the α globin promoter is severely reduced (although not completely absent) compared with normal chromosomes, and no α globin mRNA expression is detected (Figure 4 and Supplementary Figure S4B). Interestingly, in the absence of just the major regulatory element (HS-40, C40 hybrid; Figure 4 and Supplementary Figure S4B), the recruitment of some components of the PIC (e.g. TFIIB) was not changed at the promoter, whereas the recruitment of Pol II was severely reduced. As before, the level of α globin mRNA expression was considerably reduced (∼3% of normal; Bernet et al, 1995, and data not shown). These findings show that recruitment of the complete PIC at the α globin promoter is dependent on the combined effect of the upstream elements.
It was of interest that in the absence of just the small segment of DNA, which contains HS-40 (C40 hybrid), PIC recruitment (e.g. Pol II and TFIIB) was also reduced at HS-48 (upstream) and undetectable at HS-33 (downstream) (Figure 4), suggesting that recruitment of the PIC to each of the upstream elements depends on the presence of one or more of the other upstream elements (i.e. there is a cooperative effect). In summary, therefore, it appears that whereas the upstream elements can recruit the PIC independently in what seems to be a cooperative manner, efficient recruitment of the full PIC to the promoter is dependent on the upstream elements.
The α globin promoter and the remote, conserved non-coding sequences physically interact when the PIC is recruited
To investigate the mechanism by which the upstream elements recruit Pol II to the α globin promoters, we analysed the spatial organisation of the α globin regulatory domain throughout erythropoiesis using chromosome capture conformation (called 3C; Dekker et al, 2002). In these experiments, formaldehyde is used to crosslink protein/DNA interactions in intact nuclei. The crosslinked chromatin is then digested by a restriction enzyme, followed by ligation. If there is apposition between a remote regulatory sequence and a promoter, new, hybrid fragments containing these two elements are generated and carefully designed PCR reactions can be used to detect and quantify these newly combined elements. This approach was thus used here to analyse physical interactions between cis-acting chromosomal elements during erythropoiesis.
We applied 3C analysis to multipotent mouse embryonic stem (ES) cells , erythroid progenitors (uninduced MEL cells) and mature erythroblasts (induced MEL cells and primary mouse Ter119+ erythroblasts). Digesting nuclei with HindIII allowed us to evaluate interactions between the α globin gene (fixed) and a variety of points, including some of the upstream elements (Figure 5A). Appropriate controls were performed for each 3C experiment (Figure 5 and Supplementary Figure S5), including those previously described (Palstra et al, 2003; Drissen et al, 2004). All PCR fragments analysed were shown to be ligation dependent; neither samples of undigested crosslinked chromatin, nor chromatin that was digested without subsequent ligation generated PCR products (Supplementary Figure S5B).
Figure 5.
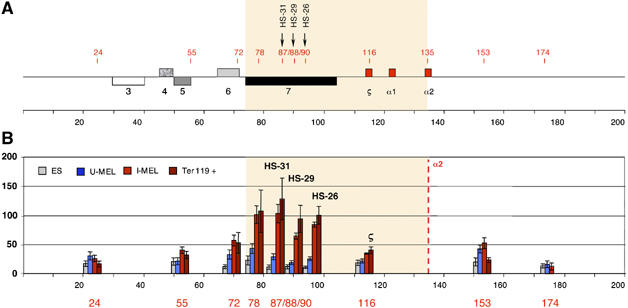
Intrachromosomal interactions involving mouse remote regulatory sequences and the mouse α2 globin promoter. (A) Chromosomal organisation of the mouse α globin locus. The following genes are annotated: 3, IL9RP3; 4, m-99; 5, Dist; 6, MPG; 7, c16orf35 gene. These are shown as boxes transcribed from the top (above the line) or bottom (below the line) DNA strand. Red numbers indicate the points (coordinates) analysed by 3C. 3C assays were performed using HindIII-digested, fixed chromatin from ES, uninduced MEL (U-MEL), induced MEL (I-MEL) and primary erythroblasts (Ter119+). The shaded area corresponds to the region containing all sequences that interact with the α globin gene (B). The bar chart (y-axis) shows the enrichment of PCR product (%) normalised to the enrichment within the Ercc3 gene (=100%). This provides an internal, genomic control for the crosslinking procedure and any general changes in nuclear or chromatin structure (de Laat and Grosveld, 2003). This, in turn, is a measure of the association between the points indicated to a fixed point (the mouse α2 globin promoter) in cells representing different stages of differentiation, from embryonic stem cells (ES) to mature erythroblasts (I-MEL and primary Ter119+ cells). We have not considered the α1 gene in this study because the HindIII fragment containing this gene is too large (14 kb) for appropriate 3C analysis. Data shown represent the average of at least two independent experiments using Taqman/real-time PCR. Error bars denote s.e.m. Each PCR was performed several times and averaged. Signals were normalised to the total amount of DNA used, estimated with an amplicon located within a HindIII fragment. Coordinates of the points analysed are indicated on the x-axis. As an illustration of the specificity of these interactions, the interaction between α globin and HS-26 (∼40 kb apart) is enriched in erythroid cells, whereas the interaction between α globin and a region at +174 (∼40 kb in the opposite direction) is not enriched.
By observing the presence or absence of PCR products following the 3C protocol, we might have concluded that intrachromosomal interactions in the α globin locus take place in uninduced MEL cells (Supplementary Figure S5B) and even in uncommitted ES cells (data not shown). However, as noted by others using this technique (Palstra et al, 2003; Drissen et al, 2004; Osborne et al, 2004; Spilianakis et al, 2005; Zhou et al, 2006), a background level of interaction, associated with specific PCR products, is nearly always detected. We therefore considered it essential to quantitate (and also stringently control the specificity) all interactions and hence designed and calibrated Taqman probes for real-time PCR analysis. Using this approach, it became clear that there is an increase in interaction between the upstream regulatory elements and the α globin gene in cells that actively produce globin (I-MEL and Ter119+ cells; Figure 5B) compared with those that do not (ES and U-MEL cells; Figure 5B). For example, the interaction between HS-26 and α globin is increased 11-fold when comparing Ter119+ cells (erythroid) with ES cells (non-erythroid). Parenthetically, these findings demonstrate that it is essential to quantitate 3C analyses accurately rather than simply base interpretations on the presence or absence of PCR products.
We initially evaluated 3C interactions by analysing the interactions between the α2 gene and various points along the α globin cluster. However, interactions were subsequently confirmed using other fixed regions upstream and outside of the cluster (coordinates 55, 87 (HS-31) and 90 (HS-26); Figure 5 and Supplementary Figure 6). Using this approach, we concluded that in ES cells and proerythroblasts (uninduced MEL), there appeared to be relatively little interaction between elements along the α globin locus (Figure 5B), which we interpret as showing that the cluster predominantly adopts a linear conformation in these cells. By contrast, in activated cells (I-MEL and Ter119+), we could readily detect increased interactions showing that the region of the cluster spanning coordinates 78–90, including HS-31 and HS-26, which also bind the PIC, comes into close spatial proximity with each other and the α2 gene, whereas other regions outside (24, 55, 72, 153 and 174) and between (e.g. ζ globin) do not interact (Figure 5B). Thus, it appears that late in erythroid differentiation, when Pol II is being recruited to the upstream elements (HS-31 and HS-26) and the α globin promoter, the chromosome conformation changes, such that these elements are now physically interacting (Figure 5B). As Pol II is specifically bound to the upstream elements and the promoter but not to the regions in between, we have found no evidence to support a model in which Pol II tracks along the chromosome from the upstream elements to the promoter (Hatzis and Talianidis, 2002; Wang et al, 2005). Rather it appears that these regions are interacting via one or more looping mechanisms (de Laat and Grosveld, 2003; Osborne et al, 2004), presumably allowing the exchange and/or interaction of prebound multiprotein complexes.
Discussion
The aim of this study was to provide a detailed understanding of how cis-acting elements controlling mammalian genes might activate transcription at the appropriate time during differentiation, using expression of the α globin gene cluster during erythropoiesis as a model. We have built on our previous studies showing that the α globin regulatory domain is first activated by TF binding and chromatin modifications at the remote upstream elements in uncommitted haematopoietic cells (Anguita et al, 2004). Such changes then extend along the chromatin domain from the upstream elements to the promoter, producing a fully poised state at each of the conserved elements (enhancers and promoters) throughout the regulatory domain in early, committed erythroid progenitors. We have shown here and elsewhere that establishing this poised state occurs independently at each of the regulatory elements; for example, histone modifications, TF binding and appearance of DHS occur at the promoter whether or not the enhancer element(s) are intact. Similar observations have been made at some (Bender et al, 2000) but not all (Spicuglia et al, 2002) other loci studied. However, for the very few genes that have been studied in sufficient detail, the development of the poised state often appears to proceed in a unidirectional manner from the remote enhancer to the promoter, and any model should account for this (Figure 6).
Figure 6.
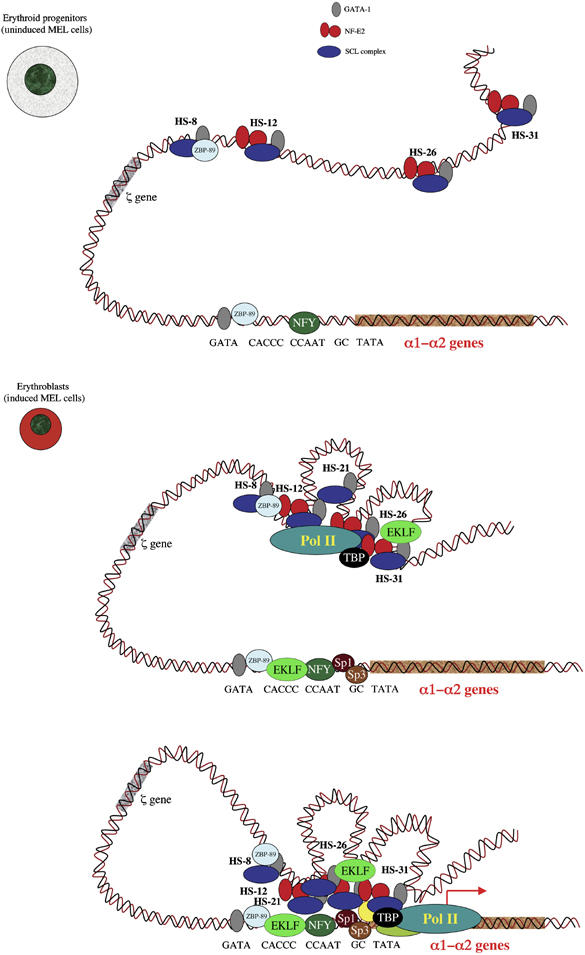
A model proposing how complexes form and interact at the mouse α globin locus during erythropoiesis. In committed erythroid progenitors (U-MEL, proerythroblast stage), the remote regulatory sequences (HS-31, HS-26, HS-12 and HS-8) are bound by multiprotein complexes containing various combinations of SCL, NF-E2 and GATA1. At this stage, the α globin promoter is also occupied by GATA1 in combination with the ubiquitous transcription factors ZBP-89 and NFY and is best poised for expression. In differentiating erythroid cells (I-MEL and primary erythroblasts), the PIC, including Pol II, is recruited to the enhancers in a cooperative manner, but independently of the promoter. Sp/X-Kruppel-like transcription factors (e.g. Sp1 and Sp3) are also recruited independently of the upstream elements, to the promoter. At this final stage, the α globin promoter is now occupied by a multiprotein complex including GATA1, ZBP-89, EKLF, NFY, Sp1 and Sp3 that represents a docking site for the recruitment of PIC, which is entirely dependent on the presence of the upstream elements.
Clearly, the multistep process leading to the poised state is an important aspect of being able to delay the timing of expression. During the early stage of differentiation when inappropriate expression of a specialised protein like globin might be deleterious to the cell, a multistep process is driving the silent gene towards a state in which it could be easily and safely switched on by a final, late switch such as a post-translational modification, removal of a repressor protein or the binding of an additional, activating TF. In this context, it was interesting that we have shown that Sp/X-KLF factors, known to behave as activators in erythroid cells (Bieker, 2001; Van Loo et al, 2003), bind to the promoter only when the genes are activated and could provide part of the final trigger for globin expression.
Here, we have concentrated on understanding when and how the general transcriptional machinery (GTFs, Pol II, mediator and elongation factors) is recruited to the α globin regulatory elements during differentiation. We found, as in most (e.g Sawado et al, 2003; Szutorisz et al, 2005a; Levings et al, 2006) but not all (Hatzis and Talianidis, 2002; Soutoglou and Talianidis, 2002) genes that have been studied in this way, the PIC is recruited to the promoter only late in differentiation. We also found that the PIC is recruited to the upstream elements independently of the promoter, most likely via its interaction with prebound TFs. Interestingly, in contrast to all previous genes studied, including the β globin cluster (Szutorisz et al, 2005a; Levings et al, 2006), recruitment occurs only at the α globin upstream elements late in differentiation.
Recruitment of the PIC at the upstream elements occurs at what appears to be relatively low levels compared to the promoter. It seems unlikely that this observation was due to inappropriate choice of amplicons, which can affect the degree of enrichment measured, as similar low levels of enrichment were seen at all of the upstream elements in both mouse and human. It is therefore possible that components of the PIC are recruited to the upstream elements via protein/protein interactions rather than directly to DNA and consequently its component proteins are not so effectively crosslinked to DNA in the ChIP assay. This could also explain why, despite careful analysis using nuclear run on assays and strand-specific PCR (unpublished observations on normal but not abnormal chromosomes), to date, we have not observed non-genic transcripts originating at these sites, as documented at some other regulatory elements (Ashe et al, 1997; Plant et al, 2001; Masternak et al, 2003; Rogan et al, 2004; Ling et al, 2005; Szutorisz et al, 2005a). Such transcripts might have been expected if the PIC were directly bound to DNA. Whatever the reason for this difference in enrichment, the nature of the interaction of the PIC appears different at the upstream sites from the promoters.
How do the upstream elements facilitate recruitment of the PIC to the promoter? By studying mutant chromosomes derived from patients with natural deletions of the α genes and their regulatory elements, we have shown that recruitment of the complete PIC to the promoter is strictly dependent on the presence of these upstream elements, and our data imply but do not prove that during differentiation and maturation, the PIC will be first recruited to the upstream elements and then to the promoter (Figure 6). The time lapse separating these two events may be very short and we have not been able to separate recruitment at the upstream elements and the promoter in time-course experiments. The newly modified 3C analysis described here, using Taqman technology, supports a model in which there is direct physical interaction between the upstream elements and the promoter. ChIP analyses (distribution of Pol II and histone modifications) suggest that this does not involve the intervening chromatin, arguing against previously described tracking (Hatzis and Talianidis, 2002; Wang et al, 2005) or linking mechanisms (Bulger and Groudine, 1999; Dorsett, 1999). However, these findings are consistent with the previously described looping model of enhancer/promoter interaction (de Laat and Grosveld, 2003; Osborne et al, 2004). Applying 3C to cells from different stages of differentiation showed that looping coincides with the onset of transcription (Figure 6).
The molecular basis for looping is not yet clear, although interactions between structural proteins (Kurukuti et al, 2006), TFs (Drissen et al, 2004; Vakoc et al, 2005) or components of the PIC within a transcription factory (Bartlett et al, 2006) are all candidates. It is of interest that in the absence of the major regulatory element (HS-40), the PIC is not recruited to all of the other upstream elements, suggesting that the upstream elements in some way act together to recruit the PIC. Again, deletion studies suggest that this structure forms first and then, subsequently facilitates recruitment of the PIC to the α globin promoters, with looping occurring between the upstream elements and the α globin genes (Figure 6). Presumably, the directional order of this looping is dictated by the location of the regulatory elements and the order in which they recruit TFs, cofactors and the PIC during differentiation. Clearly, this will also be influenced by which TFs are available to bind. For example, the first two upstream elements to be activated in mouse (HS-26 and HS-12) are bound by GATA2, which is expressed exclusively in early haemopoietic progenitors. By contrast, these and all other upstream elements (including HS-31, -21 and -8), and the α globin promoters are bound by GATA1 when it replaces GATA2 expression later in erythropoiesis. This simple switch in TF expression could explain the appearance of sequential activation of elements along the chromosome. It is of interest that the last event described here (recruitment of the PIC to the promoter) occurs at the same time as the recruitment of Sp/X-KLF factors to the promoter.
It would seem that the purpose of the interaction between the upstream elements and the promoters is either to deliver the PIC (Johnson et al, 2003; Szutorisz et al, 2005a, 2005b) or relocate the promoter into a nuclear sub-compartment containing components of the PIC (so-called transcription factory). Currently, there is no way to distinguish between these models. Once the looped protein/DNA complex (remote regulatory elements, promoter and PIC) has formed, it is possible that the upstream elements also influence later stages in transcription (promoter melting, clearance, elongation and termination), as suggested for other genes (Boehm et al, 2003; Sawado et al, 2003).
We demonstrate here a comprehensive, dynamic picture of the sequential events resulting in transcriptional activation of a mammalian locus during differentiation and terminal maturation by evaluating the orthologous human and mouse α globin clusters. Our observations are in keeping with the general models of a temporal, stepwise orchestration of promoter assembly in lower organisms (Cosma et al, 1999). However, in mammalian systems, additional control of the promoters occurs via remote regulatory elements, which appear to play a specialised role within the hierarchical order of events leading to chromatin remodelling and transcriptional initiation, possibly regulating the timing and efficiency of expression. Very few mammalian genes have been studied in such depth as the globin clusters. As more examples become available, it should be possible to distinguish the principles governing the timing and levels of expression from the inevitable differences in detail between different mammalian genes.
Materials and methods
Primary cells and cell lines
MEL cell line 585 and interspecific MEL hybrids were grown and induced as described (Higgs et al, 1990; Bernet et al, 1995; Craddock et al, 1995). ES cells were grown as reported (Anguita et al, 2004). Mature primary erythroid cells were obtained from phenylhydrazine-treated mice (Spivak et al, 1973) and Ter119+ cells were magnetically sorted using MACS. Human erythroid progenitors are derived from peripheral blood mononuclear cells of normal subjects and grown in two-step cultures (Brown et al, 2006). Human activated T lymphocytes were obtained from buffy coat by culture for 3 days in RPMI/20% fetal calf serum in the presence of 2% phytohaemagglutinin M (GIBCO BRL) and 20 U/ml interleukin-2.
Chromatin immunoprecipitation assay
ChIP was performed with the ChIP assay kit and protocol from Upstate with the following modifications: formaldehyde was added to the culture medium at a final concentration of 0.5% for 15 min at room temperature and samples were sonicated for 2 × 4 min at 4°C to cleave genomic DNA to an average of 500 bp. Antibodies Pol II (N20, H224), TFIIA (FL109), TFIIB (SI-1), TBP (SI-1), TFIIE (C17, C21), TFIIF (C18), TFIIH (S19), SSRP1 (H300), SPT16 (H300), NF-E2 (C19, C16), GATA1 (N6), Sp3 (D20, H225), Sp1 (PEP2), NFY (H209), cdk7 (FL346), cdk8 (H139), cdk9 (H169) and TFII-I (H58) were purchased from Santa Cruz. ZBP-89 antibody was purchased from Rockland. Real-time PCRs using primers and probes (5′FAM-3′TAMRA) for murine and human α globin locus were described previously (Anguita et al, 2004; De Gobbi et al, in preparation). PCR master mix was purchased from Eurogentec. Additional primers and probes used in this study are available on request.
Chromosome conformation capture
PCR reactions, primers and probes were optimised by digesting and ligating equimolar amount of BAC templates containing the entire mouse α globin and the Ercc3 loci. PCR efficiency was measured by amplifying 100–0.1 ng of mixed BACs with a fixed amount (200 ng) of digested genomic DNA. All primer combinations showed a linear correlation between BAC template and PCR product. The endogenous Ercc3 locus has been reported to adopt the same spatial conformation in different tissues (de Laat and Grosveld 2003; Palstra et al, 2003; Drissen et al, 2004). Thus, all 3C results were corrected by data from Ercc3 analysis, controlling for changes in nuclear size, chromatin density and crosslinking efficiency. Primers and probes were designed as follows: a universal sequence-specific Taqman probe and reverse primer on a fixed restriction fragment in combination with different forward primers specific of other restriction fragments. 3C templates (200 ng) were used for Taqman/PCR reaction using normal PCR condition with ABI prism 7000. Mouse primers and probes used are available on request.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
3ta
Acknowledgments
We are grateful to Merlin Crossley, Sjaak Philipsen and Jim Bieker for providing EKLF antibodies and Catherine Porcher for SCL antibody, Boris Guyot for advice and Richard Gibbons, Paresh Vyass, Catherine Porcher and David Garrick for critically reading the manuscript. MDG is a PhD student in Pharmacology and Experimental and Clinical Therapy at the University of Turin, Italy. This work was funded by the Medical Research Council.
References
- Anguita E, Hughes J, Heyworth C, Blobel GA, Wood WG, Higgs DR (2004) Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J 23: 2841–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguita E, Johnson CA, Wood WG, Turner BM, Higgs DR (2001) Identification of a conserved erythroid specific domain of histone acetylation across the alpha-globin gene cluster. Proc Natl Acad Sci USA 98: 12114–12119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ (1997) Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev 11: 2494–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett J, Blagojevic J, Carter D, Eskiw C, Fromaget M, Job C, Shamsher M, Trindade IF, Xu M, Cook PR (2006) Specialized transcription factories. Biochem Soc Symp 73: 67–75 [DOI] [PubMed] [Google Scholar]
- Bender MA, Mehaffey MG, Telling A, Hug B, Ley TJ, Groudine M, Fiering S (2000) Independent formation of DnaseI hypersensitive sites in the murine beta-globin locus control region. Blood 95: 3600–3604 [PubMed] [Google Scholar]
- Bernet A, Sabatier S, Picketts DJ, Ouazana R, Morle F, Higgs DR, Godet J (1995) Targeted inactivation of the major positive regulatory element (HS-40) of the human alpha-globin gene locus. Blood 86: 1202–1211 [PubMed] [Google Scholar]
- Bieker JJ (2001) Kruppel-like factors: three fingers in many pies. J Biol Chem 276: 34355–34358 [DOI] [PubMed] [Google Scholar]
- Boehm AK, Saunders A, Werner J, Lis JT (2003) Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol 23: 7628–7637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Leach J, Reittie JE, Atzberger A, Lee-Prudhoe J, Wood WG, Higgs DR, Iborra FJ, Buckle VJ (2006) Coregulated human globin genes are frequently in spatial proximity when active. J Cell Biol 172: 177–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M (1999) Looping versus linking: toward a model for long-distance gene activation. Genes Dev 13: 2465–2477 [DOI] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K (1999) Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97: 299–311 [DOI] [PubMed] [Google Scholar]
- Craddock CF, Vyas P, Sharpe JA, Ayyub H, Wood WG, Higgs DR (1995) Contrasting effects of alpha and beta globin regulatory elements on chromatin structure may be related to their different chromosomal environments. EMBO J 14: 1718–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Laat W, Grosveld F (2003) Spatial organization of gene expression: the active chromatin hub. Chromosome Res 11: 447–459 [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N (2002) Capturing chromosome conformation. Science 295: 1306–1311 [DOI] [PubMed] [Google Scholar]
- Dorsett D (1999) Distant liaisons: long-range enhancer–promoter interactions in Drosophila. Curr Opin Genet Dev 9: 505–514 [DOI] [PubMed] [Google Scholar]
- Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W (2004) The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev 18: 2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RC, Taxman DJ, Seshasayee D, Kensinger MH, Bieker JJ, Wojchowski DM (1996) Functional interaction of GATA1 with erythroid Kruppel-like factor and Sp1 at defined erythroid promoters. Blood 87: 1793–1801 [PubMed] [Google Scholar]
- Hatzis P, Talianidis I (2002) Dynamics of enhancer–promoter communication during differentiation-induced gene activation. Mol Cell 10: 1467–1477 [DOI] [PubMed] [Google Scholar]
- Higgs DR, Wood WG, Jarman AP, Sharpe J, Lida J, Pretorius IM, Ayyub H (1990) A major positive regulatory region located far upstream of the human alpha-globin gene locus. Genes Dev 4: 1588–1601 [DOI] [PubMed] [Google Scholar]
- Huang DY, Kuo YY, Lai JS, Suzuki Y, Sugano S, Chang ZF (2004) GATA-1 and NF-Y cooperate to mediate erythroid-specific transcription of Gfi-1B gene. Nucleic Acids Res 32: 3935–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JR, Cheng JF, Ventress N, Prabhakar S, Clark K, Anguita E, De Gobbi M, de Jong P, Rubin E, Higgs DR (2005) Annotation of cis-regulatory elements by identification, subclassification, and functional assessment of multispecies conserved sequences. Proc Natl Acad Sci USA 102: 9830–9835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KD, Grass JA, Park C, Im H, Choi K, Bresnick EH (2003) Highly restricted localization of RNA polymerase II within a locus control region of a tissue-specific chromatin domain. Mol Cell Biol 23: 6484–6493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabe Y, Yamada J, Uga H, Yamaguchi Y, Wada T, Handa H (2005) NF-Y is essential for the recruitment of RNA polymerase II and inducible transcription of several CCAAT box-containing genes. Mol Cell Biol 25: 512–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan DA, van Heyningen V (2005) Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet 76: 8–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurukuti S, Tiwari VK, Tavoosidana G, Pugacheva E, Murrell A, Zhao Z, Lobanenkov V, Reik W, Ohlsson R (2006) CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci USA 103: 10684–10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levings PP, Zhou Z, Vieira KF, Crusselle-Davis VJ, Bungert J (2006) Recruitment of transcription complexes to the beta-globin locus control region and transcription of hypersensitive site 3 prior to erythroid differentiation of murine embryonic stem cells. FEBS J 273: 746–755 [DOI] [PubMed] [Google Scholar]
- Ling J, Baibakov B, Pi W, Emerson BM, Tuan D (2005) The HS2 enhancer of the beta-globin locus control region initiates synthesis of non-coding, polyadenylated RNAs independent of a cis-linked globin promoter. J Mol Biol 350: 883–896 [DOI] [PubMed] [Google Scholar]
- Masternak K, Peyraud N, Krawczyk M, Barras E, Reith W (2003) Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nat Immunol 4: 132–137 [DOI] [PubMed] [Google Scholar]
- Merika M, Orkin SH (1995) Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF. Mol Cell Biol 15: 2437–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P (2004) Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet 36: 1065–1071 [DOI] [PubMed] [Google Scholar]
- Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W (2003) The beta-globin nuclear compartment in development and erythroid differentiation. Nat Genet 35: 190–194 [DOI] [PubMed] [Google Scholar]
- Plant KE, Routledge SJ, Proudfoot NJ (2001) Intergenic transcription in the human beta-globin gene cluster. Mol Cell Biol 21: 6507–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogan DF, Cousins DJ, Santangelo S, Ioannou PA, Antoniou M, Lee TH, Staynov DZ (2004) Analysis of intergenic transcription in the human IL-4/IL-13 gene cluster. Proc Natl Acad Sci USA 101: 2446–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawado T, Halow J, Bender MA, Groudine M (2003) The beta -globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes Dev 17: 1009–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu YC, Wen SC, Lee TL, Chen X, Hsu CT, Chen H, Chen RL, Hwang JL, Shen CK (2006) Chromatin-binding in vivo of the erythroid kruppel-like factor, EKLF, in the murine globin loci. Cell Res 16: 347–355 [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Talianidis I (2002) Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295: 1901–1904 [DOI] [PubMed] [Google Scholar]
- Spicuglia S, Kumar S, Yeh JH, Vachez E, Chasson L, Gorbatch S, Cautres J, Ferrier P (2002) Promoter activation by enhancer-dependent and -independent loading of activator and coactivator complexes. Mol Cell 10: 1479–1487 [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA (2005) Interchromosomal associations between alternatively expressed loci. Nature 435: 637–645 [DOI] [PubMed] [Google Scholar]
- Spivak JL, Toretti D, Dickerman HW (1973) Effect of phenylhydrazine-induced hemolytic anemia on nuclear RNA polymerase activity of the mouse spleen. Blood 42: 257–266 [PubMed] [Google Scholar]
- Szutorisz H, Canzonetta C, Georgiou A, Chow CM, Tora L, Dillon N (2005a) Formation of an active tissue-specific chromatin domain initiated by epigenetic marking at the embryonic stem cell stage. Mol Cell Biol 25: 1804–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szutorisz H, Dillon N, Tora L (2005b) The role of enhancers as centres for general transcription factor recruitment. Trends Biochem Sci 30: 593–599 [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA (2005) Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell 17: 453–462 [DOI] [PubMed] [Google Scholar]
- Van Loo PF, Bouwman P, Ling KW, Middendorp S, Suske G, Grosveld F, Dzierzak E, Philipsen S, Hendriks RW (2003) Impaired hematopoiesis in mice lacking the transcription factor Sp3. Blood 102: 858–866 [DOI] [PubMed] [Google Scholar]
- Vyas P, Vickers MA, Picketts DJ, Higgs DR (1995) Conservation of position and sequence of a novel, widely expressed gene containing the major human alpha-globin regulatory element. Genomics 29: 679–689 [DOI] [PubMed] [Google Scholar]
- Wang Q, Carroll JS, Brown M (2005) Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell 19: 631–642 [DOI] [PubMed] [Google Scholar]
- Woo AJ, Moran TB, Choe S-K, Schindler YL, Sullivan MR, Fujiwara Y, Paw BH, Cantor AB (2005) Identification of zfp148 (ZBP-89) as a novel GATA-1 associated transcription factor involved in megakaryopoiesis and definitive erythropoiesis. Blood 106: 828a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou GL, Xin L, Song W, Di LJ, Liu G, Wu XS, Liu DP, Liang CC (2006) Active chromatin hub of the mouse alpha-globin locus forms in a transcription factory of clustered housekeeping genes. Mol Cell Biol 26: 5096–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Figure S6
3ta


