Abstract
The translocon is a membrane-embedded protein assembly that catalyzes protein movement across membranes. The core translocon, the SecYEG complex, forms oligomers, but the protein-conducting channel is at the center of the monomer. Defining the properties of the SecYEG protomer is thus crucial to understand the underlying function of oligomerization. We report here the reconstitution of a single SecYEG complex into nano-scale lipid bilayers, termed Nanodiscs. These water-soluble particles allow one to probe the interactions of the SecYEG complex with its cytosolic partner, the SecA dimer, in a membrane-like environment. The results show that the SecYEG complex triggers dissociation of the SecA dimer, associates only with the SecA monomer and suffices to (pre)-activate the SecA ATPase. Acidic lipids surrounding the SecYEG complex also contribute to the binding affinity and activation of SecA, whereas mutations in the largest cytosolic loop of the SecY subunit, known to abolish the translocation reaction, disrupt both the binding and activation of SecA. Altogether, the results define the fundamental contribution of the SecYEG protomer in the translocation subreactions and illustrate the power of nanoscale lipid bilayers in analyzing the dynamics occurring at the membrane.
Keywords: membrane, Nanodiscs, SecA, SecYEG, translocon
Introduction
The Sec translocon is a membrane protein complex that cooperates with cytosolic partners to drive polypeptide substrate translocation into and across membranes. The translocon may be comprised of multiple components, but the core is made of a conserved heterotrimer known as SecYEG in bacteria, SecYEβ in archaea and Sec61p in eukaryotes (Cao and Saier, 2003). The remarkable fact that the translocon forms a protein-conducting channel is now rationalized through structural analysis of the core complex: cryo-electron microscopy and high-resolution crystal structures recently unraveled the location of the protein channel path at the center of the SecY subunit (Van den Berg et al, 2004; Mitra et al, 2005). Three other key structural elements were identified following structural determinations: the ‘plug' domain formed by a small helix blocking the channel on its periplasmic side; the ‘pore ring' made by a few residues forming a constriction point at the center of the channel; and the ‘lateral gate' created by the juxtaposition of two SecY transmembrane segments forming an opening toward the lipid bilayer. Recent biochemical experiments along with computer-based simulations support this model, including the widening of the pore and the displacement of the plug away from the center of the channel by the incoming polypeptide substrate (Cannon et al, 2005; Tam et al, 2005; Gumbart and Schulten, 2006; Tian and Andricioaei, 2006; Maillard et al, 2007).
Despite this tremendous progress, there are major problems that still remain in the field. One of the major questions, complicated by the observation that the channel is contained within a single SecY subunit, is that the SecYEG complex spontaneously forms oligomers of two or more copies, both in the membrane or in detergent solution (Manting et al, 2000; Collinson et al, 2001; Bessonneau et al, 2002; Mori et al, 2003; for review see Rusch and Kendall, 2007). One original hypothesis assumed that the channel was formed at the center of these oligomers, but experimentation now supports the structural prediction that a single SecYEG complex forms the translocation pathway (Duong, 2003; Cannon et al, 2005). Nevertheless, additional evidence indicates that the SecYEG dimer and also probably the tetramer are essential and active oligomeric assemblies. Cryo-electron microscopy of a translocon engaged with a ribosome-nascent chain complex shows electron density representative of a SecYEG dimer (Mitra et al, 2005). Similarly, at the endoplasmic reticulum membrane, the active channel bound to ribosomes consists of four copies of Sec61p (Beckmann et al, 2001; Ménétret et al, 2005). However, the significance of SecYEG complex oligomerization remains puzzling. Defining the underlying function of monomer and oligomers in the translocation subreaction is thus crucial for the mechanistic understanding of translocon.
Other current questions concern the interaction of the SecYEG complex with the motor protein SecA. SecA is an ATPase that binds to substrate proteins and delivers them post-translationally to the translocon (for review see Veenendaal et al, 2004). Once bound, SecA undergoes cyclic conformational changes that ultimately result in the ATP-dependant stepwise translocation of the unfolded polypeptide (Schiebel et al, 1991). Despite the fact that SecA has been extensively studied and also crystallized in various conformational states (Hunt et al, 2002; Sharma et al, 2003; Vassylyev et al, 2006; Zimmer et al, 2006), little is known about its association and stoichiometry with the SecYEG complex. SecA exists mostly as a dimer in solution (Woodbury et al, 2002), but its oligomeric state during membrane binding and preprotein translocation is a topic of intense debate; it is proposed to be monomer or dimer depending on the study (Driessen, 1993; Or et al, 2002, 2005; de Keyzer et al, 2005; Jilaveanu et al, 2005). Here again, defining the association between the SecYEG complex and its effector SecA has deep implications for the understanding of the inner working of translocon.
Characterizing the interaction and dynamic taking place at a membrane-associated or membrane-embedded protein complex is notoriously difficult. Reconstitution into large lipid bilayer vesicles or analysis in detergent micelles often limits the experimentation or introduces bias in the interpretation of results. Here, we made use of Nanodiscs (Bayburt and Sligar, 2003), an emerging technology that allowed us to prepare nanometer-sized soluble particles containing only one SecYEG complex (Figure 1). Nanodiscs faithfully recreate a lipid bilayer and thus permit investigating the reactivity and interactive nature of SecA with the SecYEG translocon in a near native environment, without use of detergent.
Figure 1.
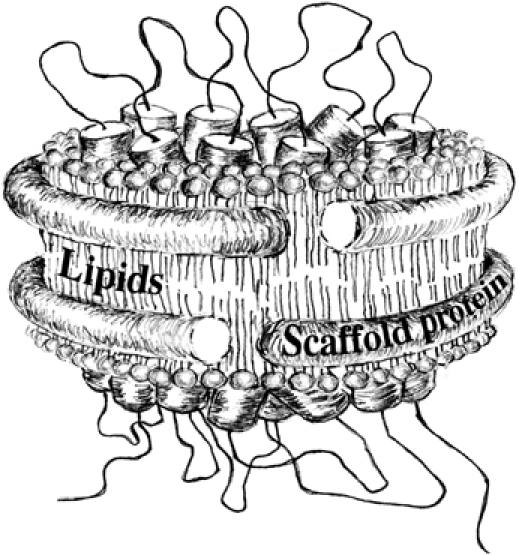
Schematic view of a membrane protein complex embedded into the Nanodisc structure (adapted from Bayburt and Sligar, 2003). The image was kindly provided by Ms Kailun Jiang.
Results
Incorporation of the SecYEG complex into Nanodiscs
Nanodisc incorporates membrane proteins into soluble, 10-nm-wide, lipid bilayer structures of controlled size (Bayburt and Sligar, 2003; Denisov et al, 2004). Integral to this technology is a bioengineered 200-amino-acid amphipathic and multihelical membrane scaffold protein (hereafter referred to as MSP), whose hydrophobic faces circumscribe the edges of small lipid bilayer (∼130–160 lipid molecules) and whose polar faces interact with polar aqueous environment (Figure 1). The principle of the self-assembly process and the ratio lipids:MSP forming the Nanodisc structure have been well studied (Denisov et al, 2004), and pioneering studies for proteins containing various seven-transmembrane segments have been reported (Bayburt et al, 2006; Leitz et al, 2006). We used this system to incorporate the SecYEG heterotrimer, which consists of 15 transmembrane helices. Purified SecYEG complex was incubated with an optimized amount of MSPs and total Escherichia coli phospholipids. Following detergent removal, the resulting mixture was fractionated on a calibrated gel filtration column (Figure 2A). The majority of the proteins eluted as two peaks, whereas a minimal amount eluted as larger aggregates in the void volume (V0). SDS–PAGE analysis shows that the larger peak corresponds to an association between the MSP and the SecYEG complex, whereas the second peak corresponds to the MSP alone (Figure 2B). Omission of the MSP from the self-assembly mixture resulted in total aggregation of the SecYEG complex. Thus, the SecYEG complex has been associated with the MSP to form a water-soluble structure characteristic of Nanodiscs containing incorporated target protein (referred to as Nd–SecYEG complex or SecYEG–Nanodisc hereafter).
Figure 2.

Reconstitution of the SecYEG complex into Nanodiscs. (A) Typical protein elution profile obtained after size-exclusion chromatography of a SecYEG–Nanodisc preparation. (B) The fractions corresponding to elution volume 10–16 ml in (A) were analyzed by SDS–PAGE, followed by Coomassie blue staining of the gel. For molecular size comparison, the SecYEG complex and the MSPs were loaded on thetwo lanes on the left. (C) Sedimentation velocity analysis and c(s) distribution of the Nd–SecYEG complex at 5.6 μM (gray lane) and 9.6 μM (black lane). A small peak is present at lower and higher S values, which represent small amount of free MSP and MSP bound to a dimeric SecYEG complex, respectively. The insets represent the c(M) distribution of the 9.6-μM sample (f/f0=1.4).
Stoichiometry and stability of the Nanodisc-reconstituted SecYEG complex
To determine the number of SecYEG complex incorporated per Nanodisc, the Nd–SecYEG complex was analyzed by analytical ultracentrifugation (Figure 2C). Sedimentation velocity data obtained at 5.6 and 9.6 μM show a major peak (>90%) that sediments at ∼5.3S. The c(M) distribution (f/f0=1.4; Figure 2C, inset) indicates that the peak corresponds to a molecular weight (MW) of ∼123 kDa, consistent with particles made of two MSPs (∼50 kDa) and one SecYEG complex (∼75 kDa). Such a stoichiometry is also confirmed by blue native PAGE (BN-PAGE) analysis (Figure 3A). The purified SecYEG complex migrates as a population of monomer and dimer (Bessonneau et al, 2002; Figure 3A, lane 1), whereas the Nd–SecYEG complex migrates predominantly as a single band (lane 2), between the SecYEG monomer and dimer. Reconstitution in the absence of the SecYEG complex leads to the formation of empty discs (labeled Nd, lane 3) that migrate slightly above the dimeric MSP, probably owing to the contribution of lipid molecules to the molecular weight of the discs. Depending on the quality of the separation by size-exclusion chromatography, the SecYEG–Nanodisc preparation was sometimes contaminated with free MSP (e.g. Figures 2C and 3A, lane 2).
Figure 3.
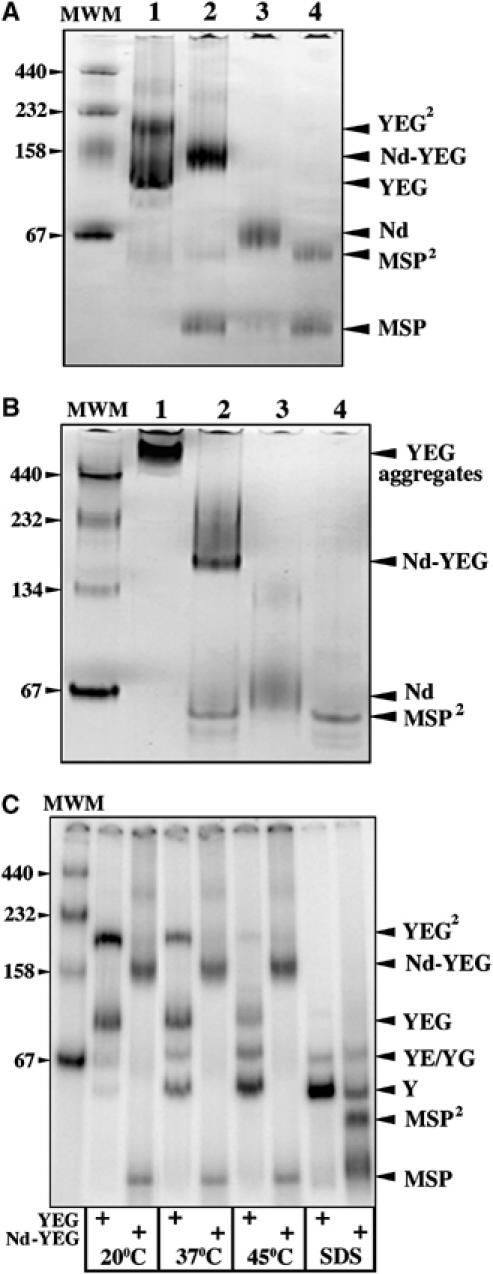
The SecYEG monomer is stably incorporated into the Nanodisc structure. (A) The Nd–SecYEG complex (4 μg, lane 2) was analyzed by BN-PAGE followed by Coomassie blue staining. For comparison, the detergent-soluble SecYEG complex (4 μg, lane 1), the empty discs (2 μg, lane 3) and the MSPs (2 μg, lane 4) were loaded on the same gel. Molecular weight markers: ferritin, 440 kDa; catalase, 232 kDa; aldolase, 158 kDa and BSA 66/132 kDa. (B) The protein samples described in (A) were analyzed by CN-PAGE, followed by Coomassie blue staining of the gel. (C) The [125I]SecYEG and [125I]Nd–SecYEG complexes (each ∼20 000 c.p.m.; ∼20 ng) were incubated at the indicated temperature for 5 min before analysis by BN-PAGE and phosphorimaging. In the presence of 0.1% SDS, both complexes are dissociated.
When analyzed by colorless native PAGE (CN-PAGE; Schägger et al, 1994 and Figure 3B), the Nd–SecYEG complex migrates into the gel (lane 2), whereas the detergent-maintained SecYEG complex readily aggregates and remains at the top of the gel (lane 1). The MSP dimers (∼50 kDa, lane 3) and the empty discs (∼50 kDa+lipids, lane 4) are also detected at the expected positions. We note that the sharpness of the bands seems sensitive to the presence of lipids; both the Nd–SecYEG complex and the empty discs, but not the MSPs alone, migrate as smeary bands on native gel. Altogether, the results show that a single SecYEG complex is incorporated in a water-soluble Nanodisc structure.
When embedded in the membrane, the SecYEG heterotrimer naturally forms a stable complex (Joly et al, 1994). When extracted from the lipid bilayer with detergents, the SecYEG complex easily dissociates into single subunits (Brundage et al, 1992). This instability can be detected when the purified SecYEG complex is prewarmed before analysis by BN-PAGE (Figure 3C). Temperature leads to the progressive conversion of the SecYEG monomer and dimer into dissociated subunits. The same incubation conditions do not, however, dissociate the Nanodisc-reconstituted SecYEG complex, and the migration pattern of the preparation remains unchanged (Figure 3C). Thus, the disc structure has created a protective environment around the SecYEG complex that mimics that of the natural membrane lipid bilayer.
Binding of monomeric SecA onto the monomeric SecYEG complex
The SecA protein migrates on CN-PAGE as a single band around 200 kDa, as expected because SecA forms dimers at micromolar concentrations (Woodbury et al, 2002; Figure 4A, lane 1). However, upon incubation with increasing amounts of SecYEG–Nanodisc, the SecA band shifts to a higher position on the gel (lanes 2–5). The same size shift is also observed when the Nd–SecYEG complex is incubated with increasing amounts of SecA (Figure 4B). Identical results were obtained when incubation was performed at room temperature or at 37°C (data not shown). In the control experiment, the SecA protein was incubated with empty discs, but a size shift is not observed either for SecA or for the empty discs (Figure 4A, lanes 6 and 7). Thus, the Nanodisc-reconstituted SecYEG monomer forms a complex with SecA (referred to as Nd–SecYEG–SecA complex hereafter). Formation of the complex is efficient as almost all SecA molecules associate with the SecYEG–Nanodiscs when equimolar amounts are mixed together (Figure 4A, lane 5 or B, lane 6).
Figure 4.
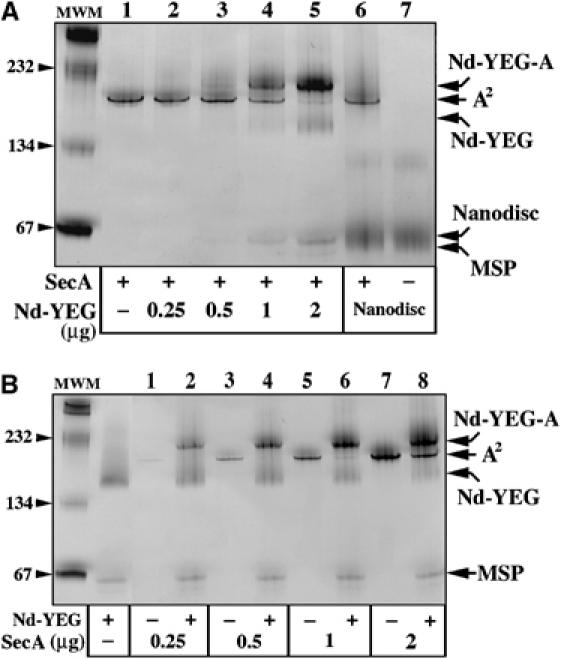
Binding of the SecA protein onto the SecYEG–Nanodisc. (A) The SecA protein (1 μg, lanes 1–5) was incubated for 5 min at room temperature with the indicated amount of SecYEG–Nanodiscs. As control, the empty discs (2 μg, lanes 6 and 7) were incubated with or without SecA (0.5 μg). The protein samples were separated by CN-PAGE followed by Coomassie blue staining of the gel. (B) The indicated amounts of SecA proteins were incubated with (even lanes) or without (odd lanes) the Nd–SecYEG complex (2 μg) and analyzed by CN-PAGE followed by Coomassie blue staining.
The apparent molecular weight of the Nd–SecYEG–SecA complex (i.e. migrating just below the 232 kDa marker, Figure 4) suggests that only one SecA molecule associates per SecYEG–Nanodisc (calculated MW ∼225 kDa). This is a surprising result as SecA is mostly dimeric in aqueous solution and indeed migrates as a dimer on native gels. To ascertain the stoichiometry, the SecA dimeric state was stabilized by oxidation with Cu2+(phenanthroline)3, resulting in the formation of an intra-dimeric SecA disulfide bond (de Keyzer et al, 2005). Incubation with an increasing amount of oxidizing agent converts about half of SecA into covalently linked dimers (Figure 5A, lanes 1–4). On CN-PAGE, both the crosslinked SecA dimers and the native dimers migrate at the same position, around 200 kDa (Figure 5B, lanes 1–4). Next, the preparations of oxidized SecA molecules, made of a mixed population of native and undissociable SecA, were incubated with the SecYEG–Nanodiscs and then separated by CN-PAGE (lanes 5–8). In addition to the Nd–SecYEG–SecA complex, an additional band of higher molecular weight is apparent on the gel. As the intensity of the band increases with the quantity of undissociable SecA dimers (compare Figure 5A, lanes 1–4 with B, lanes 5–8), we conclude that it represents a complex between the SecYEG–Nanodisc and dimeric SecA. Thus, the Nanodisc-reconstituted SecYEG complex binds exclusively monomeric SecA, unless dimeric SecA has been artificially stabilized.
Figure 5.
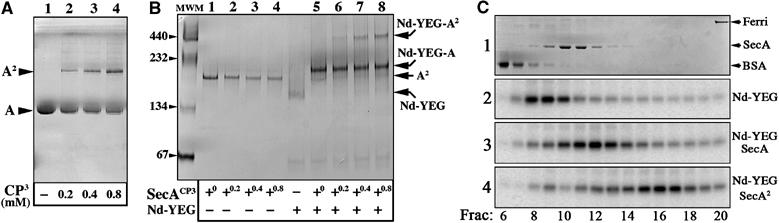
Binding of disulfide-linked SecA onto the SecYEG–Nanodisc. (A) The SecA protein was oxidized with Cu2+(phenanthroline)3 at the indicated concentration and aliquots (1 μg) were analyzed by non-reducing SDS–PAGE, followed by Coomassie blue staining of the gel. (B) The same SecA aliquots (labeled SecACP3 on the figure) were analyzed by CN-PAGE, either alone (lanes 1–4) or after incubation with the Nd–SecYEG complex (2 μg; lanes 5–8). (C) Separation of the Nd–SecYEG–SecA complexes by sucrose density centrifugation. The Nd–SecYEG complex was reconstituted in the presence of trace amounts of 125I-labeled SecYEG. The samples contained (1) 70 μg BSA, 70 μg SecA and 30 μg ferritin, (2) [125I]Nd–SecYEG, (3) [125I]Nd–SecYEG+70 μg SecA and (4) [125I]Nd–SecYEG+70 μg of purified cysteine-linked SecA dimer. Samples from fractions 6–20 were analyzed by SDS–PAGE, followed by Coomassie blue staining (panel 1) or phosphorimaging (panels 2–4). The band detected by phosphorimaging is [125I]SecY. Crosslinked SecA dimer and unmodified SecA dimer sediment at the same position on these sucrose gradients (data not shown).
This conclusion is also reached by sucrose gradient analysis (Figure 5C). On its own, the [125I]Nd–SecYEG complex sediments in-between BSA and dimeric SecA (fractions 8–10). In the presence of SecA, the [125I]Nd–SecYEG complex sediments in higher-molecular-weight fractions (fractions 11–13), just after the position of dimeric SecA. When the purified cysteine-linked SecA dimer is incubated with the [125I]Nd–SecYEG complex, the peak is further shifted toward higher-molecular-weight positions (fractions 15–17).
Dissociation of the SecA dimer onto the monomeric SecYEG complex
The above observations strongly suggest that the SecYEG complex causes disassembly of the SecA dimer. To provide addtional evidences, the SecYEG–Nanodiscs and SecA were incubated together with an amine-reactive crosslinking reagent (Figure 6A). In these experiments, either the SecYEG complex reconstituted in Nanodiscs (lanes 1 and 2) or the SecA protein (lanes 3–6) were 125I-radiolabeled. The results show that in both cases, the crosslinking products are identical and migrate at positions in-between the SecA monomer and dimer. Thus, the crosslinker reagent captures the SecYEG complex only with the SecA monomer. Incubation of 125I-labeled SecA with higher amounts of SecYEG–Nanodiscs not only increases the yield of crosslinking products, but also decreases the yield of crosslinked SecA dimers (Figure 6B). This last observation is consistent with the dissociation of the SecA dimer during incubation with the Nd–SecYEG complex.
Figure 6.

Crosslinking analysis of the Nd–SecYEG–SecA complex. (A) About 2 μg of [125I]Nd–SecYEG (lane 1) was mixed with 1 μg SecA (lane 2) and then incubated with the crosslinker reagent EGS as described in Materials and methods. In the reverse experiment, [125I]SecA (0.5 μg, lanes 3–6) was incubated with unlabeled Nd–SecYEG (2 μg, lane 5) or empty Nanodiscs (2 μg, lane 6) before crosslinking with EGS. Proteins were dissolved in 0.1% SDS and analyzed by BN-PAGE followed by Coomassie blue staining (left panel) and phosphorimaging (right panel). (B) [125I]SecA (0.5 μg) was incubated with the indicated amounts of Nd–SecYEG or empty Nanodiscs. The crosslinking reaction was performed with EGS as described in Materials and methods.
The same conclusion is also reached using fluorescence resonance energy transfer (FRET) assays. Two populations of SecA were independently labeled at their carboxy-terminal cysteine residues with either coumarin (DACIA, donor fluorophore) or fluorescein (5-IAF, acceptor fluorophore). The labeled proteins were denatured with urea, mixed together at a 1:2 molar ratio and renatured. The recorded spectra for the heterodimer (Figure 7A, black trace) shows that the fluorescence emission of the donor (dashed black trace) is decreased by 15%, whereas that of the acceptor is increased correspondingly (dashed gray trace). This FRET reflects the dimeric association of the SecA proteins (see also Or et al, 2002). Taking into account the heterodimer formation during refolding (i.e. ∼33% if Poisson distributed at a 1:2 molar ratio), the FRET efficiency was calculated to be 45%. As the SecA monomers and dimers exist in dynamic equilibrium, addition of a five-fold molar excess of unlabeled SecA reduces the energy transfer efficiency (Figure 7B, gray trace). Similarly, addition of a five-fold molar excess of SecYEG–Nanodiscs (Figure 7C, gray trace) strongly abolishes FRET (∼12% increase in donor fluorescence and corresponding decrease in acceptor fluorescence). The intensity of FRET is not changed upon addition of empty discs (Figure 7D).
Figure 7.
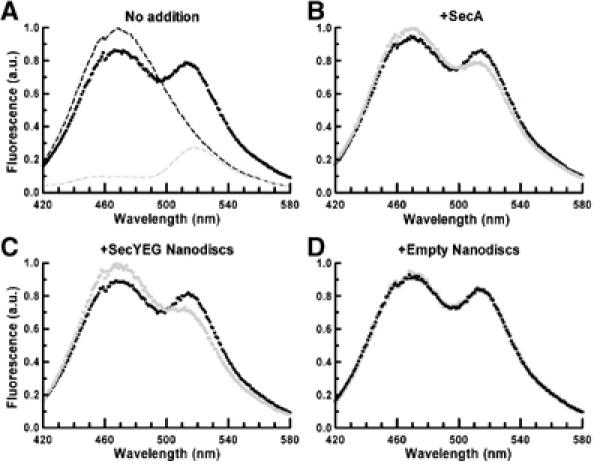
Analysis of the SecA dimer dissociation by steady-state FRET. Excitation was set at 390 nm and emitted light was recorded from 420 to 580 nm. (A) Coumarin/fluorescein-labeled SecA heterodimer (9 μg) (dark trace) compared with coumarin/unlabeled (dashed dark trace) or fluorescein/unlabeled SecA heterodimer (9 μg) (dashed gray trace). The emitted light for coumarin/fluorescein-labeled SecA heterodimer (9 μg, dark trace) was again recorded after the following addition (gray traces). (B) 45 μg of unlabeled SecA; (C) 45 μg of Nd–SecYEG; and (D) 18 μg of empty Nanodiscs.
Altogether, the results from native gel electrophoresis, crosslinking analysis, sucrose gradient centrifugation and FRET experiments support the conclusion that the SecYEG complex embedded in Nanodiscs binds the SecA monomer and shifts the SecA monomer–dimer equilibrium toward the dissociated state.
The SecY residue R357 is essential for the binding of SecA
Previous genetic and biochemical studies indicate that mutations at the conserved residue Arg357, located in the largest cytosolic loop of SecY, prevents SecA ATPase activation (Mori and Ito, 2001; van der Sluis et al, 2006). To test whether the mutation also results in defective SecA–SecYEG association, the SecYEG (R357E) mutant complex was incorporated into Nanodiscs. The results show that the efficiency of complex formation is indeed reduced (Figure 8A, middle panel). Compared with the wild type (right panel), lesser amounts of Nd–SecYEG–SecA complex are formed and, accordingly, higher amounts of SecA remain uncomplexed. Furthermore, when the amino-acyl residues adjacent to R357 are also substituted (sequence R357G358D359 replaced by E357D358P359), the binding of SecA is totally abolished (left panel). Thus, the residue R357 (and the adjacent positions) constitutes a major binding site for SecA. The results also prove that the interaction between the Nd–SecYEG complex and SecA is specific and, as expected, occurs on a SecY loop normally located on the cytosolic face of the membrane.
Figure 8.

The SecY residue R357 and acidic lipids contribute to the binding of SecA. (A) Wild-type or mutant SecYEG Nanodiscs (1 μg) were incubated for 5 min at room temperature with the indicated amounts of SecA. Samples were analyzed by CN-PAGE followed by Coomassie blue staining of the gel. Incubation at 37°C did not change the results (data not shown). (B, C) The SecYEG complex reconstituted into Nanodisc (1 μg) with the indicated phospholipids was incubated for 5 min at room temperature with the indicated amount of SecA protein and then analyzed by CN-PAGE and Coomassie blue staining. Given the smeary aspect of the Nd–SecYEG–SecA complex on native gel, densitometry analysis was performed on unbound SecA obtained at saturating binding concentration (i.e. 2 μg of SecA in this experiment). The densitometry values were compared with SecA concentration standards run on the same gel.
Acidic lipids contribute to the binding of SecA at Nanodisc-reconstituted SecYEG
Binding of SecA to inner membrane vesicles depends on phosphatidylglycerol (Lill et al, 1990; Hendrick and Wickner, 1991; Breukink et al, 1992), but the contribution of these lipids to the formation and/or stability of the SecYEG–SecA associations remains unknown. The SecYEG complex was thus reconstituted in Nanodiscs containing different phospholipids; either dioleoylphosphatidylcholine (PC) only or supplemented with 30% dioleoylphosphatidylglycerol (PG). Control experiments included reconstitution without lipids or with total E. coli lipids (made of ∼70% neutral and ∼30% acidic phospholipids). When analyzed by CN-PAGE, these various preparations migrate at different positions depending on the nature of the lipid incorporated (Supplementary Figure S1). The efficiency of reconstitution and the stoichiometry SecYEG:MSP is however not affected by the nature or the absence of lipids (Supplementary Figure S1). In the latter case, it is possible that a few lipid molecules are brought from the purified SecYEG complex itself. Next, the SecYEG–Nanodisc preparations were incubated with SecA and analyzed by CN-PAGE (Figure 8B and C). The results show that the binding capacity of the Nd–SecYEG complex is increased when it contains acidic lipids (Figure 8B, compare left and right panels). When 2 μg of the SecA protein (i.e. saturating concentration) is added to the SecYEG–Nanodiscs containing acidic lipids, the fraction of bound SecA represents ∼55%. This fraction is reduced to ∼18% when SecA is incubated with the SecYEG–Nanodisc containing only neutral lipids. Similarly, the formation of the Nd–SecYEG–SecA complex appears facilitated when the SecYEG–Nanodisc structure includes E. coli lipids (Figure 8C). At saturating concentration (i.e. 2 μg of SecA), the fraction of SecA bound to the Nd–SecYEG complex is ∼62% (right panel), but decreases to ∼20% when the SecYEG–Nanodisc is prepared without additional acidic lipids (left panel). Altogether, the results show that acidic lipids located in the surroundings of the SecYEG complex contribute significantly to the binding of SecA.
Activation of SecA at Nanodisc-reconstituted SecYEG
The SecA ATPase is stimulated by membrane-embedded or detergent-solubilized SecYEG (Lill et al, 1990; Duong, 2003). Thus, SecA is able to sense the SecYEG complex as an activating ligand. Accordingly, the results show that the endogenous SecA ATPase is stimulated ∼5-fold during incubation with the SecYEG–Nanodiscs (Figure 9A). The stimulation is comparable to the values previously reported when SecA and the purified SecYEG complex are incubated together in detergent solution (Duong, 2003). Neither the MSPs nor the empty discs can activate SecA to the same extent (Figure 9A). Furthermore, the SecYEG–Nanodiscs carrying the SecY mutations analyzed above (i.e. E357 and E357D358P359) do not stimulate the SecA ATPase (Figure 9B), suggesting that the activation requires binding to the SecYEG complex.
Figure 9.

Activation of the SecA ATPase at Nanodisc-reconstituted SecYEG. (A) SecA (1 μg) was incubated at 37°C in TKA buffer (50 mM KCl, 1 mM DTT, 1 mM ATP and 25 mM Tris–HCl pH 8.8) alone or in the presence of 4 μg of either the Nd–SecYEG complex, the empty discs or the MSPs. The amount of inorganic phosphate released during incubation was measured every 5 min by photocolorimetric method. (B) SecA (1 μg) was incubated at 37°C in TKA buffer in the presence of 4 μg of the indicated Nanodisc preparation. The amount of inorganic phosphate was measured after 20 min of incubation. (C) 125I-labeled SecA protein (∼20 000 c.p.m., ∼20 ng, lane 1) was incubated for 5 min at 37°C in TSG buffer with 1 μg of SecYEG–Nanodiscs (lanes 2–11) in the presence or absence of 1 mM ATP. Increasing amounts of unlabeled SecA were then added as indicated for an additional 5 min at 37°C. Protein samples were separated by CN-PAGE and analyzed by phosphorimaging.
As the SecA ATPase is stimulated by acidic lipid-containing membrane (Hendrick and Wickner, 1991), we measured the ATP hydrolysis occurring during incubation with the SecYEG–Nanodiscs reconstituted with various lipids. The results show that the SecA ATPase is stimulated 4- to 5-fold either with the SecYEG–Nanodiscs containing E. coli total lipids or PC lipids supplemented with PG lipids (Figure 9B). In contrast, the stimulation obtained in the absence of lipids or in the presence of PC lipids only is less significant (∼2). Thus, acidic lipids contribute significantly to the SecYEG-dependent stimulation of the SecA ATPase.
Finally, we measured the effect of nucleotides on the formation and/or stability of the Nd–SecYEG–SecA complex. The SecYEG–Nanodiscs were incubated with 125I-labeled SecA followed by the addition of unlabeled SecA (Figure 9C). On its own, SecA migrates as a monomer when a radiochemical amount is loaded on the gel (lane 1), and this monomer associates with the Nd–SecYEG complex (lane 2). In the presence of increasing amounts of unlabeled SecA, 125I-labeled SecA is displaced from (or prevented from interacting with) the SecYEG complex and reforms dimeric associations with unlabeled SecA (lanes 3–6). In the presence of ATP in the incubation buffer (lanes 7–11), the binding efficiency of 125I-labeled SecA is unchanged and the competition with unlabeled SecA remains the same. Identical results are obtained when using a non-hydrolyzable ATP analog (ATPγS) or when ADP is present in the incubation buffer (data not shown). Thus, the binding and/or dissociation of the SecA protein onto the SecYEG–Nanodisc seems to be independent of ATP binding and hydrolysis.
Discussion
Strategies for characterizing membrane proteins such as the SecYEG complex usually involve lipid bilayer vesicles or membrane mimetics such as detergent. Lipid vesicles are compromised by micron-sized dimensions, heterogeneity and inherent difficulties to measure protein–protein interactions, whereas detergent micelles often affect protein activity and readily destabilize protein associations. Study of the translocon is further challenged by its oligomeric nature, which cannot be controlled in conventional lipid vesicle or detergent micelles systems. Thus, the ability of Nanodiscs to produce a monodisperse population of soluble membrane-like structures allowed us to investigate the specific contribution of a SecYEG protomer to the translocation subreactions. Indeed, several observations reported here demonstrate that the SecYEG complex embedded in Nanodiscs has faithfully conserved its structural and functional characteristics. First, the disc structure preserves the thermostability of the SecYEG complex, much like when it is embedded in a membrane lipid bilayer. Second, the SecYEG complex conserves the capacity to bind its cytosolic binding partner SecA. The association is not merely binding onto the disc but involves a defined site on the SecY subunit. Third, much like when it is embedded in liposomes, the SecYEG complex embedded in Nanodiscs activates the SecA ATPase. Fourth, acidic lipids, which facilitate the translocation reaction, also contribute to the binding and activation of SecA at the SecYEG–Nanodisc. Our capacity to reproduce some fundamental aspects of the SecYEG–SecA interaction in near-native conditions allows additional observations and conclusions.
Previous results from hydrodynamic studies, crosslinking analysis and X-ray crystallography indicate that various forms of SecA dimer exist in solution that differing in their conformation and associations (Hunt et al, 2002; Vassylyev et al, 2006; Zimmer et al, 2006). Here, our analysis reveals that the SecYEG complex shifts the SecA equilibrium toward the dissociated state and forms associations with only the SecA protomer. This conclusion is in agreement with Or et al (2002), who reported that crosslinking of the SecA dimer cannot be obtained in the presence of SecYEG-containing liposomes. In fact, the predisposition of SecA for dissociation is supported by earlier analysis. Small changes in the primary sequence of SecA easily destabilize its dimeric state (Or et al, 2002, 2005), whereas incubation with high-salt concentration shifts the association equilibrium (kd∼0.1 μM at low-salt concentration) toward the monomer (Woodbury et al, 2002). Furthermore, comparison of the SecA crystal structures indicates different dimeric associations (parallel, antiparallel and interwined), suggesting the absence of strong and defined surface of dimerization (Hunt et al, 2002; Vassylyev et al, 2006; Zimmer et al, 2006). In our study, the SecA protein is present at concentrations (e.g. ∼2 μM) well above the dissociation constant, but the entire population of SecA becomes dissociated and trapped as a monomer with the SecYEG complex. Binding of the SecA dimer onto the SecYEG–Nanodisc can be observed, but this requires artificial stabilization of SecA by cysteine covalent linkage (Figure 5B and C). As the SecYEG complex triggers the dissociation of the SecA dimer, the most likely explanation is that the crosslinked SecA dimer binds to the SecYEG complex as tethered monomers, remaining together because of the covalent linkage. Our study does not exclude, however, the possibility that oligomerization of the SecYEG complex generates binding sites for additional SecA. Accordingly, covalently linked or antibody-stabilized SecYEG dimers were found to associate with both one and two SecA molecules (Duong, 2003; Tziatzios et al, 2004). It should be stressed that these previous studies were all performed in detergent solution and abnormal interactions cannot be excluded.
Another conclusion reached here is that residues around position R357, located between transmembrane segments TM8 and TM9 on the SecY subunit, form a major determinant for SecA association. The equivalent residue on the Sec61p complex (i.e. R406) is also critical for efficient binding of the ribosome (Cheng et al, 2005), and elegant in vivo crosslinking analysis shows that other SecY residues in the same region (i.e. 347 and 358) directly contact SecA (Mori and Ito, 2006). In fact, a large number of cold-sensitive mutants and export-defective mutations map in this cytosolic loop and among them, residue R357 was shown to be critical in supporting the SecA catalytic activity (Mori and Ito, 2001; van der Sluis et al, 2006). However, binding assays such as Scatchard analysis (F Duong, unpublished results) and surface plasmon resonance (van der Sluis et al, 2006) did not detect a reduction of the SecA binding affinity to membrane enriched for the R357E mutant complex. Although we show here that a maximal disruption of SecA–SecYEG complex is obtained when the residues adjacent to R357 are also substituted (Figure 8A), the above methods should have been sensitive enough to detect some decrease in the SecA binding affinity or capacity. Here again, it is possible that oligomerization of the SecYEG complex in the membrane provides additional binding points for the SecA protein, so that the effect of the R357E mutation is minimized and therefore difficult to detect. Nevertheless, our observations clearly indicate that the translocation defect observed with the mutation R357E finds its root in a defective SecA–SecYEG binding association. An attractive possibility is that the loop containing these residues forms a structure important for promoting the dissociation of the SecA dimer and therefore binding onto the SecYEG complex.
Reconstitution of SecYEG in Nanodiscs also allowed us to test the influence of surrounding phospholipids during the binding of SecA. This is an important issue because acidic phospholipids are required for efficient SecA-mediated preprotein translocation, both in vivo and in vitro, but a clear mechanistic interpretation is lacking (van Dalen and de Kruijff, 2004). Earlier biochemical studies showed that SecA inserts into lipid monolayers in a reaction that is facilitated by acidic phospholipids (Breukink et al, 1992). The contribution of these lipids to SecA conformational changes and activity is also supported by several other experiments using membrane vesicles (Hendrick and Wickner, 1991; Ulbrandt et al, 1992; van der Does et al, 2000). Most importantly, FRET and crosslinking experiments indicate that acidic lipids cause dissociation of the SecA dimer into monomers (Or et al, 2002). Here, using SecYEG-Nanodiscs reconstituted in different lipids, we are providing direct evidence that acidic lipids enhance the binding of SecA to the SecYEG complex. It is possible that acidic lipids may stabilize a conformation of the SecYEG channel, more prone to bind and activate SecA. Alternatively, acidic lipids surrounding the SecYEG complex may form a microenvironment favorable for the dissociation of SecA dimer and thus favorable for the binding to the SecYEG complex. These lipids may have additional roles in protein translocation, but increasing the binding of SecA to SecYEG would certainly contribute to facilitation of the translocation reaction.
Finally, our results demonstrate that the SecYEG protomer is sufficient for activation of the SecA ATPase. Although this activity may represent a futile cycle of ATP hydrolysis, as it occurs in the absence of protein substrate, the amount of ATP hydrolyzed is negligible compared to the ATPase activity supported by SecA during protein translocation (over 1000 mol/min; Bassilana et al, 1992). The SecYEG-dependent ATPase we observe may represent some sort of ‘preactivation' of SecA, and our results suggest that it depends on the monomerization of the SecA dimer. First, the activation of the SecA ATPase requires binding of SecA to the SecYEG complex, as both are abolished by the mutation R357E into SecY. Second, the dissociation of the SecA dimer triggered by the SecYEG complex can occur in the absence of ATP. Third, acidic lipids promote the dissociation of SecA dimer (Or et al, 2002) and also facilitate binding and activation of the SecA ATPase at the SecYEG–Nanodisc. Combined together, these observations suggest a model in which the dissociation of the SecA dimer provoked by the SecYEG complex is followed by activation of the SecA ATPase. In contrast, dimeric SecA on its own is prevented from such an activation and may represent the dormant state of the protein.
The oligomeric associations of the SecYEG complex and its effector SecA still raise important questions and it will likely remain a topic of intense debate. The capacity of Nanodiscs to isolate a single SecYEG protomer has allowed us to define the fundamental binding characteristics of the SecYEG complex and its contribution to the dissociation and activation of the SecA dimer. Future studies involving larger Nanodiscs (Denisov et al, 2004) should allow analysis of the properties of the SecYEG oligomer and thus further contribute to the understanding of the functional role of oligomerization during protein translocation.
Materials and methods
Biological reagents
Plasmids encoding His-tagged SecA, His-tagged SecYEG and SecY-R357 mutant versions were previously described (Duong, 2003; Tam et al, 2005). His-tagged SecA was isolated by Ni2+-chelating chromatography (Duong, 2003) and further purified on a Superdex 200 HR10/30 column (Amersham Biosciences) equilibrated in TSG buffer (50 mM Tris pH 7.9, 50 mM NaCl, 10% glycerol and 1 mM DTT). Purification of the His-tagged SecYEG complex was as previously described (Collinson et al, 2001). 125I-labeling of SecYEG and SecA was performed using iodogen-coated tubes (Pierce) as previously described (Duong, 2003).
Reconstitution of the SecYEG complex in Nanodiscs
Lyophilized powder of scaffold protein MSP1D1 (Denisov et al, 2004) was dissolved at 7.75 mg/ml (314 μM) in 20 mM Tris–HCl, pH 7.4, 100 mM NaCl and 0.5 mM EDTA. Manufacturer-weighted phospholipids (Avanti Polar Lipids) were dissolved in chloroform and dried under a stream of nitrogen. The lipids were resuspended in TSG buffer containing 0.5% dodecyl-β-D-maltopyranoside (DDM, Anatrace). A typical reconstitution experiment involved mixing together the SecYEG complex, the MSPs and the solubilized lipids at a molecular ratio of 1:4:100. For reconstitution of empty discs, the molecular ratio MSP:lipids was increased to 1:60. After 1 h incubation on ice, the self-assembly process was initiated by adding half volume of BioBeads (Bio-Rad) followed by gentle rocking (6 h at 4°C). Beads were removed by sedimentation and the resulting self-assembled mixture was centrifuged for 30 min at 200 000 g (Beckman TLA 110 rotor) and then injected onto a Superdex 200 HR10/30 column equilibrated in TSG buffer. The fractions containing the Nd–SecYEG complex were pooled, concentrated and stored at −80°C.
Oxidation of SecA and crosslinking assays
SecA in CL buffer (50 mM NaCl, 10% glycerol and 50 mM K-Hepes pH 7.0) was incubated with the indicated amount of Cu2+(phe)3 for 30 min at room temperature. The reaction was terminated by the addition of 5 mM neocuproine and desalted onto a Sephadex G-25 column. For sucrose gradient centrifugation, the oxidized SecA dimer was purified from unmodified SecA onto a Superdex 200 HR10/30 column equilibrated in 25 mM Tris pH 7.9 and 500 mM NaCl. Chemical crosslinks between SecA and Nd–SecYEG were obtained with 0.75 mM ethylene glycol bis-succinimidylsuccinate (EGS; Sigma) for 20 min at room temperature in CL buffer containing 2 mM DTT. Reactions were stopped by adding 1/10 volume of 250 mM Tris–HCl pH 7.5 and 1.5% SDS.
Analytical ultracentrifugation analyses
Sedimentation velocity experiments were conducted at 20°C with a Beckman Optima XL-I analytical ultracentrifuge. The partial specific volume of MSP2–SecYEG was 0.7505 ml/g using the program SEDNTERP (Laue et al, 1992). Standard epoxy double-sector centerpieces (12 mm) were filled with protein solution (400–450 μl) and reference buffer (25 mM Tris pH 7.9, 300 mM NaCl and 2 mM DTT). Velocity experiments were run at 40 000 r.p.m. and no time interval was set between scans. Data were analyzed with a c(s) distribution of the Lamm equation solutions calculated with the program SEDFIT (Schuck et al, 2002), assuming the regularization parameter P to be 0.95 (high confidence level). Sedimentation coefficient increments of 200 were used in the appropriate range for each sample. Sucrose gradient centrifugations were conducted at 40 000 r.p.m. (Beckman SW41 rotor) for 16 h at 4°C. Protein samples in 400 μl of buffer C (25 mM Tris pH 7.9 and 50 mM NaCl+2 mM DTT when required) were layered on top of 10 ml 8–13% sucrose gradients prepared in the same buffer. Twenty-one fractions of 500 μl were collected and aliquots (20 μl) were analyzed by SDS–PAGE.
Steady-state FRET experiments
SecA was labeled with iodoacetamido derivatives of coumarin (DACIA; Molecular Probes) or fluorescein (5-IAF; Sigma) as described by Or et al (2002). Briefly, 5.5 nm of reduced and purified SecA (in TSG buffer containing 5 mM EDTA) was incubated with 24 nmol of 5-IAF or 20 nm of DACIA (2 h at room temperature) and then desalted on Sephadex G25 column equilibrated in TSG buffer containing 1 mM DTT. The extent of modification was determined at 498 nm for SecA-5IAF and 393 nm for SecA-DACIA, using extinction coefficients of 24 100 and 75 500 cm−1 M−1, respectively. For both fluorophores, the labeling obtained was ∼1 mol dye/mol of SecA. In preparation for FRET experiments, 648 μg SecA–DACIA was mixed in a 2:1 molar ratio with SecA–5IAF. As controls, SecA–DACIA and SecA–5IAF were mixed in a 2:1 or 1:2 molar ratio with unlabeled SecA, respectively. The SecA mixtures were denaturated with urea (5 M final) and renatured onto a Sephadex G25 column equilibrated in TSG buffer. The SecA preparations were diluted to 0.45 μM in 50 mM Tris pH 7.4, 10% glycerol, 50 mM NaCl, 50 mM KCl, 1 mM DTT and 2 mM EDTA. Steady-state FRET measurements were performed on a Cary Eclipse spectrofluorometer at 25°C. Excitation was at 390 nm and emitted light was recorded from 420 to 580 nm (slit width 10 nm, three scans at the rate of 100 nm/min). Spectra were corrected for the reduction of the DACIA emission owing to absorbance or scattering at 390 nm by SecYEG–Nanodiscs or empty Nanodiscs (not shown). This reduction was calculated by subtracting the SecA–DACIA/SecA-unlabeled emission in the presence of Nanodiscs from the emission without Nanodiscs. All spectra were normalized using the maximal emission of the SecA–DACIA/SecA-unlabeled at an excitation wavelength of 390 nm.
Other methods
Blue native gels, colorless native gels and electrophoresis conditions were performed as described by Schägger et al (1994). All gels used during this study were linear 5–13% gradient gels. Detection of 125I-labeled proteins was performed using a Phosphoimager scanner. Densitometry was performed using ImageQuant software. Protein concentrations were determined using the Bradford reagent (Bio-Rad). The SecA ATPase activity was measured using the colorimetric method of Lanzetta et al (1979). The ATPase results represent an average of three independent experiments.
Supplementary Material
Supplementary Figure S1
Acknowledgments
We would like to express our gratitude to Dr Grant Mauk for access to the UBC Laboratory of Molecular Biophysics and to the laboratory members for stimulating discussions. BL-G is recipient of a Roman Babicki Fellowship for Medical Research, SGS is an IC Gunsalus Professor and University Scholar and FD is a Canada Research Chair Tiers II. This work was supported by the National Institutes of Health GM33775 (to SGS), the Canada Foundation for Innovation, the Canadian Institutes of Health Research and the Natural Sciences and Engineering Research Council of Canada (to FD).
References
- Bassilana M, Arkowitz RA, Wickner W (1992) The role of the mature domain of proOmpA in the translocation ATPase reaction. J Biol Chem 267: 25246–25250 [PubMed] [Google Scholar]
- Bayburt TH, Grinkova YV, Sligar SG (2006) Assembly of single bacteriorhodopsin trimers in bilayer nanodiscs. Arch Biochem Biophys 450: 215–222 [DOI] [PubMed] [Google Scholar]
- Bayburt TH, Sligar SG (2003) Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci 12: 2476–2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann R, Spahn CM, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G (2001) Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107: 361–372 [DOI] [PubMed] [Google Scholar]
- Bessonneau P, Besson V, Collinson I, Duong F (2002) The SecYEG preprotein translocation channel is a conformationally dynamic and dimeric structure. EMBO J 21: 995–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breukink E, Demel RA, de Korte-Kool G, de Kruijff B (1992) SecA insertion into phospholipids is stimulated by negatively charged lipids and inhibited by ATP: a monolayer study. Biochemistry 31: 1119–1124 [DOI] [PubMed] [Google Scholar]
- Brundage L, Fimmel CJ, Mizushima S, Wickner W (1992) SecY, SecE, and band 1 form the membrane-embedded domain of Escherichia coli preprotein translocase. J Biol Chem 267: 4166–4170 [PubMed] [Google Scholar]
- Cannon KS, Or E, Clemons WM Jr, Shibata Y, Rapoport TA (2005) Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J Cell Biol 169: 219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao TB, Saier MH (2003) The general protein secretory pathway: phylogenetic analyses leading to evolutionary conclusions. Biochim Biophys Acta 1609: 115–125 [DOI] [PubMed] [Google Scholar]
- Cheng Z, Jiang Y, Mandon EC, Gilmore R (2005) Identification of cytoplasmic residues of Sec61p involved in ribosome binding and cotranslational translocation. J Cell Biol 168: 67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinson I, Breyton C, Duong F, Tziatzios C, Schubert D, Or E, Rapoport T, Kuhlbrandt W (2001) Projection structure and oligomeric properties of a bacterial core protein translocase. EMBO J 20: 2462–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Keyzer J, van der Sluis EO, Spelbrink RE, Nijstad N, de Kruijff B, Nouwen N, van der Does C, Driessen AJ (2005) Covalently dimerized SecA is functional in protein translocation. J Biol Chem 280: 35255–35260 [DOI] [PubMed] [Google Scholar]
- Denisov IG, Grinkova YV, Lazarides AA, Sligar SG (2004) Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J Am Chem Soc 126: 3477–3487 [DOI] [PubMed] [Google Scholar]
- Driessen AJ (1993) SecA, the peripheral subunit of the Escherichia coli precursor protein translocase, is functional as a dimer. Biochemistry 32: 13190–13197 [DOI] [PubMed] [Google Scholar]
- Duong F (2003) Binding, activation and dissociation of the dimeric SecA ATPase at the dimeric SecYEG translocase. EMBO J 22: 4375–4384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbart J, Schulten K (2006) Molecular dynamics studies of the archaeal translocon. Biophys J 90: 2356–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrick JP, Wickner W (1991) SecA protein needs both acidic phospholipids and SecY/E protein for functional high-affinity binding to the Escherichia coli plasma membrane. J Biol Chem 266: 24596–24600 [PubMed] [Google Scholar]
- Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J (2002) Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science 297: 2018–2026 [DOI] [PubMed] [Google Scholar]
- Jilaveanu LB, Zito CR, Oliver D (2005) Dimeric SecA is essential for protein translocation. Proc Natl Acad Sci USA 102: 7511–7516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly JC, Leonard MR, Wickner WT (1994) Subunit dynamics in Escherichia coli preprotein translocase. Proc Natl Acad Sci USA 91: 4703–4707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA (1979) An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem 100: 95–97 [DOI] [PubMed] [Google Scholar]
- Laue TM, Shah BD, Ridgeway TM, Pelletier SL (1992) Analytical ultracentrifugation in biochemistry and polymer science. In: Harding S, Rowe A, Horton J (eds), pp 90–125. Cambridge, UK
- Leitz AJ, Bayburt TH, Barnakov AN, Springer BA, Sligar SG (2006) Functional reconstitution of Beta2-adrenergic receptors utilizing self-assembling Nanodisc technology. Biotechniques 40: 601–602 [DOI] [PubMed] [Google Scholar]
- Lill R, Dowhan W, Wickner W (1990) The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell 60: 271–280 [DOI] [PubMed] [Google Scholar]
- Maillard AP, Lalani S, Silva F, Belin D, Duong F (2007) Deregulation of the SecYEG translocation channel upon removal of the plug domain. J Biol Chem 282: 1281–1287 [DOI] [PubMed] [Google Scholar]
- Manting EH, van der Does C, Remigy H, Engel A, Driessen AJ (2000) SecYEG assembles into a tetramer to form the active protein translocation channel. EMBO J 19: 852–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménétret JF, Hegde RS, Heinrich SU, Chandramouli P, Ludtke SJ, Rapoport TA, Akey CW (2005) Architecture of the ribosome-channel complex derived from native membranes. J Mol Biol 348: 445–457 [DOI] [PubMed] [Google Scholar]
- Mitra K, Schaffitzel C, Shaikh T, Tama F, Jenni S, Brooks CL, Ban N, Frank J (2005) Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature 438: 318–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Ito K (2001) An essential amino acid residue in the protein translocation channel revealed by targeted random mutagenesis of SecY. Proc Natl Acad Sci USA 98: 5128–5133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Ito K (2006) Different modes of SecY–SecA interactions revealed by site-directed in vivo photo-cross-linking. Proc Natl Acad Sci USA 103: 16159–16164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori H, Tsukazaki T, Masui R, Kuramitsu S, Yokoyama S, Johnson AE, Kimura Y, Akiyama Y, Ito K (2003) Fluorescence resonance energy transfer analysis of protein translocase. SecYE from Thermus thermophilus HB8 forms a constitutive oligomer in membranes. J Biol Chem 278: 14257–14264 [DOI] [PubMed] [Google Scholar]
- Or E, Boyd D, Gon S, Beckwith J, Rapoport T (2005) The bacterial ATPase SecA functions as a monomer in protein translocation. J Biol Chem 280: 9097–9105 [DOI] [PubMed] [Google Scholar]
- Or E, Navon A, Rapoport T (2002) Dissociation of the dimeric SecA ATPase during protein translocation across the bacterial membrane. EMBO J 21: 4470–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch SL, Kendall DA (2007) Oligomeric states of the SecA and SecYEG core components of the bacterial Sec translocon. Biochim Biophys Acta 1768: 5–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, Cramer WA, von Jagow G (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem 217: 220–230 [DOI] [PubMed] [Google Scholar]
- Schiebel E, Driessen AJ, Hartl FU, Wickner W (1991) Delta mu H+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell 64: 927–939 [DOI] [PubMed] [Google Scholar]
- Sharma V, Arockiasamy A, Ronning DR, Savva CG, Holzenburg A, Braunstein M, Jacobs WR Jr, Sacchettini JC (2003) Crystal structure of Mycobacterium tuberculosis SecA, a preprotein translocating ATPase. Proc Natl Acad Sci USA 100: 2243–2248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P, Perugini MA, Gonzales NR, Howlett GJ, Schubert D (2002) Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys J 82: 1096–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam PC, Maillard AP, Chan KY, Duong F (2005) Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J 24: 3380–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian P, Andricioaei I (2006) Size, motion, and function of the SecY translocon revealed by molecular dynamics simulations with virtual probes. Biophys J 90: 2718–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tziatzios C, Schubert D, Lotz M, Gundogan D, Betz H, Schagger H, Haase W, Duong F, Collinson I (2004) The bacterial protein-translocation complex: SecYEG dimers associate with one or two SecA molecules. J Mol Biol 340: 513–524 [DOI] [PubMed] [Google Scholar]
- Ulbrandt ND, London E, Oliver DB (1992) Deep penetration of a portion of Escherichia coli SecA protein into model membranes is promoted by anionic phospholipids and by partial unfolding. J Biol Chem 267: 15184–15192 [PubMed] [Google Scholar]
- van Dalen A, de Kruijff B (2004) The role of lipids in membrane insertion and translocation of bacterial proteins. Biochim Biophys Acta 1694: 97–109 [DOI] [PubMed] [Google Scholar]
- Van den Berg B, Clemons WM Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA (2004) X-ray structure of a protein-conducting channel. Nature 427: 36–44 [DOI] [PubMed] [Google Scholar]
- van der Does C, Swaving J, van Klompenburg W, Driessen AJM (2000) Non-bilayer lipids stimulate the activity of the reconstituted bacterial protein translocase. J Biol Chem 275: 2472–2478 [DOI] [PubMed] [Google Scholar]
- van der Sluis EO, Nouwen N, Koch J, de Keyzer J, van der Does C, Tampe R, Driessen AJ (2006) Identification of two interaction sites in SecY that are important for the functional interaction with SecA. J Mol Biol 361: 839–849 [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Mori H, Vassylyeva MN, Tsukazaki T, Kimura Y, Tahirov TH, Ito K (2006) Crystal structure of the translocation ATPase SecA from Thermus thermophilus reveals a parallel, head-to-head dimer. J Mol Biol 364: 248–258 [DOI] [PubMed] [Google Scholar]
- Veenendaal AK, van der Does C, Driessen AJ (2004) The protein-conducting channel SecYEG. Biochim Biophys Acta 1694: 81–95 [DOI] [PubMed] [Google Scholar]
- Woodbury RL, Hardy SJ, Randall LL (2002) Complex behavior in solution of homodimeric SecA. Protein Sci 11: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer J, Li W, Rapoport TA (2006) A novel dimer interface and conformational changes revealed by an X-ray structure of B. subtilis SecA. J Mol Biol 364: 259–265 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1


