Abstract
DNA damage causes genome instability and cell death, but many of the cellular responses to DNA damage still remain elusive. We here report a human protein, PALF (PNK and APTX-like FHA protein), with an FHA (forkhead-associated) domain and novel zinc-finger-like CYR (cysteine–tyrosine–arginine) motifs that are involved in responses to DNA damage. We found that the CYR motif is widely distributed among DNA repair proteins of higher eukaryotes, and that PALF, as well as a Drosophila protein with tandem CYR motifs, has endo- and exonuclease activities against abasic site and other types of base damage. PALF accumulates rapidly at single-strand breaks in a poly(ADP-ribose) polymerase 1 (PARP1)-dependent manner in human cells. Indeed, PALF interacts directly with PARP1 and is required for its activation and for cellular resistance to methyl-methane sulfonate. PALF also interacts directly with KU86, LIGASEIV and phosphorylated XRCC4 proteins and possesses endo/exonuclease activity at protruding DNA ends. Various treatments that produce double-strand breaks induce formation of PALF foci, which fully coincide with γH2AX foci. Thus, PALF and the CYR motif may play important roles in DNA repair of higher eukaryotes.
Keywords: AP endonuclease, CYR motif, double-strand breaks, PARP1, single-strand breaks
Introduction
DNA damage affects transcription and DNA replication and causes genome instability and cell death. Base damage and DNA strand breaks are the most frequent types of DNA damage in living human cells, and cellular responses to these forms of damage are the subjects of extensive analysis. Base damage is recognized and removed by base damage-specific DNA glycosylases. Apurinic and apyrimidinic (AP) sites created during this process or as a result of base modification are nicked by AP endonuclease for further repair synthesis. In higher eukaryotes, homologs of bacterial AP endonuclease, exonuclease III, are widely distributed. By direct attack of reactive oxygen species on bases or on the DNA sugar–phosphate backbone, DNA single-strand breaks (SSBs) are produced. Closely opposed SSBs on opposite strands or SSBs encountered at replication forks give rise to DNA double-strand breaks (DSBs), which have much more serious consequences for cells than SSBs. Therefore, SSBs must be repaired effectively to minimize the production of DSBs. In mammalian cells, it has been shown in vitro and in vivo that SSBs activate poly(ADP-ribose) polymerases (PARP), which poly(ADP-ribosyl)ate proteins near SSBs as well as PARP itself. This is followed by the accumulation of XRCC1 at poly(ADP-ribose) and SSBs, and recruitment of other proteins necessary for further repair of SSBs (Caldecott, 2003; Lan et al, 2004).
DSBs are repaired either by nonhomologous end joining (NHEJ) or by homologous recombination. NHEJ is a major mode of repairing DSBs in mammals, in which a KU70 and KU86 heterodimer binds to a DSB end and initiates a direct rejoining process (Burma and Chen, 2004; Lieber et al, 2004). The kinase activity of DNA-PK regulates further steps in NHEJ by binding and phosphorylating other proteins, including ARTEMIS, which has endo- and exonuclease activities, and the XRCC4–ligase IV complex. DSBs produced at replication forks or at the G2–M phase of the cell cycle are repaired by homology-mediated recombination between DNA strands. DSBs activate cell-cycle checkpoints and initiate DSB repair processes that include phosphorylation of histone H2AX (γH2AX) and mobilization of proteins to the sites of damage. Although many proteins involved in the cellular responses to SSBs and DSBs have been identified, most of the processes occurring in living cells still remain to be elucidated.
Here, we report a novel human protein, designated as PALF (PNK and APTX-like FHA protein), with an FHA (forkhead-associated) domain and a unique zinc-finger-like domain, which was identified in the databases and characterized by in situ analysis, proteomics and in vitro assays. The protein has AP endonuclease and 3′–5′ exonuclease activities and was shown to respond to SSBs as well as to DSBs. Furthermore, the unique zinc-finger-like CYR (cysteine–tyrosine–arginine) motif may represent a novel functional domain of various proteins involved in DNA repair and DNA metabolism in higher eukaryotes.
Results
Identification of a human protein with a PALF domain revealed a novel zinc-finger-like motif conserved in higher eukaryotes
We initially searched the human databases for proteins harboring domains with sequence similarity to the FHA domains of polynucleotide kinase (PNK) and aprataxin (APTX), both of which bind to XRCC1 and are involved in SSB repair. We found one gene (C2orf13) encoding a protein that matched the criteria and designated this protein as PALF (Figure 1A). PALF is a protein of 57 kDa and, in addition to the amino-terminal FHA domain, there are unique tandem zinc-finger-like CYR sequences at the C terminus of the protein (Figure 1B, top). A search of the databases revealed that proteins harboring the CYR sequence are widely distributed in vertebrates and higher eukaryotes (Figure 1B). The strictly conserved sequence among various proteins harboring the CYR motif seems to contain two cysteine and two histidine residues within the sequence C(5x)C(6x)H(5x)H, as indicated in Figure 1C. In human PALF and its homologs in vertebrates, there is a tandem repeat of this sequence (Figure 1C). In Drosophila melanogaster, a protein encoded by CG6171 has tandem repeats of this sequence but no FHA domain. In Drosophila, there is another protein called Glaikit (CG8825-PA), which has one CYR motif at the amino terminus of a putative tyrosine phosphodiesterase protein. In Caenorhabditis elegans, a single CYR motif is found in the carboxyl terminus of a putative DNA ligase III (gi: 17562294), which has a PARP-type zinc-finger domain at the amino terminus, whereas in Dictyostelium a putative uracil-DNA glycosylase (gi: 66802188) replaces DNA ligase III (Figure 1B). Thus, in contrast to the tandem-repeat type of CYR motifs, a single CYR motif is found in proteins involved in DNA repair in various eukaryotic organisms, suggesting a role for single CYR motifs in DNA repair. Strikingly, all the single CYR motifs are found either at the amino terminus or the carboxyl terminus (Figure 1B). Here, we further analyze human and Drosophila proteins with tandem CYR motifs.
Figure 1.
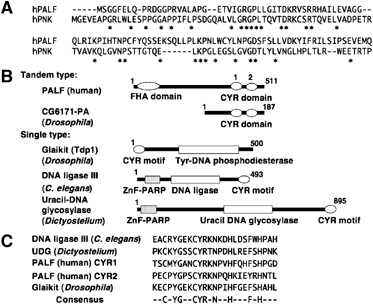
PALF and distribution of CYR motif in eukaryotes. (A) Amino-acid sequence alignment of FHA domains of human PALF and human PNK. (B) Proteins with CYR motif. Domains and number of amino acids in proteins with tandem CYR motifs (tandem type), PALF (human) and CG6171-PA (D. melanogaster) and with a single copy of the motif (single type), Glaikit (Tdp1) protein (D. melanogaster), DNA ligase III (C. elegans) and uracil DNA glycosylase (Dictyostelium discoideum) are shown. (C) Sequence alignment and conserved amino-acid residues in CYR motifs. CYR1 and CYR2 are the first and the second motifs in human PALF as shown in (B).
PALF has endo- and exonuclease activities
Because PALF has FHA domain and unique zinc-finger like motifs, we reasoned that it might possess DNA-binding and/or enzymatic activities against damaged DNA. We, therefore, prepared purified His-tagged recombinant PALF protein from wild-type Escherichia coli host cells, shown in Figure 2A, by silver staining of the protein. In order to exclude bacterial contaminants, we purified His-PALF by passing it through five different chromatography columns (see Materials and methods). Purified recombinant PALF may be a dimer (see below) and migrates at around 90 kDa in SDS gels, possibly for structural reasons. His-PALF was mixed with double-stranded 30-mer DNA molecules containing various types of DNA damage at the 14th nucleotide on one strand, which was labeled with 32P at its 5′ end. To our surprise, PALF efficiently introduced nicks at AP sites and at 5-hydroxyuracil and, to a lesser extent, at hydroxycytosine and thymine glycol (Figure 2B). There is one band below the major band at the nicked AP site, which presumably resulted from the exonuclease activity of His-PALF on the nicked AP site (indicated by the lower arrow in Figure 2B and thereafter). In addition, there are several bands below the major bands (see below). Figure 2C shows the purified His-PALF after gel filtration by silver staining (upper panel) and its nicking activity against an AP site (lower panel). There are two bands as well as a lower band indicated with arrows in the figure. These data suggest that His-PALF may be a homodimer with AP-endonuclease and -exonuclease activities in this preparation. To further confirm the nicking activity of PALF and to exclude any possibility of bacterial contamination, we expressed PALF as a GST fusion protein in an E. coli strain RPC501 (nfo xth double mutant), which lacks AP endonuclease activity, and purified the fusion protein through a glutathione Sepharose column alone. The activity was compared with that of GST alone, and the nicking activity was obtained only in the fusion protein (Figure 2D). GST-PALF from RPC501 was then further purified through three different chromatography columns (see Materials and methods), and its metal-dependent AP endonuclease activity as well as nicking activity at 5-hydroxyuracil was identified (Figure 2E). From these data, we concluded that PALF possesses nicking activities at AP sites and at some types of base damage.
Figure 2.
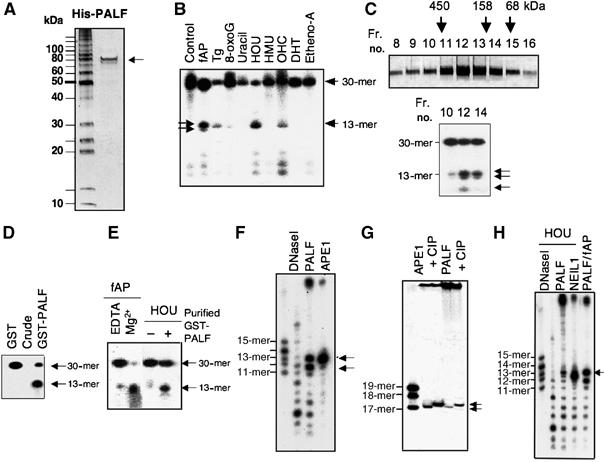
Nicking activity of PALF against various types of DNA damage. (A) Silver staining of His-tagged PALF (His-PALF) purified from BL21 host E. coli cells. (B) Nicking activity of His-PALF against various types of damaged base on one strand in 30-mer double-stranded DNA. fAP (AP site with furan ring), Tg (thymine glycol), 8-oxoG (8-oxoguanine), uracil, HOU (5-hydroxyuracil), HMU (5-hydroxymethyluracil), OHC (5-hydroxycytosine), DHT (5,6-dihydrothymine) and Etheno-A (1, N6-ethenoadenine) are located at the 14th base of the 5′ 32P-labeled 30-mer strand, which was treated with His-tagged PALF. Two arrows in fAP lane show the major nicked sites. (C) Purified His-PALF after gel filtration shown by silver staining (upper panel) and nicking activity of fractions 10, 12 and 14 against AP site (lower panel). (D) Nicking activity of GST-tagged PALF (GST-PALF) and GST alone at the fAP site. Recombinant proteins were prepared from E. coli PRC501 strain and purified simply through a glutathione Sepharose column. (E) Nicking activity of GST-PALF purified through glutathione Sepharose, HiTrap Heparin, HiTrap Q and HiTrap S columns. Nicking activity against fAP with EDTA or Mg2+ (left) or against HOU (right) is shown. (F) Nicking activity of His-PALF against an AP site compared with that of human AP endonuclease (APE1) in sequencing gel. (G) Determination of nicked sites using 3′-end-labeled oligos with an AP site. Oligos were treated with His-APE1 or His-PALF with or without subsequent CIP phosphatase treatment. (H) Nicking activity of His-PALF against 5-hydroxyuracil (HOU) is compared with nicking activity of NEIL1 and fAP digested by His-PALF shown on sequencing gel.
To gain an understanding of how AP sites are nicked by PALF, we analyzed the nicked fragments using sequencing gels. Figure 3F shows that PALF introduces a nick, against 30-mer double-stranded oligonucleotides with an fAP site at the 14th base and that PALF possesses a strong exonuclease activity at the 3′ end, preferentially excising one single nucleotide (indicated by the lower arrow) as compared with human AP endonuclease (APE1). By labeling the 3′ end of a strand harboring fAP and by phosphatase (CIP) treatment after nicking with PALF, we could show retarded migration of the phosphatase-treated fragments (the rightmost lane in Figure 2G). This was also the case for the same substrate nicked by APE1 (third lane from the left in Figure 2G). We thus confirmed that the nicked 5′ end of the 17-mer had a phosphate and that PALF nicked the AP site just as did APE1. Using 5-hydroxyuracil as the damaged base in the 32P-labeled 30-mer, the site that was nicked was determined on a sequencing gel (Figure 2H). PALF nicked 5′ to the 14th (damaged) base creating a 5′-labeled 13-mer DNA, which migrated at the same speed as the 32P-labeled fragment generated by digestion of an AP site with PALF (indicated by an arrow). The substrate nicked with NEIL1 showed a faster migration rate than that nicked by PALF, owing to the δ-elimination activity of NEIL1 (Takao et al, 2002). There was also extensive exonuclease activity. Thus, PALF possesses AP endonuclease activity, nicking activity to other lesions and 3′–5′ exonuclease activity.
Figure 3.
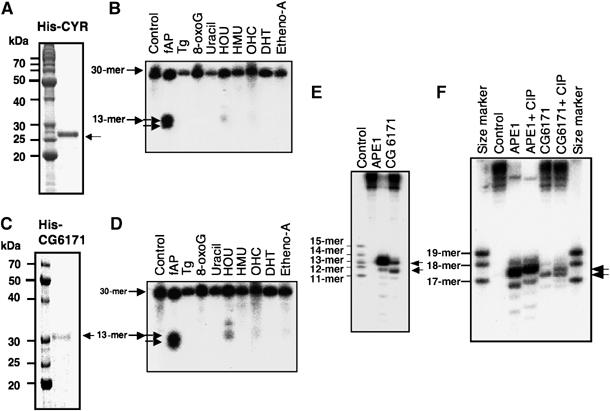
Nicking activity of CYR domain of hPALF and Drosophila CG6171-PA against various types of DNA damage. (A) Purified His-tagged CYR domain (His-CYR from amino-acid residues 361–551, Coomassie blue staining) of PALF. (B) Nicking activity of His-CYR of PALF against various substrates. Abbreviations are the same as in (B). (C) Purified His-tagged Drosophila CG6171-PA (His-CG6171) shown by Coomassie blue staining. (D) Nicking activity of His-CG6171 against various substrates. Abbreviations are the same as in (B). (E) Nicking activity of His-CG6171 against an AP site in sequencing gel. Nicking activity is compared with that of APE1. (F) Determination of nicked site by 3′-end-labeled oligos containing an AP site. Oligos were treated with APE1 or His-CG6171 with or without subsequent CIP phosphatase treatment, which partially removed the phosphate from the substrates.
The activity of the CYR domain of PALF and its homolog, Drosophila CG6171-PA
The nicking activity of PALF may well be derived from the novel CYR domain. Therefore, we examined the activity of the CYR domain. A protein containing the His-tagged CYR domain (from aa 361 to 511) of PALF was purified (Figure 3A). As expected, we found that the recombinant CYR domain exhibited a strong nicking activity at AP sites (Figure 3B). There was again one further band, lower down the gel, but there was almost no nicking activity at other lesions, nor were any lower bands seen. These data indicate that the CYR domain alone has nicking activity at AP sites with exonuclease activity for one base and suggest that the full-length PALF has a wider spectrum of substrates than does the CYR domain.
In order to determine the activities of PALF homologs in other organisms, we isolated the D. melanogaster gene CG6171, which encodes a protein that has tandem CYR sequences but no FHA domain (Figure 1B). We expressed the His-tagged protein in E. coli and purified the recombinant protein (Figure 3C). The protein proved to have the same nicking activity at AP sites as the CYR domain of PALF with the same substrate specificities as well as an exonuclease activity for one base against AP sites (Figure 3D). The nicking characteristics of CG6171 are just the same as those of hPALF with 5′-phosphate end (Figure 3E and F). This indicates that the nicking activity of the tandemly repeated CYR motif is conserved in humans and Drosophila, and that in higher eukaryotes there are at least two types of AP endonuclease, which are homologs of ExoIII (APE1) and PALF.
In situ analysis of the DNA damage response of PALF
Having established the enzymatic activities of PALF, we wanted to know the mobilization of PALF in response to DNA damage produced locally in cell nuclei. HeLa cells were irradiated with a laser micro-beam through a microscope lens. Figure 4A (left panel) depicts accumulation of endogenous PALF determined with anti-PALF antibody and immunohistochemistry at the lines irradiated with 405 nm laser light after 250 scans. These lines overlap exactly with those of XRCC1 (Figure 4A, middle and right panels), indicating that PALF accumulates at DNA damage within human cell. To characterize the kinetics and target of the response of PALF on DNA damage, we expressed GFP-tagged PALF in human or mouse cells. GFP-PALF accumulates very rapidly at sites irradiated either with a single pulse of 365 nm laser light passed through an F20 filter or with five scans of 405 nm laser light in HeLa cells (Figure 4B, middle and right panel). GFP alone does not move at all (not shown). Under these conditions, cells are exposed to 0.5–0.75 μJ/μm2 of laser light and SSBs are the predominant lesion produced by each of the irradiation protocols (Lan et al, 2004, 2005). We predicted, therefore, that PALF would probably accumulate at SSBs as well. To confirm this, we produced SSBs by an alternative method, in which UV light is passed through a porous filter to irradiate XPA-UVDE cells established from a nucleotide excision repair-deficient XPA cell line by expressing the Neurospora crassa UV endonuclease, UVDE (Okano et al, 2003). UVDE introduces a nick immediately 5′ to various types of UV-induced lesions (Yajima et al, 1995; Kanno et al, 1999), and, therefore, SSBs are produced at UV-irradiated sites by this method. In contrast to cells transformed with vector alone (Figure 4C, far left panel), GFP-PALF accumulates in XPA-UVDE cells at irradiated sites (Figure 4C, second panel), where XRCC1 also accumulates (Figure 4C, third panel and merged figure), indicating that PALF accumulates at UV-induced lesions nicked by UVDE. As purified GFP-tagged PALF (Figure 4D, left panel) possesses AP endonuclease activity (Figure 4D, right panel), the GFP-tagged PALF may well be involved in the repair pathway with its enzymatic activity.
Figure 4.
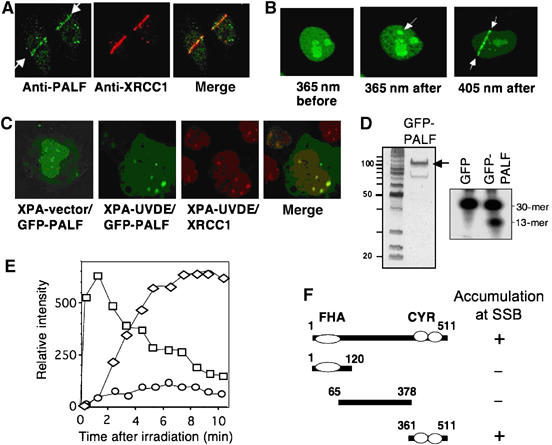
Accumulation of PALF at locally produced DNA damage. (A) Accumulation of endogenous PALF after irradiation with laser. PALF (left indicated with arrows) and XRCC1 (middle) were identified with antibodies and were merged together (right) in HeLa cells after 250 scans of 405-nm laser light. (B) Accumulation of GFP-PALF at sites of laser micro-irradiation. Immediately after one pulse of 365-nm laser light to a HeLa cell through an F20 filter (left before and middle after irradiation) or five scans of 405-nm laser light (right), GFP-tagged PALF has accumulated at the irradiated site as indicated by the arrows. (C) Accumulation of PALF at UVDE-induced SSBs in XPA-UVDE cell. After UV irradiation of XPA-UVDE cell through a porous filter, the spots of accumulated GFP-PALF (second from left) are merged (far right) with those of XRCC1 (third from left) stained with antibody. No SSB was produced in XPA-vector cells (far left). (D) Enzymatic activity of GFP-PALF. Purified GFP-PALF (left panel) shows AP endonuclease activity (right). (E) Accumulation and dissociation kinetics of GFP-PALF after 5 scans of 405-nm laser light in mouse cells. Intensity of fluorescence caused by accumulated GFP-PALF is measured in mouse wild-type cells (squares), wild-type cells treated with PARP inhibitor DIQ (diamonds) and PARP1 KO cells (circles). Data presented are the mean values of six independent experiments. (F) Minimum accumulation domain of PALF at SSB. GFP was tagged to the amino terminus of each construct. + and – indicate accumulation or no accumulation, respectively.
Figure 4E shows the kinetics of GFP-PALF accumulation in mouse cells at sites irradiated with five scans of 405 nm laser light. In wild-type mouse cells, GFP-PALF accumulates immediately after irradiation at SSBs and starts to dissociate 2 min after irradiation (squares in Figure 4E), suggesting that PALF may be active in an early step of the response to SSBs in these cells. While a PARP inhibitor, DIQ, totally suppresses the accumulation of XRCC1 at SSBs (Okano et al, 2003), it only caused a delay in the post-irradiation accumulation of PALF and suppressed its dissociation in wild-type mouse cells (diamonds in Figure 4E). In contrast to the effect of DIQ in wild-type cells, the accumulation of GFP-PALF after laser irradiation is significantly suppressed in PARP1-KO cells (circles in Figure 4E). As GFP-PALF accumulates at SSBs in XRCC1-deficient mouse or Chinese hamster ovary cells as rapidly and to as great an extent as in wild-type cells (not shown), PALF must work upstream of XRCC1. Thus, in situ analysis suggests that PALF is involved in an early step of the SSB response, and functions in a PARP1-dependent manner.
Next, we analyzed which region of PALF accumulates at the laser-irradiated sites by tagging GFP to the amino terminus of three deletion fragments—the FHA, internal and CYR domains—and expressing the tagged fragments in HeLa cells. Figure 4F shows that the fragment necessary for accumulation at SSBs after five scans of 405 nm laser irradiation is the C-terminal region containing the CYR motif. We found furthermore that after irradiation with 500 scans, which produces both DSBs and base damage in addition to SSBs, the FHA and the internal domains also accumulated at irradiated sites, even in PARP1-deficient cells, indicating that these domains are able to respond to DSB and/or base damage produced by higher dose of laser light (data not shown; see below).
Physical and functional interaction of PALF with PARP1
Because of the significant influence of PARP1 on the accumulation of PALF at SSB, shown in Figure 4E, we decided to analyze the possible physical and functional interactions between PALF and PARP1. Having prepared GST- or His-tagged PARP1 and PALF as recombinant proteins, we found that full-length GST-PARP1 is able to pull down His-PALF (Figure 5A, middle panels, left). PARP1 consists of four functional domains (Figure 5A, top panel), among which only the GST-zinc-finger domain was able to pull down His-PALF (Figure 5A, middle panels). In the reverse experiment, GST-full-length PALF pulled down His-PARP1 (Figure 5A, bottom panels, left) and the GST-CYR domain of PALF pulled down His-PARP1 (Figure 5A, bottom panels, right). These data indicate a physical interaction between PARP1 and PALF via the zinc-finger and the CYR domain, respectively, which explains nicely the in situ accumulation of the CYR domain at SSBs (Figure 4F).
Figure 5.
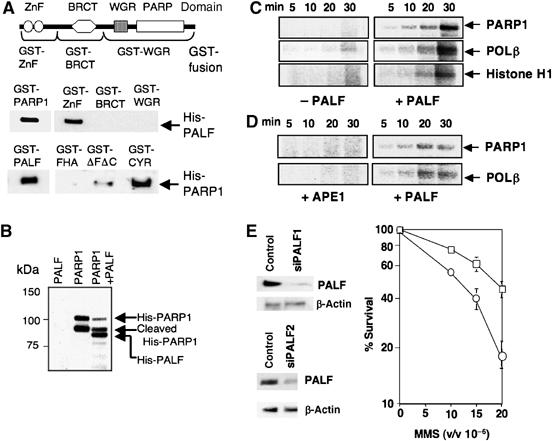
Physical and functional interactions of PALF with PARP1. (A) Direct interaction of PARP1 with PALF. Four domains of PARP1 and GST-tagged regions are shown in the upper figure. GST-tagged full-length PARP1 as well as its zinc-finger domain are able to pull down His-PALF (middle panel). GST-tagged full-length PALF and the CYR domain (see Figure 4F) pull down His-PARP1 (bottom panel). ΔFΔC domain is the region between FHA and CYR domains (Figure 4F). (B) Poly(ADP) ribosylation of PALF and PARP1 by PARP1. Salmon testis DNA (0.2 μg) was sonicated and added to the reaction. Transfer of 32P from NAD to each of the proteins was identified by autoradiography. Recombinant His-PARP1 is cleaved during its preparation from E. coli cells. (C) PALF activates PARP1 in the presence of plasmids harboring SSBs. Plasmids prepared by UV irradiation and UVDE treatment were mixed without (three left-hand panels) or with His-PALF (three right-hand panels) and ribosylation of PARP1, POLβ and Histone H1 by PARP1 was measured by autoradiography of 32P transferred from NAD to the proteins. (D) PALF but not APE1 activates PARP1 at AP sites. Plasmids treated with glyoxal were mixed with APE1 (two left-hand panels) or His-PALF (two right-hand panels) and ribosylation of PARP1 and POLβ was measured. (E) Influence of knock-down of PALF expression in HeLa cells on MMS sensitivity. Colony-forming ability after MMS treatment of HeLa cells transfected with siRNA designed to suppress PALF expression is shown. Mean values of three independent cell survival experiments for siPALF1 and siPALF2 (open circles) and control (open squares) are shown. Expression of PALF was determined by Western blotting using anti-PALF antibody (β-actin as loading control).
Having established the physical interaction between PALF and PARP1, we further questioned whether PALF is modified by PARP1. Purified His-PARP1, which is partly cleaved during its preparation, and His-PALF were mixed with sonicated salmon testis DNA and 32P-labeled NAD. Phosphate transferred from the NAD can be detected in the poly(ADP-ribosyl)ated protein by autoradiography. Label was detected in the full-size His-PARP1 and the cleaved His-PARP1 (Figure 5B, middle lane) and in His-PALF when it was added to the reaction mix (Figure 5B, right lane). Thus, PARP1 interacts with PALF and ribosylates it.
Although it was thought that PARP1 is directly activated by SSBs, it has recently been reported that the activation of PARP1 could be achieved by binding of PARP1 to gapped double-stranded DNA, enabling dimerization of PARP1, which is necessary for its activation (Dantzer et al, 2006). As the creation of base gaps at SSBs by PALF exonuclease activity (Figure 2) may influence PARP1 activity, we wanted to know whether PALF influences the activation of PARP1 at SSBs. As DNA ends activate PARP1, we used here, instead of double-stranded oligos, plasmids harboring SSBs as substrates for the activation of PARP1. We prepared plasmids with SSBs in two different ways. In the first method, plasmids were irradiated with UV and treated with UVDE. The resulting plasmids containing SSBs were mixed with PARP1, POLβ, histone and 32P-labeled NAD. Labeled phosphates, transferred from NAD to these proteins by ribosylation, can be detected by autoradiography. While none of the proteins was ribosylated in the absence of PALF (Figure 5C, left panels), significant phosphate transfer was observed if we added PALF to the reaction mixture (Figure 5C, right panels). SSBs can also be produced by AP endonuclease. We have developed a method for producing plasmids containing abasic sites by treating plasmids with glyoxal so that depurinated bases are produced. Plasmids are then either nicked with APE1 or with PALF, thus examining the difference between SSBs produced by either of the AP endonucleases, APE1 and PALF. As shown in Figure 5D, only plasmids treated with PALF were able to activate PARP1, and PARP1 and POLβ were poly(ADP-ribosyl)ated. Thus, we concluded that PALF, but not APE1, is able to activate PAPR1 at AP sites in plasmids.
To determine the biological significance of PALF in human cells, we knocked down PALF expression using siRNA in HeLa cells. Among 10 siRNA sequences tested, two sequences (siPALF1 and siPALF2) reduced the expression of PALF to 20% of the wild-type level (Figure 5E). In both cases, the HeLa cells became mildly but consistently sensitive to methyl methane sulfonate (MMS) (Figure 5E). These data indicate that PALF is involved in the resistance of human cells to SSBs.
The FHA domain and involvement of PALF in responses to DSBs
As it was the presence of an FHA domain that enabled us to identify PALF, we wanted to know the function of FHA domain. As the FHA domain is the domain interacting with phosphorylated proteins, its presence in PALF prompted us to identify proteins interacting with PALF. Using purified His-tagged PALF (Figure 2A) and affinity chromatography, we identified, in nuclear extracts prepared from HeLa cells, proteins capable of binding to His-PALF. Figure 6A shows the putative binding proteins so far determined by mass spectrometry. To our surprise, except ribosomal protein S3a, all the other proteins are involved in damage response processes. In addition to proteins involved in SSB responses like PARP1, XRCC1 and LIGASEIIIα, most of the other proteins identified by this assay are those involved in NHEJ of DSB. A GST-pull-down assay using recombinant proteins showed that GST-tagged KU86 and LIGASEIV interact directly with PALF, but GST-tagged KU70 and XLF (Cernunnos) failed to pull down His-PALF in this assay (Figure 6B). As GST-XRCC4 is shown to have a weak interaction with His-PALF (Figure 6B) and XRCC4 is known to be phosphorylated and to interact with the FHA domain of APTX (Clements et al, 2004), we phosphorylated GST-XRCC4 either by CK-II or by DNA-PKcs and mixed it with His-PALF. As shown in Figure 6C (upper panels), GST-XRCC4 phosphorylated by either of the kinases pulled down His-PALF. Furthermore, the GST-XRCC4 phosphorylated by CK-II pulled down the FHA domain of PALF (Figure 6C, lower panels), indicating that phosphorylated XRCC4 interacts with the FHA domain of PALF. The interaction of PALF with KU86 was also analyzed by a pull-down assay (Figure 6D). To our surprise, neither the FHA nor the CYR domain of PALF interacts with KU86, but the region between the two domains (indicated as ΔFΔC) does interact. These data strongly suggest that PALF responds to DSBs by interacting with proteins involved in NHEJ.
Figure 6.
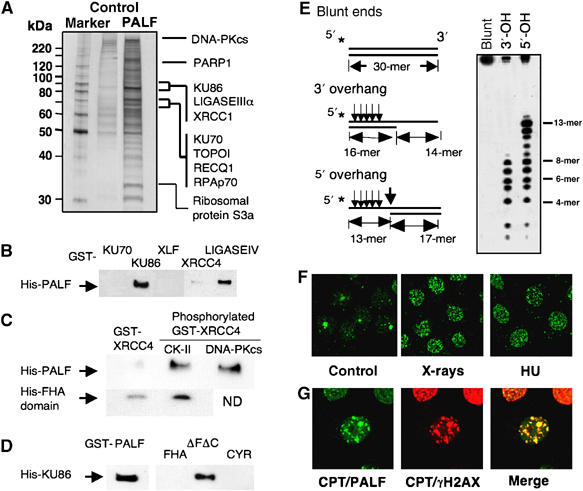
Involvement of PALF in DSB responses. (A) Putative PALF-binding proteins identified by affinity chromatography and mass spectrometry. His-PALF was attached to a nickel agarose column for binding of proteins prepared from HeLa nuclear extracts. Control lane shows the proteins obtained by column attached with a control protein (GST). All the proteins identified so far by this method are shown. (B) GST-tagged KU86 and LIGASEIV, but not KU70, XLF and XRCC4, are able to pull down His-PALF. (C) GST-XRCC4 phosphorylated either by CK-II or by DNA-PKcs pulls down His-PALF (upper panels); the FHA domain of PALF is pulled down more effectively by phophorylated GST-XRCC4 (lower panels). (D) GST-PALF and GST-tagged domain, ΔΦΔX, between FHA and CYR domains pull down His-tagged KU86. (E) Nicking activities of PALF against various double-strand ends. Three substrates used for analysis of the nicking activity of PALF at DSBs are shown (left). Stars indicate the sites for 32P-labeling. Nicked DNA fragments were identified in sequencing gel after treatment of the three substrates (right). Determined nicked sites are indicated by arrows on the left. (F) Formation of PALF foci after treatment of HeLa cells with various agents. Cells were treated with X-rays (6 Gy) and hydroxyurea (HU, 10 μM); cells were treated with anti-PALF antibody 30 min after X-ray treatment or 2 h after HU treatment. (G) Colocalization of PALF and γH2AX foci induced by CPT treatment of HeLa cells. Two hours after 2 μM CPT treatment, cells were double-stained with antibodies against PALF and γH2AX.
Because of the endo- and exonuclease activities of PALF and its interaction with proteins involved in NHEJ of DSBs, we reasoned that PALF might have endo- and exonuclease activities at DSB ends as does ARTEMIS (Ma et al, 2005). Figure 6E (left lane) shows two test substrates with protruding 3′ and 5′ ends in addition to a substrate with blunt ends. Substrates were labeled with 32P at the 5′ ends and treated with His-PALF. The right panel in Figure 6E depicts the results from the substrates nicked by His-PALF. The substrate with blunt ends was not nicked (left lane). However, the 3′-overhang substrate was strongly nicked at the 8th, 7th, 6th, 5th and 4th bases from the 5′ ends, whereas there was no nicking at all at the 3′ end (middle lane; see arrows in Figure 6E, left panel). The protruding 5′ end was strongly nicked at the 13th and 12th bases in addition to the nicking from the 8th to 4th bases, which results in a blunt end. It is interesting to note that the nicking activity of PALF against fAP (Figure 2F) is similar to that of PALF against the 5′ overhang. Thus, PALF is able to introduce nick at double-stranded DNA ends with an overhang.
Many proteins involved in DSB repair relocate within the nucleus to form foci in response to DSB-producing treatments. Although the mechanisms and the meanings of the foci formation are not well understood, accumulation at foci by a protein suggests that the protein is involved in DSB responses and repair. Using antibody raised against PALF, we analyzed the formation of foci by endogenous PALF in HeLa cells after various treatments that produce DSB. Whereas in control cells without treatment there are only a few PALF foci in the nucleus, many nuclear foci are observed after treatment with hydroxyurea (HU; 2 h after treatment) and X-rays (30 min after irradiation), which are known to produce DSBs in different ways (Figure 6F). We compared the distribution of PALF foci with those of γH2AX after camptothecin (CPT) treatment. CPT treatment also produces PALF foci, and these completely coincide with γH2AX foci (Figure 6G), further suggesting that PALF is involved in the response to DSBs in cells. As most proteins involved in NHEJ do not show formation of foci, these data suggest that the response of PALF to DSBs is not limited to NHEJ, but that it may also be involved in other cellular responses to DSBs including homologous recombination.
Discussion
We have identified a novel human protein with AP endonuclease and exonuclease activities. The protein, designated as PALF, has two domains—a domain with sequence similarity to the FHA domains of PNK and APTX, and another domain containing CYR motifs. A database search revealed that the CYR sequence motif is widely conserved in eukaryotes from Dictyostelium, C. elegans and Drosophila to vertebrates including mammals. A CYR motif contains two cysteine and two histidine residues, which are perfectly conserved in the proteins derived from eukaryotes (Figure 1C). However, we have not yet found a protein with this motif in yeast, plants or bacteria. If a major role of this protein is to activate PARP, as shown in this paper, it is reasonable to suppose that yeast and bacteria do not need this protein because of the absence of PARP activity in these organisms. In Figure 1B, two types of CYR domains are presented. Humans, mammals and other vertebrates have a tandem repeat of the CYR motif together with an FHA domain, whereas Drosophila CG6171 and proteins from other insects (e.g., proteins in the mosquito databases, GI:55240892 from Anopheles gambiae and GI: 108872456 from Aedes aegypti) possess only tandem CYR motifs without an FHA domain. Drosophila Glaikit, C. elegans Ligase III and Dictyostelium UNG have only one CYR motif without an FHA domain. As, in contrast to PALF and CG6171, the proteins so far identified with one CYR motif have other domains for putative DNA repair enzymes, and the sequences within the CYR motif are quite well conserved, the single CYR motif may also be involved in the enzymatic activities of the proteins. The absence of a CYR motif in yeast, plants or bacterial proteins suggests a relatively late creation of this motif in eukaryotic evolution, and proteins with tandem CYR motifs may have been produced much later for DNA repair in higher eukaryotes.
PALF possesses a significant AP endonuclease activity (Figure 2). Two types of AP endonucleases are known in living organisms, the endoIV and exoIII types, both of which are present in E. coli. We, as well as another group, have found that UVDE, a UV damage endonuclease found in some fungi and bacteria, also possesses AP endonuclease activity (Avery et al, 1999; Kanno et al, 1999). Proteins with tandem CYR motifs are, therefore, a fourth type of AP endonuclease. In addition to its AP endonuclease activity, PALF is able to introduce nicks at hydroxyuracil and other types of pyrimidine base damage (Figure 2B and H), an activity that is not significant in the CYR domain alone or in Drosophila CG6171 (Figure 3B and D). These data indicate that tandem CYR motifs alone recognize and introduce nicks at AP sites and that domains other than the CYR domain contribute to the recognition of damage other than AP sites. Whereas the yeast Saccharomyces cerevisiae AP endonuclease, Apn, has a 3′–5′ exonuclease activity and may provide an alternative repair pathway for AP sites (Ishchenko et al, 2005), the 3′–5′ exonuclease activity of human APE1 is a very weak activity as indicated in Figure 3F. Furthermore, APE1 inhibits PARP1 from binding to the nicked site (Peddi et al, 2006). In contrast to APE1, PALF possesses extensive 3′–5′ exonuclease activity and interacts with and activates PARP1 for SSB repair. This exonuclease activity, in addition to the AP endonuclease activity, is able to activate PARP1 at abasic sites, which was not achieved by APE1 as shown in Figure 5D. This interaction between PALF and PARP1 may represent not only the repair of SSBs but also an alternative way of repairing AP sites or base damage in human cells, although the extent of this repair remains to be determined. At SSBs created at AP sites or base damage, PARP1 is able to be activated by PALF and recruits XRCC1 for further repair processes. This may be an example of the nucleotide incision repair process recently proposed for higher eukaryotes (Ishchenko et al, 2006).
In situ analysis using micro-irradiation revealed that PALF accumulates at SSBs via the CYR domain in an early step of the SSB response. This accumulation of PALF at SSBs is dependent on PARP1 (Figure 4E). This is in contrast to AP site or base damage, which are recognized and processed by PALF itself. In situ analysis showed that poly(ADP-ribosyl)ation accelerates both the accumulation and the dissociation of PALF at SSBs (Figure 4E). Thus, the presence of PARP1 and poly(ADP-ribose) contributes to the accumulation of PALF at SSB. These data indicate that, besides AP site and base damage, SSB is a substrate for PALF. While accumulation of PALF at SSBs is dependent on PARP1, PALF is necessary for the activation of PARP1 at SSBs produced by UVDE (Figure 5C). Then, poly(ADP-ribosyl)ated proteins around the SSBs recruit XRCC1, the matchmaker in SSB repair (Lan et al, 2004). From the data obtained during purification of His-PARP1 and His-PALF, we believe that both PARP1 and PALF function as homodimers. It has been reported that WRN, the protein involved in Werner's syndrome, binds to PARP1 and influences activation of PARP1 (von Kobbe et al, 2003; Li et al, 2004). Therefore, WRN and PALF may share some roles in SSB responses, although we have not detected accumulation of WRN at SSBs (Lan et al, 2005). The enhanced sensitivity of HeLa cells to MMS by suppression of PALF expression (Figure 5E) indicates that PALF is indeed important in protecting human cells against SSBs.
In addition to base damage and SSBs responses, PALF may be involved in the cellular response to DSBs. Besides the PARP1-dependent response of PALF to SSBs via the CYR domain, PALF responds to a high dose of laser radiation in PARP1-deficient cells via its FHA domain and the internal domain (data not shown). This may be explained by the interactions of the domains with XRCC4, LIGASEIV and KU86 as shown in Figure 6B–D. Binding of PALF to XRCC4 is enhanced by the phosphorylation of XRCC4 and the FHA domain binds to phosphorylated XRCC4 (Figure 6C). Although we do not know the exact roles of PALF in the NHEJ complex, the endonuclease activity coupled with the 3′–5′ exonuclease activity of PALF at protruding ends in double-stranded DNA (Figure 6E) suggests a role in the processing of DNA ends that is similar to that of ARTEMIS, which interacts with DNA-PKcs and possesses endonuclease and exonuclease activities (Pannicke et al, 2004). Because of the intensive endonuclease activity at the 5′ overhang, one product resulting from the activity of PALF will be blunt ends. It is tempting, therefore, to suppose that PALF may be needed to cleave the ends at DSBs in NHEJ as well as for homologous recombination. However, the role of PALF and its relationship to the role of ARTEMIS remains to be determined.
The involvement of PALF in the DSB response is further suggested by its localization at foci after various treatments that produce DSBs (Figure 6F). We also found PALF foci after treatment of human cells with bleomycin, mitomycin C and VP-16 (not shown). The perfect overlap of camptothecin-induced PALF foci with γH2AX foci (Figure 6G) suggests that PALF is involved in the response to DSBs. Formation of PALF foci by HU treatment suggests its involvement in DSBs at blocked replication forks. Knock-down of PALF expression did not have any influence on cell survival after X-irradiation in our experimental conditions (not shown). Thus, PALF may be involved in various DSB responses, but its real function in DSB responses remains to be further elucidated.
Here, we have described a novel protein with DNA processing activity at DNA damage. As homologs of PALF and especially the CYR motifs are widely distributed in higher eukaryotes, this motif may play important roles in the different stages of the responses to DNA damage of higher eukaryotes.
Materials and methods
Identification and cloning of C2orf13 and other genes
Proteins were searched for in the NCBI human protein databases using PSI-BLAST and 3D-PSSM. cDNAs were prepared from HeLa cells or from D. melanogaster by reverse transcription of mRNA and PCR and were identified by sequencing. XhoI and NotI sites were attached at the start and stop codons, respectively, of the full-length or deletion fragments by PCR and introduced, after sequencing, into various expression plasmids for GFP (pEGFP-C1 or -N1 vectors, Clontech), GST or His-tag.
Laser micro-irradiation
Laser micro-irradiation was carried out as reported in our previous papers (Lan et al, 2004, 2005). We used both a 405-nm scanning laser (Olympus, Tokyo) and a 365-nm pulse laser (photonic Instruments, St Charles, IL) micro-irradiation systems combined with a confocal microscope (Olympus). Cells were incubated with Opti-medium (Gibco) in glass-bottomed dishes, which were placed on a 37°C warming plate in chambers to prevent evaporation. Each experiment was performed at least three times and representative data are presented here.
Cell lines, culture conditions and transfection
The following cell lines were used: HeLa, EM9 (Xrcc1-deficient CHO cell line), AA8 (wild-type CHO cell line), Parp1 (a cell line from Parp1 KO mouse embryonic fibroblasts, a generous gift of Mitsuko Masutani), WTB (mouse embryonic fibroblasts from wild-type mouse), XPA-UVDE (SV40-transformed human cells from the fibroblasts of a xeroderma pigmentosum A patient stably expressing UVDE (Okano et al, 2000) and XPA-Vector (XPA cells transfected with vector plasmid)). Cells were grown in Eagle's minimal essential medium (Nissui) supplemented with 10% fetal bovine serum. These plasmids were introduced into cells with Fugene 6 (Roche).
Local UV irradiation
Local UV irradiation was performed essentially as described previously (Okano et al, 2003). Cell monolayers in 35-mm glass-bottomed culture dishes were covered with a polycarbonate isopore membrane filter with pores of 3 μm in diameter (Millipore) and UV irradiated with a germicidal lamp (GL-10; Toshiba; predominantly 254-nm UV) at a dose rate of 1.82 J/m2/s.
Purification of tagged PALF, CYR and CG6171 and other enzymes
The E. coli strain BL21(DE3) was used for expression of His-tagged proteins. Purification of His-tagged proteins was performed according to the manufacturer's protocols. Partially purified His-tagged recombinant proteins were further purified by HiTrap Heparin, HiTrap Q, HiTrap S and Superose12 (GE Healthcare). The E. coli strain RPC501 (from Dr RP Cunningham), lacking both Exo III and Endo IV AP endonucleases, was used to purify GST-tagged PALF from the AP endonuclease-free host cells. GST-tagged recombinant proteins, partially purified through a glutathione Sepharose column, were further purified by HiTrap Heparin, HiTrap Q and HiTrap S. GFP-tagged PALF was purified by a similar method. Purified recombinant proteins were checked by gel electrophoresis and stained with Coomassie brilliant blue or silver staining. Casein kinase II (CK-II) was purified from mouse liver by modification of previously reported procedures (Carmichael et al, 1982). DNA-PKcs was a generous gift of Dr David J Chen (Burma and Chen, 2004).
Incision assay using synthetic oligonucleotides
Oligonucleotides containing a damaged site were 5′- or 3′-labeled with [γ-32P]ATP by T4 PNK or terminal deoxytransferase (TdT) (NEB) and immediately annealed to the complementary oligomer. A 10 fmol portion of 5′- or 3′-labeled double-strand oligonucleotides was incubated with 1 pmol of purified His-tagged PALF or CG6171 in reaction buffer (50 mM HEPES (pH 7.5), 2 mM DTT, 1 mM MgCl2 (or 10 mM) and 0.1 M NaCl) for 30 min at 37°C. In the case of GST-PALF and GST, 2 mM of each protein, purified through a glutathione Sepharose column, was used in the reaction. The reaction was stopped by adding sodium acetate (pH 3.4) and oligonucleotides were precipitated with ethanol. After centrifugation, precipitates were dissolved in 90% formamide loading buffer, heated at 70°C for 3 min and analyzed on 15% denaturing (7 M urea) polyacrylamide gels. Migration patterns of DNA fragments were analyzed using a BAS 2000 Image Analyzer (Fuji Film). As size markers, 5′-end labeled 11- to 19-mer oligonucleotides with the same sequences as the substrates were used. hAPE1 and hNEIL1 were prepared as previously reported (Kanno et al, 1999; Takao et al, 2002).
Oligonucleotide substrates
The following were used: 30-mer containing a damaged site: 5′-CTCGTCAGCATCT-X-CATCATACAGTCAGTG (X=fAP, U, Tg, 8-oxoG, HOU, HMU, OHC, DHT, etheno-A). For 3′ and 5′ overhangs, 30-mer: 5′-CACTGACTGTATGATGAAGATGCTGACGAG, 16-mer: 5′-CATCATACAGTCAGTG, 14-mer: CACTGACTGTATGA, 17-mer: ATGAAGATGCTGACGAG.
PALF affinity column and nanoLC/MS/MS
The PALF affinity column was prepared by coupling purified His-tagged PALF to Affi-gel 10 (Bio-Rad) (200 mg/ml). HeLa nuclear extracts were prepared as follows: HeLa cell pellets (1 g) were homogenized in SHE buffer (10 mM Hepes (pH 7.4), 0.21 M mannitol, 0.07 M sucrose, 0.1 M EDTA, 0.1 M EGTA, 0.15 mM spermine, 0.75 mM spermidine) and centrifuged for 10 min at 3000 r.p.m. The pellets were rinsed with SHE buffer and centrifuged once more. The pellets were then extracted in 1 ml of extraction buffer by sonication and the extracts were clarified by centrifugation at 12 000 r.p.m. for 30 min at 4°C. A 300 ml volume of His-tagged PALF-beads was incubated in the 1 ml HeLa nuclear extracts for 2 h at 4°C. After centrifugation for 3 min at 3000 r.p.m., the beads were washed three times with buffer A (50 mM Tris–HCl (pH 7.5), 1 mM DTT) containing 0.15 M NaCl and eluted with 1 ml buffer A containing 1 M NaCl. The eluates were concentrated to 50 μl by centrifugal filter devices Amicon Ultra (Millipore) and 2 × SDS sample buffer was added. The samples were resolved on a 12.5% SDS–PAGE gel and stained using a Wako Mass silver stain kit. Gel slippage was reduced by 100 mM of DTT and alkylated by 100 mM idoacetamide. After washing, the gels were incubated with trypsin overnight at 30°C. Recovered peptides were desalted by Ziptip c18 (Millipore). Samples were analyzed by nanoLC/MS/MS systems (DiNa HPLC system KYA TECH Corporation/QSTAR XL Applied Biosystems). Mass data acquisitions were piloted by Mascot software.
GST pull-down assay
GST-recombinant proteins for the GST-pull-down assay were expressed in BL21(DE3). The bacterial cell pellets were lysed by sonication in extraction buffer (50 mM phosphate buffer (pH 8.0), 0.3 M NaCl, 0.1% NP-40). The lysate was clarified by centrifugation at 4°C. One milliliter of the resulting supernatant was incubated with 100 μl of glutathione Sepharose (GS) beads (50% v/v) for 1 h at 4°C. The beads were washed three times with 1 ml of extraction buffer. For binding experiments, GS beads bound with GST-fusion protein (10 μg) were incubated for 1 h at 4°C with 100 μg of purified His-tagged recombinant protein in buffer A containing 0.15 M NaCl. The beads were subsequently washed three times with buffer A and protein eluted by boiling in 50 μl of Laemmli buffer. Proteins were electrophoresed on 10–20% gradient polyacrylamide SDS gels and transferred to PVDF membranes and detected by Western blotting.
PARP1 activation assay
Nicked plasmids were prepared by two different methods. In one method, plasmids were UV-irradiated (100 J/m2) and exposed to UVDE, which was derived from Neurospora crassa and prepared as previously reported (Kanno et al, 1999). PARP1 activity was assayed in a final volume of 50 μl containing 10 pmol of His-tagged PARP1, 100 mM NAD+ spiked with 32P-NAD+ (Amersham Biosciences), 50 mM HEPES (pH 7.5), 1 mM DTT, 1 mM MgCl2, 0.5 μg His-tagged pol β, 0.5 μg histone H1, UV (100 J/m2) irradiated pBluescript 10 μg and 10 pmol UVDE in the presence or absence of 10 pmol His-tagged PALF. At the indicated time, a 10 μl aliquot of reaction mixture was added to 2 times the volume of SDS buffer. The samples were resolved on a 12.5% SDS–PAGE gel and the radioactivity in the gel was analyzed on BAS 2000 (Fuji). In an alternative method, we used chemical treatment of plasmids. A 300 μg portion of pBluescript plasmid was incubated with 1% glyoxal on ice for 1 h and then was purified by agarose gel electrophoresis. Under these conditions, glyoxal adducts were not generated, but AP sites were produced by a deamination-like reaction. A 10 pmol portion of APE1 or 10 pmol of His-PALF was used to treat the plasmids.
siRNA
Two siRNA oligos (sense RNA sequences, siPALF1: GGA AGA GUC UAC CAU UUCA; siPALF2: GGC AAC UGA UUC AGU UCU ACTT) were able to suppress the expression of PALF effectively. Two days after HeLa cells were transfected with siRNA (100 nM of siPALF1 or 200 nM of siPALF2 or control nonspecific siRNA), cells were treated with MMS. Ten days after treatment, cells were stained with crystal violet and colonies were counted.
Chemicals
1,5-Isoquinolinediol (DIQ), hydroxyurea and MMS were purchased from Sigma. 40% glyoxal solution, iodoacetamide and camptothecin were from Wako Pure Chemical Industries. Anti-His antibody was purchased from GE Healthcare and anti-XRCC1 from Neomakers. Antibodies were used after 1000 times dilution.
Preparation of antibodies
Rabbit polyclonal antibodies were raised against purified recombinant His-tagged PALF and affinity purified on a PALF affinity column.
In situ detergent extraction and immunofluorescence
Cells grown for 48 h in glass-bottomed dishes to about 50% confluence were treated or mock-treated with DNA-damaging agents or X-rays. After 2 or 0.5 h, cell extraction was carried out in situ by incubation in detergent solution (0.5% Triton X-100, 5 mM EDTA, 1% BSA in PBS) for 15 min on ice. Cells were rinsed twice with PBS and fixed in 4% formaldehyde for 10 min at room temperature (RT). Staining of cells was performed by standard methods.
Acknowledgments
We thank Dr SJ McCready for editing the text. We thank Drs M Masutani, S Iwai and DJ Chen for providing us with PARP1 cells, furan-AP oligo and DNA-PKcs, respectively. Technical assistance by Ms Mikiko Hoshi is acknowledged. This work was supported in part by Grant-in-Aid for Scientific Research (nos. 17710042, 12143201 and 13480162) and by a genome network project grant from the Ministry of Education, Science, Sports and Culture of Japan.
References
- Avery AM, Kaur B, Taylor JS, Mello JA, Essigmann JM, Doetsch PW (1999) Substrate specificity of ultraviolet DNA endonuclease (UVDE/Uve1p) from Schizosaccharomyces pombe. Nucleic Acids Res 27: 2256–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burma S, Chen DJ (2004) Role of DNA-PK in the cellular response to DNA double-strand breaks. DNA Repair (Amst) 3: 909–918 [DOI] [PubMed] [Google Scholar]
- Caldecott KW (2003) XRCC1 and DNA strand break repair. DNA Repair (Amst) 2: 955–969 [DOI] [PubMed] [Google Scholar]
- Carmichael DF, Geahlen RL, Allen SM, Krebs EG (1982) Type II regulatory subunit of cAMP-dependent protein kinase. Phosphorylation by casein kinase II at a site that is also phosphorylated in vivo. J Biol Chem 257: 10440–10445 [PubMed] [Google Scholar]
- Clements PM, Breslin C, Deeks ED, Byrd PJ, Ju L, Bieganowski P, Brenner C, Moreira MC, Taylor AM, Caldecott KW (2004) The ataxia-oculomotor apraxia 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair (Amst) 3: 1493–1502 [DOI] [PubMed] [Google Scholar]
- Dantzer F, Ame JC, Schreiber V, Nakamura J, Menissier-de Murcia J, de Murcia G (2006) Poly(ADP-ribose) polymerase-1 activation during DNA damage and repair. Methods Enzymol 409: 493–510 [DOI] [PubMed] [Google Scholar]
- Ishchenko AA, Deprez E, Maksimenko A, Brochon JC, Tauc P, Saparbaev MK (2006) Uncoupling of the base excision and nucleotide incision repair pathways reveals their respective biological roles. Proc Natl Acad Sci USA 103: 2564–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishchenko AA, Yang X, Ramotar D, Saparbaev M (2005) The 3′ → 5′ exonuclease of Apn1 provides an alternative pathway to repair 7,8-dihydro-8-oxodeoxyguanosine in Saccharomyces cerevisiae. Mol Cell Biol 25: 6380–6390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno S, Iwai S, Takao M, Yasui A (1999) Repair of apurinic/apyrimidinic sites by UV damage endonuclease; a repair protein for UV and oxidative damage. Nucleic Acids Res 27: 3096–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L, Nakajima S, Komatsu K, Nussenzweig A, Shimamoto A, Oshima J, Yasui A (2005) Accumulation of Werner protein at DNA double-strand breaks in human cells. J Cell Sci 118: 4153–4162 [DOI] [PubMed] [Google Scholar]
- Lan L, Nakajima S, Oohata Y, Takao M, Okano S, Masutani M, Wilson SH, Yasui A (2004) In situ analysis of repair processes for oxidative DNA damage in mammalian cells. Proc Natl Acad Sci USA 101: 13738–13743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Navarro S, Kasahara N, Comai L (2004) Identification and biochemical characterization of a Werner's syndrome protein complex with Ku70/80 and poly(ADP-ribose) polymerase-1. J Biol Chem 279: 13659–13667 [DOI] [PubMed] [Google Scholar]
- Lieber MR, Ma Y, Pannicke U, Schwarz K (2004) The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst) 3: 817–826 [DOI] [PubMed] [Google Scholar]
- Ma Y, Schwarz K, Lieber MR (2005) The Artemis:DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair (Amst) 4: 845–851 [DOI] [PubMed] [Google Scholar]
- Okano S, Kanno S, Nakajima S, Yasui A (2000) Cellular responses and repair of single-strand breaks introduced by UV damage endonuclease in mammalian cells. J Biol Chem 275: 32635–32641 [DOI] [PubMed] [Google Scholar]
- Okano S, Lan L, Caldecott KW, Mori T, Yasui A (2003) Spatial and temporal cellular responses to single-strand breaks in human cells. Mol Cell Biol 23: 3974–3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannicke U, Ma Y, Hopfner KP, Niewolik D, Lieber MR, Schwarz K (2004) Functional and biochemical dissection of the structure-specific nuclease ARTEMIS. EMBO J 23: 1987–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peddi SR, Chattopadhyay R, Naidu CV, Izumi T (2006) The human apurinic/apyrimidinic endonuclease-1 suppresses activation of poly(ADP-ribose) polymerase-1 induced by DNA single strand breaks. Toxicology 224: 44–55 [DOI] [PubMed] [Google Scholar]
- Takao M, Kanno S, Kobayashi K, Zhang QM, Yonei S, van der Horst GT, Yasui A (2002) A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J Biol Chem 277: 42205–42213 [DOI] [PubMed] [Google Scholar]
- von Kobbe C, Harrigan JA, May A, Opresko PL, Dawut L, Cheng WH, Bohr VA (2003) Central role for the Werner syndrome protein/poly(ADP-ribose) polymerase 1 complex in the poly(ADP-ribosyl)ation pathway after DNA damage. Mol Cell Biol 23: 8601–8613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima H, Takao M, Yasuhira S, Zhao JH, Ishii C, Inoue H, Yasui A (1995) A eukaryotic gene encoding an endonuclease that specifically repairs DNA damaged by ultraviolet light. EMBO J 14: 2393–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]


