Abstract
The residues in tRNA that account for its tertiary fold and for its specific aminoacylation are well understood. In contrast, relatively little is known about the residues in tRNA that dictate its ability to transit the different sites of the ribosome. Yet protein synthesis cannot occur unless tRNA properly engages with the ribosome. This study analyzes tRNA gene sequences from 145 fully sequenced bacterial genomes. Grouping the sequences according to the anticodon triplet reveals that many residues in tRNA, including some that are distal to the anticodon loop, are conserved in an anticodon-dependent manner. These residues evade detection when tRNA genes are grouped according to amino acid family. The conserved residues include those at positions 32, 38, and 37 of the anticodon loop, which are already known to influence tRNA translational performance. Therefore, it seems likely that the newly detected anticodon-associated residues also influence tRNA performance on the ribosome. Remarkably, tRNA genes that belong to the same amino acid family and therefore share identical residues at the second and third anticodon positions have diverged, during bacterial evolution, into highly conserved groups that are defined by the residue at the first (wobble) anticodon position. Current ideas about the properties of tRNA and the translation mechanism do not fully account for this phenomenon. The results of the present study provide a foundation for studying the adaptation of individual tRNAs to the translation machinery and for future studies of the translation mechanism.
Keywords: tRNA, translation, ribosome, evolution, bacteria, anticodon, wobble position
INTRODUCTION
Transfer RNA (tRNA) is the workhorse of translation. It is responsible for accepting amino acids and for donating them to the polypeptide chain that is synthesized at the ribosome. The sequence and structural features of tRNA that enable it to participate in the many biochemical steps of the translation pathway are of longstanding interest (Saks et al. 1994; RajBhandary and Soll 1995; Hoagland 1996; Giege et al. 1998; Zamecnik 2005). Yet the determinants of these diverse biochemical functions have not been studied with equal intensity. Previous studies have provided information on the residues that dictate the tertiary fold of tRNA and that thereby account for the L-shaped structure that is characteristic of all tRNAs (Rich and RajBhandary 1976; Dirheimer et al. 1995; Steinberg et al. 1997). They have also elucidated how the sequence and structure of tRNA influence recognition by the cognate aminoacyl-tRNA synthetase (McClain 1993; Martinis and Schimmel 1995; Pallanck et al. 1995; Giege et al. 1998) and how these tRNA attributes, in combination with the nature of the esterified amino acid, influence binding to elongation factor Tu (EF-Tu) (LaRiviere et al. 2001). In contrast, relatively little attention has focused on the problem of whether tRNA has features, other than the anticodon triplet, that influence its decoding function. Three residues of the anticodon loop, a base pair in the anticodon stem, and a base pair in the D-stem have received the most attention, and they have been studied more frequently within the context of nonsense suppressor as opposed to wild-type tRNAs (Yarus and Smith 1995). Therefore, a more comprehensive analysis is needed to assess whether collaboration between the anticodon triplet and residues in the tRNA body is universally important to the translational performance of tRNA.
Bioinformatics has proven to be a useful tool for identifying residues that are important to molecular function. An implicit and powerful assumption of bioinformatics is that when genes are properly grouped the conserved sites reflect the imprint of natural selection, making it likely that these sites influence the molecular properties of the group members. The proper way to group genes is self-evident in cases where the encoded product performs a single function in the cell. However, grouping becomes problematic for molecules, such as tRNA, which perform several functions and that interact with several molecular partners. Bioinformatics studies of these sorts of molecules can be misleading if the genes are grouped in a manner that reveals variation that would otherwise be explained by a different grouping parameter. Several bioinformatics studies of tRNA have appeared (McClain and Nicholas 1987; Steinberg and Kisselev 1993; Marck and Grosjean 2002; Ardell and Andersson 2006). Most of these studies have focused on the determinants of tRNA amino acid identity. By grouping tRNA gene sequences according to amino acid family, these studies have successfully identified residues in tRNA that are recognized by the cognate synthetase. However, a substantial amount of the variation among tRNA gene sequences remains unexplained, even when the results of the tRNA identity studies are considered along with the results of studies that have identified residues that influence the tRNA tertiary fold.
The unexplained genetic variation may be simply random noise due to the accumulation of neutral mutations. However, the genes for the molecular partners of tRNA, such as EF-Tu and 16S ribosomal RNA, are highly conserved (Gutell et al. 1985; Baldauf et al. 1996; Keeling and Inagaki 2004), and there is no a priori reason to think that tRNA would be exempt from strong selective pressure. Therefore, perhaps the variation is symptomatic that tRNA gene sequences have not been grouped properly. The convention is to group tRNA gene sequences by amino acid identity, without regard to the subtly different anticodon triplets of isoacceptors. However, if tRNAs are subjected to selection pressures on the basis of their anticodon triplet, then lumping the sequences by amino acid would inadvertently inflate estimates of the variation that exists among tRNA genes. Moreover, it would hinder the detection of residues that are conserved in an anticodon-dependent manner and that might therefore pertain to tRNA performance on the ribosome.
Here we explore the problem of whether collaboration between the anticodon triplet and residues in the tRNA body is universally important to the translational performance of tRNA. Only one bioinformatics study has focused on identifying sites in tRNA that pertain to translational performance, and this analysis was performed nearly 25 yr ago when the gene sequences for only some Escherichia coli and phage tRNAs were available (Yarus 1982). In the present study, tRNA gene sequences were extracted from 145 fully sequenced bacterial genomes that represent distantly related bacteria. This large data set makes it possible (1) to determine whether some residues in tRNA genes are conserved in an anticodon-dependent manner; (2) to determine the location of such residues; and (3) to explore whether coevolution between the anticodon triplet and residues in the tRNA body is a general or an idiosyncratic aspect of tRNA gene evolution.
RESULTS
Bacteria have been attractive candidates for genome sequencing projects due to their relatively small genome sizes (less than ∼10 Mb) and their relevance to medical and biotechnological problems. Complete genome sequences are now available for bacteria that are distantly related, have evolved different lifestyles, and have adapted to a variety of habitats. Thus the breadth of this sequence coverage provides an opportunity to identify robust and universal attributes of tRNA gene sequences.
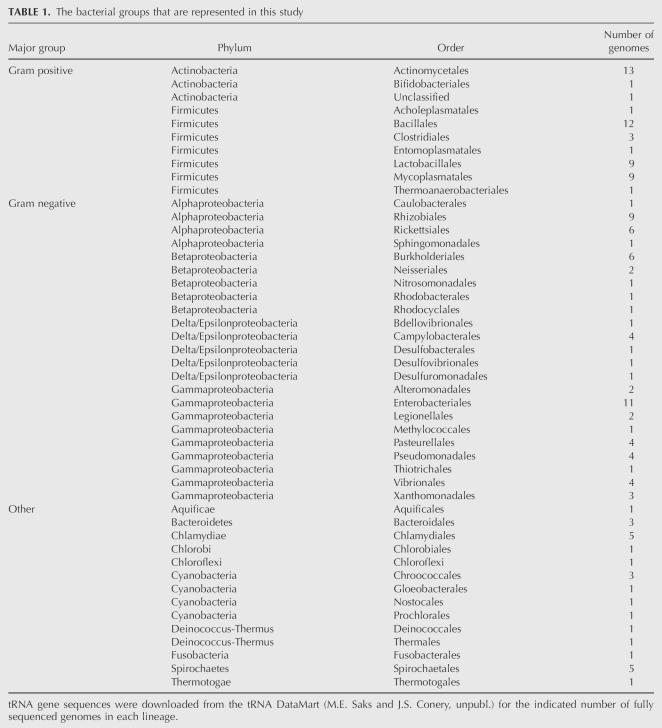
tRNA gene sequences were obtained from the tRNA DataMart (M.E. Saks and J.S. Conery, unpubl.). This publicly available Web site (http://trnamart.uoregon.edu/) was developed to facilitate both the formulation and testing of hypotheses that pertain to protein synthesis in bacteria. The Web site houses the tRNA gene sequences of the bacterial genomes that have been entirely sequenced thus far. In addition, it houses information on the physical location of each tRNA gene; characteristics of the associated genome such as its taxonomic affiliation, size, and GC content; and codon usage in the protein-encoding genes. The tRNA gene sequences that are used in this study were extracted from 145 bacterial genomes, which represent each of the bacterial lineages that have been sequenced to date (for details, see Materials and Methods; Table 1).
TABLE 1.
The bacterial groups that are represented in this study
The effect of grouping parameters on estimates of tRNA gene conservation
The first set of analyses focused on the problem of whether the choice of a grouping parameter (amino acid identity versus anticodon triplet) has any effect on the detection of conserved residues in tRNA gene sequences. For these analyses, the extent of sequence conservation across the entire tRNA sequence was calculated for each group as the number of conserved sites relative to the total number of sites in tRNA.
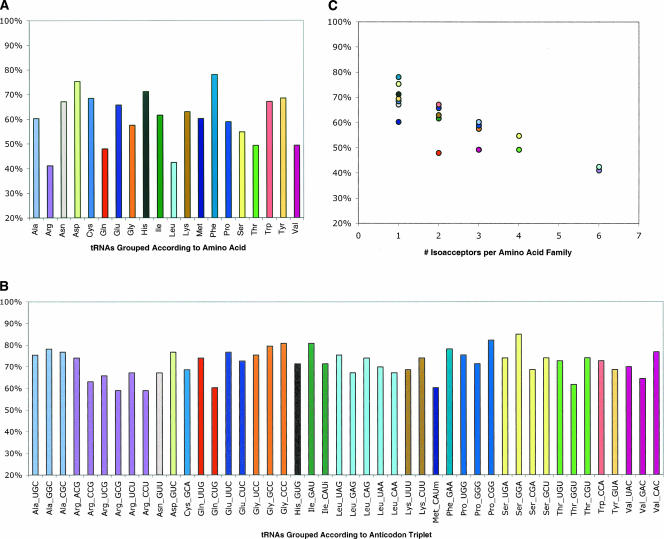
Figure 1A presents the percentage of residues that are conserved in tRNA when the sequences are grouped according to amino acid identity. The results indicate that the extent of sequence conservation differs markedly across the 20 amino acid families. For example, whereas only ∼40% of the positions in leucine tRNAs are conserved (excluding the variable arm), ∼80% of the positions in phenylalanine tRNAs are conserved. The extent of the variation is surprising. The aminoacyl-tRNA synthetases and EF-Tu impose selection pressure on all tRNAs to maintain a proper correspondence between their sequence and their amino acid identity (Giege et al. 1998; LaRiviere et al. 2001). If this is the principle selection pressure to which tRNAs are exposed, then the sequences of tRNAs in each of the 20 amino acid families ought to be conserved to similar extents.
FIGURE 1.
A comparison of the extent to which tRNA gene sequences are conserved when they are grouped according to the amino acid identity versus the anticodon triplet of the encoded tRNA. (A) The sequences are grouped by the amino acid identity of the encoded tRNA. (B) The sequences for the tRNAs in each amino acid family are separated according to their encoded anticodon triplet. (C) The extent of sequence conservation as a function of the number of different anticodon triplets (isoacceptors) in each amino acid family; Pearson's product moment correlation coefficient, r = −0.851, p < 0.01. Different colors are assigned to each amino acid family, and the colors are consistent across the three panels.
The explanation for the unexpected differences in the extent of sequence conservation across the 20 amino acid families becomes apparent when the tRNA sequences for each family are separated on the basis of the anticodon triplet. In all cases where more than one anticodon is used to translate a set of cognate codons, the extent of sequence conservation is higher for each anticodon-triplet group as compared to when the sequences are lumped by the corresponding amino acid family (Fig. 1A,B, cf. similarly colored bars). For example, leucine tRNAs share only ∼40% of their residues; but this number jumps to between 65% and 75% when each leucine tRNA is analyzed separately on the basis of its anticodon triplet. Moreover, as a consequence of this alternative criterion for grouping sequences, the extent of conservation becomes very similar for all tRNAs, regardless of their amino acid family (Fig. 1B). This uniformity is indicative of the anticodon triplet, rather than the amino acid, being the appropriate criterion to use for grouping tRNA gene sequences.
The above results indicate that tRNA genes are subjected to two types of selection pressures. The need to maintain a correct correspondence between a tRNA's sequence and its amino acid identity accounts for why tRNAs in the same amino acid family share some conserved residues. In addition, a need to maintain a correct correspondence between a tRNA's sequence and the nature of its anticodon triplet seems to account for the divergent evolution of isoacceptors that belong to the same amino acid family. According to these ideas, the extent of conservation and the number of anticodon triplets within each family ought to be inversely related. Indeed, Figure 1C shows that the extent of sequence conservation decreases with an increase in the number of anticodon triplets per amino acid family (Pearson's product moment correlation coefficient, r = −0.851, p < 0.01). These results indicate that the tRNA genes within each amino acid family comprise an admixture of solutions to at least two different sorts of selection pressures. Consequently, the joint influence of these pressures can be captured only if the sequences are grouped according to the anticodon triplet.
Conservation of anticodon-associated residues
A second set of analyses used a subset of tRNAs to identify anticodon-associated residues and to distinguish them from residues that are conserved on the basis of tRNA amino acid identity. The analyses are based on tRNA genes that correspond to the eight amino acid families (Ala, Arg, Gly, Leu, Pro, Ser, Thr, and Val) that are specified by four or more codons. This allows the sequences for the tRNAs in each amino acid family to be subdivided into at least three anticodon triplet (isoacceptor) groups that comprise replicate samples for each amino acid family. If tRNA gene sequences have diverged on the basis of the entire anticodon triplet, then the isoacceptors within each amino acid family should conserve different sets of sequence elements. It is not possible to apply this approach to cases where an amino acid is specified by only NNU and NNC codons. In these cases, the two synonymous codons are translated by a single tRNA species, making it impossible to treat the amino acid identity and anticodon triplet as separable parameters.
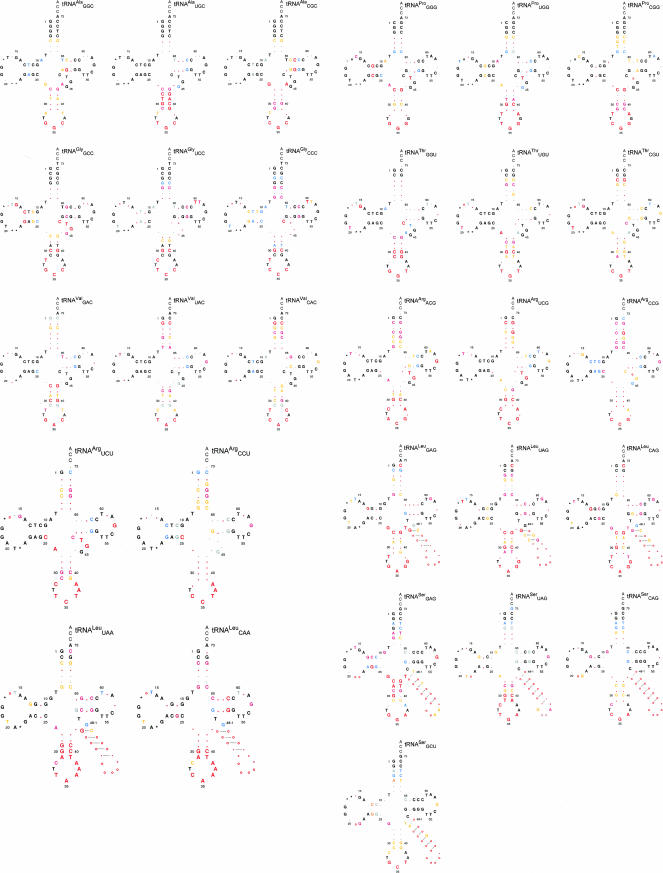
Cloverleaf diagrams for each isoacceptor are given in Figure 2. Nucleotides are colored and shaded to reflect the basis and extent of sequence conservation when the tRNAs from the eight amino acid families are grouped according to their amino acid identity and their distinctive anticodon triplet. The residues that are 90%–100% conserved on the basis of tRNA amino acid identity are indicated by nucleotide symbols in black, and for lesser degrees of conservation, in blue and gray (hereafter, black residues). Any additional residues that are conserved when the tRNAs in each family are separated according to their anticodon triplet are indicated by nucleotide symbols in shades of red, depending on their extent of conservation (hereafter, red residues). Note that the anticodon triplets are indicated in red to emphasize their distinctive nature and their association with the red residues.
FIGURE 2.
Sequence conservation among tRNA genes from 145 species, which represent the major lineages of the bacteria. The conserved residues are displayed on cloverleaf diagrams of alanine, arginine, glycine, leucine, proline, serine, threonine, and valine tRNA gene sequences that have been grouped according to the encoded anticodon triplet. The standard single-letter abbreviations are used to indicate the residues that are conserved at each position: (G) guanine, (A) adenine, (C) cytosine, (T) thymine; periods indicate that no residue is conserved. Nucleotide frequencies at positions that are conserved on the basis of amino acid family are indicated as: (black) 90%–100%; (blue) 80%–89%; and (gray) 70%–79%. Nucleotide frequencies at positions that are detected as conserved once the tRNAs for each amino acid family are separated according to the anticodon triplet are indicated as (red, large font) 90%–100%; (dark pink) 80%–89%; and (orange) 70%–89%. Symbols at the variable positions (17, 17a, 20a, 20b, and 47, the variable arm of leucine and serine tRNAs) indicate whether the position is conserved as (o) occupied; (*) unoccupied; or (period, .) neither. The 3′-terminal CCA was not included in the analyses.
A substantial number of residues are conserved when tRNAs are grouped by amino acid identity. Among these are residues that are known to influence the tRNA tertiary fold and/or recognition by the aminoacyl synthetases. Figure 2 shows that the extent of sequence conservation is often 90%–100% in the case of structural elements (Rich and RajBhandary 1976; Dirheimer et al. 1995; Steinberg et al. 1997), such as the residues in the D stem and D loop (positions 9–25), the variable loop (positions 45–48), and the T loop with its associated closing base pair (positions 53–61). Subtle differences in tRNA structure are usually conserved among tRNAs that belong to the same amino acid family, indicating that these differences have probably evolved due to selective pressures that are imposed by the aminoacyl-tRNA synthetases and perhaps the ribosome (Westhof et al. 1985; Steinberg et al. 1997; Nissan et al. 1999; Nissan and Perona 2000; Tukalo et al. 2005). The imprint of synthetase-imposed selection is also evident in other ways. Studies using the Escherichia coli and Thermus thermophilus systems show that synthetases typically recognize the residues at the top of the acceptor stem and the “discriminator” base at position 73 (Shimizu et al. 1992; McClain 1993; Saks et al. 1994; Pallanck et al. 1995; Giege et al. 1998). In addition, residues of the tRNA core are known to influence the recognition of arginine (A20) and leucine (A14-U48, A20a, G21, and the size of the D loop) tRNAs by their cognate synthetases (McClain and Foss 1988; Asahara et al. 1993; Tocchini-Valentini et al. 2000; Tukalo et al. 2005). Remarkably, most of the recognition elements that have been identified for each amino acid family are highly conserved across the bacteria (Fig. 2). Thus, the synthetases for the amino acid families in the present study have apparently imposed strong and consistent selective pressures on their cognate tRNAs throughout the course of bacterial evolution.
A remarkable number of residues in tRNA are conserved according to the sequence of the anticodon triplet rather than the amino acid family (Fig. 2, red residues). The total number of red residues ranges from 9 (tRNAGGGPro and tRNAGGUThr) to 25 (tRNAUAGLeu). Thus, ∼12%–32% of the residues in tRNA are devoted to functions that are associated with the anticodon triplet. The sequences of tRNAs that belong to the same amino acid family but that have different anticodon triplets (isoacceptors) have diverged in two ways. Isoacceptors sometimes conserve different residues at the same position in tRNA (Fig. 2, cf. [32–38] pair of valine isoacceptors). In other cases, isoacceptors have conserved residues at different positions in tRNA. For example, the [2–71] pair is conserved in >98% of the sequences for tRNAUACVal and tRNACACVal, whereas this pair is only conserved in 58% of the sequences for tRNAGACVal. On the other hand, the [3–70] pair is highly conserved in tRNAGACVal and tRNACACVal, but not in tRNAUACVal.
Conserved anticodon-associated residues are located throughout the tRNA (Fig. 2). Many of these residues are proximal to the anticodon triplet. This includes the residues at positions 32 and 38, which form a pseudo-base pair at the top of the anticodon loop (Auffinger and Westhof 1999) and the residue at position 37, which is adjacent to the third anticodon nucleotide. In addition, at least two of the five base pairs of the anticodon stem are conserved in nearly all of the anticodon triplet groups. Although most of the residues that influence the tRNA fold are conserved on the basis of tRNA amino acid identity (above), a few of the residues in this functional class are conserved in an anticodon-dependent manner. These red residues are usually located at positions 26 and 44 at the top of the anticodon stem, and in the D, T, and variable loops (Fig. 2). For example, isoacceptors in the same amino acid family sometimes differ with respect to the size of the α or the β regions of the D loop; the presence of a residue at position 47 of the variable loop; or the identity of the residues at positions 57, 59, or 60 of the T loop (Fig. 2). The ubiquity of anticodon-associated residues in the coaxial acceptor-TΨC stem (ATS) is striking. All but three of the 29 tRNAs in the study have at least one red residue in the ATS. Moreover, in some instances, the number of red residues in the ATS is greater than the number in the anticodon stem and loop. Arginine tRNAs, particularly tRNACCGArg and tRNACCUArg provide striking examples of this phenomenon.
DISCUSSION
Early on, phylogenetic studies of E. coli and Haemophilus influenzae tRNA gene sequences hinted that the isoacceptors in some amino acid families may have diverged during the course of evolution (Squires and Carbon 1971; Cedergren et al. 1980; Fitch and Upper 1987; Staves et al. 1987, 1988; Saks et al. 1998). However, the significance of these observations was unclear because a full complement of tRNA gene sequences was available for only a limited number of species. Thus it was impossible to discern whether the observed isoacceptor divergence was indicative of a general phenomenon or was simply a peculiarity of E. coli and H. influenzae. The results of the present study settle this issue. By using tRNA gene sequences from distantly related bacteria and from bacteria that have a variety of lifestyles, the present study unambiguously shows that isoacceptors in the same amino acid family have evolved distinctive and highly conserved signature sequences in association with their different anticodon triplets.
The evolution of anticodon-associated residues must arise from a need to compensate for a property(s) of the anticodon that is intimately related to an aspect of tRNA performance. The possibility that the association has evolved in response to synthetase-imposed selection pressures is ruled out because the isoacceptors within the alanine, leucine, and serine families have evolved different sequence signatures even though their cognate synthetases do not recognize any anticodon residues (Fig. 2; Pallanck et al. 1995). On the other hand, several lines of evidence support the idea that anticodon-associated residues have evolved in response to selective pressures that are related to tRNA performance on the ribosome.
The idea that anticodon-associated residues influence the translational performance of tRNA originated with Yarus's observation of an association between residues of the anticodon stem and loop and the “cardinal” (3′) nucleotide of the anticodon triplet (Yarus 1982). Subsequent in vivo studies of the translation efficiency of E. coli suppressor tRNAs (Yarus et al. 1986; Kleina et al. 1990; Yarus and Smith 1995) and in vitro biochemical studies of tRNA binding to the ribosome decoding center (A site) (Lustig et al. 1993; Claesson et al. 1995; Konevega et al. 2004; Olejniczak et al. 2005) have chiefly focused on the residues of the anticodon loop. When taken together, the results of these studies show that the residue at position 37, which is adjacent to the third anticodon nucleotide, and the residues of the [32–38] pseudopair, at the top of the anticodon loop, can affect translation efficiency and binding to the cognate codon.
The correspondence between the available experimental data and the bioinformatics presented here is quite good. The residue at position 37 is conserved in all of the tRNAs in the present study; and at least one member of the [32–38] pseudopair is conserved in an anticodon-dependent manner in all but two of the 29 anticodon triplet groups (Fig. 2). Thus the residues at positions 37, 32, and 38 seem to comprise universal “tuning” elements with regard to decoding. In addition, the [27–43] pair, which is known to influence codon recognition (Komine and Inokuchi 1990; Schultz and Yarus 1994; Yarus et al. 2003), and/or the adjacent [26–44] pair are conserved in an anticodon-dependent manner in 22 of the 29 tRNAs in the study (Fig. 2). The [26–44] pair and most of the additional red residues have yet to be studied in detail. However, given the general agreement between the bioinformatics results and the results of previous experimental studies, it seems likely that most of the red residues collaborate with the anticodon during translation.
Why are so many of the residues in tRNA conserved in an anticodon-dependent manner? The residues at positions 37, 32, and 38 are proximal to the anticodon triplet, and they comprise part of the anticodon loop. These residues, including their post-transcriptional modifications (Agris 2004), have probably coevolved with the anticodon triplet due to a need to convert an idiosyncratic property of the anticodon, such as its binding energy or the conformation of the anticodon loop, into a property that conforms to the universal interaction rules that are set by the ribosome (Olejniczak et al. 2005). In principle, residues that are distal to the anticodon loop can also provide compensatory binding energy. X-ray crystallographic studies of the T. thermophilus 70S ribosome containing bound messenger RNA and tRNA show that the ribosome contacts the acceptor, TΨC, and D stems, as well as the residues at the junction between the D and T loops (Yusupov et al. 2001; Korostelev et al. 2006; Selmer et al. 2006). These contacts may be especially important when tRNA passes through hybrid states where its anticodon and aminoacyl ends must, respectively, remain stably bound to adjacent sites on the 30S and 50S subunits (Moazed and Noller 1989).
During translation, the stability of the codon–anticodon helix and of the tRNA–ribosome complex is challenged by both EF-Tu- and ribosome-imposed forces. The existence of these forces is evidenced by the different conformations that tRNA adopts as it interacts with its molecular partners. For example, in complex with EF-Tu, aminoacyl-tRNA binds to the ribosome in the A/T state, where its anticodon binds to the cognate codon while its 3′-aminoacyl end remains bound to EF-Tu. Cryoelectron microscopy studies reveal that this hybrid state induces a kink in tRNAGAAPhe near the junction of the D and anticodon stems that is relieved when the 3′-aminoacyl end of tRNA moves into the A site and becomes properly oriented in the peptidyl-transferase center of the 50S subunit (Stark et al. 2002; Valle et al. 2002, 2003). Similarly, molecular dynamics simulations indicate the transition from the A/T to the A/A state involves changes in the acceptor stem conformation, rearrangements of the variable loop, and changes in the base-stacking interactions between the D and T loops (Sanbonmatsu et al. 2005). These observations clearly show that the deformability of the tRNA structure is central to the translation mechanism. Moreover, studies of the [27–43] pair at the top of the anticodon stem (Komine and Inokuchi 1990; Schultz and Yarus 1994; Yarus et al. 2003) and the [11–24] pair of the D stem (Hirsh 1971; Schultz and Yarus 1994; Cochella and Green 2005) indicate that codon recognition is influenced by tRNA deformability. In the case of E. coli tRNACCATrp, a G24A mutation seems to alter the deformability of the tRNA in a manner that increases the forward rate by which tRNA is accommodated into the ribosome A site, thereby facilitating the translation of near-cognate UGA codons through a normally prohibited C-A pair (Cochella and Green 2005).
Given the above observations, it stands to reason that the residues that account for tRNA deformability ought to coevolve with the anticodon triplet. Yet this specific idea has yet to be explored. Here we detect an association between anticodon triplets and residues that is likely to influence the deformability of tRNA and thus its translational performance. These anticodon-associated residues include members of the [27–43] and the [26–44] pairs at the junction between the D- and anticodon-stem residues of the D and T loops, as well as residues of the ATS (Fig. 2). It is tempting to speculate that tRNA deformability plays a role in translation that is analogous to the role already described with regard to the contribution that the [32–38] pair makes to uniform ribosome binding (Olejniczak et al. 2005). In principle, deformability could enable individual tRNAs to convert the uniform forces that are imposed by EF-Tu and the ribosome into forces that are compatible with the particular properties of the corresponding codon–anticodon helix. Accordingly, in addition to proofreading the codon–anticodon helix by monitoring its constellation of hydrogen bonds (Ogle et al. 2001), the ribosome may also discriminate tRNAs on the basis of the match between the deformability of a tRNA and its anticodon triplet.
A subtle but important question that emerges from the present study is whether anticodon-associated residues have evolved due to a need to compensate for a property that is associated with a particular position within the anticodon triplet rather than a property of the entire anticodon. Because the anticodons of isoacceptors that belong to the same amino acid family usually differ at only the first position (position 34, the “wobble” position) (Fig. 2), the design of the present study provides an opportunity to explore whether selective pressures are imposed on tRNA as a consequence of properties of the wobble nucleotide. If the observed divergence in isoacceptor gene sequences constitutes a response to selective pressures that arise due to the inherent differences in the overall binding energies of related anticodon triplets, then the sequences of cognate tRNAs that have G34 and C34 should be more similar to each other than either is to the cognate tRNA that has U34. However, this sort of relationship is not evident in the data (Fig. 2). Moreover, studies of threonine tRNAs show that while a C34U substitution in tRNACGUThr results in a tRNA that translates cognate ACA codons with an efficiency similar to that of a normal tRNAUGUThr, the analogous mutation (G34U) in tRNAGGUThr results in a variant that has a substantially reduced efficiency (W. Liu and M.E. Saks, unpubl.). Parallel studies using glycine tRNAs show an identical pattern (M.E. Saks and A.S. Barnett, unpubl.), and in both cases, translational performance is improved by second-site mutations in the tRNA body. Therefore, some residues in tRNA seem to have evolved in association with the identity of the wobble nucleotide. Perhaps the conformation of the anticodon loop or attributes of the codon–anticodon helix are differentially affected by purine versus pyrimidine at the wobble position to an extent that warrants the evolution of compensatory residues in the tRNA body. In any event, the results of the present study raise the possibility that the first nucleotide of the anticodon or the third codon–anticodon pair participates in one or more steps in translation that remain to be clarified.
MATERIALS AND METHODS
The tRNA gene sequences in this study were obtained from the tRNA DataMart using the options associated with its search engine (M.E. Saks and J.S. Conery, unpubl.; http://trnamart.uoregon.edu/). The data set includes the tRNA genes that are encoded in 145 bacterial genomes, which represent all of the major bacterial lineages that have been identified to date (Table 1). To minimize bias, only one strain of each bacterial species is used for the analyses. The tRNA gene copy number varies extensively across the bacteria. Sometimes the copies have identical sequences, whereas in other cases, the copies encode slightly different variants of a tRNA gene. To normalize the data set while preserving the natural variation, we directed the search engine of the tRNAMart to assemble a data set that comprises only the distinctive tRNA gene sequences in each genome. A sequence is defined as distinctive if at least one of its residues (including those of the anticodon) differs from those of the other tRNA genes in the same genome. The gene sequences of the tRNAs in all 20 amino acid families were downloaded using the tRNAMart subroutine that aligns them according to the conventional cloverleaf structure of tRNA, and the alignments were subsequently checked by hand.
The anticodon unambiguously specifies the amino acid identity of most, but not all, tRNA genes. In the exceptional cases, tRNA amino acid identity is specified by the anticodon in conjunction with other sequence elements in the tRNA body, making it possible to infer the amino acid identity of these tRNAs with some confidence. Here we use the established criteria to distinguish tRNAUCATrp from tRNAUCASec, and to distinguish among tRNACAUMetf(the bacterial initiator tRNA), tRNACAUMetm (the methionine elongator tRNA), and tRNACAUIle (an isoleucine tRNA in which C at the first anticodon position is post-transcriptionally modified to lysidine) (Marck and Grosjean 2002). The resulting data set comprises ∼6000 tRNA gene sequences.
The aligned tRNA gene sequences were grouped according to the encoded anticodon triplet; and the nucleotide frequency at each position in tRNA was determined for each anticodon triplet group. A position is classified as conserved if at least 70% of the sequences within a group have the same residue at the position. This cutoff was chosen in light of the fact that the genomic GC content of the bacteria in the study ranges from ∼25% to 75% (data not shown). An analysis of the entire data set shows that on average the nucleotide frequency is >90% for about half of the positions in tRNA. The results of the analyses of the anticodon triplet groups were used to identify the positions that are conserved in each amino acid family. A position is defined as conserved within an amino acid family if all of the anticodon triplet groups for the amino acid conserve (as defined above) the same residue. The results of the analyses for alanine, arginine, glycine, leucine, proline, serine, threonine, and valine tRNAs are presented, in detail, in Figure 2. Although the residues of the 3′-terminal CCA were not scored, they are represented in Figure 2, using shadowed text, to indicate that –CCA3′ is a universal feature of transcribed and processed tRNAs.
The extent of sequence conservation within a group (Fig. 1) is defined as the number of conserved positions divided by the total number of positions in tRNA. The 11 positions of the D loop and positions 45, 46, and 47 of the variable loop are included in the analyses; the anticodon residues and the 3′-terminal CCA are excluded. The variable positions in tRNA (positions 17, 17a, 20a, 20b, and 47) are scored in a manner that detects structural variation. If a single residue was not conserved (as defined above) at one of these sites, then a determination was made of whether the position is conserved as occupied: there is a residue at the position in at least 70% of the sequences within a group; or unoccupied: at least 70% of the sequences within a group have no residue at the position. Positions that failed to meet one of these criteria are scored as variable.
ACKNOWLEDGMENTS
We gratefully acknowledge O.C. Uhlenbeck and S. Ledoux for thoughtful comments on the manuscript and O.C. Uhlenbeck for stimulating discussions about tRNA and the ribosome. This work was supported by NIH grant GM059779 to M.E.S.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.345907.
REFERENCES
- Agris, P.F. Decoding the genome: A modified view. Nucleic Acids Res. 2004;32:223–238. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardell, D.H., Andersson, S.G. TFAM detects co-evolution of tRNA identity rules with lateral transfer of histidyl-tRNA synthetase. Nucleic Acids Res. 2006;34:893–904. doi: 10.1093/nar/gkj449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahara, H., Himeno, H., Tamura, K., Hasegawa, T., Watanabe, K., Shimizu, M. Recognition nucleotides of Escherichia coli tRNALeu and its elements facilitating discrimination from tRNASer and tRNATyr. J. Mol. Biol. 1993;231:219–229. doi: 10.1006/jmbi.1993.1277. [DOI] [PubMed] [Google Scholar]
- Auffinger, P., Westhof, E. Singly and bifurcated hydrogen-bonded base pairs in tRNA anticodon hairpins and ribozymes. J. Mol. Biol. 1999;292:467–483. doi: 10.1006/jmbi.1999.3080. [DOI] [PubMed] [Google Scholar]
- Baldauf, S.L., Palmer, J.D., Doolittle, W.F. The root of the universal tree and the origin of eukaryotes based on elongation factor phylogeny. Proc. Natl. Acad. Sci. 1996;93:7749–7754. doi: 10.1073/pnas.93.15.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedergren, R.J., LaRue, B., Sankoff, D., Lapalme, G., Grosjean, H. Convergence and minimal mutation criteria for evaluating early events in tRNA evolution. Proc. Natl. Acad. Sci. 1980;77:2791–2795. doi: 10.1073/pnas.77.5.2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson, C., Lustig, F., Boren, T., Simonsson, C., Barciszewska, M., Lagerkvist, U. Glycine codon discrimination and the nucleotide in position 32 of the anticodon loop. J. Mol. Biol. 1995;247:191–196. doi: 10.1006/jmbi.1994.0132. [DOI] [PubMed] [Google Scholar]
- Cochella, L., Green, R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirheimer, G., Keith, G., Dumas, P., Westhof, E. Primary, secondary, and tertiary structures of tRNAs. In: Soll D., RajBhandary U.L., editors. tRNA: Structure, biosynthesis, and function. American Society for Microbiology; Washington, DC: 1995. pp. 93–126. [Google Scholar]
- Fitch, W.M., Upper, K. The phylogeny of tRNA sequences provides evidence for ambiguity reduction in the origin of the genetic code. Cold Spring Harb. Symp. Quant. Biol. 1987;52:759–767. doi: 10.1101/sqb.1987.052.01.085. [DOI] [PubMed] [Google Scholar]
- Giege, R., Sissler, M., Florentz, C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell, R.R., Weiser, B., Woese, C.R., Noller, H.F. Comparative anatomy of 16-S-like ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- Hirsh, D. Tryptophan transfer RNA as the UGA suppressor. J. Mol. Biol. 1971;58:439–458. doi: 10.1016/0022-2836(71)90362-7. [DOI] [PubMed] [Google Scholar]
- Hoagland, M. Biochemistry or molecular biology? The discovery of “soluble RNA”. Trends Biochem. Sci. 1996;21:77–80. [PubMed] [Google Scholar]
- Keeling, P.J., Inagaki, Y. A class of eukaryotic GTPase with a punctate distribution suggesting multiple functional replacements of translation elongation factor 1α. Proc. Natl. Acad. Sci. 2004;101:15380–15385. doi: 10.1073/pnas.0404505101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleina, L.G., Masson, J.M., Normanly, J., Abelson, J., Miller, J.H. Construction of Escherichia coli amber suppressor tRNA genes. II. Synthesis of additional tRNA genes and improvement of suppressor efficiency. J. Mol. Biol. 1990;213:705–717. doi: 10.1016/S0022-2836(05)80257-8. [DOI] [PubMed] [Google Scholar]
- Komine, Y., Inokuchi, H. Importance of the G27-A43 mismatch at the anticodon stem of Escherichia coli tRNAThr2. FEBS Lett. 1990;272:55–57. doi: 10.1016/0014-5793(90)80447-q. [DOI] [PubMed] [Google Scholar]
- Konevega, A.L., Soboleva, N.G., Makhno, V.I., Semenkov, Y.P., Wintermeyer, W., Rodnina, M.V., Katunin, V.I. Purine bases at position 37 of tRNA stabilize codon–anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. RNA. 2004;10:90–101. doi: 10.1261/rna.5142404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev, A., Trakhanov, S., Laurberg, M., Noller, H.F. Crystal structure of a 70S ribosome–tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- LaRiviere, F.J., Wolfson, A.D., Uhlenbeck, O.C. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001;294:165–168. doi: 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
- Lustig, F., Boren, T., Claesson, C., Simonsson, C., Barciszewska, M., Lagerkvist, U. The nucleotide in position 32 of the tRNA anticodon loop determines ability of anticodon UCC to discriminate among glycine codons. Proc. Natl. Acad. Sci. 1993;90:3343–3347. doi: 10.1073/pnas.90.8.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck, C., Grosjean, H. tRNomics: Analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinis, S.A., Schimmel, P. Small RNA oligonucleotide substrates for specific aminoacylations. In: Soll D., RajBhandary U.L., editors. tRNA: Structure, biosynthesis, and function. American Society for Microbiology; Washington, DC: 1995. pp. 349–370. [Google Scholar]
- McClain, W.H. Rules that govern tRNA identity in protein synthesis. J. Mol. Biol. 1993;234:257–280. doi: 10.1006/jmbi.1993.1582. [DOI] [PubMed] [Google Scholar]
- McClain, W.H., Foss, K. Changing the acceptor identity of a transfer RNA by altering nucleotides in a “variable pocket”. Science. 1988;241:1804–1807. doi: 10.1126/science.2459773. [DOI] [PubMed] [Google Scholar]
- McClain, W.H., Nicholas H.B., Jr Differences between transfer RNA molecules. J. Mol. Biol. 1987;194:635–642. doi: 10.1016/0022-2836(87)90240-3. [DOI] [PubMed] [Google Scholar]
- Moazed, D., Noller, H.F. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- Nissan, T.A., Perona, J.J. Alternative designs for construction of the class II transfer RNA tertiary core. RNA. 2000;6:1585–1596. doi: 10.1017/s1355838200001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissan, T.A., Oliphant, B., Perona, J.J. An engineered class I transfer RNA with a class II tertiary fold. RNA. 1999;5:434–445. doi: 10.1017/s1355838299981827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle, J.M., Brodersen, D.E., Clemons W.M., Jr, Tarry, M.J., Carter, A.P., Ramakrishnan, V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Olejniczak, M., Dale, T., Fahlman, R.P., Uhlenbeck, O.C. Idiosyncratic tuning of tRNAs to achieve uniform ribosome binding. Nat. Struct. Mol. Biol. 2005;12:788–793. doi: 10.1038/nsmb978. [DOI] [PubMed] [Google Scholar]
- Pallanck, L., Pak, M., Schulman, L.H. tRNA discrimination in aminoacylation. In: Soll D., RajBhandary U.L., editors. tRNA: Structure, biosynthesis, and function. American Society for Microbiology; Washington, DC: 1995. pp. 371–394. [Google Scholar]
- RajBhandary, U.L., Soll, D. Transfer RNA in its fourth decade. In: Soll D., RajBhandary U.L., editors. tRNA: Structure, biosynthesis, and function. American Society for Microbiology; Washington, DC: 1995. pp. 1–4. [Google Scholar]
- Rich, A., RajBhandary, U.L. Transfer RNA: molecular structure, sequence, and properties. Annu. Rev. Biochem. 1976;45:805–860. doi: 10.1146/annurev.bi.45.070176.004105. [DOI] [PubMed] [Google Scholar]
- Saks, M.E., Sampson, J.R., Abelson, J.N. The transfer RNA identity problem: A search for rules. Science. 1994;263:191–197. doi: 10.1126/science.7506844. [DOI] [PubMed] [Google Scholar]
- Saks, M.E., Sampson, J.R., Abelson, J. Evolution of a transfer RNA gene through a point mutation in the anticodon. Science. 1998;279:1665–1670. doi: 10.1126/science.279.5357.1665. [DOI] [PubMed] [Google Scholar]
- Sanbonmatsu, K.Y., Joseph, S., Tung, C.S. Simulating movement of tRNA into the ribosome during decoding. Proc. Natl. Acad. Sci. 2005;102:15854–15859. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, D.W., Yarus, M. tRNA structure and ribosomal function. I. tRNA nucleotide 27–43 mutations enhance first position wobble. J. Mol. Biol. 1994;235:1381–1394. doi: 10.1006/jmbi.1994.1095. [DOI] [PubMed] [Google Scholar]
- Selmer, M., Dunham, C.M., Murphy F.V., IV, Weixlbaumer, A., Petry, S., Kelley, A.C., Weir, J.R., Ramakrishnan, V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Shimizu, M., Asahara, H., Tamura, K., Hasegawa, T., Himeno, H. The role of anticodon bases and the discriminator nucleotide in the recognition of some E. coli tRNAs by their aminoacyl-tRNA synthetases. J. Mol. Evol. 1992;35:436–443. doi: 10.1007/BF00171822. [DOI] [PubMed] [Google Scholar]
- Squires, C., Carbon, J. Normal and mutant glycine transfer RNAs. Nat. New Biol. 1971;233:274–277. doi: 10.1038/newbio233274a0. [DOI] [PubMed] [Google Scholar]
- Stark, H., Rodnina, M.V., Wieden, H.J., Zemlin, F., Wintermeyer, W., van Heel, M. Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat. Struct. Biol. 2002;9:849–854. doi: 10.1038/nsb859. [DOI] [PubMed] [Google Scholar]
- Staves, M.P., Bloch, D.P., Lacey J.C., Jr Evolution of E. coli tRNAIle: Evidence of derivation from other tRNAs. Z. Naturforsch. C. 1987;42:129–133. doi: 10.1515/znc-1987-1-221. [DOI] [PubMed] [Google Scholar]
- Staves, M.P., Bloch, D.P., Lacey J.C., Jr Evolution of E. coli tRNATrp. Orig. Life Evol. Biosph. 1988;18:97–105. doi: 10.1007/BF01808784. [DOI] [PubMed] [Google Scholar]
- Steinberg, S.V., Kisselev, L.L. Mosaic tile model for tRNA-enzyme recognition. Nucleic Acids Res. 1993;21:1941–1947. doi: 10.1093/nar/21.8.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg, S., Leclerc, F., Cedergren, R. Structural rules and conformational compensations in the tRNA L-form. J. Mol. Biol. 1997;266:269–282. doi: 10.1006/jmbi.1996.0803. [DOI] [PubMed] [Google Scholar]
- Tocchini-Valentini, G., Saks, M.E., Abelson, J. tRNA leucine identity and recognition sets. J. Mol. Biol. 2000;298:779–793. doi: 10.1006/jmbi.2000.3694. [DOI] [PubMed] [Google Scholar]
- Tukalo, M., Yaremchuk, A., Fukunaga, R., Yokoyama, S., Cusack, S. The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer-editing conformation. Nat. Struct. Mol. Biol. 2005;12:923–930. doi: 10.1038/nsmb986. [DOI] [PubMed] [Google Scholar]
- Valle, M., Sengupta, J., Swami, N.K., Grassucci, R.A., Burkhardt, N., Nierhaus, K.H., Agrawal, R.K., Frank, J. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J. 2002;21:3557–3567. doi: 10.1093/emboj/cdf326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle, M., Zavialov, A., Li, W., Stagg, S.M., Sengupta, J., Nielsen, R.C., Nissen, P., Harvey, S.C., Ehrenberg, M., Frank, J. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat. Struct. Biol. 2003;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- Westhof, E., Dumas, P., Moras, D. Crystallographic refinement of yeast aspartic acid transfer RNA. J. Mol. Biol. 1985;184:119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Yarus, M. Translational efficiency of transfer RNA's: Uses of an extended anticodon. Science. 1982;218:646–652. doi: 10.1126/science.6753149. [DOI] [PubMed] [Google Scholar]
- Yarus, M., Smith, D. tRNA on the ribosome: A waggle theory. In: Soll D., RajBhandary U.L., editors. tRNA: Structure, biosynthesis, and function. American Society for Microbiology; Washington, DC: 1995. pp. 443–469. [Google Scholar]
- Yarus, M., Cline, S.W., Wier, P., Breeden, L., Thompson, R.C. Actions of the anticodon arm in translation on the phenotypes of RNA mutants. J. Mol. Biol. 1986;192:235–255. doi: 10.1016/0022-2836(86)90362-1. [DOI] [PubMed] [Google Scholar]
- Yarus, M., Valle, M., Frank, J. A twisted tRNA intermediate sets the threshold for decoding. RNA. 2003;9:384–385. doi: 10.1261/rna.2184703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., Noller, H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- Zamecnik, P. From protein synthesis to genetic insertion. Annu. Rev. Biochem. 2005;74:1–28. doi: 10.1146/annurev.biochem.74.050304.091632. [DOI] [PubMed] [Google Scholar]