Abstract
Fragile X syndrome is caused by the absence or reduction of the fragile X mental retardation protein (FMRP) because FMR1 gene expression is reduced. Alleles with repeat sizes of 55–200 are classified as premutations, and it has been demonstrated that FMR1 expression is elevated in the premutation range. However, the majority of the studies reported were performed in males. We studied FMR1 expression in 100 female fragile X family members from the northern region of Spain using quantitative (fluorescence) real-time polymerase chain reaction. Of these 100 women, 19 had normal alleles, 19 were full mutation carriers, and 62 were premutation carriers. After confirming differences between the three groups of females, and increased levels of the FMR1 transcript among premutation carriers, we found that the relationship between mRNA levels and repeat size is nonlinear. These results were obtained using a novel methodology that, based on the size of the CGG repeats, allows us to find out the most probable threshold from which the relationship between CGG repeat number and mRNA levels changes. Using this approach, a significant positive correlation between CGG repeats and total mRNA levels has been found in the premutation range <100 CGG, but this correlation diminishes from 100 onward. However, when correcting by the X inactivation ratio, mRNA levels increase as the number of CGG repeats increases, and this increase is highly significant over 100 CGG. We suggest that due to skewed X inactivation, mRNA levels tend to normalize in females when the number of CGG repeats increases.
Keywords: FMR1, mRNA, FXS, FRAXA, FMR1 female carriers
INTRODUCTION
Fragile X syndrome (FXS) is the most common form of inherited mental retardation, with an estimated incidence of 1 in 4000 males and 1 in 8000 females (for review, see Crawford et al. 2001). The syndrome is nearly always caused by a large expansion of a CGG trinucleotide repeat (>200 CGG = “full mutation”) within the promoter region of the FMR1 gene, which is usually associated with hypermethylation and transcription silencing. However, this simple relationship between CGG repeats, hypermethylation, and gene silencing cannot explain all clinical manifestations described in FXS patients.
In the general population, the CGG repeat number ranges from approximately seven to 55, with no clinical or biochemical abnormalities, whereas premutation alleles (PM) present expansions of ∼55–200 CGG repeats (Fu et al. 1991). These alleles are linked to a high risk of expansion, through maternal transmission, in the next generation.
Although individuals within the premutation range do not have fragile X-related mental retardation and have normal IQ measurements, in some cases clinical involvement has been reported, including learning disabilities, anxiety, and emotional problems. Moreover, two characteristic phenotypes have been identified that only occur in the premutation range: premature ovarian failure (POF) in 13%–26% of women (for review, see Sherman 2000); and a new neurodegenerative syndrome, Fragile X-associated tremor and ataxia syndrome (FXTAS), in older premutation carrier males, with progressive tremor, ataxia, and generalized brain atrophy (Hagerman et al. 2001, 2003; Jacquemont et al. 2003).
Recent studies of FMR1 gene expression (Tassone et al. 2000c) have shown elevated FMR1 mRNA levels in cells of males with alleles in the premutation range. The increased transcriptional activity appears to be positively correlated with CGG repeat size. That is, CGG expansion in individuals in the high premutation range (≥100 CGG) is observed to be four- to 10-fold higher than usual, whereas in the lower range (55–100 CGG) the average was twofold elevation. This molecular abnormality has been reported in other studies (Tassone et al. 2000a,b, 2001; Kenneson et al. 2001; Allen et al. 2004), the majority of which were carried out on males. However, the relationship between molecular status and phenotype is much more complex in women, where the activation ratio of the X chromosome must be considered.
In females, FMR1 gene transcript levels have been studied by Tassone et al. (2000a) and Allen et al. (2004). The former reported mRNA levels in 35 female premutation carriers and in eight full mutation females and demonstrated that mRNA levels were elevated in females with premutation alleles. Allen et al. (2004) studied a larger population than Tassone et al. (2000a) and confirmed the results obtained by the former by means of different analyses.
In any event, these works revealed the need to carry out further research into transcript levels in the female population in order to better define the possible correlation between molecular parameters and clinical phenotypes associated with the premutation range. This was the purpose of our study, which is presented here.
RESULTS
Comparison analyses of FMR1 mRNA levels and X inactivation ratio
We initiated the evaluation of our results by investigating and comparing mean mRNA levels and mean X inactivation ratios (calculated using the normal X chromosome that remains active) among noncarrier females (N), female premutation carriers (PM), and females with the full mutation (FM). The analyses were carried out using only 95 females (95/100), after having removed two females with FM and three females with PM because they were considered outlier observations: The Grubbs’ statistics and the associated p-values were 3.16 (p = 0.001) and 2.69 (p = 0.020) for the FM outliers; and 3.69 (p = 0.002), 4.15 (p = 0.0001), and 3.41 (p = 0.01) for the PM outliers. Furthermore, from these 95 females, we could not determine the X inactivation ratio in five females, so they were not considered for the analyses of this ratio and the corresponding mRNA levels corrected by the activation ratio (AR). Table 1 shows the mean (±SD) values of the total observed mRNA levels, the X inactivation ratio expressed as the mean of the percentage of the observed normal active X chromosomes in Southern blot (AR), and the mean for the mRNA levels corrected by AR. Table 1 also shows the statistical results of a comparison between groups (globally and for pairs of groups). It is noteworthy that differences among the three groups are highly significant for the mRNA and mRNA corrected by AR levels.
TABLE 1.
Distribution of mRNA, mRNA corrected for AR, and AR for each group
Influence of CGG repeats in the premutation range
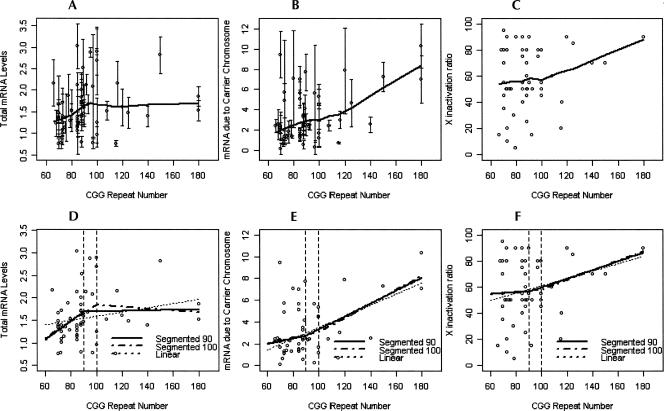
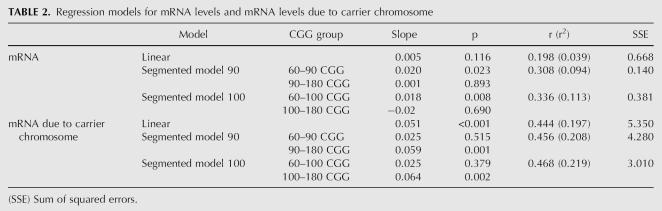
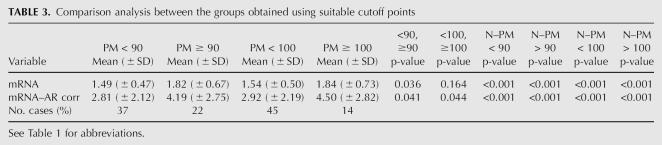
The locally ordinary weighted regression (lowess) curve obtained for females with alleles in the premutation range, using the total mRNA levels versus the CGG repeat number, shows that their relationship is nonlinear: mRNA levels appear to increase with the CGG repeat number up to ∼90–100 CGG; thereafter, this increase is stopped and a zero slope curve is observed (Fig. 1A). When mRNA from a carrier chromosome is used, the pattern of increase differs substantially. In fact, the pattern of increase for repeats <100 CGG appears to be moderate, but in contrast the slope seems to be higher from 100 onward (Fig. 1B). The explanation for this phenomenon could be a skewing toward a high proportion of normal active X that we have observed when the number of CGG repeats increases (Fig. 1C).
FIGURE 1.
mRNA versus molecular status. (A) Error bar plot and lowess curve for total mRNA levels, (B) error bar plot and lowess curve for mRNA due to carrier chromosome, (C) scatter plot and lowess curve for X inactivation ratio, (D) linear and segmented regression models with cutoffs of 90 and 100 for mRNA levels, (E) linear and segmented regression models with cutoffs of 90 and 100 for mRNA levels due to carrier chromosome, and (F) linear and segmented regression models with cutoffs of 90 and 100 for X inactivation ratio.
According to this lowess curve, the most suitable cutoff points seem to be 90 and 100 CGG repeats. Using these cutoff points, the fit of the segmented linear models for the total mRNA levels shows that there is a significant increasing trend for the first segment (slopes = 0.020 and 0.018), whereas this increase is practically zero and not significant for the second one (slopes = 0.001 and −0.02) (Table 2). These two segment models improve both the goodness of fit and the sum of squared errors (SSE) with respect to the linear model (see r2 and SSE values in Table 2), indicating that they capture the local behavior of the data better. Of the two segmented models, the lowess curve is closer to the model with a cutoff of 90 (see the SSE values and Fig. 2C), but the goodness of fit is better for the model that considers the cutoff as 100.
TABLE 2.
Regression models for mRNA levels and mRNA levels due to carrier chromosome
Looking at the second part of Table 2 (the segmented linear models for the mRNA levels corrected by AR), the results for both models (considering 90 and 100 as cutoff points) also improve those obtained by the linear model in terms of goodness of fit and SSE. In particular, the apparent increasing trend for the first segment, with a slope of 0.025 for both segment models, is not significant, whereas the increasing trend of the second segment is highly significant for both models, with a slope of ∼0.06. Both the SSE and the goodness of fit are slightly better for the segmented model with 100 CGG as the cutoff point, although both models lead to similar conclusions regarding significance and slope values, being close to each other (Fig. 2D), and indicating that both cutoff points seem plausible. Taking the segmented model with 100 CGG as a threshold, a difference of 10 CGG in the low CGG group produces on average an increment of 0.25 units in transcript levels, while an increment of 0.64 units can be expected in the high CGG group (Table 2).
Table 3 shows the comparison analysis of total mRNA levels and mRNA levels after AR correction among the subgroups derived from the segmented analysis: Taking 100 CGG as the cutoff, there are not significant differences in the mean values of mRNA levels between the low-intermediate CGG group and the high CGG group (Mann–Whitney test, p = 0.164), whereas there are significant differences between the mRNA levels corrected by AR (Mann–Whitney test, p = 0.044). Interestingly, if we use 90 as the cutoff point instead, the results differ slightly, as the differences between the two subgroups are significant even for the total mRNA levels (Mann–Whitney test, p = 0.036). Finally, when comparing each of these PM groups with the normal group, the differences are highly significant using both 90 and 100 as cutoffs (p < 0.001 for the four combinations) (Table 3).
TABLE 3.
Comparison analysis between the groups obtained using suitable cutoff points
DISCUSSION
The premutation status of FXS has aroused fresh interest since Tassone et al. (2000c) described seven carrier males who had a fivefold increase in FMR1 mRNA levels. This molecular abnormality was previously reported in FM males, whose FMR1 gene was partially unmethylated, although FMRP levels decreased (Tassone et al. 2000b). Moreover, in the low premutation range, from 60 to 100 CGG repeats, 16 males showed elevated FMR1 transcript levels in another study reported by Tassone et al. (2000a). These findings were confirmed by Kenneson et al. (2001), who found a significant positive correlation between the number of CGG repeats and FMR1 mRNA levels.
In the current work, we studied FMR1 expression in 100 female fragile X family members from the northern region of Spain using quantitative (fluorescence) real-time PCR. Of these 100 women, 19 had normal alleles, 19 were full mutation carriers, and 62 were premutation carriers. We followed exactly the methods used by Tassone et al. (2000c), and all the measurements were taken from fresh blood, normal control included, because they seemed more representative of the in vivo measurement, as Allen et al. (2004) said. With regard to the normal control, we ensured that there was no biological variation because the RNA used was the same in every reaction: a mix of RNA samples obtained from different blood extractions of the same volunteer, who was a normal woman homozygous for 29 repeats. After confirming differences between the three groups of females, our results showed that total mRNA levels are significantly elevated in females with CGG expansions in the premutation range and that these levels increase with increasing repeat size. These observations coincide with previous studies (Tassone et al. 2000b; Allen et al. 2004), showing a higher variation in mRNA levels compared with those obtained in males, due likely to the influence of the X inactivation ratio.
Furthermore, the modest trend obtained from the lowess curve in the scatter plot (Fig. 1A) suggested to us that the assumption of a constant linear trend confirmed by Allen et al. (2004) along the whole range of CGG repeats could not be adequate. Our findings seem to corroborate the presence of a threshold, above which the relationship of mRNA levels with CGG repeats is different. This molecular phenomenon could be very important, in that it would support the nonlinear relationship between CGG repeat number and the risk of having POF recently published (Sullivan et al. 2005; Ennis et al. 2006).
Following these points, we considered it very important to use an approach that could capture these changes in trend with a low degree of arbitrariness. With this aim, we have developed a novel methodology that, based on the size of the CGG repeats, allows us to find out the most probable threshold from which the relationship between CGG repeat number and mRNA levels changes. This procedure consists of fitting a robust segmented linear regression model using the cutoff points suggested by the lowess curve and has the advantage of allowing different slopes within each segment of the model. Doing so, we avoid: (1) the stiff structure of the linear model used in this context by other authors (Tassone et al. 2000c; Allen et al. 2004), which does not allow for different increasing rates depending on the repeat group; and (2) the risk of using different regression models for each of the repeat groups, which does not incorporate continuity as it assumes independent intercepts for each segment. Furthermore, the use of a robust estimation procedure instead of the classical estimation approach avoids the excessive influence that some points may have on the parameter estimates. Hence, we consider our approach useful in order to detect and evaluate threshold effects. Similar methods have also been previously used by other authors (Muggeo 2003) to take advantage of the flexibility they provide in contrast to the rigid assumptions of simple linear models. The models we use, also called piecewise regression models, may provide valuable information when the effect of a variable on the response is suspected to change, a fact that linear models ignore. They also complement the information provided by other nonlinear models such as quadratic or exponential models, as they allow for the assessment of the signification of the change.
Using this method, a significant positive correlation between CGG repeats and total mRNA levels has been found in the premutation range <100 CGG, but this correlation diminishes from 100 onward. It is noteworthy, however, that there is not much difference in the use of 90 or 100 as the cutoffs regarding the results obtained, but that a breakpoint provides a better representation and modeling of the data than a simple linear model.
However, when correcting by the X inactivation ratio, mRNA levels increase while increasing the number of CGG repeats, and this increase is highly significant over 100 CGG. To calculate these mRNA levels adjusted for the X inactivation ratio of our premutated female carriers, we followed the approach of Tassone et al. (2000a), using the X chromosome that remains active (AR), but we used the average of total mRNA for our 19 normal females instead the arbitrary number of 1 for a normal X chromosome. Although it could be considered that this quantity is not enough and could be a limitation, we decided to use normal females from the same families to rule out the bias of the normal X chromosome's repeat size. In any case, it is important to highlight the significant differences in mRNA levels corrected by AR between the low-intermediate and the high premutation range using the 100 CGG repeat number as a cutoff point. No less important are the significant differences in these levels between the normal and low-intermediate ranges, even though the correlation of CGG number of repeats and mRNA levels is not significant for this range. These results indicate that there may be a molecular abnormality in FMR1 gene expression of the alleles with the PM, which reveals altered regulation in individuals even with slightly higher repeats than the normal range. This model of FMR1 gene expression observed in our population predicts three times higher mRNA levels in females with 120 CGG than in women within the low PM range. Nevertheless, this deregulation seems to be biologically counteracted by increasing the proportion of normal active X when the number of CGG repeats increases (Fig. 1C). Skewing toward a high proportion of normal active X has also been observed in full mutation females, but in relation to age (Taylor et al. 1994). However, to our knowledge this is the first time that it is described in females with the PM. This molecular phenomenon may be very important, in that it would explain the recently published nonlinear relationship between CGG repeat number and the risk of having POF (Sullivan et al. 2005; Ennis et al. 2006), and it supports the concept of a protective effect from expression of the allele on the normal X chromosome also observed in relation with FXTAS (Berry-Kravis et al. 2005).
Finally, we wanted to mention that, throughout our research study, we have also found some signs that indicate the possible existence of an intrafamilial effect, because we observed that mRNA levels due to the carrier X chromosome were more similar among members of the same family than among unrelated females. But, due to the fact that in no family were there enough women to corroborate this observation, nor were there enough to analyze the intrafamilial versus interfamilial variation, we only mention it to attract the attention of other researchers because it would be an important topic of research for the future.
MATERIALS AND METHODS
Patients
We studied FMR1 expression in 100 female fragile X family members from the northern region of Spain. They were all diagnosed in our laboratory and belong to 40 different families. Of these 100 women, 19 had normal alleles (18–52 CGG repeats), 19 were full mutation carriers, and 62 were premutation carriers. These PM carriers belong to 31 families, 16 of them having more than one female in the premutation range. All the participants in the study were volunteers and signed an informed consent form. The Hospital Research Committee approved the protocol.
Genomic DNA analysis
Genomic DNA was isolated from 10–20 mL of peripheral blood following the “salting-out” method (Miller et al. 1998). FRAXA CGG repeats were determined by PCR analysis followed by electrophoresis in polyacrylamide gels and hybridization with a chemoluminescent oligonucleotide (CGG)n probe, as previously reported (Duran Dominguez et al. 2001). Methylation status was determined in Southern blots, hybridized with the Stb12.3 probe after digestion of DNA with EcoRI and EagI (Tejada et al. 1992), and detected with Digoxigenin and CSPD. X inactivation ratios were determined by densitometry of the normal active X chromosome in Southern blots following previously published methods (Taylor et al. 1994).
Total RNA isolation and expression level analysis
Total RNA was collected from 2.5 to 5 mL of peripheral blood in PAXgene Blood RNA tubes and isolated by the PAXgene Blood RNA Kit (Qiagen). Reverse transcription (RT) and quantitative real-time PCR were performed following exactly the method described by Tassone et al. (2000c), except for the sequence detector, since we have the 7300 Real Time PCR System (Applied Biosystems). With regard to the control reactions for FMR1 and GUS, we used a normal control female's mRNA to perform relative quantification by fluorescence quantitative PCR using the 2−ΔΔCt method (Tassone et al. 2000c). This mRNA was a mix of mRNA samples obtained from different blood extractions of the same volunteer, a normal woman homozygous for 29 repeats. It is important to note that, as was the case for Tassone's group, for each blood sample, three RNA concentrations were used and quantitative PCR reactions were performed in duplicate for each starting total RNA concentration (a total of 12 reactions). The relative abundance of FMR1 mRNA levels was also assessed by using the housekeeping gene β-glucoronidase (GUS) mRNA (GenBank accession number NM000181).
FMR1 mRNA levels due to the carrier chromosome
Following the approach of Tassone et al. (2000a) and for comparative purposes, we also derived FMR1 mRNA levels due to the carrier chromosome assuming that the total FMR1 mRNA levels observed are the sum of contributions from both normal and expanded alleles, with relative contributions determined by the X inactivation ratio. More precisely, for PM females we used the formula: mRNAobs = mRNAnormal × AR + mRNAp × (1 − AR), where: (1) mRNAobs represents the total mRNA level obtained for each sample; (2) mRNAnormal is the mean value of the normal samples (0.91 from 19 normal females in our study); (3) AR is the observed percentage of normal active X chromosomes; and (4) mRNAp is the mRNA value due to the carrier chromosome, obtained with this formula. For the normal control population, we assumed the value of 0.5 as the activation ratio for each X chromosome due to random X inactivation, and for females with the FM, we assumed that just about all mRNA observed is due to the normal X chromosome, because the X chromosome with the full mutation must be methylated.
Statistical analyses
Prior to the analyses, an outlier detection procedure was carried out based on the Grubbs’ test (Grubbs 1950). The outliers detected underwent an expert assessment, on the basis of which the decision to exclude them from the analysis was taken. We then described the observed mRNA levels, mRNA levels corrected by the influence of the AR, and the AR values using the mean and standard deviation of each of the groups considered, namely, noncarrier females (N), premutation carrier females (PM), and fully mutated females (FM) (Table 1). To compare the mean levels of these variables between the groups, we used ANOVA, t-test, Kruskal–Wallis, or Mann–Whitney tests depending on the normality assumption and the number of groups.
To analyze the relationship between the number of CGG repeats and the total mRNA levels, as well as the AR-corrected levels in female premutation carriers, we present here a two-step procedure that allows for different relationships between CGG repeats and RNA levels depending on the CGG range. In the first step, a lowess curve is fitted to the data, which is a flexible model representing the relationship between CGG repeats and RNA levels based only on the neighborhood points, in contrast to the regression models that use all points (Fig. 1A,B). Hence, it allows for detection of changes in the pattern of the relationship. Then, in the second step, and using the lowess curve to decide which are the most appropriate cutoff points, we fit a robust segmented linear regression model that allows for different slopes within each segment of the model (the repeat groups), and then we evaluate the significance of each of the slopes (Fig. 1C,D). These models (using different cutoff points), together with the linear model, were compared in terms of goodness of fit, significance of the slopes, and also using the SSE obtained from the discrepancies between each model and the lowess curve: SSE = ∑(predicted − lowess)2 (the lower the value, the better). The most suitable model was used to classify individuals, and the mean mRNA levels of these groups were then compared using appropriate statistical tests.
All the statistical analyses were performed using Statistical Package R, and the codes to carry them out are available upon request.
ACKNOWLEDGMENTS
We thank the female subjects whose participation made this work possible. We also thank Paul Hagerman and Flora Tassone for training us in the methodology and to Noelia Viguera for her technical assistance. This work was supported by FIS (ISCIII) grant No. PI02/0812.
Footnotes
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.206307.
REFERENCES
- Allen, E.G., He, W., Yadav-Shah, M., Sherman, S.L. A study of the distributional characteristics of FMR1 transcript levels in 238 individuals. Hum. Genet. 2004;114:439–447. doi: 10.1007/s00439-004-1086-x. [DOI] [PubMed] [Google Scholar]
- Berry-Kravis, E., Potanos, K., Weinberg, D., Zhou, L., Goetz, G. Fragile X-associated tremor/ataxia syndrome in sisters related to X inactivation. Ann. Neurol. 2005;57:144–147. doi: 10.1002/ana.20360. [DOI] [PubMed] [Google Scholar]
- Crawford, D.C., Acuna, J.M., Sherman, S.L. FMR1 and the fragile X syndrome: Human genome epidemiology review. Genet. Med. 2001;3:359–371. doi: 10.1097/00125817-200109000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran Dominguez, M., Molina Carrillo, M., Fernandez Toral, J., Martinez Merino, T., Lopez Aristegui, M.A., Alvarez Retuerto, A.I., Onaindia Urquijo, M.L., Tejada Minguez, M.I. Diagnóstico molecular por reacción en cadena de la polimerasa del Síndrome X frágil: Aplicación de un protocolo diagnóstico en 50 familias del norte de España. An. Esp. Pediatr. 2001;54:331–339. [PubMed] [Google Scholar]
- Ennis, S., Ward, D., Murray, A. Nonlinear association between CGG repeat number and age of menopause in FMR1 premutation carriers. Eur. J. Hum. Genet. 2006;14:253–255. doi: 10.1038/sj.ejhg.5201510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y.H., Kuhl, D.P., Pizzuti, A., Pieretti, M., Sutcliffe, J.S., Richards, S., Verkerk, A.J., Holden, J.J., Fenwick R.G., Jr, Warren, S.T., et al. Variation of the CGG repeat at the fragile X site results in genetic instability: Resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- Grubbs, F.E. Sample criteria for testing outlying observations. Ann. Math. Stat. 1950;21:27–58. [Google Scholar]
- Hagerman, R.J., Leehey, M., Heinrichs, W., Tassone, F., Wilson, R., Hills, J., Grigsby, J., Gage, B., Hagerman, P.J. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology. 2001;57:127–130. doi: 10.1212/wnl.57.1.127. [DOI] [PubMed] [Google Scholar]
- Hagerman, P.J., Greco, C.M., Hagerman, R.J. A cerebellar tremor/ataxia syndrome among fragile X premutation carriers. Cytogenet. Genome Res. 2003;100:206–212. doi: 10.1159/000072856. [DOI] [PubMed] [Google Scholar]
- Jacquemont, S., Hagerman, R.J., Leehey, M., Grigsby, J., Zhang, L., Brunberg, J.A., Greco, C., Des Portes, V., Jardini, T., Levine, R., et al. Fragile X premutation tremor/ataxia syndrome: Molecular, clinical, and neuroimaging correlates. Am. J. Hum. Genet. 2003;72:869–878. doi: 10.1086/374321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenneson, A., Zhang, F., Hagedorn, C.H., Warren, S.T. Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number intermediate-length and premutation carriers. Hum. Mol. Genet. 2001;10:1449–1454. doi: 10.1093/hmg/10.14.1449. [DOI] [PubMed] [Google Scholar]
- Miller, S.A., Dykes, D.D., Polesky, H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1998;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muggeo, V.M.R. Estimating regression models with unknown break-points. Stat. Med. 2003;22:3055–3071. doi: 10.1002/sim.1545. [DOI] [PubMed] [Google Scholar]
- Sherman, S.L. Premature ovarian failure in the fragile X syndrome. Am. J. Med. Genet. 2000;97:189–194. doi: 10.1002/1096-8628(200023)97:3<189::AID-AJMG1036>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Sullivan, A.K., Marcus, M., Epstein, M.P., Allen, E.G., Anido, A.E., Paquin, J.J., Yadav-Shah, M., Sherman, S.L. Association of FMRP1 repeat size with ovarian dysfunction. Hum. Reprod. 2005;20:402–412. doi: 10.1093/humrep/deh635. [DOI] [PubMed] [Google Scholar]
- Tassone, F., Hagerman, R.J., Chamberlain, W.D., Hagerman, P.J. Transcription of the FMR1 gene in individuals with fragile X syndrome. Am. J. Med. Genet. 2000a;97:195–203. doi: 10.1002/1096-8628(200023)97:3<195::AID-AJMG1037>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Tassone, F., Hagerman, R.J., Loesch, D.Z., Lachiewicz, A., Taylor, A.K., Hagerman, P.J. Fragile X males with unmethylated, full mutation trinucleotide repeat expansions have elevated levels of FMR1 messenger RNA. Am. J. Med. Genet. 2000b;94:232–236. doi: 10.1002/1096-8628(20000918)94:3<232::aid-ajmg9>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Tassone, F., Hagerman, R.J., Taylor, A.K., Gane, L.W., Godfrey, T.E., Hagerman, P.J. Elevated levels of FMR1 mRNA in carrier males: A new mechanism of involvement in the fragile-X syndrome. Am. J. Hum. Genet. 2000c;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone, F., Hagerman, R.J., Taylor, A.K., Hagerman, P.J. A majority of fragile X males with methylated full mutation alleles have significant levels of FMR1 messenger RNA. J. Med. Genet. 2001;38:453–456. doi: 10.1136/jmg.38.7.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, A.K., Safanda, J.F., Fall, M.Z., Quince, C., Lang, K.A., Hull, C.E., Carpenter, I., Staley, L.W., Hagerman, R.J. Molecular predictors of cognitive involvement in female carriers of fragile X syndrome. JAMA. 1994;271:552–553. [PubMed] [Google Scholar]
- Tejada, I., Mornet, E., Biancalana, V., Oberlé, I., Boué, J., Mandel, J.-L., Boué, A. Direct DNA analysis of fragile X syndrome in Spanish pedigrees. Am. J. Med. Genet. 1992;43:282–290. doi: 10.1002/ajmg.1320430144. [DOI] [PubMed] [Google Scholar]