Abstract
Association of alternative splicing (AS) with accelerated rates of exon evolution in some organisms has recently aroused widespread interest in its role in evolution of eukaryotic gene structure. Previous studies were limited to analysis of exon creation or lost events in mouse and/or human only. Our multigenome approach provides a way for (1) distinguishing creation and loss events on the large scale; (2) uncovering details of the evolutionary mechanisms involved; (3) estimating the corresponding rates over a wide range of evolutionary times and organisms; and (4) assessing the impact of AS on those evolutionary rates. We use previously unpublished independent analyses of alternative splicing in five species (human, mouse, dog, cow, and zebrafish) from the ASAP database combined with genomewide multiple alignment of 17 genomes to analyze exon creation and loss of both constitutively and alternatively spliced exons in mammals, fish, and birds. Our analysis provides a comprehensive database of exon creation and loss events over 360 million years of vertebrate evolution, including tens of thousands of alternative and constitutive exons. We find that exon inclusion level is inversely related to the rate of exon creation. In addition, we provide a detailed in-depth analysis of mechanisms of exon creation and loss, which suggests that a large fraction of nonrepetitive created exons are results of ab initio creation from purely intronic sequences. Our data indicate an important role for alternative splicing in creation of new exons and provide a useful novel database resource for future genome evolution research.
Keywords: alternative splicing, divergent exons, exon creation and loss rates, genes
INTRODUCTION
Evolution can generate new functions either by creating new genes (typically through duplication) or by introducing new functional elements into existing genes, for example, by creation of new exons. Recently, there has been widespread interest in the role of alternative splicing (AS) in evolution of eukaryotic gene structure because of evidence that alternative splicing is associated with accelerated rates of exon evolution in some organisms (Modrek and Lee 2003; Nurtdinov et al. 2003; Xing and Lee 2006). A number of groups have reported that alternatively spliced exons are more frequently divergent in comparisons of human versus mouse genomes (e.g., not conserved between these genomes) than are constitutive exons. These data suggest an important role for alternative splicing in driving exon creation. For example, one estimate reported that 87% of newly created exons in mouse were found to be alternatively spliced (versus only 13% constitutive) (Wang et al. 2005), suggesting that the majority of exon creation events may be associated with alternative splicing. This interesting hypothesis merits further investigation.
One important question that recent studies have begun to address is the problem of distinguishing exon creation versus exon loss events (Kondrashov and Koonin 2003; Wang et al. 2005). If an exon is present in one organism A but absent in another organism B, this difference could have been caused by either creation of the exon during evolution of organism A (from the most recent common ancestor [MRCA] of the two species), or loss of the exon during evolution of organism B. The only way to distinguish between these two possibilities is to establish the ancestral state of the MRCA. To do so, we can look at the conservation of the same exon in a third, more distantly related organism C, called an outgroup. If the exon is present in the outgoup, then by parsimony it must have been present in the MRCA as well (Fig. 1A), implying an exon loss event in organism B. Conversely, if the exon is absent in the outgroup, this implies an exon creation event in organism A (Fig. 1B). Thus one important goal in the field is to analyze patterns of exon creation and loss using outgroup analysis. One study has identified evidence for 25 exon creation events and 48 exon deletions, based on comparisons of alternative isoforms in human versus other vertebrates, using prokaryotic and yeast protein sequences as outgroups (Kondrashov and Koonin 2003). Another outgroup study has estimated exon creation rates in mouse versus rat (Wang et al. 2005). More recently, an analysis of eight vertebrates has estimated rates of exon creation in human and mouse (Zhang and Chasin 2006).
FIGURE 1.
Outgroup method for distinguishing between creation and loss. (A) Exon is present in the source and outgroup organism, which implies that the ancestral state is also present. This means that the exon was lost from the target organism after the split with source. (B) Exon is not present in either the target or outgroup organism; therefore, it must have been created after the split of source and target.
A second important question concerns the detailed evolutionary mechanisms for exon creation and loss. A number of plausible mechanisms that contribute to alternative splicing divergence have been reported. For example, it has been shown that some divergences are due to tandem exon duplication (Kondrashov and Koonin 2001), whereby a constitutive exon is duplicated and is converted into a mutually exclusive alternative exon. Alu exonization (Sorek et al. 2002) and other repetitive elements (Zhang and Chasin 2006) appears to be a second major mechanism. A number of systematic studies have shown that Alu repeat elements can easily mutate to create a new alternative splice site (Lev-Maor et al. 2003; Sorek et al. 2004). This mechanism can be generalized: Single nucleotide mutations can destroy or create splice sites within any intronic sequence, leading to exon loss or ab initio creation from purely intronic sequence (Modrek and Lee 2003). This last mechanism is intriguing because it opens the possibility for recruiting totally new sequences into the transcriptome and proteome.
Third, it would be interesting to measure the specific rates of exon creation and loss over time and their total impact over the course of vertebrate evolution. Is AS-associated exon creation a recent process (e.g., only in mammals), or is it a long-term pattern in vertebrate evolution? Currently, data are mainly available for mammals. For example, one study (Wang et al. 2005) has estimated that the new exon creation rate is as high as 2.71 × 10−3 in rodents. Using new genome data, it should be possible to extend such an analysis over several branches of vertebrate evolution (e.g., primates, rodents, birds, and fish) and much further back in evolutionary time. Analyses of both exon creation and loss have never been attempted in a unified framework and on such a global multigenome scale.
Fourth, is it possible to distinguish different types of alternatively spliced exons based on their evolutionary history? Several studies have reported subtypes of alternative exons that appear to have markedly different evolutionary histories. Based on EST data, Modrek measured exon inclusion levels (what fraction of a gene's transcripts include a particular exon) and showed that inclusion levels are strongly conserved between independent human versus mouse expression data sets (Modrek and Lee 2003). Moreover, major-form exons (defined as exons with inclusion level >2/3) showed low exon divergence (due to either exon creation or loss) rates, similar to constitutive exons, whereas minor-form exons (defined as exons with inclusion level <1/3) showed exon divergence rates that were many-fold higher. The same result has recently been obtained for both mouse and human (Zhang and Chasin 2006). Independent microarray data for mouse have shown a similar result (Pan et al. 2004; Pan 2005), and suggest that minor-form exons may represent tissue-specific alternative splicing events (Xing and Lee 2005). Thus it would be interesting to assess the impact of exon inclusion level on exon creation and loss rates over long-term vertebrate evolution and to ask when minor-form exons have been created.
In this article, we present a large-scale analysis of exon creation and loss in 17 complete genomes, over 360 million years of vertebrate evolution. This study brings together a variety of data: (1) independent analyses of alternative splicing in five source species; (2) analysis of exon creation and loss of both constitutive and alternatively spliced exons in mammals, fish, and birds, using outgroup analysis and genomewide multiple alignment of the 17 genomes; (3) the effects of exon inclusion level on exon creation and loss rates; and (4) detailed analysis of mechanisms by global dynamic programming alignment of introns in a separate study of mammalian genomes. Our analyses are based on well-characterized, widely used data sources: the University of California at Santa Cruz (UCSC) multigenome alignments (specifically, the 17-way conservation track) and the ASAP alternative splicing database (Modrek et al. 2001). These results provide a detailed, genomewide picture of exon creation and loss processes measured at 13 time points spanning the last 4 million to 360 million years of vertebrate evolution. These data along with the Supplemental materials for this article are available at the VEEDB Web site (http://www.bioinformatics.ucla.edu/VEEDB).
RESULTS
Global alignment analyses of splice site conservation versus exon conservation in mammals
Splice site conservation is a widely used criterion in comparative genomics for functional exon conservation (i.e., whether a sequence region orthologous to a known exon is likely to be also expressed as an exon in spliced transcripts). Given the poor EST coverage for many organisms of interest, EST data would have far too high a false negative rate, especially for alternative exons that are only expressed in certain tissues or in a minor fraction of a gene's transcripts. For this reason, splice site conservation has been widely adopted as a criterion for exon conservation (see, e.g., Ovcharenko et al. 2004; Hsieh et al. 2006): If its GT/AG splice sites are conserved, the exon is scored as conserved (in the absence of a valid splice site, the sequence is unlikely to be expressed as an exon).
To test this criterion empirically, we have compared splice site conservation against exon conservation, using detailed genomic sequence alignments. We performed this analysis on a set of internal exons, by matching their flanking exons to the target genome and generating full dynamic programming global alignment of the regions bounded by the flanking exons in the source and target genomes (see Materials and Methods for details). Whereas BLAST may lack adequate sensitivity to reliably detect intron homologies, the flanking exons’ alignment forces global alignment to align whatever regions of homology are present between them, even if individually they are too weak to be detected by BLAST. Global alignment has been shown to produce superior quality intron alignment (Pollard et al. 2004). Since intronic sequence accumulates mutations rapidly, accurate intron alignment is only possible for recently diverged genomes. We therefore focused on mammalian genomes: human versus chimp (average intron identity 97%, using mouse as an outgroup); and mouse versus rat (average intron identity 86%, using human as an outgroup). This analysis produced a data set containing a total of 167 alternative and 1999 constitutive human exons and 207 alternative and 4546 constitutive mouse exons (Table 1).
TABLE 1.
Summary for exons included in in-depth study
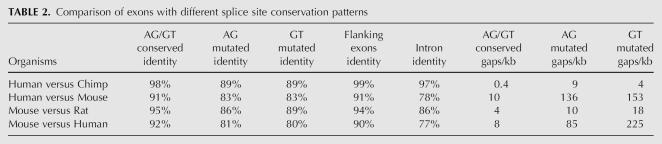
Using these data, we compared splice site conservation against two different metrics of exon conservation: percent sequence identity (much higher in exons than in introns) and the frequency of insertions and deletions (indels; much lower in exons than in introns). The sequence alignments divided into two clearly distinct groups (Table 2). In the first group, both splice sites (GT, AG) were conserved, the level of sequence percent identity was similar to that in the flanking constitutive exons, and the frequency of indels was very low (0.4/kb human versus chimp; 4/kb mouse versus rat) (Fig. 2A). In the second group, one or both of the splice sites were mutated (splice site motif not conserved), the level of sequence percent identity was similar to that of the surrounding intron, and the frequency of indels was high (4–10/kb human versus chimp; 10–18/kb mouse versus rat), again like the surrounding intron (Fig. 2B). We observed the same patterns consistently in all four genome–genome comparisons. Thus, both of the independent conservation metrics indicate that if the splice sites are conserved, the sequence region displays conservation patterns characteristic of an exon, whereas if either splice site is mutated, it becomes intronic in its characteristics. These results validate previous studies’ use of splice site conservation as a marker of exon conservation, and we therefore also used this criterion throughout this study. We will comment on possible shortcomings of this criterion in the Discussion.
TABLE 2.
Comparison of exons with different splice site conservation patterns
FIGURE 2.
Typical alignments of exons to the flanked region of other organisms. In A, a skipped exon of the cdc37l gene provides an example of a well-conserved class of exons showing very good sequence conservation (mismatches underlined) with no insertions and the AG/GT splice signals (bold) are conserved, while in B, a skipped exon of the dnajc7 gene shows an alignment typical of the second class, with conservation similar to the surrounding intron including a large insertion (italic) and splice-sites mutated.
Large-scale analysis of exon creation and loss over 17 vertebrate genomes
To analyze exon creation over a wide range of evolutionary timescales, we have taken advantage of massive data sets from recent comparative genomics studies of vertebrate genomes. Haussler and colleagues have generated whole-genome multiple alignments of 17 vertebrate genomes, including mammals (e.g., human, mouse, dog, and cow), birds (e.g., chicken), and fish (e.g., zebrafish) (Blanchette et al. 2004). We therefore used these data to analyze the conservation of splice sites for AS exons and constitutive exons. The UCSC alignments are well characterized and widely used for such applications (see, e.g., Siepel et al. 2005; Washietl et al. 2005; Zhang and Chasin 2006).
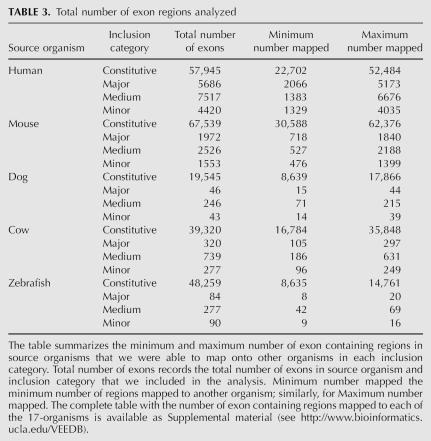
We have analyzed gene structure and alternative splicing via genomewide studies in five organisms and performed outgroup analysis over 17 vertebrate genomes to construct the Vertebrate Exon Evolution Database (VEEDB), containing 258,404 exons. This genomewide database identifies exon creation and exon loss events during vertebrate evolution of primate, rodent, other mammalian, and fish exons (Table 3). VEEDB actually consists of five parallel studies, each one based on a complete exon data set from a given organism's EST data: human, mouse, dog, cow, and zebrafish. For each study, the exons were compared against each of the other genomes—starting from its closest relative and moving to progressively more distant relatives—to detect exon creation versus loss events with respect to an outgroup genome. For example, when comparing human exons versus the chicken genome, the frog genome was used as the outgroup (see Supplemental Materials for the full list of the source, target, and outgroup triples of genomes used: http://www.bioinformatics.ucla.edu/VEEDB). VEEDB provides detailed information about (1) gene, gene structure, and splicing; (2) evidence of alternative splicing in different organisms and tissues; (3) when a specific exon creation or loss event occurred during vertebrate evolution, based on outgroup analysis; and (4) detailed multigenome alignment of the exon. VEEDB contains independent analysis of alternative splicing from EST and mRNA data in 15 different organisms, including human, mouse, cow, dog, zebrafish, and others, and thus has many potential future applications, including comparison of alternative splicing patterns between different vertebrate species.
TABLE 3.
Total number of exon regions analyzed
Exon creation and loss trends over vertebrate evolution
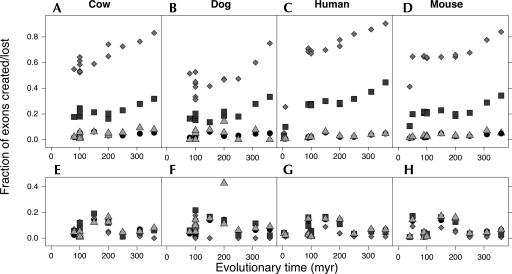
VEEDB provides a detailed time course for exon creation and loss over vertebrate evolution, with time points approximately every 50 million years (my) (Fig. 3). These data show a progressive process of exon creation of minor-form AS exons, whose slope appears to be significantly higher during mammalian evolution (last 100 my), compared with the previous 250 my of vertebrate evolution. Independent AS data sets from different species (cow, dog, human, and mouse; each generated from expression data exclusively from that species) showed the same trend. This pattern was corroborated by the data for medium-form exons, which also showed a steep increase in exon creation during mammalian evolution. In human, for instance, the fraction of created exons rapidly rose from 7% (5 my) to 33% (100 my), followed by a slow increase to 49% over the period 100 my–360 my ago. Major-form exons and constitutive exons, in contrast, did not show significant growth in exon creation over this period.
FIGURE 3.
Time course of exon creation and loss. The fraction of minor (light diamonds), medium (dark squares), major (light triangles), and constitutive (dark circles) created in cow (A), dog (B), human (C), and mouse (D) or lost (E–H, respectively) within the evolutionary distance. The amount of loss and creation in constitutive and major-form exons is the same, indicated by strong overlap of points on the graph. In the alternative set, the fraction of created exons is anticorrelated with the inclusion level. The amount of loss is similar for all times and inclusion categories. Similar trends are observed in data from different source organisms.
A strong association between the exon-inclusion level (frequency of alternative splicing of a given exon) and the exon-creation rate was observed consistently in all species and at all timescales (Fig. 3A–D). In general, major-form exons behaved like constitutive exons, both with very low exon creation rates (see also Supplemental materials: http://www.bioinformatics.ucla.edu/VEEDB). In contrast, medium-form and minor-form AS exons displayed dramatically higher exon creation rates. Indeed during the course of vertebrate evolution, these two categories represent almost opposite evolutionary histories: Whereas >90% of major-form and constitutive exons are older than 360 my, it appears that ∼90% of existing minor-form exons were created in the last 360 my.
We performed regression analysis to test the statistical significance of these trends (see Supplemental materials: http://www.bioinformatics.ucla.edu/VEEDB). We have designed this analysis to account for two sources of variation: (1) rate variations associated with different exon inclusion levels and (2) different forms of time-dependent autocorrelation, which may affect the overall predictions of the model (see Supplemental materials: http://www.bioinformatics.ucla.edu/VEEDB). The results of this analysis were completely consistent with our previous conclusions. These data showed that exon creation rates were different for alternative exons with different inclusion levels, and these differences were statistically significant (p-value <10−4). The exon creation rate was highest for minor-form exons and lowest for constitutive and major-form exons. Our analysis indicated no significant difference between creation rates in major-form and constitutive exons. The fraction of lost exons appeared to be relatively constant over all timescales and exon types. The amount of loss (Fig. 3E–H) never exceeded 20% and was consistent with Figure 4 (see below) at the same timescales. Our analysis showed no association of the loss rate with exon inclusion level (see Supplemental materials: http://www.bioinformatics.ucla.edu/VEEDB). Therefore, these data suggest that not only is the degree of conservation different in general between different inclusion categories, but the specific creation rate also varies accordingly, while exon loss remains approximately constant for exons in all inclusion categories.
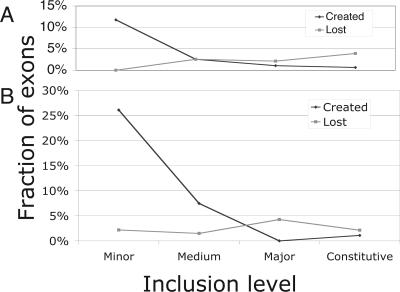
FIGURE 4.
Fraction of exons created and lost by inclusion category. The fraction of exons created (dark diamonds) or lost (light squares) after the split between (A) human–chimp and (B) mouse–rat in each of the four inclusion categories. The amount of loss is almost constant within time (5 my [A], 40 my [B]) and inclusion category. The amount of creation is anticorrelated with the inclusion level.
Global alignment analyses of exon creation and loss in mammals
To validate these results, and to investigate the detailed mechanism of exon creation, we analyzed exon creation and loss using full dynamic programming global alignment of introns (see Materials and Methods for details). Distinguishing different mechanisms (such as single nucleotide mutation versus exon duplication or insertion of exogenous sequences) requires detailed alignment across the whole intron (up to the flanking constitutive exons) in the two genomes being compared and the outgroup genome. Since intronic sequence accumulates mutations rapidly, accurate intron alignment is only possible for recently diverged genomes, so again we focused on mammalian genome evolution. Since we first identified the homologous intron and only then establish exon conservation, we effectively control for the presence of the gene in the organism in question, which allows us to filter out gene creation and loss effects from the analysis. Because repeat elements (such as Alu) are well known to cause unusually high rates of exon creation (Sorek et al. 2002, 2004; Lev-Maor et al. 2003), we excluded exons containing known repeats from this analysis; the filtered data set statistics are summarized in Table 1.
Global alignment analyses of exon creation and loss trends
We analyzed the outgroup sequences in these intron alignments to distinguish exon creation versus loss events (Fig. 4). In rodents (mouse versus rat; 40 my divergence), exon creation rates were low for constitutive and major-form exons (1%) but increased dramatically for medium-form (7%) and minor-form (26%) AS exons. Exon loss rates appeared to be constant over all categories, at ∼2%–4%. In primates (human versus chimp; 4 my divergence), exon creation rates were lower but followed a similar pattern: 0.5%–1% for constitutive and major-form exons and higher in medium-form (2.5%) and minor-form AS exons (12%). Exon loss rates appeared to be similar (2%–4%) to those observed in the mouse versus rat comparison.
These data corroborate the results of our large-scale studies. First, the VEEDB mouse versus rat results (Fig. 3D; 40 my time point) agree with our intron alignment studies (exon creation rates highest for minor-form > medium-form > major-form, constitutive), despite the fact that they used a different alternative splicing data set, genome sequence data, and completely different alignment method. Similarly, the VEEDB human versus chimp results (Fig. 3C; 5 my time point) are similar to our intron alignment results, again using a new AS data set and alignment data set.
Global alignment analyses of exon creation mechanisms in mammals
We next analyzed possible mechanisms of exon creation and loss, based on the detailed intron alignment. In the vast majority of cases (>92%), alternative exons were found to be aligned to a homologous region in the corresponding intron. Indeed, even in cases where the splice sites GT/AG were not conserved, we found that the intron showed aligned homology with the whole exon in 90% of cases for alternative exons and 88% of cases for constitutive exons (see Materials and Methods for details). Because these sequences still showed clear homology with the AS exon, they are consistent with a mechanism of single nucleotide mutation (creating or destroying a valid splice site) rather than a mechanism of inserting a functional exon sequence from elsewhere in the genome. These data suggest that exon creation events were primarily derived from intron sequences via mutation (introducing new splice sites). In < 8% of cases, the lack of alignment of the exon versus any portion of the intronic sequence suggests a translocation event (i.e., insertion or deletion of an entire exon).
Large-scale analysis of recent exon creation: Species-specific exons
Since the role of repeat elements in exon creation has been analyzed thoroughly by previous studies (Sorek et al. 2002, 2004; Lev-Maor et al. 2003; Zhang and Chasin 2006), we did not include repeats in our primary analysis. However, we have examined one specific category: their role in the most recent exon creation events. Specifically, we screened species-specific alternative exons (i.e., exons that are present only in one source organism) for the presence of repetitive sequences using RepeatMasker (Table 4). We found that up to 45% of these exons are associated with known repeats. Moreover, an additional 26% of human-specific exons have homologous sequences in other areas of the human genome, suggesting that they are either novel repeat sequences or duplicated regions. Therefore, up to 71% of human-specific exons are repeats (including Alu) or duplications. A large fraction of the rest of the exons shows significant sequence conservation, suggesting that they arose via mutations creating new splice sites.
TABLE 4.
Results for genomewide searches of created alternative exons
DISCUSSION
These analyses provide a large expansion in the available data set of exon creation and loss events during vertebrate evolution and further understanding of the role of alternative splicing in these processes. By performing computationally intensive global alignment of introns in primates and rodents, this study found clear evidence that the majority of exon creation events in nonrepetitive sequences were caused by mutations introducing new splice sites (as opposed to exon duplication), resulting in recruitment of formerly intronic sequence. Our genomewide studies of 17 genomes extending over 360 million years of vertebrate evolution also provide evidence of recruitment of formerly intronic sequences by mutations creating splice sites in nonduplicated and nonrepetitive regions. This complements our current knowledge of mechanisms involved in exon creation, which also include duplication (Kondrashov and Koonin 2001) and mutations within repetitive elements (Sorek et al. 2002, 2004; Lev-Maor et al. 2003). Our analysis also indicates that exon creation has been strongly associated with alternative splicing not only during mammalian evolution (Kondrashov and Koonin 2001, 2003; Sorek et al. 2002; Modrek and Lee 2003; Nurtdinov et al. 2003; Wang et al. 2005), but during previous ages of vertebrate evolution as well, which is also supported by previous analysis of eight vertebrate genomes (Zhang and Chasin 2006).
We have constructed the Vertebrate Exon Evolution Database (VEEDB) to make these results available (online at http://www.bioinformatics.ucla.edu/VEEDB). VEEDB provides extensive information (gene, gene structure, splicing, alternative splicing analyses in multiple vertebrate genomes, and comparative genomics analysis of splice site conservation and exon sequence conservation) for exon creation and loss events in human, mouse, dog, cow, and zebrafish exons. This database can be a valuable resource for other researchers interested in comparative genomics of alternative splicing or exon evolution.
Evolutionary studies based on modern genome sequences can underestimate certain types of events because they cannot accurately measure the kinetics of features whose average “lifetime” is shorter than the evolutionary branch lengths of the phylogenetic tree under study. For example, if a type of exon is lost within 5 my of its initial creation, comparison of two genomes separated by 40 my will significantly underestimate both creation and loss rates for this type of exon. It seems likely that this and other comparative genomics studies of exon creation will underestimate exon creation and loss rates for this reason. The common use in comparative genomics of splice site conservation as a criterion for exon conservation (see, e.g., Ovcharenko et al. 2004; Hsieh et al. 2006) is another potential source of false negatives (e.g., a GT→GC mutation may still function as a splice site; loss of a splice site might be compensated by a nearby cryptic splice site) and false positives (other mutations might inactivate an exon even if splice sites are conserved). Our in-depth studies of intron alignments showed that splice site conservation correlates strongly with other criteria for exon conservation, such as sequence identity and indel frequency; however, splice site conservation is clearly an imperfect criterion.
Another source of possible underestimation is the use of UCSC multigenome alignments generated by MULTIZ (Blanchette et al. 2004), since the alignment method involves sensitivity thresholds that can lead to false negatives (failure to report regions of homology that fall below the method's sensitivity threshold). Such alignment false negatives could cause underestimation of exon loss rates since failure to detect homology in the outgroup would convert a putative “exon creation” event to be categorized as an “exon loss” event. Another potential problem would be if MULTIZ aligned paralogs instead of orthologs. This method was designed to ensure “that different projects present consistent predictions of which genomic positions are orthologous,” by seeking the subset of alignments that represent genuine synteny (Blanchette et al. 2004). To validate the UCSC results, we performed an independent study of exon creation in rodents and primates using full dynamic programming global alignment of intron sequences (which does not suffer the same sensitivity threshold problem, because global alignment of the intron is constrained to find the best homology within the intron, regardless of its level of conservation). We also designed this analysis to avoid paralog errors: in cases where the flanking exons showed multiple hits in the target genome (indicating possible paralogs), we performed full dynamic programming global alignment versus all of the target hits, and only scored the exon as nonconserved if it was not conserved in all of the target hits. Overall, the results strongly corroborated the MULTIZ-based results for both exon creation and loss trends and detailed results on individual exons. In general, the alignments extracted from the UCSC 17-genome alignment and the full dynamic programming global alignment of intron sequences were similar.
Our results indicate very different evolutionary histories of minor-form alternative exons versus major-form and constitutive exons. First, these data indicate that most minor-form exons evolved during vertebrate evolution (younger than 360 my), whereas most major-form exons (and constitutive exons) are ancient (older than 400 my). It should be noted that “medium-form” exons (inclusion level >1/3 and <2/3) show a similar pattern as minor-form exons: a strong slope of increasing exon creation with increasing evolutionary time, unlike major-form and constitutive exons, which show no such increase. Second, these results confirm the apparent importance of alternative splicing (and minor-form exons in particular) to exon creation during vertebrate evolution. Fifty-six percent of “young” vertebrate exons (created <410 my ago) are alternatively spliced, whereas this number drops to 11% for “ancient” exons (older than 410 my).
These data raise intriguing questions about the function of the alternatively spliced exons that have been created during vertebrate evolution. Our data show that many of these are quite old: for minor-form exons, >20% are older than 300 my; for medium-form exons, >66% are older than 300 my. Thus (1) they clearly seem to have important biological functions and (2) they represent the bulk of new exonic material added during vertebrate evolution. What functions are associated with such minor-form exons? One recent study of microarray data from 10 mouse tissues found that one important category of conserved minor-form exons was tissue-specific exons: regulated by alternative splicing to only be included in the transcript in certain tissues (Xing and Lee 2005). It is possible that many “minor-form” exons (as identified by whole-body EST analyses) actually have tissue-specific alternative splicing and may be expressed as the major-form in just one tissue or cell type. Further microarray studies are needed, with finer resolution, to assess whether alternative splicing of such exons is regulated at the level of whole organs or at the level of individual cell types or finer cellular differentiation or activation states.
MATERIALS AND METHODS
To give an overview of the two independent analyses performed in this study (large-scale analysis of 17 vertebrate genomes using UCSC multigenome alignments; global alignment of introns using full dynamic programming), we provide a detailed dataflow schematic of our analyses (Fig. 5), which we will refer to throughout this section.
FIGURE 5.
Dataflow of the analyses. Analyses of two data sets are schematically depicted: (A) in-depth study, (B) 17-way exon conservation analysis. Roughly, the analysis consisted of the following steps. Source data collection included downloading and arranging data. In the preprocessing step, we identified the regions that should contain the exons in question by mapping the adjacent (flanking) exons onto the target genomes. Then we aligned (in A) introns using full dynamic programming and (in B) just the exons and splice sites using UCSC conservation track. Next, the alignments were scored using various metrics of conservation. The resulting conservation data sets make up the VEEDB database and were used for further outgroup analyses.
Alternative splicing data sets
We identified alternative and constitutive exons in human, mouse, dog, cow, and zebrafish as our source organisms using UniGene builds #188 Homo sapiens, #151 Mus musculus, #13 Canis familiaris, #74 Bos taurus, and #89 Danio rerio, respectively, as previously described (Modrek et al. 2001). We mapped the expressed sequence data (UniGene) to the same genome assemblies as are used in the UCSC Genome Browser 17-way conservation track: human, May 2004 (hg17); mouse, May 2004 (mm7); dog, May 2005 (canFam2); cow, Mar 2005 (bosTau2); and zebrafish, May 2005 (danRer3) (Fig. 5B, source data). The data for these splicing calculations are available at the ASAP2 website: http://bioinfo.mbi.ucla.edu/ASAP2/.
Genome alignment data sources
The estimates of divergence times of vertebrate organisms were obtained from Hedges and Kumar (2002). The phylogenetic tree for the 17 organisms was obtained from the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgTrackUi?g=multiz17way). We have also downloaded all the genomes (Fig. 5B, source data) that were used to construct this alignment database (see above link for full list).
Exon conservation scoring
One important issue we tried to distinguish in our analysis is the difference between conservation of an exon and conservation of a gene ortholog. To control for gene presence when establishing exon conservation, we limit our analysis to cases where we are able to map onto the genome of interest a pair of exons flanking (on the right and on the left) the region where the exon is located. Specifically, in our source organisms (human, dog, cow, mouse, and zebrafish) we look for all alternatively spliced exons and flanking exons that are immediately adjacent to these exons in the isoform that includes this exon and are both present in an isoform that skips the alternative exon. The left and right flanking exons define the region where we look for conservation of the alternative exon in the target and outgroup organisms in the UCSC Genome Browser Database (Hinrichs et al. 2006) 17-way conservation track (Fig. 5B, preprocessing; Blanchette et al. 2004). To map this region in other organisms we look for conservation of splice sites of the flanking exons (GT/AG) internal to the region. In order to draw inference about creation or loss, we have to require that we are able to map both left and right exons to both target and outgroup organisms.
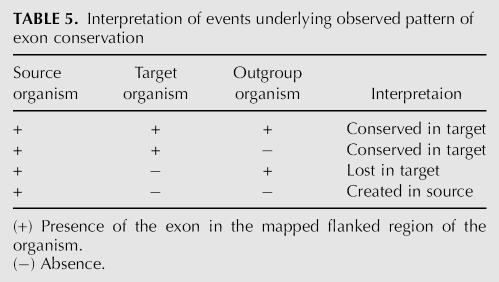
After locating putative homologous regions in target and outgroup genome we record presence (+) or absence (−) of the splice sites around the exon (see Table 5). In order to deal with possible paralogs, for a given exon we consider all possible positions in the target genome where it may be present. We call it an absence (−) only if no putative region contains evidence for the presence of the exon. Specifically, we look for conservation of GT/AG sequence around the exon sequence. Conservation of the splice site motif, as is evident from our results (Table 2), was highly predictive of conservation (% identity, number of indels) relative to surrounding exons and introns (Fig. 5, scoring). We record whether these di-nucleotides are aligned and whether they match in the target and outgroup organisms. If both conditions are met we consider the exon conserved (+); otherwise it is not conserved (−). Note that the source organism always has a “plus” as its state, since we are observing splicing of the exon in this organism in the first place. We select an outgroup that is the closest to the source and target organisms (see Supplemental Materials, http://www.bioinformatics.ucla.edu/VEEDB), in order to maximize the number of homologous regions. To compute the fraction of lost exons we divide the number of exons that have a (+,−,+) conservation pattern by the total number of number of regions that are mapped to both target and outgroup (Fig. 5, analysis). To compute the fraction created we do the same for the (+,−,−) conservation pattern.
TABLE 5.
Interpretation of events underlying observed pattern of exon conservation
Global alignment analysis using full dynamic programming
For this independent study, we used UniGene builds #160 Homo sapiens and #122 Mus musculus and genomes from the Human Genome Sequencing Consortium (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens), July 2003, and the NCBI Mouse Genome Assembly (ftp://ftp.ensembl.org/pub/current_mouse/), July 2003. We used the Rat Genome Project (ftp://ftp.hgsc.bcm.tmc.edu/pub/analysis/rat/), December 2002, and Chimp (ftp://ftp.ensembl.org/pub/current_pan_troglodytes), June 2004 (Fig. 5A, source data). The exon and alternative splicing data are available online at http://www.bioinformatics.ucla.edu/ASAP.
The goal of this analysis was to obtain detailed alignment across the whole intron (up to the flanking constitutive exons) in the two genomes being compared and the outgroup genome. We therefore used full dynamic programming global alignment (using the program POA; Lee et al. 2002) to align the entire intron as defined by the flanking exons. Full dynamic programming is a computationally intensive procedure compared with simply running a BLAST search for the exon sequence, but it is more robust than a BLAST search, which can have substantial false negative rates in introns. Due to sensitivity limitations, BLAST only reports local regions of homology that are above a certain level of score, leaving out less conserved regions, and thus it cannot be relied on for a global alignment of an intron. Full dynamic programming global alignment avoids this potential risk of losing important regions of alignment by requiring alignment of all homologies within the region bounded by the flanking exons.
Specifically, (1) the genomes in this analysis were masked for species-specific repeats using RepeatMasker and (2) for each exon, we searched for conservation of its flanking exon sequences using BLASTN with a 10−4 expectation cutoff and required conservation of the left and right exons’ GT/AG splice sites (Fig. 5A, preprocessing). In cases where multiple hits were found in the target genome, we analyzed all of them in the subsequent steps. (3) We extracted the complete sequence bounded by the flanking exons from both source and target genomes and performed full dynamic programming global alignment using POA (Lee et al. 2002; Fig. 5A, alignment). (4) We analyzed the resulting alignments by a variety of criteria including splice-site conservation, percent sequence identity, and indel frequency. In cases with multiple hits in the target genome, an exon was only scored as absent (−) if its splice sites were not conserved even in the most similar hit (based on analyzing all the hits’ global alignments).
ACKNOWLEDGMENTS
We thank Yi Xing and Barmak Modrek for useful suggestions on this article. A.V.A. was supported through a UCLA-IGERT bioinformatics traineeship (NSF DGE-998764) and by the NIH under Ruth L. Kirschstein National Research Service Award GM008185 from the National Institutes of General Medical Sciences. C.J.L. was supported through a National Institutes of Health Grant (U54-RR021813), a Department of Energy Grant (DE-FC02-02ER63421), and a Dreyfus Foundation Teacher-Scholar Award.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.325107.
REFERENCES
- Blanchette, M., Kent, W.J., Riemer, C., Elnitski, L., Smit, A.F., Roskin, K.M., Baertsch, R., Rosenbloom, K., Clawson, H., Green, E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges, S.B., Kumar, S. Genomics. Vertebrate genomes compared. Science. 2002;297:1283–1285. doi: 10.1126/science.1076231. [DOI] [PubMed] [Google Scholar]
- Hinrichs, A.S., Karolchik, D., Baertsch, R., Barber, G.P., Bejerano, G., Clawson, H., Diekhans, M., Furey, T.S., Harte, R.A., Hsu, F., et al. The UCSC Genome Browser Database: Update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, S.J., Lin, C.Y., Liu, N.H., Chow, W.Y., Tang, C.Y. GeneAlign: A coding exon prediction tool based on phylogenetical comparisons. Nucleic Acids Res. 2006;34:W280–W284. doi: 10.1093/nar/gkl307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, F.A., Koonin, E.V. Origin of alternative splicing by tandem exon duplication. Hum. Mol. Genet. 2001;10:2661–2669. doi: 10.1093/hmg/10.23.2661. [DOI] [PubMed] [Google Scholar]
- Kondrashov, F.A., Koonin, E.V. Evolution of alternative splicing: Deletions, insertions and origin of functional parts of proteins from intron sequences. Trends Genet. 2003;19:115–119. doi: 10.1016/S0168-9525(02)00029-X. [DOI] [PubMed] [Google Scholar]
- Lee, C., Grasso, C., Sharlow, M.F. Multiple sequence alignment using partial order graphs. Bioinformatics. 2002;18:452–464. doi: 10.1093/bioinformatics/18.3.452. [DOI] [PubMed] [Google Scholar]
- Lev-Maor, G., Sorek, R., Shomron, N., Ast, G. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- Modrek, B., Lee, C.J. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat. Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- Modrek, B., Resch, A., Grasso, C., Lee, C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurtdinov, R.N., Artamonova, I.I., Mironov, A.A., Gelfand, M.S. Low conservation of alternative splicing patterns in the human and mouse genomes. Hum. Mol. Genet. 2003;12:1313–1320. doi: 10.1093/hmg/ddg137. [DOI] [PubMed] [Google Scholar]
- Ovcharenko, I., Boffelli, D., Loots, G.G. eShadow: A tool for comparing closely related sequences. Genome Res. 2004;14:1191–1198. doi: 10.1101/gr.1773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, Q., Shai, O., Misquitta, C., Zhang, W., Saltzman, A.L., Mohammad, N., Babak, T., Siu, H., Hughes, T.R., Morris, Q.D., et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Pan, W. Incorporating biological information as a prior in an empirical Bayes approach to analyzing microarray data. Stat. Appl. Genet. Mol. Biol. 2005;4 doi: 10.2202/1544-6115.1124. Article12, PMID: 16646829. [DOI] [PubMed] [Google Scholar]
- Pollard, D.A., Bergman, C.M., Stoye, J., Celniker, S.E., Eisen, M.B. Benchmarking tools for the alignment of functional noncoding DNA. BMC Bioinformatics. 2004;5:6. doi: 10.1186/1471-2105-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel, A., Bejerano, G., Pedersen, J.S., Hinrichs, A.S., Hou, M., Rosenbloom, K., Clawson, H., Spieth, J., Hillier, L.W., Richards, S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek, R., Ast, G., Graur, D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorek, R., Lev-Maor, G., Reznik, M., Dagan, T., Belinky, F., Graur, D., Ast, G. Minimal conditions for exonization of intronic sequences: 5′ splice site formation in alu exons. Mol. Cell. 2004;14:221–231. doi: 10.1016/s1097-2765(04)00181-9. [DOI] [PubMed] [Google Scholar]
- Wang, W., Zheng, H., Yang, S., Yu, H., Li, J., Jiang, H., Su, J., Yang, L., Zhang, J., McDermott, J., et al. Origin and evolution of new exons in rodents. Genome Res. 2005;15:1258–1264. doi: 10.1101/gr.3929705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl, S., Hofacker, I.L., Lukasser, M., Huttenhofer, A., Stadler, P.F. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat. Biotechnol. 2005;23:1383–1390. doi: 10.1038/nbt1144. [DOI] [PubMed] [Google Scholar]
- Xing, Y., Lee, C.J. Protein modularity of alternatively spliced exons is associated with tissue-specific regulation of alternative splicing. PLoS Genet. 2005;1:e34. doi: 10.1371/journal.pgen.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing, Y., Lee, C. Alternative splicing and RNA selection pressure–evolutionary consequences for eukaryotic genomes. Nat. Rev. Genet. 2006;7:499–509. doi: 10.1038/nrg1896. [DOI] [PubMed] [Google Scholar]
- Zhang, X.H., Chasin, L.A. Comparison of multiple vertebrate genomes reveals the birth and evolution of human exons. Proc. Natl. Acad. Sci. 2006;103:13427–13432. doi: 10.1073/pnas.0603042103. [DOI] [PMC free article] [PubMed] [Google Scholar]