Abstract
The 5′ untranslated region (UTR) of the mouse hepatitis virus (MHV) genome contains cis-acting sequences necessary for transcription and replication. A consensus secondary structural model of the 5′ 140 nucleotides of the 5′ UTRs of nine coronaviruses (CoVs) derived from all three major CoV groups is presented and characterized by three major stem–loops, SL1, SL2, and SL4. NMR spectroscopy provides structural support for SL1 and SL2 in three group 2 CoVs, including MHV, BCoV, and HCoV-OC43. SL2 is conserved in all CoVs, typically containing a pentaloop (C47-U48-U49-G50-U51 in MHV) stacked on a 5 base-pair stem, with some sequences containing an additional U 3′ to U51; SL2 therefore possesses sequence features consistent with a U-turn-like conformation. The imino protons of U48 in the wild-type RNA, and G48 in the U48G SL2 mutant RNA, are significantly protected from exchange with solvent, consistent with a hydrogen bonding interaction critical to the hairpin loop architecture. SL2 is required for MHV replication; MHV genomes containing point substitutions predicted to perturb the SL2 structure (U48C, U48A) were not viable, while those that maintain the structure (U48G and U49A) were viable. The U48C MHV mutant supports both positive- and negative-sense genome-sized RNA synthesis, but fails to direct the synthesis of positive- or negative-sense subgenomic RNAs. These data support the existence of the SL2 in our models, and further suggest a critical role in coronavirus replication.
Keywords: coronavirus replication, U-turn motif, SARS, MHV, 5′ untranslated region
INTRODUCTION
Mouse hepatitis virus (MHV), a member of the family Coronaviradae, contains a positive-sense, single-stranded RNA genome 32 kb long. MHV infects cells via MHV-specific receptors (Dveksler et al. 1991; Williams et al. 1991), or in a receptor-independent fashion (Nakagaki et al. 2005). After infection and uncoating, MHV releases its genomic RNA into the cytoplasm and the viral genomic RNA serves directly as a messenger RNA directing the synthesis of two large polyproteins, ORF1a and ORF1ab (Leibowitz et al. 1982; Baker et al. 1989); the latter polypeptide is synthesized by a −1 ribosomal frameshift mechanism (Denison and Perlman 1986, 1987; Brierley and Dos Ramos 2006). The resulting 740 kDa polypeptide contains a conserved array of functional domains, which upon proteolytic processing, results in 16 nonstructural proteins, the majority of which are thought to be required for RNA synthesis (Snijder et al. 2003; Masters 2006).
MHV infected cells contain 7–8 plus sense RNA species, with the longest RNA being the intracellular counterpart of the virion RNA (Lai et al. 1981; Leibowitz et al. 1981; Spaan et al. 1981; Wege et al. 1981; Weiss and Leibowitz 1981). The subgenomic RNAs form a nested set with common 3′ ends. The 5′ ends of the subgenomic RNAs contain a 72 nucleotide (nt) leader sequence identical to that present in the 5′ end of the genome (Spaan et al. 1982, 1983; Lai et al. 1984). The genomic RNA serves as a template for the synthesis of full-length and subgenomic negative-sense RNAs, the latter through a discontinuous transcription mechanism (Sawicki and Sawicki 1990, 1998; Zuniga et al. 2004; Sola et al. 2005). In turn, these negative-strand RNAs serve as templates for the synthesis of genomic RNA and subgenomic messenger RNAs.
Many cis-acting sequences required for coronaviruses transcription and replication have been defined by studying defective interfering (DI) RNAs. Coronavirus DI RNAs are extensively deleted genomic remnants that replicate by using the RNA synthesis machinery of a helper virus, often interfering with viral genomic RNA replication. Cis-acting sequence elements for virus transcription and replication have been defined for several coronaviruses such as MHV (Kim et al. 1993; Lin and Lai 1993; Liao and Lai 1994; Hsue and Masters 1997; Hsue et al. 2000; Liu et al. 2001), bovine coronavirus (BCoV) (Chang et al. 1994, 1996; Brian and Spaan 1997; Williams et al. 1999; Raman et al. 2003; Raman and Brian 2005), porcine transmissible gastroenteritis virus (TGEV) (Mendez et al. 1996), and infectious bronchitis virus (IBV) (Penzes et al. 1996). Kim et al. (1993) demonstrated that ∼470 nt at the 5′ terminus of MHV, 469 nt at the 3′ terminus, and ∼135 nt in an internal position ∼0.9 kb from the 5′ end of DI RNA were necessary for replication of an MHV-JHM DI RNA. The requirement for the internal sequence element is specific to MHV-JHM DI RNAs (Koetzner et al. 1992; Chang et al. 1994). Lin and Lai (1993) demonstrated that 859 nt from the 5′ end and 436 nt from the 3′ end of the MHV RNA genome were necessary for DI RNA replication. More recent studies have shown that the MHV 3′ UTR, containing 301 nt, plus the poly(A) tail provide all of the 3′ cis-acting signals needed for viral replication (de Haan et al. 2002; Goebel et al. 2004).
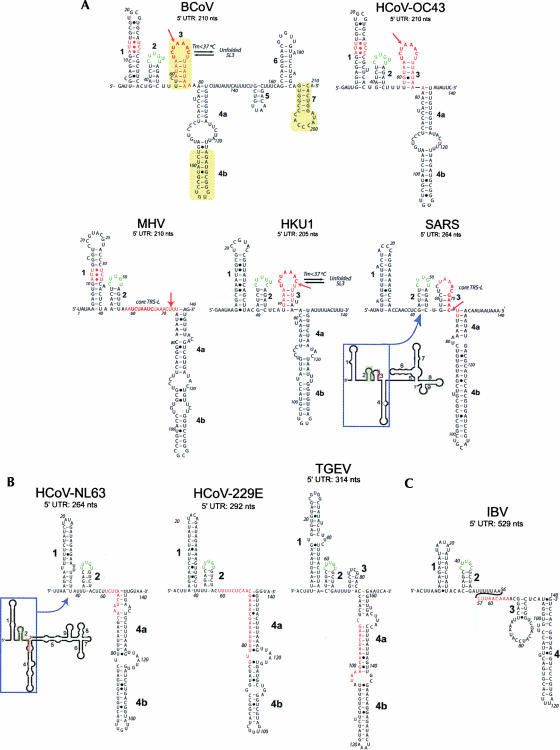
Although secondary structure models of the 3′ UTRs of group 2 coronaviruses are available and well supported by a variety of functional data (Williams et al. 1999; Liu et al. 2001; Nanda and Leibowitz 2001; Goebel et al. 2004; Nanda et al. 2004; Johnson et al. 2005), the 5′ UTR of only one group 2 coronavirus, BCoV, has been extensively studied to date. Four stem–loops, denoted I, II, III, and IV, that map within the 210-nt 5′ UTR of BCoV have been predicted, and their existence is supported by RNase probing and functional studies (Chang et al. 1994, 1996; Wang and Zhang 2000; Raman et al. 2003; Raman and Brian 2005). The predicted stem–loop I (nt 11–42; see Fig. 1A) contains just three contiguous Watson–Crick base pairs and a large 16-nt loop and is not conserved among group 2 coronaviruses. In addition, a mutational study designed to examine the requirement for stem–loop I was not definitive, in that all of the mutations in the DREP1 DI RNA construct were rapidly replaced by wild-type (WT) sequences (presumably derived from helper virus by recombination) (Chang et al. 1994) irrespective of whether they were predicted to maintain or disrupt the stem–loop. The predicted stem–loop II (nt 51–84) is an A-U base-pair-rich hairpin with a low free energy that folds the transcription regulatory sequence (TRS, the core motif at the RdRp template switch site) into the terminal loop (Raman et al. 2003; Raman and Brian 2005). In contrast, stem–loop III is phylogenetically conserved among group 2 coronaviruses and appears to have homologs in coronavirus groups 1 and 3, and enzymatic structure probing and DI RNA replication assays support its existence (Raman et al. 2003). The fourth predicted stem–loop, stem–loop IV, mapped to nucleotides 186–215, and is also predicted to be conserved among group 2 coronaviruses (Raman and Brian 2005). RNase mapping supports the existence of this stem–loop and DI RNA replication assays indicate that this structure likely plays a functional role in RNA replication, perhaps as a target for the binding of cellular proteins (Raman and Brian 2005).
FIGURE 1.
(A) Predicted secondary structure models for the entire 5′ UTR of BCoV compared with the 5′ 140 nt of selected group 2 coronaviruses. (B) Predicted secondary structure models for three group 1 coronaviruses. (C) Predicted secondary structure model of a group 3 coronavirus, avian infectious bronchitis virus (IBV). (Bold numbers) Predicted stem–loops SL1, SL2, SL3, and SL4 (4a and 4b), (bold red letters) leader TRS-L sequences, (yellow) SL-II, SL-III, and SL-IV of Raman et al. (2003) and Raman and Brian (2005). Nucleotide positions are numbered according to GenBank accession numbers (BCoV-LUN: AF391542; HCoV-OC43: NC_005147; MHV-A59: NC_001846; HKU1: NC_006577; SARS-CoV: NC_004718; HCoV-NL63: NC_005831; HCoV-229E: NC_002645; TGEV: NC_002306); IBV: NC_001451). All models except one represent mfe structures, and are predicted by Mfold, PKNOTS, and ViennaRNA. The lone exception is MHV, which represents a structure within 1.7 kcal mol−1 of the mfe structure. If SL2, the strongest secondary structure in a covariation analysis (not shown), is forced to pair as indicated, the structure shown represents the mfe structure.
Here, we present consensus secondary structure predictions of the 5′-most 140 nt in the 5′ UTR regions of nine group 1 and group 2 coronaviruses, including five human coronaviruses. All nine coronaviral genomes are predicted to fold into similar secondary structures containing three or four stem–loops in this region, including a highly conserved 5-nt hairpin loop (SL2) that possesses sequence features consistent with a U-turn motif containing a UNR sequence (Gutell et al. 2000). NMR studies of RNAs corresponding to SL1, SL2, and SL1/SL2 fragments from MHV, HCoV-OC43, and BCoV provide structural support for the general features of the model. A mutational analysis of SL2 in the context of the complete MHV genome supports the existence of this stem–loop structure and further reveals that SL2 has an essential role in MHV replication.
RESULTS
RNA secondary structure prediction for group 1 and 2 coronaviruses
Vienna RNA 1.5 (Hofacker et al. 2004) was used to examine the secondary structures of nine coronavirus 5′ UTRs, including five group 2 CoVs, BCoV, and the closely related human coronavirus HCoV-OC43, MHV-A59, HKU1, and SARS-CoV (Fig. 1A), as well as three representative group 1 CoVs, HCoV-NL63, HCoV-229E, and TGEV (Fig. 1B). A similar secondary structure was predicted for the avian coronavirus IBV, a group 3 CoV (Fig. 1C). Minimum free energy (mfe) secondary structural models of the 5′ 140 nt of all CoVs are remarkably similar, and all contain three major helical stems, denoted SL1, SL2, and SL4. Some sequences are predicted to contain a fourth stem–loop, SL3, which folds the leader TRS (TRS-L) sequence into a hairpin loop. Only for OC43 and SARS-CoV is SL3 predicted (at 37°C, ΔG37 = 1.5 kcal mol−1 and ΔG37 = 2.2 kcal mol−1, respectively); BCoV is capable of adopting the analogous SL3 stem–loop, corresponding to stem–loop II of (Raman and Brian 2005; Fig. 1A), although its Tm is predicted to be ≤37°C. (Note: Preliminary investigations of an RNA encompassing SL2 and SL3 of SARS-CoV [nt 42–72] by NMR spectroscopy and thermal denaturation experiments are consistent with a double hairpin conformation as indicated [see Fig. 1A; L. Li and D. Giedroc, unpubl.]). The SARS-CoV 5′ UTR differs from the other classical group 2 coronaviruses, in that SL2 is more distal to SL1.
Extending these predictions to encompass the entire 5′ UTR typically adds a few additional stem–loops to the mfe structure, as in the case of BCoV and OC43 (SL5–7), or another long multibranched stem structure in the 3′ region as shown for SARS-CoV (Fig. 1A) and NL63 (Fig. 1B), leaving the fold of the 5′ ∼140 nt intact; this strongly suggests that our predictions are meaningful. We note that SL4b and SL7 in our complete BCoV prediction (Fig. 1A) correspond to phylogenetically conserved and functionally important stem–loops III (Raman et al. 2003) and IV (Raman and Brian 2005).
SL2 is absolutely conserved and strongly predicted to form in all coronaviruses examined. Except for the core TRS-L, the (C/U)UUG(U/C) sequence encompassing the predicted SL2 loop is the most conserved contiguous run of nucleotides in the entire 5′ UTR of all coronaviruses examined. This conserved sequence only appears three or five times in the entire MHV or SARS-CoV genomes. Secondary structure analysis shows that this pentaloop is always stacked on a predicted 5 base-pair (bp) helix. This sequence conservation and the constancy of the predicted SL2 suggest an important functional role in coronavirus replication.
The (C/U)UUG(U/C) sequence of SL2 contains features of a canonical U-turn motif, in which the middle 3 nt of the loop, UNR (U0•N+1•R+2), forms a triloop that stacks on a Y:Y, Y:A, or G:A noncanonical base pair (Gutell et al. 2000). U-turn motifs are widely distributed in transfer (Quigley and Rich 1976; Lescrinier et al. 2006), ribosomal (Lebars et al. 2003), catalytic (Stallings and Moore 1997), and viral (Puglisi and Puglisi 1998) RNAs and often mediate RNA–RNA interactions between helical elements (Campbell and Legault 2005). The basic structural feature of the canonical U turn is a sharp turn in the phosphate backbone between U0 and N+1, with U0 stacked on the noncanonical base pair and engaged in two critical hydrogen bonds: the U0 imino proton donates a hydrogen bond to the nonbridging phosphate oxygen following R+2, and the U0 2′-OH proton donates a hydrogen bond to the N7 of R+2. Single or multiple nucleotide insertions in a U-turn motif are not uncommon, and in some cases the polynucleotide strand is diverted in a different direction (Gutell et al. 2000); in most cases, these breaks occur exclusively 3′ to R+2 (G) like that in the hammerhead ribozyme (Feig et al. 1998). This is, in fact, where one additional uridine is inserted in the SL2s of BCoV, OC43, and HKU1 (Fig. 1A). However, it is important to recognize that recent structural data suggest that U-turn motifs are conformationally diverse in solution, and may be lacking one or more of the key structural features associated with canonical U-turn motifs (Campbell and Legault 2005).
NMR spectroscopy of SL1- and SL2-containing RNAs
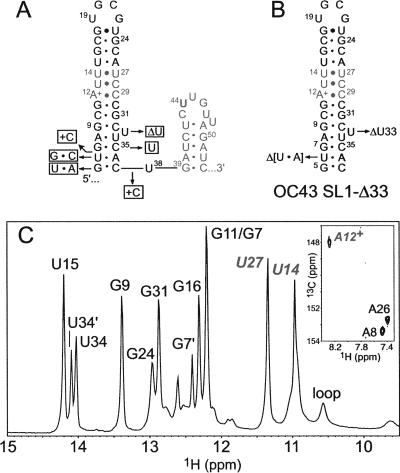
SL1s from BCoV/HCoV-OC43 and MHV are predicted to contain 13–14 bp capped by a 4-nt UGCG (YNMG) (Proctor et al. 2002; Theimer et al. 2003) (BCoV/OC43) or 8-nt (MHV) hairpin loop. Both SL1s are predicted to contain 2–3 noncanonical base pairs in the middle of the stem. To determine whether SL1 forms in solution, several RNAs were prepared and characterized by NMR spectroscopy. Figure 2A shows a sequence comparison of BCoV and OC43 SL1s, and as can be seen, all sequence differences are localized to the base of SL1. The ID imino proton region of an RNA corresponding to HCoV-OC43 SL1 termed OC43 SL1-Δ33 (see Fig. 2B) is shown in Figure 2C, with resonance assignments obtained from analysis of a 200-msec Watergate NOESY spectrum acquired at 10°C at pH 6.0 (not shown). The most notable feature of this spectrum is a U14•U27 base pair as predicted by the model, with both uridine imino protons strongly protected from exchange with solvent (Theimer et al. 2003; Du et al. 2004; Ohlenschlager et al. 2004). In addition, analysis of a 1H-13C HSQC experiment (Fig. 2C, inset) clearly shows the presence of an adenosine residue protonated at N1, since the 13C2 chemical shift is strongly shifted upfield (Huppler et al. 2002). A8 and A26 H2 protons were assigned by virtue of the intense NOE to the U H3 imino proton of the A8-U34 and U15-U27 Watson–Crick base pairs, respectively, with the A12+ H2 assigned by elimination. Consistent with this, analysis of thermal melting profiles for OC43 SL1-Δ33 acquired at pH 5.5 and 8.0 reveal an ∼3°C shift in Tm upon protonation, consistent with previous studies of A+•C base pairs (data not shown; Huppler et al. 2002). These studies establish that SL1 forms in BCoV/OC43 as predicted by the model.
FIGURE 2.
(A) Predicted secondary structures of SL1 and SL2 of HCoV-OC43. Nucleotide substitutions, insertions, and deletions at the base of SL1 that correspond to the BCoV-Lun sequence are indicated in the adjacent boxes. (B) HCoV-OC43 SL1 construct used for NMR studies, denoted OC43 SL1-Δ33. The U6-A36 base pair was excised to enable transcription by T7 RNA polymerase; the extrahelical U33 was also deleted. (C) Imino proton region of a 1D jump–return echo spectrum acquired at 10°C, pH 6.0 for OC43 SL1-Δ33. Resonance assignments were obtained from analysis of a homonuclear Watergate NOESY spectrum (τm=150 msec). The U14-U27 base pair was verified by the presence of a strong crosspeak in a NOESY spectrum acquired at a short mixing time (τm=50 msec). (Inset) Region of a natural-abundance 1H-13C HSQC spectrum acquired for OC43 SL1-Δ33, with assigned adenosine 13C2-1H2 cross-peaks indicated.
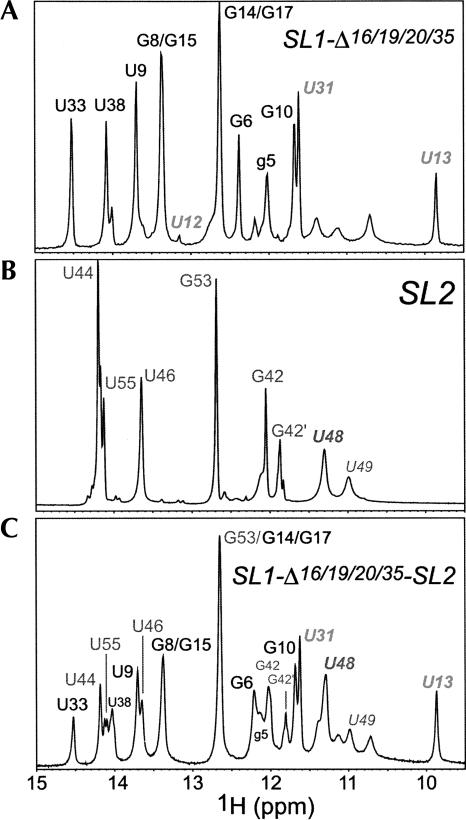
We next prepared a series of SL1 RNAs corresponding to SL1 from MHV-A59. In contrast to BCoV/HCoV-OC43 SL1, MHV SL1 contains two unpaired nucleotides in the stem, C16 and A35, and is capped by an 8-nt loop (Fig. 3A). Interestingly, if C19 and C20 are looped out, the MHV loop sequence becomes structurally identical to that of BCoV/HCoV-OC43; i.e., a YNMG (U22-G25) tetraloop (Proctor et al. 2002) capped on a closing G21•U26 base pair (Fig. 3B). ID imino proton spectra are shown for three different variants of SL1, one with all four “looped-out” nucleotides deleted (SL1-Δ16/19/20/35, Fig. 3C), SL1-Δ16/19/20/35 with A35 added back (SL1-Δ16/19/20, Fig. 3D), and SL1-Δ16/19/20/35 with A35 and C16 added back (SL1-Δ19/20, Fig. 3E). The spectra of all three RNAs are substantially identical, and all are characterized by a noncanonical U13•U31 base pair as predicted by the model (Fig. 3A). A comparison of SL1-Δ16/19/20/35 (Fig. 3C) with SL1-Δ16/19/20 (Fig. 3D) reveals that the major change in the spectrum is dynamic in nature, with the imino protons corresponding to U9, G10, and U33, i.e., those closest to the introduced A35, substantially broadened (at 10°C). Indeed, examination of the pyrimidine H5/H6 regions of homonuclear TOCSY spectra acquired for SL1-Δ16/19/20/35 and SL1-Δ16/19/20 (at 25°C) suggest that the two RNAs adopt virtually identical structures (data not shown); these results collectively suggest that A35 is extrahelical and that alternative pairing of U9 with A35 or A36 introduces a local destabilization of this region of SL1. The same appears true of SL1-Δ19/20, except that the spectra degrade substantially in overall quality. The broader linewidths in this sample suggest that this is attributable to dimerization of the RNA.
FIGURE 3.
(A) Predicted secondary structure of MHV-A59 SL1. (B) Representation of the SL1, SL2 (boxed regions), and SL1-SL2 chimeras characterized in this study. All MHV SL1 constructs have a nonnative g5-c40 base pair (native sequence shown in brackets) at the base of SL1 to facilitate transcription by T7 RNA polymerase, and incorporate a nonnative base G-C pair base of SL2 and invert the MHV A44-U54 pair in MHV to the U-A pair present in SARS-coronavirus (the native MHV sequence is shown in brackets, see text for details). Imino proton regions of 1D jump–return echo spectra acquired at 10°C unless otherwise indicated for SL1-Δ16/19/20/35 (C), SL1-Δ16/19/20 (D), and SL1-Δ19/20 (E) (5°C). Resonance assignments were obtained from analysis of a homonuclear Watergate NOESY spectra (τm=150 msec) acquired for each RNA (300 msec for SL1-Δ19/20). Imino resonances for the U13-U31 base pair are indicated. (*) 14.2 ppm, the expected absence of the imino resonance for the A7-U38 base pair due to substitution of a nonnative g7-c38 base pair in this construct.
We next prepared a 16-nt SL2 RNA that corresponds to the SARS-CoV SL2 (see Fig. 3B). This SL2 is more efficiently transcribed by SP6 RNA polymerase than the equivalent MHV SL2, and the stem will be more stable. Relative to the MHV sequence, SL2 contains A41-U56 to G41-C56 and A44-U54 to U44-A54 substitutions. A 3′ adenosine cap was also added in an attempt to stabilize the adjacent stem (Theimer et al. 1998). We note that this SL2 stem sequence is also found in BCoV/HCoV-OC43 and HKU1 (Figs. 1A, 2A), and replacement of the MHV SL2 with the SARS-CoV SL2 fully supports MHV replication (Kang et al. 2006). The 1D imino proton region of SL2 is shown in Figure 4B (pH 6.0, 10°C, no added salt), with the spectrum of SL1-Δ16/19/20/35 also reproduced here (Fig. 4A) to facilitate comparison with the spectrum derived from a 49-nt RNA encompassing MHV SL1-Δ16/19/20/35 and SL2 (Fig. 4C). The most notable feature of the SL2 spectrum is a relatively intense upfield shifted uridine imino proton, assigned to U48 (U0 in the U0-N+1-R+2 nomenclature, see below). This is consistent with the U48 H3 proton donating a hydrogen bond and thereby stabilizing a U-turn-like conformation. In addition, the imino proton of the closing U46-A52 base pair of the stem is also observable, suggesting the loop is structured and stacked on the U46-A52 base pair; this contrasts with a previous solution structure of a noncanonical U-turn motif (Campbell and Legault 2005). In any case, the spectrum obtained for SL1-Δ16/19/20/35-SL2 RNA (Fig. 4C) is essentially identical to that expected for a superposition of component SL1 and SL2 hairpins. These spectra are consistent with the idea that SL1 and SL2 are independently folded and do not substantially interact with one another.
FIGURE 4.
Imino proton regions of 1D jump–return echo spectra acquired at 10°C, pH 6.0 for SL1-Δ16/19/20/35 (A), SL2 (B), and SL1-Δ16/19/20/35-SL2 (C) RNA. See Fig. 3B for sequences for these RNA constructs. Note that the spectra for SL2 are characterized by slow conformational heterogeneity at the base of SL2 (G42-C56 and A43-U55 base pairs). The imino protons corresponding to U48 and U49 in the SL2 pentaloop are also indicated. The assignment of U12 is based on a weak NOE to a C32 amino proton.
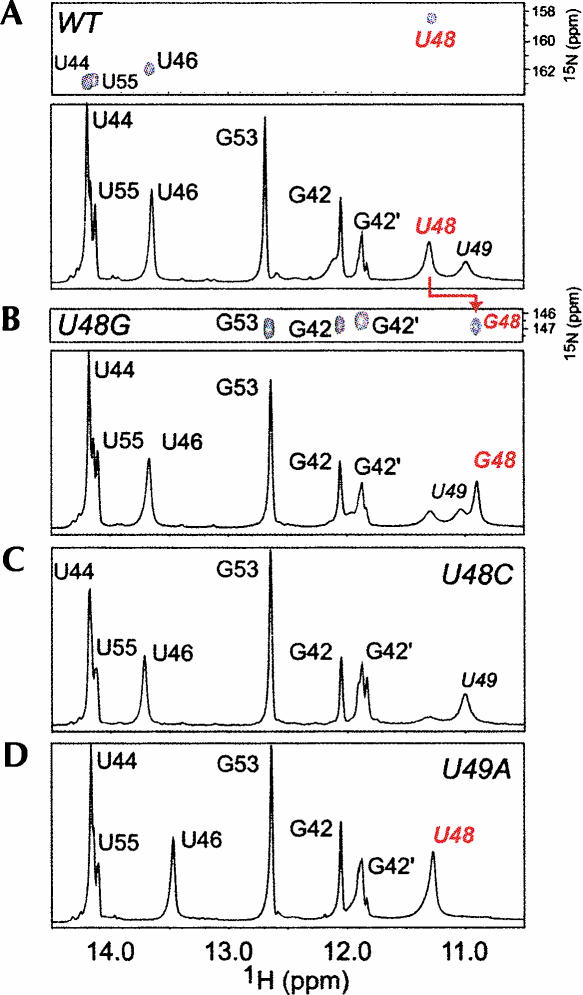
We next determined the extent to which the SL2 was amenable to substitution, since the U-turn hypothesis predicts that substitutions of U48 may be deleterious to the structure, in contrast to U49. 1D imino proton spectra are shown for WT (Fig. 5A), U48G (Fig. 5B), U48C (Fig. 5C), and U49A (Fig. 5D) SL2 RNAs acquired at low salt. We also show 1H-15N HSQC spectra that derive from 13C,15N-[U]-labeled WT SL2 (Fig. 5A) and 13C,15N-[G]-labeled U48G (Fig. 5B) RNAs. These spectra establish that the most intense upfield-shifted slowly exchanging imino proton corresponds to U48 and G48 in WT and U48G RNAs, respectively, and analysis of an 1hJ-HNN-COSY experiment acquired for these two RNAs suggest non-U or non-G nitrogen acceptors in the WT and U48G RNAs, respectively, as well (data not shown). These data, coupled with an analysis of the nonexchangeable NOE data support the contention that the G48 imino proton conserves the basic structure of the wild-type pentaloop, in a manner analogous to U48 in the WT sequence. The U48C loop structure may be disrupted due to the loss of U48 imino proton (Fig. 5C). In contrast, as expected from the consensus UNR sequence, the U49A RNA appears to maintain a wild-type-like loop structure, given the identical chemical shift of the U48 H3 proton in both RNAs (Fig. 5D).
FIGURE 5.
Imino proton regions of 1D jump–return echo spectra acquired at 10°C, pH 6.0 for SL2 variants, with 1H-15N-HSQC spectra shown for 13C,15N-[U]-labeled WT SL2 and 13C,15N-[G]-labeled U48G SL2, as well. (A) WT SL2 (see Fig. 3B); (B) U48G SL2; (C) U48C SL2; and (D) U49A SL2. G42 and G42′ represent alternative conformations for the terminal G42-C56 base pair. The immediately adjacent U55 resonance is also doubled, indicative of heterogeneity at the base of SL2.
We could find no evidence in either the WT or U48G SL2 RNAs in support of a C47•U51 base pair that would be expected for a canonical U-turn motif. In fact, the data suggest that U51 is extruded from the loop with G50 in close proximity to A52 (L. Li and D. Giedroc, unpubl.); these data suggest a noncanonical U-turn conformation in CoV SL2. Although U-turn structures are capable of forming in the absence of divalent cations and at low salt (Puglisi and Puglisi 1998), they can be stabilized by high concentrations of Mg2+ or Co(NH3)63+ (Cabello-Villegas et al. 2004). We find that the basic structural features of the pentaloop are unchanged in the presence of 0.1 M KCl, 5.0 mM MgCl2, while the addition of mM Co3+(NH3)6 leads to significant dimer formation (data not shown).
Mutational analysis of the SL2 stem
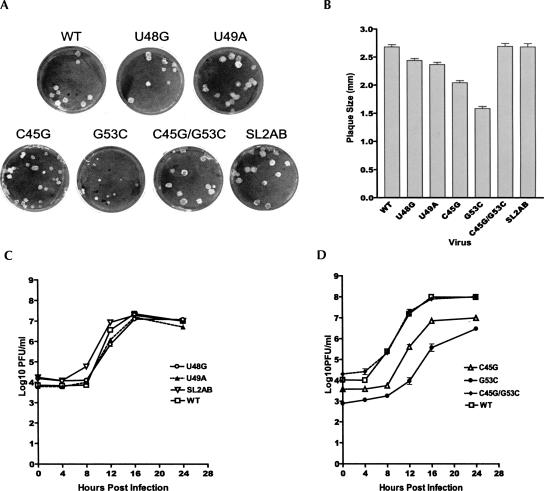
Having established the structural features of the SL2 hairpin, we next investigated the functional importance of SL2 for viral replication. Three mutations were introduced into the helical stem of MHV SL2 to either open up the stem by introducing multiple transversion mutations into either the left (5′, nt 44–46, mutant SL2A; see Fig. 3B for numbering of nucleotides) or right (3′, nt 52–54, mutant SL2B) side of the stem, or to maintain base pairing by introducing the transversion mutations into both sides of the stem (mutant SL2AB). When these mutations were introduced into the MHV-A59 genome, mutants SL2A and SL2B were not viable, whereas mutant SL2AB was viable and had a replication phenotype that was similar to wild-type MHV-A59 1000. Mutant SL2AB formed similar-sized plaques (Fig. 6A,B) and reached a titer virtually identical to that achieved by the wild-type virus (Fig. 6C). These results strongly support the existence of SL2 and indicate that the stem structure, rather than its nucleotide sequence, is required for viral replication, a result consistent with the viability of an SL2 SARS-MHV chimeric genome (Kang et al. 2006).
FIGURE 6.
Growth phenotypes of SL2 mutant viruses. (A) Plaque morphologies of mutant and wild-type viruses. (B) Averaged plaque size of mutant and wild-type viruses. Plaque sizes were measured after DBT cell monolayers were stained with crystal violet. Plaque sizes were determined as described previously (Johnson et al. 2005). (C) One-step growth curves of viable loop and stem clustered point mutants. (D) One-step growth curve of stem point and compensatory mutations. Triplicate wells of DBT cells in 96-well plates were infected with mutant or wild-type viruses at a MOI of 3 and harvested at 0, 4, 8, 12, 16, and 24 h post-infection. Virus titers were determined by plaque assays. Error bars represent the standard errors of the mean.
To further characterize the SL2 helical stem, point mutations predicted to destabilize the stem were introduced into the MHV-A59 1000 genome at position 45 (mutant C45G) or 53 (mutant G53C). The double point mutant C45G/G53C introduces both of these changes and is predicted to restore base pairing in the stem. All three mutants were viable, although they have different phenotypes. As shown in Figure 6, A and B, mutants C45G and G53C, predicted to destabilize SL2, both form smaller plaques than wild-type MHV-A59 1000, with mutant G53C having a much greater effect on plaque size. As expected, the double mutant C45G/G53C forms plaques identical to or slightly larger than wild-type MHV-A59 1000. One-step growth curves confirm the growth phenotypes of these viruses (Fig. 6C,D). Under one-step growth conditions, mutants C45G and G53C grow much more slowly and achieve lower final titers than wild-type MHV-A59 1000. However, the G53C mutation induces a more severe effect on virus replication than does the C45G mutation, consistent with the greater effect of this mutation on plaque size. Under multistep growth conditions (M.O.I.=0.01, data not shown), the G53C mutant appears much more severely impaired in its ability to replicate, reaching a final titer ∼105-fold less than wild-type virus. The effect of the C45G mutation is much less severe (titer decreased ∼10-fold compared with wild type), while the C45G/G53C mutation replicates almost as well as wild-type virus (titer decreased about threefold compared with wild type).
The SL2 U-turn motif-containing pentaloop is crucial for virus viability
As covariation analysis, NMR data, and mutational studies supported the existence of the SL2 stem–loop, we made a series of mutations at positions 48 and 49 to characterize the functional importance of the proposed UNR (U48-U49-G50) in the CUUGU pentaloop in viral replication. Replacing uridine 48 with either cytidine (mutant U48C) or adenosine (mutant U48A) resulted in genomes that were not viable, consistent with a requirement for a UNR loop structure for SL2. In contrast, the U48G mutant was viable, resulting in a virus that produced near normal-sized plaques, reached a final titer virtually identical to the wild-type virus, and had growth kinetics that were very similar to the wild-type virus (Fig. 6A–C). These findings reveal that the major groove side of the Watson–Crick face, involving U O4/H3 or G O6/H1 hydrogen bond acceptors/donors in WT and U48G RNAs, can functionally substitute for one another to maintain virus viability. The same is true of viruses harboring a nonconservative replacement of uridine 49 with adenosine (mutant U49A), which leads to recovery of virus with near normal-sized plaques (Fig. 6A,B) and growth kinetics nearly identical to the U48G and WT viruses (Fig. 6C). This result is consistent with the NMR studies that suggest that U48G SL2 folds into a wild-type-like noncanonical U-turn motif and further suggest that nt 49 marks the apex of the loop structure and simply stacks on G50 (L. Li and D. Giedroc, unpubl.), similar to other UNR-loop-containing U-turn motifs; in such a structure, any nucleotide would be tolerated at position 49 (Fig. 5D).
Mutations in SL2 affect RNA synthesis
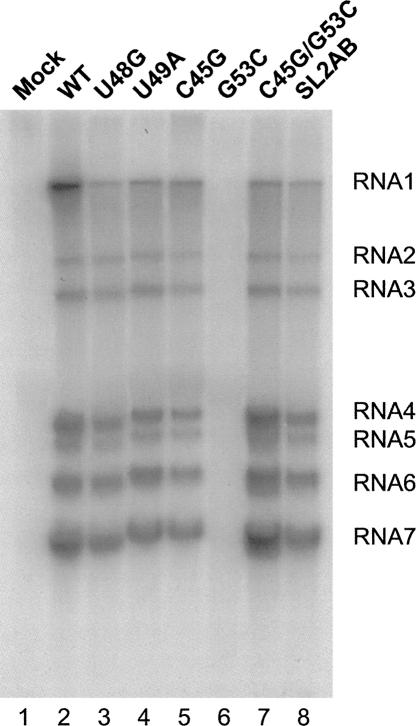
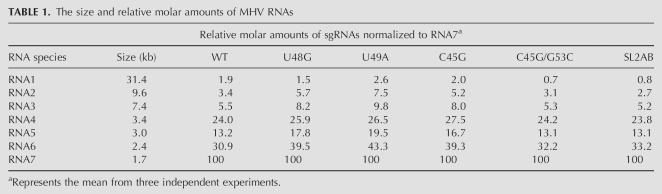
To determine if mutations in SL2 affected RNA synthesis, we infected replicate cultures of DBT cells with mutant and wild-type MHV-A59 1000, and metabolically labeled MHV-specific RNAs with 32P-orthophosphate for 6 h. At the end of the labeling period, the intracellular RNAs were extracted and resolved by electrophoresis on a formaldehyde agarose gel. As shown in Figure 7, virus-specific RNAs synthesized by the mutant G53C were barely detectable relative to other mutants as well as the wild-type virus; this is consistent with the severe defect of this mutant in growth kinetics (Fig. 6). For the other mutants, quantitation of the total amount of label incorporated into MHV-specific RNAs reveals that the amount of RNA synthesized by each mutant generally correlates with the virus growth phenotypes, with the next most functionally compromised mutant, C45G, containing approximately half of wild-type levels of total RNA (Fig. 6). Examination of the molar ratios of genomic and subgenomic RNA species revealed only small or no significant differences, with no obvious trends in the data (Table 1).
FIGURE 7.
Analysis of MHV specific RNAs synthesis. DBT cells infected with wild-type and mutant viruses RNAs were metabolically labeled from 6–12 h post-infection in the presence actinomycin D. Total RNAs were extracted and resolved on a formaldehyde agarose gel. (Lane 1) Mock infected cells, (lanes 2–8) cells infected with wild-type MHV A59–1000 (100%), U48G (72%), U49A (88%), C45G (57%), G53C (<1%), C45G/G53C (109%), and SL2AB (94%) genomes, respectively. The total amount of RNA synthesized relative to wild-type MHV A59–1000 is indicated in parentheses. (RNA 1–7) MHV-specific RNA bands. Molar ratios of indicated RNAs are compiled in Table 1.
TABLE 1.
The size and relative molar amounts of MHV RNAs
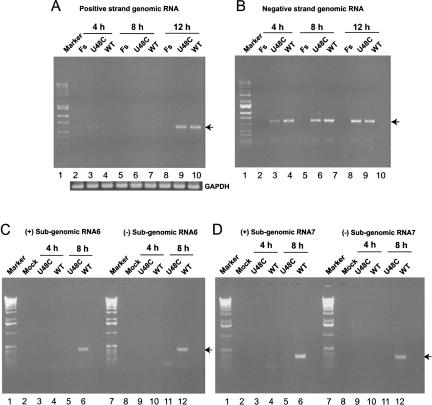
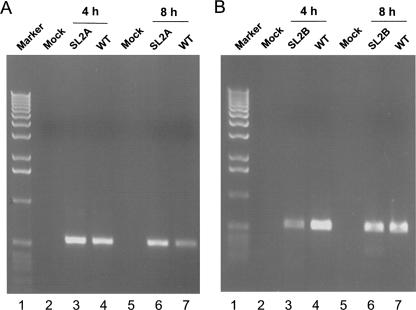
We next investigated the RNA phenotypes of the nonviable mutant genomes. Poly(A)+ mRNAs were extracted at 4, 8, and 12 h post-electroporation with wild-type, Fs (a frameshift mutant incapable of synthesizing RNA), and U48C genomes and analyzed by RT-PCR. As shown in Figure 8A, input positive genomic RNA can be detected 4 h post-electroporation. By 8 h post-electroporation the input RNA is virtually completely degraded. Newly synthesized positive genomic RNA can be detected at 12 h post-electroporation for U48C and wild type, but as expected, not for the Fs mutant.
FIGURE 8.
Analysis of genomic and subgenomic RNAs of nonviable mutant U48C. RNAs were extracted 4, 8, and 12 h after cells were electroporated with in vitro assembled and transcribed Fs, wild-type MHV A59–1000, or U48C genomes and analyzed by RT-PCR or nested RT-PCR. (A) Positive-strand genomic RNA synthesis. (Marker, lane 1) 1-Kb DNA ladder, (Fs) RdRp frameshift mutant. (B) Negative-strand genomic RNA synthesis, with lanes marked as in panel A. (C) Positive-strand (left) and negative-strand (right) synthesis of RNA6; (Mock) Mock-infected cells. (D) Positive-strand (left) and negative-strand (right) synthesis of RNA7, with lanes marked as in panel C.
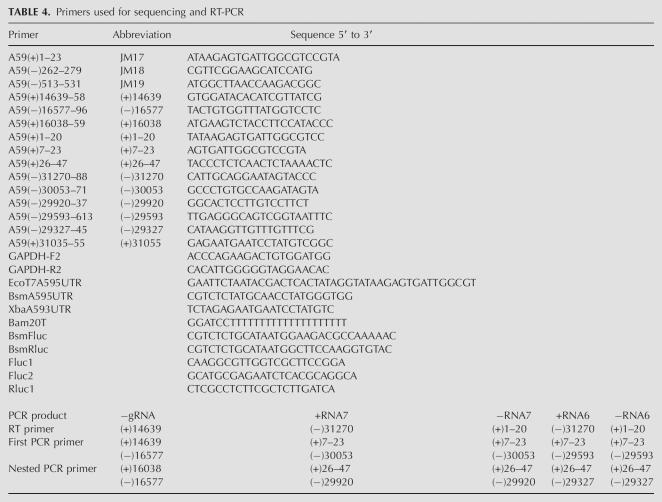
To detect negative-strand genomic RNA, total RNAs were extracted at 4, 8, and 12 h post-electroporation. Nested RT-PCR results showed that negative-sense genome-sized RNAs were present in cells electroporated with U48C genomes (Fig. 8B); however, neither positive- nor negative-sense subgenomic RNA6 and RNA7 were detected (Fig. 8C,D). Negative-sense subgenomic RNA3 was also undetectable (data not shown). An identical analysis performed with cells electroporated with SL2A or SL2B genomes gave results similar to those obtained with the U48C mutant; negative-sense genome-sized RNAs were synthesized (Fig. 9) but no subgenomic RNAs could be detected (data not shown). For each sample, parallel RT-PCR reactions in which the RT step was omitted were performed to ensure that residual DNA transcription templates were not producing spurious signals (not shown). Taken together, these results are consistent with a critical role for SL2 in MHV RNA replication and transcription.
FIGURE 9.
RT-PCR analysis of negative-strand genomic RNA of nonviable stem clustered point mutations SL2A and SL2B. (A) Full-length negative-strand genomic RNA synthesis of SL2A versus wild-type A59–1000. (B) Full-length negative-strand genomic RNA synthesis of SL2B versus wild-type A59–1000.
Mutations in SL2 affect in vitro translation
To determine if mutations in SL2 affected translation, the MHV-A59 wild-type 5′ UTR and the U48C mutant 5′ UTR were each cloned immediately upstream of the Renilla and firefly luciferase coding sequences fused to the MHV-A59 3′ UTR. Capped RNAs corresponding to WT-5′UTR-Renilla luciferase-3′UTR (WT-Ren) and U48C-5′UTR-Renilla luciferase-3′UTR (U48C-Ren) were synthesized by in vitro transcription with T7 RNA polymerase and translated in reticulocyte lysates programmed with a subsaturating amount of RNA. The RNA containing the U48C 5′ UTR was translated to yield ∼19% of Renilla luciferase product as RNAs containing the wild-type 5′ UTR (Table 2, Experiment 1). To be certain that this decrease in translation was not due to a different interaction of the mutant and wild-type UTRs with the Renilla sequence, and to better control for small differences between individual in vitro translation reactions, we performed a series of ratiometric assays with in vitro translation reactions programmed with equal molar amounts of WT-Ren, U48C-Ren, WT-FF (WT-5′UTR-firefly luciferase-3′UTR), and U48C-FF (U48C-5′UTR-firefly luciferase-3′UTR) in the combinations shown in Table 2 (Experiment 2). These ratiometric assays confirmed that RNAs containing the wild-type 5′ UTR are translated in vitro about fivefold more efficiently than those containing a 5′ UTR with a U48C mutation.
TABLE 2.
Effect of the SL2 U48C mutation on in vitro translation
DISCUSSION
In this study, we present a consensus RNA secondary structure model for the most 5′ 140 nt of nine representative coronaviruses derived from all three major coronavirus groups. We also provide structural and functional support for the model, focusing here on the functional role that a pentaloop-containing SL2 plays in MHV replication. This work complements previous studies of chimeric MHV/SARS-CoV viruses in which portions of the MHV 5′ UTR have been replaced by the corresponding SARS-CoV stem–loop structures (Fig. 1A; Kang et al. 2006). Although the SL1s of SARS-CoV and MHV have only 47.7% sequence identity, SL1 of SARS-CoV is capable of forming a stem–loop structure of similar length to that of MHV SL1. Substitution of the SARS-CoV SL1 for the MHV SL1, which also increases the spacing between the predicted MHV SL1 and SL2 by 2 nt, does not strongly affect viral viability but yields a virus with a smaller plaque size, impaired RNA synthesis, and which replicated to lower titer than wild-type MHV. Replacing the MHV SL4 with the SARS-CoV SL4 structure also generated a viable chimeric virus with a similar phenotype. SL2 is the most conserved secondary structure in the coronavirus 5′ UTR; replacing the SL2 of MHV with SARS SL2 resulted in a viable chimeric virus with a replication phenotype very similar to wild-type MHV.
SL1 from MHV and BCoV/OC43 is uniquely characterized by 2–3 noncanonical U•U, U•C, or A+•C base pairs in the middle of the stem (Figs. 2–4). Their functional significance remains to be tested; however, they are obviously not absolutely required for replication since SARS-CoV SL1 can substitute, albeit weakly, for MHV SL1 (Kang et al. 2006). We point out, however, that the specific structural features of the upper two-thirds of BCoV/OC43 SL1, including the noncanonical base pairs and a predicted UNCG (YNMG; M=A or C) tetraloop stacked on a G•U wobble pair, are strikingly reminiscent of stem–loop D (SLD) in the 5′ UTRs of picornoviruses that forms a binding site for the viral chymotrypsin-like protease (3CLpro) (Du et al. 2004; Ohlenschlager et al. 2004; Ihle et al. 2005). A C•U Watson–Crick base pair in the triple pyrimidine mismatch widens the major groove of the stem by shortening the C1′-C1′ distance across the helix (Theimer et al. 2003; Ohlenschlager et al. 2004). Our NMR experiments are consistent with the basic stem–loop structure, including the noncanonical pairing in the helical stem. However, it is clear that the anticipated 5′-GUGCGU tetraloop, where the closing base pair is 5′ G•U, is destabilized with respect to the UUNCGG tetraloop in picornoviral SLD since characteristic imino proton resonances associated with the 3′ guanosine in the loop and the closing G•U base pair are absent or solvent-exchange broadened (Du et al. 2003; Ohlenschlager et al. 2004); this is consistent with previous thermodynamic studies that reveal that inversion of the 5′ C-G base pair to 5′ G-C destabilizes a UUCG tetraloop by 2.3 kcal mol−1 (Antao et al. 1991). Efforts are underway to solve the solution structure of SL1 and test the possibility that CoV 3CLpro, which adopts a similar tertiary structure to picornovirus 3Cpro(Matthews et al. 1999; Yang et al. 2003), forms a functionally significant interaction with SL1.
Our structural and functional data provide strong evidence in support of the formation of SL2. Ablation of the stem and introduction of compensatory mutations that restore SL2 base pairing argue strongly for the functional importance of SL2. In addition, we also show that U48, which is predicted to stabilize the pentaloop conformation by virtue of formation of one or more hydrogen bonds, is a key determinant for SL2 function. This is in contrast to U49, where a nonconservative adenosine substitution is tolerated, in contrast to the U48A virus, which, like the U48C virus, is nonviable. The precise structural basis for these findings is currently under investigation (L. Li and D. Giedroc, unpubl.), as is a more comprehensive mutagenesis analysis of the pentaloop; however, we note a 1:1 correspondence between the presence of a hydrogen bond donor at position 48 (U48 H3 or G48 H1) and MHV viability. The mutagenesis and structural studies reported here coupled with the evolutionary conservation of the pentaloop (Fig. 1) suggest a novel, noncanonical U-turn motif conforming to the sequence 5′-YY(U/G)NR(Y)R, where a Y-R base pair closes a four-base Y(U/G)NR tetraloop with the (Y) nucleotide extruded from the structure.
Our secondary structural model leaves open the possibility that SL3 is single stranded or folded into a weakly paired helical structure (Fig. 1A). (As mentioned above, preliminary investigations of an RNA encompassing SL2 and SL3 of SARS-CoV [nt 42–72] by NMR spectroscopy and thermal denaturation experiments are consistent with a double hairpin conformation as indicated [see Fig. 1A; L. Li and D. Giedroc, unpubl.].) In the latter case, the TRS sequence, which is absolutely required for template switching and discontinuous subgenomic RNA synthesis, would be found in the terminal loop of SL3 in BCoV/OC43, HKU1, and SARS-CoV (Fig. 1A). Studies of the structure of the 5′ UTR in a more distantly related member of the Nidovirus family, equine arteritis virus (EAV, an arterivirus), suggest that the EAV leader TRS sequence is incorporated into the terminal loop of a stem–loop structure (stem–loop G), and this prediction is supported by enzymatic probing and functional data (Van Den Born et al. 2004; van den Born et al. 2005). The extent to which this is the case in any coronavirus has yet to be firmly established (see Chang et al. 1996); this is in contrast to sequence requirements of both the core and flanking nucleotides of the TRS, which have been extensively investigated (Zuniga et al. 2004; Sola et al. 2005).
Interestingly, the U48C substitution in SL2 also decreases the translational efficiency of heterologous RNAs bearing the MHV 5′ UTR (Table 2). The precise mechanism by which SL2 influences translational efficiency is not clear. This could occur through interactions with proteins present in the in vitro translation reactions, by providing a structure that facilitates interactions between the 5′ and 3′ UTRs, favoring repetitive rounds of translation, or by promoting the folding of the UTR into a structure that is more easily traversed by ribosomes scanning for the initiating AUG. Our findings are consistent with earlier work implicating the 5′ leader sequence containing SL2 in enhancing in vitro translational efficiencies of heterologous transcripts in lysates prepared from MHV-infected cells (Tahara et al. 1994).
All mutations in SL2 affect virus-specific RNA synthesis, and similar findings characterize nonviable SL1 mutants as well (H. Kang, L. Li, N. Makkinje, P. Liu, S. Williamson, D. Giedroc, and J. Leibowitz, in prep.). Cells electroporated with nonviable mutants in SL2 all have defects in subgenomic RNA synthesis, although genome replication occurs. These results are consistent with those observed by Wang and Zhang (2000) in a DI RNA system. It is currently unknown how SL2 would directly affect subgenomic RNA synthesis, but there are several possibilities that can be distinguished from one another on the basis of additional structural and functional studies. For example, it is often the case that U-turn motifs mediate long-range RNA–RNA interactions, and prominent examples of this occur within the tRNA anticodon loop–mRNA interaction, ribosomal RNA–RNA interactions (Gutell et al. 2000), and within the VS ribozyme, where a noncanonical U-turn motif base pairs with a nonadjacent hairpin loop in forming three consecutive loop–loop Watson–Crick base pairs (Campbell and Legault 2005). This might be expected if, for example, the Watson–Crick edges of the bases within the loop are exposed to solvent. Another possibility is that SL2 mediates a specific interaction with a viral-encoded or cellular protein. Previous studies suggest that polypyrimidine-tract binding (PTB) protein (hnRNP I) binds to the MHV leader to a region containing UCUAA repeats in the leader TRS, where it is thought to function as a regulator of viral transcription (Li et al. 1999; Choi et al. 2002). Recent structural studies reveal the molecular basis by which PTB interacts with short pyrimidine-rich (UCU, CUCU) sequences (Oberstrass et al. 2005), and these data suggest the possibility that that SL2 might provide a specific binding site for PTB. This interaction might play a role in mediating the circularization of the genome and/or directly facilitate template switching in discontinuous subgenomic RNA synthesis. Studies along these lines aimed at testing this hypothesis are currently in progress in our laboratories.
MATERIALS AND METHODS
Cells and viruses
Baby hamster kidney cells expressing MHV receptors (BHK-R) (Dveksler et al. 1991; Yount et al. 2002) were kindly provided by Ralph Baric (University of North Carolina at Chapel Hill) and maintained at 37°C and 5% CO2 in DMEM supplemented with 10% calf serum, 10% tryptose phosphate broth, 800 μg/mL Geneticin (G418 sulfate; Sigma), and 0.25 μg/mL kanamycin. DBT cells were maintained at 37°C and 5% CO2 in DMEM supplemented with 10% calf serum. L2 cells were maintained at 37°C and 3% CO2 in DMEM supplemented with 10% calf serum. Wild-type MHV A59–1000 (NC_001846) and all mutant viruses were propagated in DBT cells.
Sequence alignment and RNA structure prediction
Nine coronavirus 5′ UTR sequences were obtained from GenBank with accession numbers listed in the legend to Figure 1. RNA nucleotide sequences were first aligned using Clustal W and manually inspected for the presence of strongly predicted covariations that would support the presence of conserved terminal hairpin loop sequences. Each RNA sequence was then subjected to secondary structure prediction using ViennaRNA 1.5 (Hofacker 2003; Hofacker et al. 2004), Mfold 3.1 (Zuker 2003), and PKNOTS (Rivas and Eddy 1999). Mfold 3.1 predicts both a minimum free-energy structure as well as all structures within a defined energy from the minimum. PKNOTS is a dynamic programming algorithm that incorporates pseudoknot pairing. ViennaRNA uses updated nearest-neighbor thermodynamic parameters that incorporate the energetics of stabilizing tetraloops and unpaired dangling ends and generates both a minimum free-energy (mfe) structure as well as equilibrium base-pairing probabilities using the partition function (pf) algorithm of McCaskill (1990). With the exception of MHV, the mfe and thermodynamic ensemble predictions corresponded in every case, taken as good evidence for one major lowest-energy secondary structural conformer that speaks to the robustness of the prediction. For MHV, the structure shown is within 1.7 kcal mol−1 of the mfe structure; if SL2 is forced to pair as indicated, the structure shown represents the mfe structure.
RNA preparation and NMR spectroscopy
RNAs were prepared by run-off transcription using SP6 or T7 RNA polymerase, purified by denaturing PAGE, and subjected to multiple cycles of ethanol precipitation as described previously (Nixon et al. 2002; Cornish et al. 2005). 13C,15N-[U]-labeled WT SL2 and 13C,15N-[G]-labeled U48G RNAs were also prepared using 13C,15N-[UTP] and 13C,15N-[GTP] in transcription reactions, respectively. The final NMR buffer was 10 mM potassium phosphate, pH 6.0, unless otherwise indicated. The RNA concentration in each case ranged from 0.8 to 2.0 mM. All RNAs were checked for the presence of a monomer–dimer equilibrium by nondenaturing PAGE prior to extensive NMR analysis. All NMR experiments were performed on a Varian Inova 500- or 600-MHz spectrometer in the Texas A&M University Biomolecular NMR Laboratory. The proton resonances were referenced to an internal standard (100 μM DSS). The jump–return echo 1D and Watergate 1H-1H NOESY spectra (τmix = 200 msec) were acquired to obtain imino proton resonance assignments, while 1H-1H D2O NOESY (τmix = 250 msec) and TOCSY experiments were performed to obtain nonexchangeable proton resonance assignments using standard methodologies (Furtig et al. 2003). A natural-abundance 2D 1H-13C CT-HSQC spectrum was acquired to obtain 13C and 1H assignments for the adenosines in HCoV-OC43 SL1. Through-bond 2D HNCCCH (Simorre et al. 1995) H(CCN)H-TOCSY (Sklenar et al. 1996) experiments were used to correlate imino protons with uridine H6 and guanosine H8 protons, respectively, in 13C,15N-[U]-labeled WT and 13C,15N-[G]-labeled U48G SL2 RNAs, to provide unique starting points for sequential base–ribose connectivities in a D2O NOESY spectrum (τm = 150–250 msec for SL1- and SL2-containing RNAs, respectively). A combination of sensitivity-optimized HCN-HMQC (ribose moiety, H1′-C1′-N1/9), HCN-TROSY (base moiety, H6/8-C6/8-N1/9), and HCNCH experiments provided unambiguous, through-bond sugar to base connectivities (Fiala et al. 2000) with complete ribose proton connectivities obtained with a 1H-13C HCCH-TOCSY experiment. Additional NOE restraints were obtained from analysis of a 13C-edited NOESY spectra, a 2D 1H-15N CPMG-NOESY (60 and 250 msec) (Mueller et al. 1995), and 2JHN-HSQC experiments (Simorre et al. 1996). Adenosine H2 protons were correlated with H8 protons using a HCCH-TOCSY experiment (Sklenar et al. 1996). All of the NMR data were processed using nmrPipe and analyzed using Sparky (Goddard and Kneller 2001; Delaglio et al. 1995).
Assembly of full-length MHV-A59 cDNAs and recovery of infectious viruses
The reverse genetic system for MHV-A59 used in this study was initially described by Yount et al. (2002). cDNAs representing the entire MHV-A59 1000 (defined as wild type for this work) genome with either the wild-type sequence or containing mutations in SL2 (see below) were constructed by sequential ligation of the A-G cDNAs as described previously (Yount et al. 2002; Johnson et al. 2005). Chimeric and wild-type MHV genomes were transcribed and electroporated into cells as previously described (Johnson et al. 2005). Cultures were observed for up to 72 h for the development of cytopathic effect (cell fusion) and harvested by freezing at −70°C. Cultures that did not develop cytopathic effect were sequentially blind passed three times in DBT cells in a further attempt to recover infectious virus. At least three independent experiments, including at least one experiment in which electroporated cells were incubated at 34°C and 39.5°C, were performed before a mutant genome was considered nonviable. Recovered viruses were plaque purified and expanded once or twice in DBT cells. The sequence of mutant viruses was confirmed by sequencing the entire 5′ and 3′ UTRs.
Mutant virus construction
Plasmid A of the reverse genetic system described above (Yount et al. 2002) was used as a substrate for mutagenesis. An oligonucleotide assembly approach was used to construct plasmids containing the following mutations: C45G, U48A, U48C, U49A, G53C, and C45G/G53C. An isogenic control plasmid containing the wild-type MHV-A59 1000 sequence was constructed, as well. To introduce these mutations into wild-type MHV A59–1000, we took advantage of two conveniently located restriction sites in plasmid A: an MluI site just 5′ to the T7 promoter, and a SacII site located at position 106 in the MHV-A59 sequence (GenBank Accession Number NC 001846). To reconstruct the wild-type sequence, six partially overlapping oligonucleotides, P1A-F, P1A-R, P2A-F, P2A-R, P3A-F, and P3A-R, spanning the sequence between the MluI and SacII sites in plasmid A were synthesized (Table 3) such that an MluI site was present at the 5′ end and a SacII site was present at the 3′ end. Oligonucleotides were diluted to 10 μM, and 10 μL of each oligonucleotide were mixed together and heated to 90°C for 1 min; the oligonucleotides were then annealed by reducing the temperature by 5°C every 10 min until room temperature was reached. The annealed 135-bp DNA fragment was ligated into MluI- and SacII-digested plasmid A and cloned into TOP10 Escherichia coli. To construct viruses with the desired mutations in SL2, oligonucleotides P2A-F and P2A-R were replaced by oligonucleotides containing the mutations shown in Table 3. Plasmids were sequenced with primer JM18 [A59(−)262–279] (Table 4) to confirm the presence of the desired mutations.
TABLE 3.
Oligonucleotides used to introduce mutations into SL2
TABLE 4.
Primers used for sequencing and RT-PCR
Mutant U48G, the triple mutants SL2A (A44U/C45G/U46A) and SL2B (A52U/G53C/U54A), and mutant SL2AB (A44U/C45G/U46A/A52U/G53C/U54A) were constructed using the Quick Change II site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions with wild-type plasmid A as the substrate for mutagenesis. The sequences of the mutagenic oligonucleotides are shown in Table 3. Mutagenized plasmids were sequenced to confirm that the sequences between the MluI and SacII sites contained the desired sequence. The mutant MluI–SacII fragment was excised and religated into fresh, unmutagenized MluI-SacII–digested plasmid A, and the ligation products were then transformed into the TOP10 strain of E. coli. The region of the recovered clones between the MluI and SacII sites was sequenced to verify that the desired mutation was recovered.
To introduce frameshift mutations in the RdRp gene, plasmid F was cut with MunI at position 16152, the overhang was filled in by Klenow fragment DNA polymerase, and then the blunt-ended plasmid was religated, tranformed into E. coli, and the cloned plasmid sequenced to verify the frame shift. This frame-shifted F fragment was then used in the assembly of nonviable Fs mutant genomes.
One-step growth curves
DBT cells were grown in 96-well plates, and triplicate wells were infected with MHV-A59 1000 or mutant virus at a M.O.I of 0.01 or 3. Plates were frozen at −80°C at 0, 4, 8, 12, 16, and 24 h post-infection for cultures infected at M.O.I=3 and at 0, 8, 12, 16, 24, and 36 h post-infection for cultures infected at M.O.I.=0.01. Virus production was quantitated by plaque assay in L2 cells.
Metabolic labeling of mutant viruses
2.25 × 105 of DBT cells were seeded into each well of 24-well plates. Ten hours later, the cells were infected at an M.O.I of 1 with wild-type or mutant viruses. At 6 h post-infection, the cells were washed twice with phosphate-free DMEM without serum, and 0.4 mL of DMEM medium supplemented with 2% dialyzed phosphate free calf serum and 5 μg/mL actinomycin D were added to the cells. The cultures were incubated for 15 min, and 200 μCi of 32P-orthophosphate (MP Biomedicals) were added to the medium and incubated for 6 h, by which time 90% of the cells in the cultures infected with wild-type virus had undergone cell fusion. Cultures were washed twice with cold PBS, and total RNAs were extracted and purified using RNeasy Mini kits (Qiagen). Total RNAs were quantitated using the RediPlate 96 RiboGreen RNA Quantitation Kit (Molecular Probes). Equal amounts of RNA were denatured in formaldehyde gel loading buffer containing ethidium bromide (20 μg/mL) at 65°C for 15 min, and electrophoresed in a 0.8% agarose formaldehye RNA gel. Following electrophoresis, the gel was illuminated with UV light, the image captured with a FluorChem 8900 (AlphaInotech) imaging system, and the relative amount of 18S rRNA in each sample was determined by densitometry. The gel was then fixed with 70% methanol, dried over vacuum, and exposed to X-ray film to visualize radiolabeled MHV-specific RNAs. The relative amount of radiolabeled RNA in each sample was determined by exposing the dried gel to a Molecular Dynamics PhosphorImager equipped with Storm 8.2 software. The amount of 18S rRNA in each sample was used to normalize the PhosphorImager signals to account for small differences in total RNA loaded per sample. The relative amount of virus-specific RNA in each sample was determined by adding the pixel volumes RNAs1–7. The most abundant RNA, RNA7, was set to 100, and the relative molar amount of each individual subgenomic RNA was calculated relative to RNA7.
Detection of positive-strand genomic RNA
Total RNAs of mutants Fs, U48C, and WT were extracted 4, 8, and 12 h post-electroporation, and poly(A)+ mRNAs were extracted from these total RNAs using the QIAGEN Oligotex mRNA Midi kit according to the manufacturer's protocol. One hundred eighty nanograms of poly(A)+ mRNAs were used as a template for the synthesis of genomic cDNA and GAPDH cDNA with primers A59(−)16577–16596 and GAPDH-R2 with the Invitrogen SuperScript II RNase H- Reverse Transcription kit. Positive-strand genomic RNA was amplified by PCR with primers A59(+)16038–16059. GAPDH was also amplified with primers GAPDH-F2 and GAPDH-R2. PCR reactions with poly(A)+-selected RNA demonstrated that samples were free of detectable DNA.
Detection of negative-strand genomic RNA and subgenomic mRNAs of nonviable mutants by RT-PCR
For mutants that were nonviable, total cellular RNAs from 2.6 × 106 BHK-R cells were extracted 4 h and 8 h post-electroporation (a 12-h time point RNA sample was also collected for negative-strand genomic RNA analysis) using RNeasy Mini kits (QIAGEN). Each 50-μL RNA sample was treated with 2 U of RNase-free DNase I at 37°C for 20 min, followed by heating to 70°C for 15 min to inactivate the DNase I. The total cellular RNAs were assayed by nested RT-PCR to detect negative-sense genome-sized RNA, and positive- and negative-sense subgenomic RNAs, as described previously (Yount et al. 2002; Johnson et al. 2005). Briefly, to detect negative-strand RNA, oligo A59(+)14639–14658 was used as the RT primer for cDNA synthesis. Primers A59(+)14639–14658 and A59(−)16596–16577 were used in the first PCR reaction, and primers A59(+)16038–16059 and A59(−)16596–16577 in the nested PCR reaction. To detect positive- and negative-sense subgenomic RNA6, primers A59(−)31270–31288 and A59(+)1–20 were used as RT primers, respectively, primers A59(+)7–23 and A59(−)29593–29613 were used in the first PCR, and primers A59(+)26–47 and A59(−)29327–29345 were used in the nested PCR. The same RT primers were used to synthesize cDNA to detect positive and negative subgenomic RNA7 as for subgenomic RNA6, primers A59(+)7–23 and A59(−)30053–30071 were used in the first PCR, and primers A59(+)26–47 and A59(−)29920–29937 were used in the nested PCR. Parallel reactions in which reverse transcriptase was omitted from the cDNA step were always performed to ensure that the PCRs did not detect residual DNA transcription templates that entered the cells during electroporation (see Table 4).
Luciferase constructs
Firefly luciferase and Renilla luciferase genes were cloned between the MHV-A59 5′ and 3′ UTRs. Wild-type or U48C mutant 5′ UTRs were PCR amplified using primers EcoT7A595UTR (a T7 promoter was placed in front of the 5′ UTR sequence) and BsmA595UTR, cloned into pGEM-T Easy, and sequenced. The MHV-A59 3′ UTR was PCR amplified using primers XbaA593UTR and Bam20T, cloned into pGEM-T Easy, and sequenced. The 3′ UTR fragment was excised with XbaI and BamHI and cloned downstream of the firefly luciferase coding sequence in plasmid pGL3 using these two restriction sites or cloned downstream of the Renilla luciferase coding sequence in plasmid phRL cut with the same enzymes (both plasmids were generously provided by Dr. Lori Bernstein, Texas A&M University Health Science Center). The firefly luciferase-MHV-A59 3′ UTR fusion was PCR amplified using primers BsmFluc and Bam20T, cloned into pGEM-T Easy, and sequenced. The insert was released from pGEM-T Easy by digestion with BsmBI and BamHI and then ligated to the wild-type MHV 5′ UTR fragment or the U48C mutant 5′ UTR fragment after excision from pGEM-T Easy with EcoRI and BsmBI. The ligated fragments were then cloned into pUC18 cut with EcoRI and BamHI, producing the final plasmid constructs pWFluc and pU48CFluc. The Renilla luciferase coding sequence was fused to the MHV-A59 5′UTR (WT or the U48C mutant) and the MHV-A59 3′ UTR using the identical strategy to produce the plasmid constructs pWRluc and pU48CRluc in a pUC18 backbone.
Luciferase assay
The plasmids pWFluc, pU48CFluc, pWRluc, and pU48CRluc were linearized with BamHI. One microgram of linearized plasmid was in vitro transcribed in a total of 20 μL transcription reaction using the Ambion mMESSAGE mMACHINE High Yield Capped RNA Transcription Kit (AM1344) according to the manufacturer's instructions. The transcribed RNA was treated with 2 U of DNase I at 37°C, for 30 min and the DNase I was heat inactivated at 70°C for 15 min. The treated RNA was purified with Ambion NucAway column, quantitated by absorbance at 260 nm with a ND-1000 Spectrophotometer (NanoDrop), and diluted to 100 fmol/μL. Each transcribed RNA (100 fmol) was in vitro translated at 30°C for 30 min, individually or in different combinations (Table 2) using rabbit reticulocyte lysates (Promega). Translation reactions were stopped by the addition of cycloheximide, and 5 μL of the translation reaction was assayed for luciferase activity using the Promega Dual-Luciferase Reporter Assay System. Each translation reaction was performed in triplicate, and luciferase assays were performed for each translation reaction. Relative translation efficiencies of RNAs were calculated by an adaptation of the method of Tahara et al. (1994) to the dual luciferase system. The formula used for calculating the relative translational efficiencies is shown in the notes to Table 2.
ACKNOWLEDGMENTS
This work was supported by NIH grants AI040187 (to D.P.G.) and AI051493 (to J.L.L.). We thank Lindsay Patty for constructing the frame-shifted F plasmid used in this work.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.261807.
REFERENCES
- Antao, V.P., Lai, S.Y., Tinoco I., Jr A thermodynamic study of unusually stable RNA and DNA hairpins. Nucleic Acids Res. 1991;19:5901–5905. doi: 10.1093/nar/19.21.5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, S.C., Sheh, C.-K., Soe, L.H., Chang, M.-F., Vannier, D.M., Lai, M.M.C. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. J. Virol. 1989;63:3693–3699. doi: 10.1128/jvi.63.9.3693-3699.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian, D.A., Spaan, W.J.M. Recombination and coronavirus defective interfering RNAs. Semin. Virol. 1997;8:101–111. doi: 10.1006/smvy.1997.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley, I., Dos Ramos, F.J. Programmed ribosomal frameshifting in HIV-1 and the SARS-CoV. Virus Res. 2006;119:29–42. doi: 10.1016/j.virusres.2005.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello-Villegas, J., Tworowska, I., Nikonowicz, E.P. Metal ion stabilization of the U-turn of the A37 N6-dimethylallyl-modified anticodon stem–loop of Escherichia coli tRNAPhe. Biochemistry. 2004;43:55–66. doi: 10.1021/bi0353676. [DOI] [PubMed] [Google Scholar]
- Campbell, D.O., Legault, P. Nuclear magnetic resonance structure of the Varkud satellite ribozyme stem-loop V RNA and magnesium-ion binding from chemical-shift mapping. Biochemistry. 2005;44:4157–4170. doi: 10.1021/bi047963l. [DOI] [PubMed] [Google Scholar]
- Chang, R.Y., Hofmann, M.A., Sethna, P.B., Brian, D.A. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J. Virol. 1994;68:8223–8231. doi: 10.1128/jvi.68.12.8223-8231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, R.Y., Krishnan, R., Brian, D.A. The UCUAAAC promoter motif is not required for high-frequency leader recombination in bovine coronavirus defective interfering RNA. J. Virol. 1996;70:2720–2729. doi: 10.1128/jvi.70.5.2720-2729.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, K.S., Huang, P., Lai, M.M. Polypyrimidine-tract-binding protein affects transcription but not translation of mouse hepatitis virus RNA. Virology. 2002;303:58–68. doi: 10.1006/viro.2002.1675. [DOI] [PubMed] [Google Scholar]
- Cornish, P.V., Hennig, M., Giedroc, D.P. A loop 2 cytidine-stem 1 minor groove interaction as a positive determinant for pseudoknot-stimulated −1 ribosomal frameshifting. Proc. Natl. Acad. Sci. 2005;102:12694–12699. doi: 10.1073/pnas.0506166102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haan, C.A., Volders, H., Koetzner, C.A., Masters, P.S., Rottier, P.J. Coronaviruses maintain viability despite dramatic rearrangements of the strictly conserved genome organization. J. Virol. 2002;76:12491–12502. doi: 10.1128/JVI.76.24.12491-12502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Denison, M.R., Perlman, S. Translation and processing of mouse hepatitis virus virion RNA in a cell-free system. J. Virol. 1986;60:12–18. doi: 10.1128/jvi.60.1.12-18.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison, M.R., Perlman, S. Identification of a putative polymerase gene product in cells infected with murine coronavirus A59. Virology. 1987;157:565–568. doi: 10.1016/0042-6822(87)90303-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Z., Yu, J., Andino, R., James, T.L. Extending the family of UNCG-like tetraloop motifs: NMR structure of a CACG tetraloop from coxsackievirus B3. Biochemistry. 2003;42:4373–4383. doi: 10.1021/bi027314e. [DOI] [PubMed] [Google Scholar]
- Du, Z., Yu, J., Ulyanov, N.B., Andino, R., James, T.L. Solution structure of a consensus stem-loop D RNA domain that plays important roles in regulating translation and replication in enteroviruses and rhinoviruses. Biochemistry. 2004;43:11959–11972. doi: 10.1021/bi048973p. [DOI] [PubMed] [Google Scholar]
- Dveksler, G.S., Pensiero, M.N., Cardellichio, C.B., Williams, R.K., Jiang, G., Holmes, K.V., Dieffenbach, C.W. Cloning of the mouse hepatitis virus (MHV) receptor: Expression in human and hamster cell lines confers susceptibility to MHV. J. Virol. 1991;65:6881–6891. doi: 10.1128/jvi.65.12.6881-6891.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig, A.L., Scott, W.G., Uhlenbeck, O.C. Inhibition of the hammerhead ribozyme cleavage reaction by site-specific binding of Tb. Science. 1998;279:81–84. doi: 10.1126/science.279.5347.81. [DOI] [PubMed] [Google Scholar]
- Fiala, R., Czernek, J., Sklenar, V. Transverse relaxation optimized triple-resonance NMR experiments for nucleic acids. J. Biomol. NMR. 2000;16:291–302. doi: 10.1023/a:1008388400601. [DOI] [PubMed] [Google Scholar]
- Furtig, B., Richter, C., Wohnert, J., Schwalbe, H. NMR spectroscopy of RNA. ChemBioChem. 2003;4:936–962. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- Goddard, T.D., Kneller, D.G. Sparky 3 NMR software. University of California; San Francisco: 2001. [Google Scholar]
- Goebel, S.J., Hsue, B., Dombrowski, T.F., Masters, P.S. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 2004;78:669–682. doi: 10.1128/JVI.78.2.669-682.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell, R.R., Cannone, J.J., Konings, D., Gautheret, D. Predicting U-turns in ribosomal RNA with comparative sequence analysis. J. Mol. Biol. 2000;300:791–803. doi: 10.1006/jmbi.2000.3900. [DOI] [PubMed] [Google Scholar]
- Hofacker, I.L. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker, I.L., Stadler, P.F., Stocsits, R.R. Conserved RNA secondary structures in viral genomes: A survey. Bioinformatics. 2004;20:1495–1499. doi: 10.1093/bioinformatics/bth108. [DOI] [PubMed] [Google Scholar]
- Hsue, B., Masters, P.S. A bulged stem–loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J. Virol. 1997;71:7567–7578. doi: 10.1128/jvi.71.10.7567-7578.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsue, B., Hartshorne, T., Masters, P.S. Characterization of an essential RNA secondary structure in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 2000;74:6911–6921. doi: 10.1128/jvi.74.15.6911-6921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppler, A., Nikstad, L.J., Allmann, A.M., Brow, D.A., Butcher, S.E. Metal binding and base ionization in the U6 RNA intramolecular stem–loop structure. Nat. Struct. Biol. 2002;9:431–435. doi: 10.1038/nsb800. [DOI] [PubMed] [Google Scholar]
- Ihle, Y., Ohlenschlager, O., Hafner, S., Duchardt, E., Zacharias, M., Seitz, S., Zell, R., Ramachandran, R., Gorlach, M. A novel cGUUAg tetraloop structure with a conserved yYNMGg-type backbone conformation from cloverleaf 1 of bovine enterovirus 1 RNA. Nucleic Acids Res. 2005;33:2003–2011. doi: 10.1093/nar/gki501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, R.F., Feng, M., Liu, P., Millership, J.J., Yount, B., Baric, R.S., Leibowitz, J.L. The effect of mutations in the mouse hepatitis virus 3′(+)42 protein binding element on RNA replication. J. Virol. 2005;79:14570–14585. doi: 10.1128/JVI.79.23.14570-14585.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H., Feng, M., Schroeder, M.E., Giedroc, D.P., Leibowitz, J.L. Putative cis-acting stem-loops in the 5′ untranslated region of the severe acute respiratory syndrome coronavirus can substitute for their mouse hepatitis virus counterparts. J. Virol. 2006;80:10600–10614. doi: 10.1128/JVI.00455-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.-N., Jeong, Y.S., Makino, S. Analysis of cis-acting sequences essential for coronavirus defective interfering RNA replication. Virology. 1993;197:53–63. doi: 10.1006/viro.1993.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koetzner, C.A., Parker, M.M., Ricard, C.S., Sturman, L.S., Masters, P.S. Repair and mutagenesis of the genome of a deltion mutant of the coronavirus mouse hepatitis virus by targeted RNA recombination. J. Virol. 1992;66:1841–1848. doi: 10.1128/jvi.66.4.1841-1848.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, M.M.C., Brayton, P.R., Armen, R.C., Patton, C.D., Pugh, C., Stohlman, S.A. Coronavirus: A jumping RNA transcription. J. Virol. 1981;39:823–834. doi: 10.1128/jvi.39.3.823-834.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, M.M.C., Baric, R.S., Brayton, P.R., Stohlman, S.A. Characterization of leader RNA sequences on the virion and mRNAs of mouse hepatitis virus, a cytoplasmic RNA virus. Proc. Natl. Acad. Sci. 1984;81:3626–3630. doi: 10.1073/pnas.81.12.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebars, I., Yoshizawa, S., Stenholm, A.R., Guittet, E., Douthwaite, S., Fourmy, D. Structure of 23S rRNA hairpin 35 and its interaction with the tylosin-resistance methyltransferase RlmAII. EMBO J. 2003;22:183–192. doi: 10.1093/emboj/cdg022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz, J.L., Wilhelmsen, K.C., Bond, C.W. The virus-specific intracellular RNA species of two murine coronaviruses: MHV-A59 and MHV-JHM. Virology. 1981;114:39–51. doi: 10.1016/0042-6822(81)90250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz, J.L., Weiss, S.R., Paavola, E., Bond, C.W. Cell-free translation of murine coronavirus RNA. J. Virol. 1982;43:905–913. doi: 10.1128/jvi.43.3.905-913.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescrinier, E., Nauwelaerts, K., Zanier, K., Poesen, K., Sattler, M., Herdewijn, P. The naturally occurring N6-threonyl adenine in anticodon loop of Schizosaccharomyces pombe tRNAi causes formation of a unique U-turn motif. Nucleic Acids Res. 2006;34:2878–2886. doi: 10.1093/nar/gkl081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.P., Huang, P., Park, S., Lai, M.M. Polypyrimidine tract-binding protein binds to the leader RNA of mouse hepatitis virus and serves as a regulator of viral transcription. J. Virol. 1999;73:772–777. doi: 10.1128/jvi.73.1.772-777.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, C.L., Lai, M.M. Requirement of the 5′-end genomic sequence as an upstream cis-acting element for coronavirus subgenomic mRNA transcription. J. Virol. 1994;68:4727–4737. doi: 10.1128/jvi.68.8.4727-4737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y.-J., Lai, M.M.C. Deletion mapping of a mouse hepatitis virus defective interfering RNA reveals the requirement of an internal and discontinous sequence for replication. J. Virol. 1993;67:6110–6118. doi: 10.1128/jvi.67.10.6110-6118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., Johnson, R.F., Leibowitz, J.L. Secondary structural elements within the 3′ untranslated region of mouse hepatitis virus strain JHM genomic RNA. J. Virol. 2001;75:12105–12113. doi: 10.1128/JVI.75.24.12105-12113.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters, P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006;66:193–292. doi: 10.1016/S0065-3527(06)66005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, D.A., Dragovich, P.S., Webber, S.E., Fuhrman, S.A., Patick, A.K., Zalman, L.S., Hendrickson, T.F., Love, R.A., Prins, T.J., Marakovits, J.T., et al. Structure-assisted design of mechanism-based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes. Proc. Natl. Acad. Sci. 1999;96:11000–11007. doi: 10.1073/pnas.96.20.11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaskill, J.S. The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers. 1990;29:1105–1119. doi: 10.1002/bip.360290621. [DOI] [PubMed] [Google Scholar]
- Mendez, A., Smerdou, A., Izeta, A., Gebauser, F., Enjuanes, L. Molecular characterization of transmissible gastroenteritis coronavirus defective interfering genomes: Packaging and heterogeneity. Virology. 1996;217:495–507. doi: 10.1006/viro.1996.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, L., Legault, P., Pardi, A. Improved RNA structure determination by detection of Noe contacts to exchange-broadened amino protons. J. Am. Chem. Soc. 1995;117:11043–11048. [Google Scholar]
- Nakagaki, K., Nakagaki, K., Taguchi, F. Receptor-independent spread of a highly neurotropic murine coronavirus JHMV strain from initially infected microglial cells in mixed neural cultures. J. Virol. 2005;79:6102–6110. doi: 10.1128/JVI.79.10.6102-6110.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda, S.K., Leibowitz, J.L. Mitochondrial aconitase binds to the 3′-untranslated region of the mouse hepatitis virus genome. J. Virol. 2001;75:3352–3362. doi: 10.1128/JVI.75.7.3352-3362.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda, S.K., Johnson, R.F., Liu, Q., Leibowitz, J.L. Mitochondrial HSP70, HSP40, and HSP60 bind to the 3′ untranslated region of the murine hepatitis virus genome. Arch. Virol. 2004;149:93–111. doi: 10.1007/s00705-003-0196-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon, P.L., Rangan, A., Kim, Y.G., Rich, A., Hoffman, D.W., Hennig, M., Giedroc, D.P. Solution structure of a luteoviral P1-P2 frameshifting mRNA pseudoknot. J. Mol. Biol. 2002;322:621–633. doi: 10.1016/s0022-2836(02)00779-9. [DOI] [PubMed] [Google Scholar]
- Oberstrass, F.C., Auweter, S.D., Erat, M., Hargous, Y., Henning, A., Wenter, P., Reymond, L., Amir-Ahmady, B., Pitsch, S., Black, D.L., et al. Structure of PTB bound to RNA: Specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- Ohlenschlager, O., Wohnert, J., Bucci, E., Seitz, S., Hafner, S., Ramachandran, R., Zell, R., Gorlach, M. The structure of the stemloop D subdomain of coxsackievirus B3 cloverleaf RNA and its interaction with the proteinase 3C. Structure. 2004;12:237–248. doi: 10.1016/j.str.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Penzes, Z., Wroe, C., Brown, T.D., Britton, P., Cavanagh, D. Replication and packaging of coronavirus infectious bronchitis virus defective RNAs lacking a long open reading frame. J. Virol. 1996;70:8660–8668. doi: 10.1128/jvi.70.12.8660-8668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor, D.J., Schaak, J.E., Bevilacqua, J.M., Falzone, C.J., Bevilacqua, P.C. Isolation and characterization of a family of stable RNA tetraloops with the motif YNMG that participate in tertiary interactions. Biochemistry. 2002;41:12062–12075. doi: 10.1021/bi026201s. [DOI] [PubMed] [Google Scholar]
- Puglisi, E.V., Puglisi, J.D. HIV-1 A-rich RNA loop mimics the tRNA anticodon structure. Nat. Struct. Biol. 1998;5:1033–1036. doi: 10.1038/4141. [DOI] [PubMed] [Google Scholar]
- Quigley, G.J., Rich, A. Structural domains of transfer RNA molecules. Science. 1976;194:796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- Raman, S., Brian, D.A. Stem-loop IV in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 2005;79:12434–12446. doi: 10.1128/JVI.79.19.12434-12446.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman, S., Bouma, P., Williams, G.D., Brian, D.A. Stem–loop III in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 2003;77:6720–6730. doi: 10.1128/JVI.77.12.6720-6730.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas, E., Eddy, S.R. A dynamic programming algorithm for RNA structure prediction including pseudoknots. J. Mol. Biol. 1999;285:2053–2068. doi: 10.1006/jmbi.1998.2436. [DOI] [PubMed] [Google Scholar]
- Sawicki, S.G., Sawicki, D.L. Coronavirus transcription: Subgenomic mouse hepatitis virus replicative intermediates function in RNA synthesis. J. Virol. 1990;64:1050–1056. doi: 10.1128/jvi.64.3.1050-1056.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawicki, S.G., Sawicki, D.L. A new model for coronavirus transcription. Adv. Exp. Med. Biol. 1998;440:215–219. doi: 10.1007/978-1-4615-5331-1_26. [DOI] [PubMed] [Google Scholar]
- Simorre, J.P., Zimmermann, G.R., Pardi, A., Farmer, B.T.N., Mueller, L. Triple resonance HNCCCH experiments for correlating exchangeable and nonexchangeable cytidine and uridine base protons in RNA. J. Biomol. NMR. 1995;6:427–432. doi: 10.1007/BF00197641. [DOI] [PubMed] [Google Scholar]
- Simorre, J.P., Zimmermann, G.R., Mueller, L., Pardi, A. Triple-resonance experiments for assignment of adenine base resonances in C-13/N-15-labeled RNA. J. Am. Chem. Soc. 1996;118:5316–5317. [Google Scholar]
- Sklenar, V., Dieckmann, T., Butcher, S.E., Feigon, J. Through-bond correlation of imino and aromatic resonances in C-13-, N-15-labeled RNA via heteronuclear TOCSY. J. Biomol. NMR. 1996;7:83–87. doi: 10.1007/BF00190460. [DOI] [PubMed] [Google Scholar]
- Snijder, E.J., Bredenbeek, P.J., Dobbe, J.C., Thiel, V., Ziebuhr, J., Poon, L.L., Guan, Y., Rozanov, M., Spaan, W.J., Gorbalenya, A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331:991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola, I., Moreno, J.L., Zuniga, S., Alonso, S., Enjuanes, L. Role of nucleotides immediately flanking the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis. J. Virol. 2005;79:2506–2516. doi: 10.1128/JVI.79.4.2506-2516.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan, W.J.M., Rottier, P.J.M., Horzinek, M.C., van der Zeijst, B.A.M. Isolation and identification of virus-specific mRNAs in cells infected with mouse hepatitis virus. Virology. 1981;108:424–434. doi: 10.1016/0042-6822(81)90449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan, W.J.M., Rottier, P.J.M., Horzinek, M.C., van der Zeijst, B.A.M. Sequence relationships between the genome and the intracellular RNA species 1, 3, 6, and 7 of mouse hepatitis virus strain A59. J. Virol. 1982;42:432–439. doi: 10.1128/jvi.42.2.432-439.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaan, W., Delius, H., Skinner, M., Armstrong, J., Rottier, P., Smeekens, S., van der Zeijst, B.A., Siddell, S.G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2:1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallings, S.C., Moore, P.B. The structure of an essential splicing element: Stem loop IIa from yeast U2 snRNA. Structure. 1997;5:1173–1185. doi: 10.1016/s0969-2126(97)00268-2. [DOI] [PubMed] [Google Scholar]
- Tahara, S.M., Dietlin, T.A., Bergmann, C.C., Nelson, G.W., Kyuwa, S., Anthony, R.P., Stohlman, S.A. Coronavirus translational regulation: Leader affects mRNA efficiency. Virology. 1994;202:621–630. doi: 10.1006/viro.1994.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer, C.A., Wang, Y., Hoffman, D.W., Krisch, H.M., Giedroc, D.P. Non-nearest neighbor effects on the thermodynamics of unfolding of a model mRNA pseudoknot. J. Mol. Biol. 1998;279:545–564. doi: 10.1006/jmbi.1998.1812. [DOI] [PubMed] [Google Scholar]
- Theimer, C.A., Finger, L.D., Feigon, J. YNMG tetraloop formation by a dyskeratosis congenital mutation in human telomerase RNA. RNA. 2003;9:1446–1455. doi: 10.1261/rna.5152303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Born, E., Gultyaev, A.P., Snijder, E.J. Secondary structure and function of the 5′-proximal region of the equine arteritis virus RNA genome. RNA. 2004;10:424–437. doi: 10.1261/rna.5174804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Born, E., Posthuma, C.C., Gultyaev, A.P., Snijder, E.J. Discontinuous subgenomic RNA synthesis in arteriviruses is guided by an RNA hairpin structure located in the genomic leader region. J. Virol. 2005;79:6312–6324. doi: 10.1128/JVI.79.10.6312-6324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Zhang, X. The leader RNA of coronavirus mouse hepatitis virus contains an enhancer-like element for subgenomic mRNA transcription. J. Virol. 2000;74:10571–10580. doi: 10.1128/jvi.74.22.10571-10580.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wege, H., Siddell, S., Sturm, M., ter Meulen, V. Coronavirus JHM: Characterization of intracellular viral RNA. J. Gen. Virol. 1981;54:213–217. doi: 10.1099/0022-1317-54-1-213. [DOI] [PubMed] [Google Scholar]
- Weiss, S.R., Leibowitz, J.L. Comparison of the RNAs of murine and human coronaviruses. Adv. Exp. Med. Biol. 1981;142:245–259. doi: 10.1007/978-1-4757-0456-3_20. [DOI] [PubMed] [Google Scholar]
- Williams, R.K., Jiang, G.S., Holmes, K.V. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc. Natl. Acad. Sci. 1991;88:5533–5536. doi: 10.1073/pnas.88.13.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, G.D., Chang, R.Y., Brian, D.A. A phylogenetically conserved hairpin-type 3′ untranslated region pseudoknot functions in coronavirus RNA replication. J. Virol. 1999;73:8349–8355. doi: 10.1128/jvi.73.10.8349-8355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, H., Yang, M., Ding, Y., Liu, Y., Lou, Z., Zhou, Z., Sun, L., Mo, L., Ye, S., Pang, H., et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. 2003;100:13190–13195. doi: 10.1073/pnas.1835675100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yount, B., Denison, M.R., Weiss, S.R., Baric, R.S. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 2002;76:11065–11078. doi: 10.1128/JVI.76.21.11065-11078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga, S., Sola, I., Alonso, S., Enjuanes, L. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 2004;78:980–994. doi: 10.1128/JVI.78.2.980-994.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]