Abstract
Ribonuclease P (RNase P) is involved in regulation of noncoding RNA (ncRNA) expression in Saccharomyces cerevisiae. A hidden-in-reading-frame antisense-1 (HRA1) RNA in S. cerevisiae, which belongs to a class of ncRNAs located in the antisense strand to verified protein coding regions, was cloned for further use in RNase P assays. Escherichia coli RNase P assays in vitro of HRA1 RNA show two cleavage sites, one major and one minor in terms of rates. The same result was observed with a partially purified S. cerevisiae RNase P activity, both at 30°C and 37°C. These latter cells are normally grown at 30°C. Predictions of the secondary structure of HRA1 RNA in silico show the cleavage sites are canonical RNase P recognition sites. A relatively small amount of endogenous HRA1 RNA was identified by RT-PCR in yeast cells. The endogenous HRA1 RNA is increased in amount in strains that are deficient in RNase P activity. A deletion of 10 nucleotides in the HRA1 gene that does not overlap with the gene coding for a protein (DRS2) in the sense strand shows no defective growth in galactose or glucose. These data indicate that HRA1 RNA is a substrate for RNase P and does not appear as a direct consequence of separate regulatory effects of the enzyme on ncRNAs.
Keywords: endogenous RNA, RNase P cleavage, deletions
INTRODUCTION
RNase P activity has been identified in all three kingdoms of life: Archaea, Bacteria, and Eukarya, as well as in cellular organelles (mitochondria and chloroplasts) that synthesize tRNAs (Gopalan et al. 2002; Hartmann and Hartmann 2003). The enzyme removes 5′-leader sequences from precursor tRNAs by a single magnesium-dependent endonucleolytic cleavage. Homologs of the RNase P RNA gene have been annotated in many microbial genomes (Li and Altman 2004; Yang et al. 2005). Besides its involvement in tRNA maturation (Chamberlain et al. 1996; Gopalan et al. 2002; Xiao et al. 2002), RNase P cleaves some regulatory riboswitches in Escherichia coli (Mandal et al. 2004; Altman et al. 2005). RNase P substrates were also found in E. coli operons in noncoding intergenic regions (Li and Altman 2003), indicating that RNase P activity might play other important, regulatory role(s).
Recently, Samanta et al. (2006) successfully employed high-density genome tiling microarrays to check global Saccharomyces cerevisiae gene expression by depleting crucial RNase P enzyme activity in the host cells that were assayed. Many previously undescribed RNAs were identified in this way (Samanta et al. 2006). A group of ncRNAs was found in the antisense strand of genomic sequence to known protein-coding regions and was defined as hiding in reading-frame antisense (HRA) RNA. Although identified as possible RNase P substrate candidates by microarray analysis, the cleavage of those HRA RNAs by RNase P required further validation in vivo and in vitro. It is also necessary to show that HRA1 RNA is not found because of a peculiar regulatory effect of the loss of RNase P activity.
The HRA1 gene is located in the antisense strand of the DRS2 gene in S. cerevisiae chromosome 1 and partially overlaps with the DRS2 open reading frame (ORF). DRS2 encodes a Golgi membrane-located transport protein (Christie et al. 2004) that is involved in 18S rRNA maturation (Ripmaster et al. 1993). We cloned the HRA1 gene and found it was indeed a substrate for various RNase P enzymes in vitro. The amount of HRA1 RNA in yeast cells was determined by RT-PCR but could not be detected by a Northern blot. The endogenous HRA1 RNA increased about 2.5-fold in an RNase P depletion yeast strain. This result confirms the results of Samanta et al. (2006) that HRA1 is an RNase P substrate.
RESULTS
Alignment of the HRA1 gene in the S. cerevisiae genome
A set of noncoding RNAs in S. cerevisiae with a deletion of RNase P activity was identified with tiling microarrays in the antisense strand of known protein-coding regions and defined as hidden-in-reading-frame antisense RNA (Samanta et al. 2006). This report focuses on HRA1 RNA.
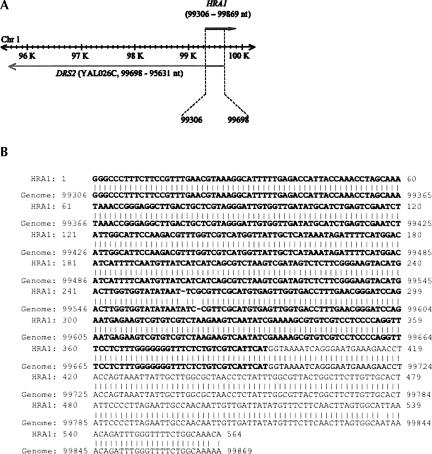
As shown in Figure 1A, the cloned HRA1 gene was located in yeast chromosome 1 from 99,306 to 99,869 (564 nucleotides [nt]) and partially overlapped from 99,306 to 99,698 (393 nt) with the sense strand DRS2 gene (YAL026C, from 99,698 to 95,631 nt; Christie et al. 2004). The upstream sense strand MAK16 gene (YAL025C) runs from 101,146 to 100,226 nt. The HRA1 gene (564 base pairs [bp]) from S. cerevisiae was amplified under the control of T7 promoter and inserted into a pUC19 vector to generate the pUC19-HRA1-3 plasmid (see Materials and Methods). Sequencing analysis of the cloned HRA1 gene shows 99% homology with S. cerevisiae strain S288C chromosome 1 sequence by BLAST2 (available at http://seq.yeastgenome.org/cgi-bin/nph-blast2sgd) and the overlapped sequence with the DRS2 gene is shown in bold letters (Fig. 1B). HRA1 RNA was tested as a substrate for RNase P.
FIGURE 1.
The HRA1 gene in S. cerevisiae Chromosome 1. (A) A cloned HRA1 gene is located in yeast chromosome I from 99,306 to 99,869 in the antisense strand of a known coding region for DRS2 (YAL026C). The upstream sense strand MAK16 (YAL025C) runs from 101,146 to 100,226 nt. (B) A yeast HRA1 gene was amplified directly from intact yeast cells by using primer oligonucleotides HRA1F3 (5′-TAATACGACTCACTATAGGCCCTTTCTTCCGTTTGAA-3′) with T7 promoter sequence (in italics) and HRA1R3 (5′-CATATGTTTGCCAGAAAACCCAAATC-3′) with the NdeI site (underlined). The overlapped sequence with the DRS2 gene is shown in bold letters. The sequence of the cloned HRA1 gene was used for WU-BLAST2 analysis with S. cerevisiae genomic sequence (http://seq.yeastgenome.org/cgi-bin/nph-blast2sgd) and shows 99% homology with the yeast genomic sequence.
Cleavage analysis of yeast HRA1 RNA
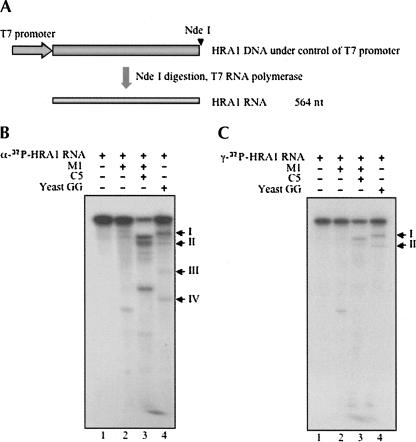
S. cerevisiae HRA1 RNA was transcribed by T7 RNA polymerase from linearized pUC19-HRA1-3 plasmid, as shown in Figure 2A. Both the internal and 5′-end-labeled HRA1 RNA substrates (Materials and Methods) were then employed for RNase P cleavage assays in vitro. The catalytic E. coli M1 RNA (Fig. 2B,C, lane 2), reconstituted E. coli RNase P holoenzymes in vitro (Fig. 2B,C, lane 3), or partially purified yeast RNase P holoenzyme (Fig. 2B,C, lane 4) could cleave HRA1 RNA, but at different sites for the various sources of the enzyme (Fig. 2B,C, cf. lane 4 and lanes 2,3).
FIGURE 2.
Cleavage analysis in vitro of yeast HRA1 RNA with various RNase P enzymes. (A) Yeast HRA1 RNA was transcribed in vitro after linearization of plasmid DNA by NdeI. Both internal labeled (B) and 5′-end-labeled (C) HRA1 RNA were prepared according to Materials and Methods, and then used for cleavage assays in vitro by catalytic E. coli M1 RNA (lane 2, plus 90 mM MgCl2), E. coli RNase P holoenzyme (lane 3), and yeast RNase P holoenzyme (lane 4) in PA buffer (50 mM Tris·HCl at pH 7.5, 10 mM MgCl2, 100 mM NH4Cl). Labeled HRA1 RNA in buffer PA was used as a negative control (lane 1). Reactions were performed at 37°C for 60 min and stopped by adding 8 M urea dye. Samples were then electrophoresed in a 5% polyacrylamide/8 M urea gel and exposed to Kodak BioMax film.
After cleavage by yeast RNase P holoenzyme at 37°C, both internal (Fig. 2B, lane 4) and 5′-end-labeled (Fig. 2C, lane 4) HRA1 RNA showed two main cleavage sites. Semi-logarithmic analysis of band mobilities in the gel (data not shown) was consistent with one cleavage in HRA1 RNA that yielded product bands I and IV and another cleavage that produced bands II and III (Fig. 2B, lane 4). No bands except the ones noted were found (Fig. 2C, lane 4), an indication that the cleavages were independent of each other. Notably, the cleavage efficiency of the yeast RNase P holoenzyme on HRA1 RNA is much lower than that on the precursor tRNA substrates (>95% in 10 min at 37°C; data not shown). Cleavage products of 5′-end-labeled HRA1 RNA were accumulated at 37°C as the reaction time increased from 5 to 60 min, as shown in Figure 3A. Another cleavage assay in vitro was also performed at 30°C. It yielded a similar accumulation of cleaved products (Fig. 3B).
FIGURE 3.
Comparison of cleavage analysis in vitro of yeast HRA1 RNA at different temperatures. (A,B) 5′-End-labeled HRA1 RNA was used for cleavage assays in vitro cleavage assays by yeast RNase P holoenzyme in PA buffer (see Materials and Methods) at 37°C (A) or at 30°C (B) and stopped at different time points by adding 8 M urea dye. Samples were then electrophoresed in 5% polyacrylamide/8 M urea gel and exposed to Kodak BioMax film. (C) Each product obtained from assays A and B was estimated by scanning films with Image Gauge (v3.3). The ratios of each product at different time points were obtained by dividing relevant product with the total amount of remaining substrate and products, as listed in the table. (Triangles) major cleavage at 37°C. (Squares) minor cleavage at 37°C. (Dashed line with triangles) major cleavage at 30°C. (Dashed line with squares) minor cleavage at 30°C. (D) 5′-End-labeled HRA1 RNA was used for cleavage assays in vitro by yeast RNase P holoenzyme in PA buffer at 30°C and stopped at time points by adding 8 M urea dye. Samples were treated as above. (E) (Dashed line with triangles) major cleavage at 30°C. (Dashed line with squares) minor cleavage at 30°C.
The cleavage results for each product are listed in a table in Figure 3C and are illustrated graphically in Figure 3C. By comparing the major and minor cleavages of the two reactions at 37°C or 30°C, yeast RNase P appears to be more specific for the major cleavage (band I) at 30°C than at 37°C. For cleavage reactions at short times (shown in Fig. 3D,E), the rate for the major cleavage is much greater than that for the minor cleavage. As the culture temperature for yeast is usually at 30°C, the yeast RNase P cleavage in vitro at 30°C should resemble results in vivo.
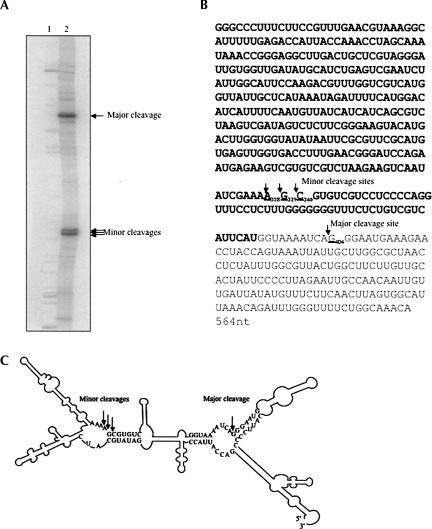
The two cleavage sites by yeast RNase P were readily confirmed (Fig. 4A) by standard methods. The major cleavage is located at the 5′ side at G404 (Fig. 4B, underlined). However, the minor cleavage seems to be a mixture of cleavages at the 5′ sides of three adjacent nucleotides from A338 to C340 (Fig. 4B, bold and underlined), which were located in the antisense region overlapped with the GRS2 gene (bold, Fig. 4B). The major cleavage site was further confirmed with 3′-end-labeled HRA1 RNA and primer extension (data not shown). Using a Zuker program (MFOLD, http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html; Zuker et al. 1999), the model structure of HRA1 RNA is shown in Figure 4C, with cleavage sites indicated by arrows. Both the major cleavage site at the 5′-end G404 and the minor cleavage sites correspond to the canonical model RNase P site, the junction of a single strand and the 5′ end of a helical region in the structure (Altman and Kirsebom 1999).
FIGURE 4.
Cleavage sites of HRA1 RNA with yeast RNase P. (A) 5′-end-labeled HRA1 RNA was a substrate for yeast RNase P cleavage in vitro. An alkali ladder and partial RNase T1 digestion of HRA1 RNA, as described in Materials and Methods, were used for comparison with the cleavage products. Yeast HRA1 RNA cleaved by yeast RNase P holoenzyme, alkali ladder of HRA1 RNA, and partial RNase T1 digestion of HRA1 RNA were individually precipitated with ethanol with glycogen as carrier, resuspended with 8 M urea dye, and loaded on to 6% polyacrylamide/8 M urea sequencing gel. After separating, the gel was dried and exposed to BioMax film (Kodak). The major and minor cleavage sites were marked with arrows. (B) The major cleavage site was located at the 5′ side of G404 (underlined), and the minor cleavage is a mixture of several cleavages at the 5′ termini of three adjacent nucleotides from A338 to G340 (bold and underlined), which were located in the antisense region that overlapped with the DRS2 gene (bold). (C) Scheme of cleavage of HRA1 RNA with major and minor cleavage sites (indicated by arrows). The secondary structure of HRA1 RNA was estimated using the MFOLD program (Zuker et al. 1999). Only individual nucleotides near the cleavage sites are shown.
Endogenous HRA1 RNA transcription
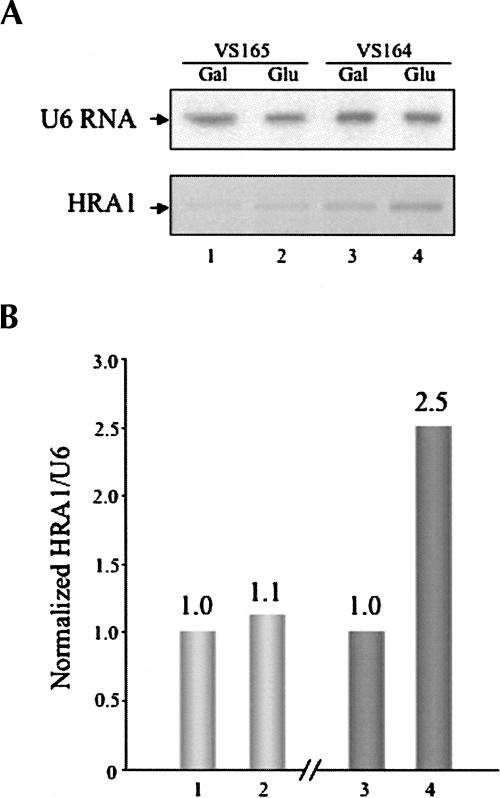
To better understand the tiling microarray result in which HRA1 gene transcription is affected by RNase P activity (Samanta et al. 2006), we checked the endogenous HRA1 RNA transcription in two yeast strains, VS165 and VS164. Unlike the control VS165 yeast strain that can grow on both plates containing galactose or glucose, the mutant VS164 yeast strain grows only on galactose plate medium, but not on glucose plates at 30°C for 72 h. The RNase P activity in VS164 on glucose is inhibited by lethal suppression of the essential Rpp1 protein subunit of yeast RNase P holoenzyme (Stolc and Altman 1997). After the phenotypes of control VS165 and mutant VS164 yeast strains were confirmed on plates, the two yeast strains were cultured in liquid medium with galactose for 4 h and split into medium with galactose or glucose for another 8 h, respectively (Materials and Methods). At 8 h, although the growth rate of the VS164 strain was similar to the control VS165 strain, the absolute amounts of some RNAs that were checked accumulated (Stolc and Altman 1997). Four total RNA samples were purified by hot phenol extraction (Materials and Methods) from VS165 with galactose (Fig. 5A, lane 1), VS165 with glucose (Fig. 5A, lane 2), VS164 with galactose (Fig. 5A, lane 3), or VS164 with glucose (Fig. 5A, lane 4), respectively. Even though 0.01 pmol transcribed HRA1 RNA in vitro was clearly detected with Northern blot analysis, the endogenous HRA1 RNA could not be observed by using two different probes (data not shown), an indication that the endogenous HRA1 RNA absolute amount might be very low. Endogenous U6 RNA was easily checked with a similar amount in all four RNA samples on the same nylon membrane (Fig. 5A, top panel), indicating the qualities of the RNA samples were good. A more sensitive RT-PCR method (Material and Methods) was then used and it successfully amplified the endogenous HRA1 RNA transcription (Fig. 5A, bottom panel). The endogenous HRA1 RNA was estimated using the RT-PCR result and then normalized with endogenous U6 RNA. The endogenous HRA1 gene transcription after normalization had little change in the control VS165 strain cultured with glucose (Fig. 5B, lane 2) or galactose (Fig. 5B, lane 1). However, the transcription of HRA1 RNA in the mutant VS164 strain cultured with glucose (Fig. 5C, lane 4) is about 2.5-fold of the cells grown with galactose (Fig. 5B, lane 3). Thus, it clearly suggests that HRA1 RNA is also a substrate in vivo for RNase P, consistent with the aforementioned cleavage assay in vitro.
FIGURE 5.
Transcription of yeast HRA1 RNA in vivo. (A) S. cerevisiae strains VS165 and VS164 (see Materials and Methods) were both cultured in liquid medium with galactose for 4 h, and then split into medium with either galactose or glucose for another 8 h, respectively. Four RNA samples were purified from VS165 with galactose (lane 1), VS165 with glucose (lane 2), VS164 with galactose (lane 3), or VS164 with glucose (lane 4) by the hot phenol method (Stolc and Altman 1997). Northern blots were performed as described (Guerrier-Takada et al. 1995). The endogenous U6 RNA was used as an internal control (top panel). The RT-PCR method was performed as descibed in Material and Methods. (B) Endogenous HRA1 RNAs from all four total RNA samples (A) were estimated by the RT-PCR result and normalized with endogenous U6 RNA.
Deletion in the yeast HRA1 gene
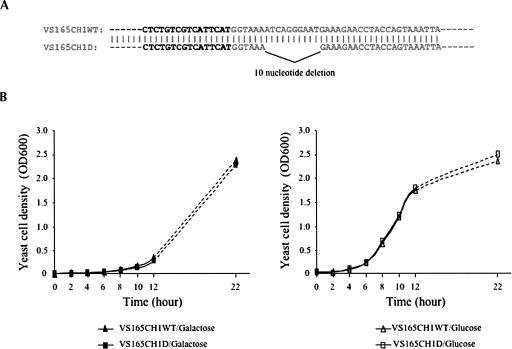
A 10-nt deletion in the yeast HRA1 gene (Fig. 6A), including the major cleavage site, G404, was introduced into the yeast genome by transformation with the integrating plasmid YIplac128SCCH1D to get the strain VS165CH1D, which had a 10-nt deletion in the HRA1 gene. A control yeast strain, VS165CH1WT, was also obtained by parallel transformation with the control integrating plasmid YIplac128SCCH1 (Materials and Methods). Both the deletion sequence in VS165CH1D and wild-type sequence in VS155CH1WT were confirmed by genomic PCR and sequence analysis. Comparison with the control wild-type yeast strain, VS165CH1WT (triangles), showed the 10-nt deletion in the HRA1 gene had no effect on the growth of the mutant strain, VS165CH1D (squares), in either galactose (left panel of Fig. 2B) or glucose (right panel of Fig. 2B) medium, when cultured at 30°C. The effect(s) of this 10-nt deletion in the HRA1 gene on global gene expression is being tested further by microarray analysis.
FIGURE 6.
Deletion analysis of the the yeast HRA1 gene. (A) The 10-nt deletion (ATCAGGGAAT, from 99,705 to 99,714) in the HRA1 gene is designed upstream of the sense DRS2 gene (bold) in yeast chromosome 1 and introduced into yeast cells (see Materials and Methods). (B) Growth of the wild-type strain, VS165CH1WT (triangles), and deletion strain, VS165CH1D (squares), after transfer from overnight galactose-containing medium to either galactose-containing (left panel) or glucose-containing (right panel) medium at T = 0 h. Yeast cells were cultured at 30°C, and cell densities were measured at times indicated.
DISCUSSION
We report the RNase P cleavage in vitro of HRA1 RNA from S. cerevisiae. HRA1 RNA could be cleaved by yeast RNase P at two sites (a major and a minor cleavage; Fig. 2). Yeast RNase P, E. coli RNase P holoenzyme, and its catalytic RNA subunit (M1 RNA) each cleaved HRA1 RNA at different sites (Fig. 2B,C). One explanation of these results is the notion that a critical function of the protein component from various species of RNase P is to alter substrate specificity and enhance catalytic efficiency besides binding the substrate (Niranjanakumari et al. 1998; Altman and Kirsebom 1999).
The endogenous transcription of HRA1 RNA was checked by RT-PCR in both control VS165 and mutant VS164 yeast strains. The HRA1 RNA amount is significantly increased in mutant VS164 yeast strain cultured with glucose in which RNase P activity is depleted.
Since the yeast HRA1 gene partially overlaps with the DRS2 ORF region on the complementary genome strand (Fig. 1), deletion of the full length of the HRA1 gene would certainly result in side effects reflected by the mutant DRS2 gene. In this study, a 10-nt deletion in the HRA1 gene, containing the cleavage G404 site, was designed upstream of the DRS2 ORF on the sense strand, but subsequent studies showed that the 10-nt deletion had no effect on yeast growth (Fig. 6). The ongoing microarray analysis will elucidate the effect(s) of the HRA1 gene on the expressions of other genes, especially DRS2 and MAK16 ORFs (Fig. 1A) on the sense strand.
RNase P in E. coli can cleave other noncoding RNAs (for example, riboswitches; Altman et al. 2005) besides tRNA and rRNA. Since almost every method of gene identification used to assume that genes encode proteins, ncRNA genes have been effectively invisible and their functions are rarely studied (for review, see Eddy 2001). Very little was known about the noncoding RNA substrate candidates for RNase P. Recently, through microarray analysis, new noncoding RNAs were identified and were reported to be regulated by RNase P in both E. coli (Li and Altman 2003) and in S. cerevisiae (Samanta et al. 2006). It is not obvious whether the function of RNase P cleaving noncoding RNAs besides tRNA (HRA1 RNA, for example) originated during the “RNA world” (Gilbert 1986) or subsequently evolved from the “RNA–protein world.” RNA genes were thought of as rare relics of a primordial “RNA world.” These genes were presumed to have largely been replaced by a more efficient “protein world.” Perhaps, now an “RNA–protein world” instead of a “protein world” is the most appropriate label for Nature as we see it.
MATERIALS AND METHODS
Strains
Saccharomyces cerevisiae strains, the control VS165 (MATα GAL1 leu2–3, 112 ura3–1+pYCpGAL [URA3]) and the mutant VS164 (MATα GAL1 leu2–3, 112 ura3–1+pYCpGAL::rpp1 [URA3]), were maintained in culture medium with galactose, and the depletion of endogenous RNase P activity in yeast VS164 strain was obtained by culturing in medium with glucose to inhibit yeast Rpp1 protein expression, but not in the VS165 control strain (Stolc and Altman 1997).
Enzymes
DNA restriction and other enzymes reacting with RNA were obtained from New England Biolabs unless described otherwise. Radiochemicals were from Amersham. Oligonucleotides were synthesized by the Keck Facility at Yale University.
Construction of DNA plasmids
The yeast HRA1 gene was amplified directly from intact S. cerevisiae cells by using primer oligonucleotides HRA1F3 (5′-TAATACGACTCACTATAGGCCCTTTCTTCCGTTTGAA-3′) with a T7 promoter sequence (in italics) and HRA1R3 (5′-CATATGTTTGCCAGAAAACCCAAATC-3′) with an NdeI site (underlined). The amplified fragment was purified and inserted into the SmaI site of the pUC19 vector, to generate plasmid pUC19HRA1-3, which contained the yeast HRA1 gene under the control of the T7 promoter. Two partially overlapped yeast genome DNA fragments were individually amplified from S. cerevisiae cells by using primer oligonucleotides SCCH1F1 (5′-AAGGCTGCAGCGCACTATACTACGCAAC-3′) with a PstI site (in italics) and SCCH1R3 (5′-CAATGCCTTTCTCTCCCGGGTCTGCGTCGCCAACGTTC-3′) with SmaI site (in italics) or SCCH1F3 (5′-GTTGGCGACGCAGACCCGGGAGAGAAAGGCATTGGTAGC-3′) with SmaI site (in italics) and SCCH1R4 (5′-AAGGGAGCTCGTGCCACTTCTTGCTCAG-3′) with SacI site (in italics), and were then fused by denaturation and annealing in a subsequent extension reaction. Finally, the fused and extended product was amplified by PCR using primer oligonucleotides SCCH1F1 and SCCH1R4. The final PCR product was digested with PstI and SacI and inserted into the PstI and SacI sites of an integrating plasmid, YIplac128, with a LEU2 selection marker to obtain plasmid YIplac128SCCH1. A 10-nt deletion (ATCAGGGAAT, from 99,705 to 99,714) in the HRA1 gene was introduced into YIplac128SCCH1 by two primer oligonucleotides Del FP (5′-CTGTCGTCATTCATGGTAAAGAAAGAACCTACCAG-3′) that did not have the sequence between A and G (underlined), normally found in HRA1 and DelRP (5′-CTGGTAGGTTCTTTCTTTACCATGAATGACGACAG-3′) without the deleted sequences between C and T (underlined) according to the protocol described by the Stratagene QuikChange Site-Directed Mutagenesis Kit to make plasmid YIplac128SCCH1D, which contains a 10-nt deletion. All plasmids were confirmed by restriction enzyme digestion and by DNA sequencing, which was performed according to the protocol described by the USB Corporation Sequencing Kit with slight modifications.
Transcription in vitro
Yeast HRA1 RNA was transcribed by T7 RNA polymerase from plasmid pUC19HRA1-3, which had been first linearized by NdeI. The transcription reactions in vitro were carried out as described (Yuan et al. 1992), and transcripts were purified by using Sephadex G-50 Quick Spin columns (Roche Diagnostics). For 5′-end labeling, the transcribed HRA1 RNA was first dephosphorylated with 0.01 U calf intestinal phosphatase (CIP)/pmol RNA, and then 5′-end labeled by T4 polynucleotide kinase with the presence of [γ-32P]ATP. For internal labeling, transcription in vitro was performed with [α-32P]GTP from linearized pUC19HRA1-3. Both the 5′-end-labeled and internally labeled HRA1 RNAs were purified on 5% polyacrylamide/8 M urea gel, and used at 5000 cpm per cleavage reaction. E. coli RNase P RNA subunit M1 was generated by transcription in vitro using T7 RNA polymerase (Promega) and purified by using Sephadex G-50 Quick Spin columns (Roche Diagnostics) (Vioque et al. 1988). The substrate RNA, E. coli precursor tRNATyr (pTyr), was transcribed in vitro with [α-32P]GTP, purified on a 5% polyacrylamide/8 M urea gel, and used at 5000 cpm per reaction.
Preparation of RNase P enzymes
E. coli RNase P protein subunit C5 (Gopalan et al. 1997) was used to reconstitute in vitro E. coli RNase P holoezyme in PA buffer (50 mM Tris·HCl at pH 7.5, 10 mM MgCl2, 100 mM NH4Cl) with E. coli M1 RNA, which was transcribed in vitro. Yeast RNase P holoenzyme was partially purified through the glycerol gradient step.
RNase P cleavage assay in vitro
RNase P cleavage assays were carried out in buffer PA (50 mM Tris·HCl at pH 7.5, 10 mM MgCl2, 100 mM NH4Cl) with E. coli M1 RNA plus an additional 90 mM MgCl2, or with reconstituted in vitro E. coli RNase P holoenzyme (M1 RNA plus C5 protein), or with purified yeast RNase P holoenzyme at 37°C or 30°C. Reactions were stopped at designated time points by adding 8 M urea dye, and the mixture was electrophoresed on 5% polyacrylamide/8 M urea gels and exposed to BioMax film (Kodak). If needed, each product from the cleavage assay was estimated quantitatively with Image Gauge (v3.3). The ratios of each product were obtained by dividing the relevant product with the total amount of remaining substrate and products.
Alkali ladder and partial RNase T1 digestion of HRA1 RNA
An alkali ladder of HRA1 RNA was obtained by incubating 5′-end-labeled HRA1 RNA in alkali buffer (50 mM NaHCO3 at pH 9.2, 1 mM EDTA) at 95°C for 3 min. Partial RNase T1 (Calbiochem) digestion of HRA1 RNA was carried out in PA buffer (50 mM Tris·HCl at pH 7.5, 10 mM MgCl2, 100 mM NH4Cl) on ice for 30 sec, and stopped by adding phenol. Yeast HRA1 RNA cleaved by yeast RNase P holoenzyme, an alkali ladder of HRA1 RNA, and partial RNase T1 digestion of HRA1 RNA were individually precipitated with ethanol and glycogen as carrier, resuspended with 8 M urea dye, and loaded on to 6% PAGE/urea sequencing gel. After electrophoretic separation, the gel was dried and exposed to BioMax film (Kodak).
Total RNA preparation, Northern blot, and RT-PCR
Two yeast strains, VS165 and VS164, were individually cultured in liquid medium with galactose for 4 h and then split into both medium with galactose or glucose for another 8 h (25 mL, OD600 = 0.5). After that, yeast pellets were obtained by centrifugation and stored at −80°C. Total RNAs were isolated from yeast cell pellets by the hot phenol method as described (Stolc and Altman 1997), followed by DNase I (Worthington Biochemical Corp.) treatment to eliminate genomic DNAs. Both Northern blot and RT-PCR were used to detect the endogenous transcription of yeast HRA1 RNA. Total RNA samples were separated in 3% agarose gel and transferred onto a positively charged nylon membrane (Roche Diagnostics) with 0.01 pmol in vitro transcribed HRA1 RNA as a positive control.
Northern blots were performed as described (Guerrier-Takada et al. 1995) with different probes NBPHRA1 (5′-CTAGGTTTGGTAATGGTC-3′) or PRIEXT1 (5′-GAATAGTGCAACAAGAAGCCAG-3′). Endogenous U6 RNA was also checked by Northern blot with oligo U6 (5′-TCATCCTTATGCAGGG-3′) as probe to be used as an internal control. Reverse transcription of yeast total RNA by superscript reverse transcriptase (Invitrogen) was carried out with gene-specific primer oligonucleotide HRA1R3, followed by PCR with oligonucleotides HRA1F3 and HRA1R3 for 30 cycles. After RT-PCR, cDNAs were separated in an agarose gel and quantified by Image Gauge (v3.3). The relative amount of endogenous HRA1 RNA was estimated by scanning the RT-PCR gels and normalized with U6 RNA as an internal control.
Yeast transformation
YIplac128SCCH1 and YIplac128SCCH1D were individually linearized with SmaI and then transformed into competent VS165 yeast cells according to the polyethylene glycol (PEG)/lithium acetate (LiAc) method (Gietz and Woods 2002) with slight modification. Briefly, an overnight culture of VS165 yeast cells was transferred to fresh media with galactose (25 mL, OD600 = 0.15) and incubated for another 5 h at 30°C (OD600 = 0.5). Yeast cells were harvested by centrifugation at 3000g for 5 min, washed with sterile water, and resuspended in 200 μL TE/LiAc (10 mM Tris HC1, 1 mM EDTA, 100 mM LiAc at pH 7.5) to obtain competent cells. For each transformation, 100 μL competent cells were added with 5 μg linearized DNA, 100 μg single-strand carrier DNA, and 600 μL PEG/LiAc (40% PEG 4000 [w/v], 10 mM Tris HCl, 1 mM EDTA, 100 mM LiAc at pH 7.5), and cultured at 30°C for 30 min. After adding 10% DMSO, the transformation mixtures were heat shocked at 42°C for 15 min and chilled on ice. The yeast cells were then centrifuged, resuspended with TE (10 mM Tris HC1, 1 mM EDTA at pH 7.5), separated onto plates that lacked leucine, and incubated at 30°C for 3–4 d. Transformants were further confirmed by genome PCR amplifying by using primer oligonucleotides HRA1F3 and HRA1R3, and were checked by DNA sequencing with HRASEQ1 (5′-TCATCAGCGTCTAAGTC-3′) at the DNA Analysis Facility at Yale University. For further study, total RNAs were isolated from yeast cell pellets by the hot phenol method as described (Stolc and Altman 1997).
ACKNOWLEDGMENTS
We thank Dr. Viktor Stolc and Dr. Hideo Tsubouchi, as well as our laboratory colleagues, especially Dr. Cecilia Guerrier-Takada, for helpful discussions and technical support. Plasmid YIplac128 was donated by Dr. Shirleen Roeder at Yale University. L.Y. is supported by a Focused Giving grant from the Johnson and Johnson Co.; Yale University assisted S.A.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.460607.
REFERENCES
- Altman, S., Kirsebom, L. Ribonuclease P. In: Gesteland R.F., et al., editors. The RNA world. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. pp. 351–380. [Google Scholar]
- Altman, S., Wesolowski, D., Guerrier-Takada, C., Li, Y. RNase P cleaves transient structures in some riboswitches. Proc. Natl. Acad. Sci. 2005;102:11284–11289. doi: 10.1073/pnas.0505271102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain, J.R., Pagan, R., Kindelberger, D.W., Engelke, D.R. An RNase P RNA subunit mutation affects ribosomal RNA processing. Nucleic Acids Res. 1996;24:3158–3166. doi: 10.1093/nar/24.16.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie, K.R., Weng, S., Balakrishnan, R., Costanzo, M.C., Dolinski, K., Dwight, S.S., Engel, S.R., Feierbach, B., Fisk, D.G., Hirschman, J.E., et al. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 2004;32:D311–D314. doi: 10.1093/nar/gkh033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy, S.R. Noncoding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., Woods, R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Gilbert, W. The RNA world. Nature. 1986;319:618. [Google Scholar]
- Gopalan, V., Baxevanis, A.D., Landsman, D., Altman, S. Analysis of the functional role of conserved residues in the protein subunit of ribonuclease P from Escherichia coli. J. Mol. Biol. 1997;267:818–829. doi: 10.1006/jmbi.1997.0906. [DOI] [PubMed] [Google Scholar]
- Gopalan, V., Vioque, A., Altman, S. RNase P: Variations and uses. J. Biol. Chem. 2002;277:6759–6762. doi: 10.1074/jbc.R100067200. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada, C., Li, Y., Altman, S. Artificial regulation of gene expression in Escherichia coli by RNase P. Proc. Natl. Acad. Sci. 1995;92:11115–11119. doi: 10.1073/pnas.92.24.11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, E., Hartmann, R.K. The enigma of ribonuclease P evolution. Trends Genet. 2003;19:561–569. doi: 10.1016/j.tig.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Li, Y., Altman, S. A specific endoribonuclease, RNase P, affects gene expression of polycistronic operon mRNAs. Proc. Natl. Acad. Sci. 2003;100:13213–13218. doi: 10.1073/pnas.2235589100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Altman, S. In search of RNase P RNA from microbial genomes. RNA. 2004;10:1533–1540. doi: 10.1261/rna.7970404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal, M., Lee, M., Barrick, J.K., Wenberg, Z., Emilsson, G.M., Ruzzo, W.L., Breaker, R.R. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- Niranjanakumari, S., Stams, T., Crary, S.M., Christianson, D.W., Fierke, C.A. Protein component of the ribozyme ribonuclease P alters substrate recognition by directly contacting precursor tRNA. Proc. Natl. Acad. Sci. 1998;95:15212–15217. doi: 10.1073/pnas.95.26.15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripmaster, T.P., Vaughn, G.P., Woolford J.L., Jr DRS1 to DRS7, novel genes required for ribosome assembly and function in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;12:7901–7912. doi: 10.1128/mcb.13.12.7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta, M.P., Tongprasit, W., Sethi, H., Chin, C.S., Stolc, V. Global identification of noncoding RNAs in Saccharomyces cerevisiae by modulating an essential RNA processing pathway. Proc. Natl. Acad. Sci. 2006;103:4192–4197. doi: 10.1073/pnas.0507669103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolc, V., Altman, S. Rpp1, an essential protein subunit of nuclear RNase P required for processing of precursor tRNA and 35S precursor rRNA in Saccharomyces cerevisiae. Genes & Dev. 1997;11:2926–2937. doi: 10.1101/gad.11.21.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vioque, A., Arnez, J., Altman, S. Protein–RNA interactions in the RNase P holoenzyme from Escherichia coli. J. Mol. Biol. 1988;202:835–848. doi: 10.1016/0022-2836(88)90562-1. [DOI] [PubMed] [Google Scholar]
- Xiao, S., Scott, F., Fierke, C.A., Engelke, D.R. Eukaryotic ribonuclease P: A plurality of ribonucleoprotein enzymes. Annu. Rev. Biochem. 2002;71:165–189. doi: 10.1146/annurev.biochem.71.110601.135352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, L., Wesolowski, D., Li, Y., Altman, S. Analysis of putative RNase P RNA from orthopoxviruses. J. Mol. Biol. 2005;354:529–535. doi: 10.1016/j.jmb.2005.09.020. [DOI] [PubMed] [Google Scholar]
- Yuan, Y., Hwang, E.S., Altman, S. Targeted cleavage of mRNA by human RNase P. Proc. Natl. Acad. Sci. 1992;89:8006–8010. doi: 10.1073/pnas.89.17.8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M., Mathews, D.H., Turner, D.H. Algorithms and thermodynamics for RNA secondary structure prediction: A practical guide in RNA biochemistry and biotechnology. In: Barciszewski J., Clark B.F.C., editors. RNA biochemistry and biotechnology. NATO ASI Series, Kluwer Academic Publishers; Dordrecht, The Netherlands: 1999. pp. 11–43. [Google Scholar]