Abstract
The previously described NMR structure of a 5′-CU-3′/5′-UU-3′ motif, which is highly conserved within the 3′-UTR Y-stem of poliovirus-like enteroviruses, revealed striking regularities of the local helix geometry, thus retaining the pseudo-twofold symmetry of the RNA helix. A mutant virus with both pyrimidine base pairs changed into Watson–Crick replicated as wild type, indicating the functional importance of this symmetry relation in viral RNA replication. Here we investigated the effect of changing only one of the two pyrimidine base pairs to Watson–Crick. We determined the NMR structures of two Y-stem variants: one containing the 5′-CU-3′/5′-AU-3′ motif, which has been found in wild-type virus isolates as well, and the other containing a 5′-CU-3′/5′-UG-3′ motif, which is not present in any enterovirus sequenced to date. Both structures show single pyrimidine mismatches with intercalated bases. In the 5′-CU-3′/5′-AU-3′ motif a C–U Watson–Crick-type base pair is formed that retains the pseudo-twofold symmetry, while in the 5′-CU-3′/5′-UG-3′ motif a single asymmetric U–U mismatch breaks the twofold symmetry. Surprisingly, for the nonnatural variant no effect of the single base-pair replacement was observed on polioviral RNA replication using an in vitro replicon assay.
Keywords: RNA, RNA structure, NMR, U–U/C–U base pair, poliovirus
INTRODUCTION
Noncanonical base pairs are essential elements in functional RNAs. Because their basic secondary structure element, the RNA helix, is dominated by Watson–Crick base pairs, the noncanonical base pairs are often the ones playing the important roles. This can be either structurally, e.g., in the formation and stabilization of higher-order structures or in protein recognition, or functionally, for example, in the mechanism of catalytic RNA (Cate et al. 1996; McKay 1996; Hermann and Westhof 1999).
For understanding how an RNA molecule works and how noncanonical base pairs contribute to function, structural knowledge is a necessity. Unfortunately, the rules that govern the folding of RNA molecules containing noncanonical base pairs are still far from understood. One complicating factor is that the structures of many noncanonical base pairs are polymorphic, i.e., that their base-pair configuration is dependent on the structure of nearby elements, e.g., adjacent base pairs or loops, or by a third interaction such as in a base triple. The current situation is that, despite the large amount of RNA structural data that have been documented, it is still hard to predict the detailed structure of even a simple structural motif, such as a helical element containing a noncanonical base pair, from sequence alone.
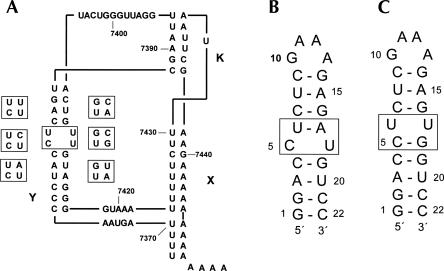
We have been interested in the structure and function of the poliovirus cis-acting element oriR, a well-characterized multidomain globular structure involving the 3′-untranslated region (3′-UTR) of the single-stranded RNA genome, thought to be involved in the initiation of the complementary strand synthesis (Pilipenko et al. 1996; Melchers et al. 1997; Wang et al. 1999; van Ooij et al. 2006). The secondary structure contains two hairpins, designated as X and Y, and a kissing loop X–loop Y interaction called the K-domain (Fig. 1A). The Y-stem is largely conserved within the polio-like viruses and contains two consecutive pyrimidine base pairs in most variants (Figs. 1A, 2). Since regulation of viral RNA synthesis by the 3′-UTR is mediated by protein binding (Harris et al. 1994; Todd et al. 1995; Mellits et al. 1998) and the number of base pairs of the Y-stem and spatial orientation with respect to the X-stem is largely restricted (Melchers et al. 2000; van Ooij et al. 2006), we envisaged that these pyrimidine base pairs could have a more specific role in viral replication, and therefore subjected them to separate investigations.
FIGURE 1.
(A) Schematic representation of the secondary structure of the 3′-UTR of poliovirus type 1. (B) RNA hairpin Ycu and (C) hairpin Yuu used for the NMR studies. Variation of the central two base pairs in the Y domain is shown on the left of the Y domain in A for the polio and polio-like coxsackie viruses, for the nonpolio-like coxsackie and ECHO viruses on the right.
FIGURE 2.
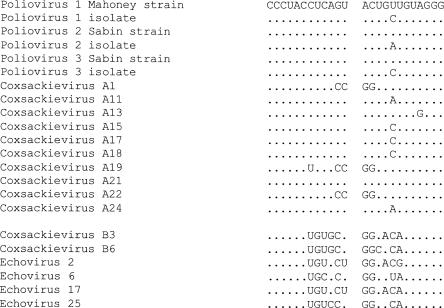
Sequence alignment of the 3′-UTR Y-stem of enteroviral RNAs. The upper section shows the sequences of the polio and polio-like coxsackie viruses, the lower section shows the nonpolio-like coxsackie and ECHO viruses.
We previously solved the structure of the poliovirus Y-stem containing the 5′-CU-3′/3′-UU-5′ tandem base-pair motif by NMR (Lescrinier et al. 2003). While the sequence of this motif is not self-complementary, and hence the secondary structure is not symmetric, the NMR structure revealed isosteric C–U and U–U base pairs retaining the typical pseudo-twofold symmetry of the A-form RNA helix. We subsequently argued that this symmetry relationship may be used to explain the specific configurations of the pyrimidine base pairs in the 5′-CU-3′/-3′UU-5′ sequence and those of other tandem pyrimidine–pyrimidine mismatches in RNA helices as well (Heus and Hilbers 2003; Lescrinier et al. 2003). Furthermore, we envisaged that the retained twofold symmetry might be important for viral RNA replication, and indeed a mutant virus with both pyrimidine base pairs changed into Watson–Crick replicated as wild type.
The pyrimidine-rich base-pair tandem is largely conserved in the 3′UTR Y-domain of polio-like enteroviruses with the 5′-CU-3′/5′-UU-3′ sequence being the most abundant, followed by the less abundant 5′-CU-3′/5′-CU-3′ (Fig. 2). However, also a single C–U pair in the context of a 5′-CU-3′/5′-AU-3′ sequence can be found in poliovirus-like enteroviruses. In the nonpolio-like enteroviruses the pyrimidine base pairs are replaced by Watson–Crick. Except for the 5′-CU-3′/5′-AU-3′ sequence, all of these sequences have been shown to possess pseudo-twofold symmetry (Saenger 1984; Lescrinier et al. 2003).
Thus, we reasoned that if the pseudo-twofold symmetry, present in regular helices containing Watson–Crick base pairs and those containing the tandem noncanonical base pairs, is important for function of the 3′-UTR, it might also be present in the wild-type 5′-CU-3′/5′-AU-3′ sequence with a single C–U mismatch. On the other hand, since single noncanonical base pairs can be notably asymmetric, introduction of a single pyrimidine base pair might break the pseudo-twofold symmetry.
To look into these possibilities we investigated two Y-stem variants either containing a 5′-CU-3′/5′-AU-3′ or a 5′-CU-3′/5′-UG-3′ motif. With respect to the wild-type 5′-CU-3′/5′-UU-3′ sequence in these variants the U–U base pair is changed into U–A or the C–U base pair is changed into C–G, respectively. The NMR structures show that pseudo-twofold symmetry is retained in the wild-type 5′-CU-3′/5′-AU-3′ sequence, but lost in the nonnatural 5′-CU-3′/5′-UG-3′ variant. These observations are corroborated by numerical analyses of helical parameters and cross-strand distances, which for the latter sequence also corresponds with local distortion of the helix at the single mismatch site.
RESULTS
Resonance assignment and structure calculations
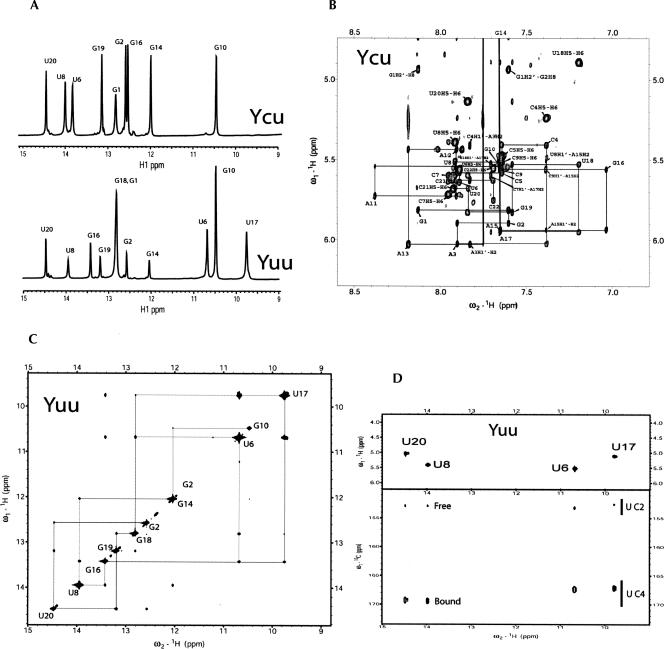
The structures of the two Y-stem variants with either a 5′-CU-3′/5′-AU-3′ or 5′-CU-3′/5′-UG-3′ sequence were determined by NMR using the 22-mer constructs shown in Figure 1, B and C. Henceforth we will refer to these variants as Ycu (Fig. 1B) and Yuu (Fig. 1C), respectively. Using the unlabeled samples, all aromatic, anomeric, and H2′ resonances of both variants could be assigned starting from a classic anomeric to aromatic proton walk (Wijmenga et al. 1993). Heteronuclear NMR experiments performed on the 13C/15N-labeled Ycu sample or U-specific 13C/15N-labeled Yuu sample, confirmed the assignments and allowed for additional identification of other ribose 1H and 13C resonances. Chemical shifts and NOE contacts in the GAAA tetraloop residues of both variants were similar to those found for hairpin structures described before (Jucker et al. 1996; Lescrinier et al. 2003) indicative of identical loop structures.
Imino protons were assigned from 2D 1H-15N HMQC experiments and classic imino-to-imino proton sequential walks in the 2D-NOESY experiments recorded in H2O. For the Ycu variant the data clearly provided evidence for the restored Watson–Crick U6–A17 base pair (Fig. 3A). However, although the upfield H5/H6 chemical shifts of the C5 and U18 bases clearly indicate intercalated stacked bases in the A-form helix (Fig. 3B), no imino proton signal could be detected for the flanking C–U pair.
FIGURE 3.
(A) One-dimensional 1H NMR spectra of the Yuu (lower trace) and Ycu (upper trace) RNA hairpins recorded at pH 6.0 at 600 MHz, showing the imino proton region. Assignments to residues are indicated. (B) Section of the 2D NOESY spectrum (300 msec mixing time) of the Ycu RNA hairpin recorded in D2O at pH 6.8 at 800 MHz, showing the anomeric to aromatic proton region. Sequential H1′-H8/H6 connectivities are indicated by lines. Intranucleotide H1′-H6/H8 NOEs are annotated according to residue type and number, e.g., G1. (C) Section of the 2D NOESY spectrum (300 msec mixing time) of the Yuu RNA hairpin recorded in 90% H2O/10% D2O at pH 6.0 at 800 MHz, showing the imino proton region. Assignments of the diagonal peaks are indicated by residue type and number. The sequential imino-to-imino proton walk is traced out. (D) Residue-specific imino proton assignment and chemical-shift dependence of the C2 and C4 chemical shifts on base pairing of the Yuu RNA hairpin. The spectra were recorded at 5°C at 600 MHz. The 2D back-and-forth H(N3C5)H5 spectrum on top shows the imino-H5 connectivities of the U-specific 13C/15N-labeled Yuu sample, used for assignnment of uridine imino protons. The 2D HNCO spectrum on the bottom was used to establish the hydrogen bonding patterns of the uridines based on the C2 and C4 chemical shifts.
For the Yuu variant all helical imino protons of the stem residues and the loop guanosine were detectable, and a complete sequential imino-to-imino proton walk could be made for these residues, including the opposite U6 and U17 pyrimidines (Fig. 3C). Uridine imino protons were unambiguously assigned using a 2D back-and-forth H(N3C5)H5 experiment (Fig. 3D). In the case of the Yuu molecule, the upfield H5/H6 chemical shifts of the U6 and U17 bases also clearly indicate intercalated, stacked bases in the A-form helix, and therefore the hydrogen bonding patterns of uridines were further assessed by inspection of the chemical-shift signatures of the uridine carbonyl groups. In this approach the 13C chemical shifts of uridine carbonyl C2 and C4 nuclei are compared to the carbonyl chemical shifts of uridines in A–U or G–U base pairs observed in a 2D H(N)CO spectrum (Fig. 3D). Earlier work showed that a downfield chemical shift of the uridine C2 and an upfield chemical shift of the C4 signal correlates with the carbonyl oxygen being hydrogen bonded (Furtig et al. 2003; Theimer et al. 2003; Du et al. 2004; Ohlenschlager et al. 2004). For the Yuu molecule, the C4 signals of U6 and U17 are shifted upfield with respect to the signals of the hydrogen-bonded C4 signals of the Watson–Crick base-paired U8 and U20, and the U6/U17 C2 signals do not show a significant downfield shift with respect to those of the free C2 carbonyls of U8 and U20. These results indicate that despite the presence of two sharp imino proton signals with a strong mutual NOE (Fig. 3A,C) the bases in the U6–U19 pair are too far apart to form stable hydrogen bonds.
Analysis of the DQF-COSY and TOCSY spectra revealed that all stem residues adopt N-type sugar conformations, except for the Ycu G19 residue that displayed a weak signal in the DQF-COSY spectrum, indicative of an N/S-type mixture.
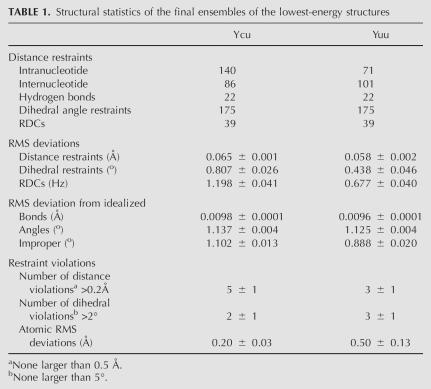
The structures of the Ycu variant were calculated using 248 distance, 175 torsion angle, and 39 RDC restraints, while those of the Yuu variant were calculated with 194 distance, 175 dihedral angle, and 39 RDC restraints (Table 1). Figures 4A and 5A show the overlays of the lowest-energy structures selected for structural analysis.
TABLE 1.
Structural statistics of the final ensembles of the lowest-energy structures
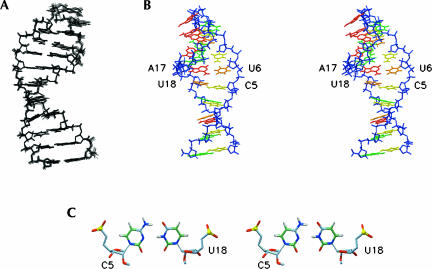
FIGURE 4.
Structure of the Ycu RNA hairpin. (A) Ensemble of the 12 lowest-energy structures. (B) Stereo view of the lowest-energy structure. The A residue bases are colored red, G residue bases are colored green, C residue bases are colored yellow, and U residue bases are colored orange. The sugar-phosphate backbone is in blue. (C) Top view down the helical axis of the lowest-energy structure, showing the C5–U18 base pair.
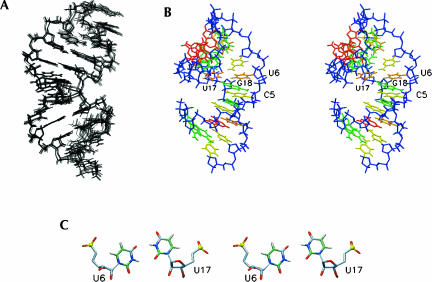
FIGURE 5.
Structure of the Yuu RNA hairpin. (A) Ensemble of the 13 lowest-energy structures. (B). Stereo view of the lowest-energy structure. Coloring is as in Figure 3. (C) Top view down the helical axis of the lowest-energy structure, showing the pseudo U6–U17 base pairs.
Structure of the Ycu RNA molecule
The structure determination of the Ycu variant reveals an A-form-type helix containing a C5–U18 Watson–Crick-type base pair (Fig. 4). Although no assumptions or explicit hydrogen bond restraints were introduced to impose the C–U base pair, for which we could not detect the imino or amino proton signals in the spectra recorded in H2O, with the implementation of RDC restraints for the sugars and bases, all structures refined with two hydrogen bonds within hydrogen-bonding range (C5N4–U18O4 2.9 ± 0.05 Å and C5N3–U18N3 3.4 ± 0.1 Å). The C5N3–U18N3 distance is substantially larger than that in canonical C–G or A–U base pairs (2.9 Å), suggesting possible fraying, which could account for the absent U18 imino proton. Chemical shifts of aromatic protons are dominated by stacking of the 5′ residue (Cromsigt et al. 2001). Enhanced stacking of A17 on U18 in the structure accounts for the very large upfield shift of the U18 H5/H6 protons (Fig. 3B). To accommodate the narrow C–U base pair in the A-form helix the minor groove width, measured by the C1′–C1′ distance is reduced from 11 Å in A-form RNA to 9.5 Å (Table 2). The cross-strand Pn–Pn′ + 1 distances, the typical measure for the major groove width remain A-form helix-like (17.5 Å). The A-form character of the incorporated C–U pair is further corroborated by the virtual bond angle λ, i.e., the angle between the C1′–C1′ line and C1′–N1 bonds, which are typical A-form-like, i.e., 55° for C5 and 58° for U18. Backbone torsion angles of all stem residues are typical for A-form RNA, except for deviating αt and γt torsion angles of the C4 and G19 residues, resulting from typical crankshaft rotations and deviating S-type sugar puckers for G19 in some structures of the ensemble. Such values for these specific torsion angles, one base-pair step down from a C–U base pair, have been found earlier (Holbrook et al. 1991; Lescrinier et al. 2003).
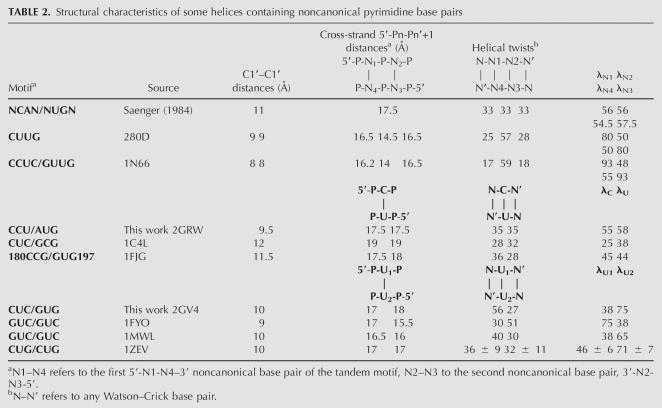
TABLE 2.
Structural characteristics of some helices containing noncanonical pyrimidine base pairs
Structure of the Yuu RNA molecule
The stem structure of the Yuu variant also forms an A-form-type helix, but contains a noncanonical pseudo U6–U17 base pair (Fig. 5). In other structures of RNA helices containing tandem U–U pairs or U–U pairs flanked by other pyrimidine base pairs the backbone is distorted to permit the formation of two hydrogen bonds with short acceptor–donor distances (Baeyens et al. 1995; Lietzke et al. 1996; Lescrinier et al. 2003). In these structures the C1′–C1′ distance is severely contracted to 8 Å–9 Å to accommodate the narrow U–U pair, and the U–U base pair is stabilized by two hydrogen bonds. To form these two hydrogen bonds the two uridine bases of the base pair have to be oriented differently within the helix. Thus, one uridine extends its O4 into the major groove and the O2 participates in a hydrogen bond with the opposite N3H. Conversely, the other uridine turns its O2 toward the minor groove and the O4 accepts a hydrogen bond from the other opposite N3H. These two different orientations are referred to as “open” and “closed” conformation, respectively (Lietzke et al. 1996).
In the Yuu molecule the interstrand C1′–C1′ distance is only slightly reduced from the normal 11 Å to 10 Å, while the cross-strand Pn–Pn′ + 1 distances are slightly increased to 18 Å–19 Å. The U6–U7 pair adopts a closed–open configuration with the bases oriented in the proper position to form two hydrogen bonds: one between U6N3H and U17O2 and the other between U6O4 and U17N3H, however, with rather large heavy atom donor–acceptor distances (3.8 and 3.4 Å, respectively). Thus, it appears that the U–U mismatch lacks hydrogen bonds, which agrees with the chemical shift signatures of the carbonyl signals (Fig. 3D) and absence of a 3hJNC′ coupling using a longer range H(N)CO experiment (results not shown). On the other hand, the distance between the two imino protons in the U–U pair is 3.6 ± 0.2 Å, which accounts for the strong mutual NOE (Fig. 3C). The virtual bond angle λ is a very useful measure in identifying the configuration of the U–U pair. Compared to regular A-form RNA this amounts to the relative smaller 38° value for U6 (closed) and larger 75° value for U17 (open). As is the case for the Ycu molecule, large upfield shifts are observed for U17 H5/H6, caused by enhanced stacking on the 5′ purine. Why U6 adopts the closed and U17 the open conformation and not vice versa, leading to the alternative possible configuration of the pseudo-U–U base pair remains an unresolved issue (vide infra).
Although we were rather surprised to find this pseudo-U–U base pair (bp) without hydrogen bonds, given the observation of the two uridine imino protons with a strong mutual NOE, this base-pair configuration was by no means unprecedented: this type of U–U mismatch has recently also been found in the crystal structure of an 18-bp RNA double helix containing six CUG repeats, which provided a structural basis for myotonic dystrophy (Mooers et al. 2005).
In vitro replication of poliovirus Y-domain mutants
In our previous study of the wild-type pyrimidine-rich internal loop 5′-CU-3′/5′-UU-3′ we assayed its functional importance by comparing growth curves of wild-type virus with a double base-pair mutant in which the C–U was changed into C–G and U–U into U–A (Lescrinier et al. 2003). Both wild-type and mutant viruses displayed similar growth characteristics, indicating that replacement of the tandem noncanonical base-pair motif by two Watson–Crick base pairs has no effect on virus viability and growth in vitro. The result also confirmed the suggestion that the pseudo-twofold symmetry of the A-form helix might be important for function.
In this study, we investigated the effect of the single base-pair replacements by looking directly at viral RNA replication in three different cell lines using a so-called viral RNA replicon (Andino et al. 1993). In this replicon the capsid coding region of the genomic RNA is replaced by the firefly luciferase gene. The expression of this reporter gene has been shown to be proportional to the amount of viral RNA present in the cell and can thus be used to efficiently and accurately quantify viral RNA replication. To discriminate between luciferase production from transfected RNA and the enhanced production of luciferase from progeny RNA replication is assayed in the absence and presence of 2 mM guanidine hydrochloride (GuHCl), an inhibitor of the poliovirus protein 2C, necessary for synthesis of minus strand RNA (Barton and Flanegan 1997).
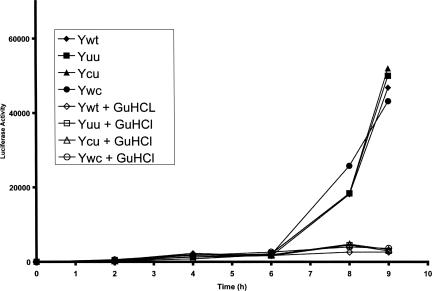
RNA replication was studied with four different constructs. The wild-type replicon with the tandem noncanonical base-pair motif 5′-CU-3′/5′-UU-3′ (Ywt), two mutants with the NMR sequences 5′-CU-3′/5′-AU-3′ (Ycu) and 5′-CU-3′/5′-UG-3′ (Yuu), and the double base-pair mutant in which the 5′-CU-3′/5′-UU-3′ motif was replaced by two Watson–Crick base pairs, 5′-CU-3′/5′-AG-3′ (Ywc). RNA replication was assayed in three different cell lines, human HeLa cells, mouse neuroblastoma CCL31, and HEK 293 cells. The accumulation of luciferase activity showed two phases in all assays. Initially, a low level of activity plateaus during 4–6 h due to expression of the transfected RNA, then a slight increase is observed for the assays in the presence of the replication inhibitor GuHCl and a strong increase for the assays in the absence of the inhibitor. The latter phase corresponds to the increase of viral RNA from replication (Fig. 6). All variants gave an enhanced luciferase production in all three cell lines, indicative of successful RNA replication, however without a significant difference in replication efficiency.
FIGURE 6.
Luciferase activity produced by wild-type and various poliovirus Y-stem mutants of the chimeric poliovirus luciferase replicon after transfection with in vitro transcribed RNA in the human HeLa cell. The symbols utilized to indicate each mutant are indicated in the inset. Closed symbols correspond to luciferase activity in the absence of GuHCL, open symbols to luciferase activity in the presence of GuHCl.
DISCUSSION
Structural implications
The poliovirus is a member of the genus of enteroviruses, which are small nonenveloped RNA viruses. A phylogenetic comparison of the enteroviral RNAs (Fig. 2) shows that in all poliovirus and human enterovirus type C (HEV-C, poliovirus-like coxsackie-A viruses) species the pyrimidine-rich base-pair tandem in the 3′-UTR Y-domain is largely conserved. The 5′-CU-3′/5′-UU-3′ tandem base-pair motif is present in most poliovirus subtypes and a number of coxsackie-A virus subtypes. A 5′-CU-3′/5′-CU-3′ tandem is found in poliovirus types 1, 3 and coxsackievirus A 15, 17, and 18 isolates. Poliovirus type 2 and coxsackie-A virus types 11 and 24 contain the 5′-CU-3′/5′-AU-3′ sequence, identical to the Ycu variant described in this work. In the other subgroup, containing nonpolio-like coxsackie type B and ECHO viruses the pyrimidine base pairs are replaced by Watson–Crick.
The structure of the wild-type poliovirus 3′-UTR Y-domain, which we described earlier, contains a tandem noncanonical 5′-CU-3′/5′-UU-3′ pyrimidine base-pair motif in which the C–U and U–U base pairs are isomorphic and related by a pseudo-twofold axis, which can be gleaned from the equal, relatively small, C1′–C1′ distances and similar λ values of the 5′ and 3′ residues, which amount to 80° and 50°, respectively (Table 2). Another regularity is observed for the 5′-CU-3′/5′-UU tandem: a very large helical twist between the two pyrimidine base pairs is compensated by small helical twists at the interface between the pyrimidine base pairs and flanking A-form helices, leading to an overall helical twist of ∼33°, which is close to the A-form. These striking regularities apply to other tandems of noncanonical base pairs as well (Heus and Hilbers 2003).
The structures of the molecules described in this paper contain only single mismatches in the stem. In the wild-type Ycu variant with the single C–U mismatch the λ angles values, cross-strand phosphorous distances, and helical twists on either side of the mismatch are closer to the A-form. In the nonnatural Yuu variant with the single U–U pair both values of the λ angles are very different, and the mismatches bear no symmetry relation to the flanking Watson–Crick base pair on either side. The twist angles are clearly deviating more from the A-form at both sides of the pseudo-U–U pair. Thus, we can conclude that the pseudo-twofold symmetry is retained in the naturally occurring Y-stem with the single C–U mismatch, but lost in the nonnatural variant with the single U–U base pair. In Figure 7 overlays of the central two base pairs of the three structures are given, providing an overall impression of the general structural differences and similarities.
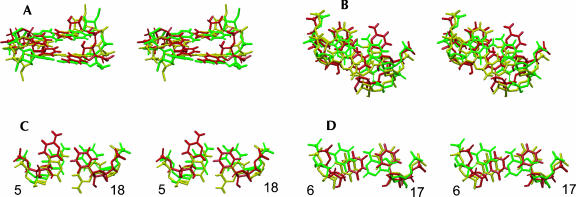
FIGURE 7.
Overlays of the lowest-energy structures of the poliovirus Y domain (Lescrinier et al. 2003) and variants (this work), superimposed on the central seven base pairs, i.e., residues 2–8 and 15–21. The poliovirus 1 structure (Lescrinier et al. 2003) is colored red, the Ycu variant green, and the Yuu variant yellow. (A) Stereo side view into the major groove of the two central residue 5–18 and 6–17 base pairs. (B) Stereo top view down the helical axes. The residue 6–17 base pairs are on top. (C) Stereo top view of the residue 5–18 base pairs. (D) Stereo top view of the residue 6–17 base pairs.
Similar to the parent 5′-CU-3′/5′-UU-3′ structure, the pseudo-U6–U17 base pair in the Yuu variant adopts the closed–open configuration and its incorporation causes similar, be it less profound distortions, on the local helical geometry. The pseudo-U–U base pair only slightly contracts the minor groove, decreasing the C1′–C1′ distance by ∼1 Å to 10 Å, in contrast with the large contraction to 8 Å in the 5′-CU-3′/5′-UU-3′ sequence (Lescrinier et al. 2003). An enlarged twist of 56° between the U6–U17 and C7–G16 base pairs is compensated by a decreased twist of 27° at the C5–G18 and U6–U17 base-pair step.
Tandem and single noncanonical base pairs or unpaired nucleotides commonly destabilize the RNA helix, and their thermodynamic stabilities depend both on the identity of the mismatch and the nature of adjacent base pairs (Xia et al. 1997; Kierzek et al. 1999; Bourdelat-Parks and Wartell 2005). For instance, it was shown that a U–U mismatch in a 5′-GUC-3′/5′-GUC-3′ context is more stable than a U–U mismatch in a 5′-CUG-3′/5′-CUG-3′ context, while a 5′-CUC-3′/5′-GUG-3′ mismatch exhibits intermediate stability. Interestingly, these differences in stability correspond to the phylogenetic distribution of single U–U base pairs in ribosomal RNA: the 5′-GUC-3′/5′-GUC-3′ sequence is 15 times more abundant than the 5′-CUG-3′/5′-CUG-3′ sequence (Kierzek et al. 1999). Similar to the noncanonical U–U base pairs, stabilities of C–U mismatches are also sequence dependent (Kierzek et al. 1999). Although less extensively investigated than the U–U base pair, its stability might also correspond to phylogenetic distribution with 5′-GCC-3′/5′-GUC-3′, being five times more abundant than 5′-CCC-3′/5′-GUG-3′ in ribosomal RNA. To assess the thermodynamic stabilities of the single and tandem noncanonical base pairs within the context of the poliovirus Y-stem sequences we also recorded melting curves of the poliovirus wild-type, Ycu, and Yuu variants (results not shown). The Tm values, measured in 1 M NaCl salt conditions, amount to 71°C (5′-CU-3′/5′-UU-3′), 78°C (Ycu), and 88°C (Yuu), respectively, in line with earlier reported values of the Turner group (Xia et al. 1997; Kierzek et al. 1999).
Thus, stabilities of single (and also tandem) noncanonical base pairs are quite sequence dependent, suggesting that rules for their folding properties can be complex. Indeed, a survey of the structural database reveals a variety of noncanonical base-pair configurations in different environments, which appears to be more diverse for C–U than for U–U mismatches. C–U base-pair configurations have been found in helical regions with a single CO2–UN3H hydrogen bond (Lescrinier et al. 2003), with a CN4H–UO4 and a water-mediated CN3–UN3H hydrogen bond (Tanaka et al. 1999; Harms et al. 2001; Lukavsky et al. 2003), with direct CN4H–UO4 and CN3–UN3H hydrogen bonds (Du et al. 2004; Ohlenschlager et al. 2004), and with a single CN4H-UO4 hydrogen bond (Ban et al. 2000; Tanaka et al. 2000; Theimer et al. 2003). In the 5′-CCU-3′/5′-AUG-3′ context described here also two hydrogen bonds are present between the C and U.
As mentioned before, U–U base pairs are able to adopt two alternative configurations, depending on which base accepts a hydrogen bond by either the O2 (open conformation) or O4 (closed conformation), and again different configurations are found in different contexts (Baeyens et al. 1995; Fourmy et al. 1996; Lietzke et al. 1996; Wimberly et al. 2000; Lynch and Puglisi 2001; Vicens and Westhof 2001, 2002, 2003; Lescrinier et al. 2003; Theimer et al. 2003; D'Souza and Summers 2004; Du et al. 2004; Ohlenschlager et al. 2004; Mooers et al. 2005). For instance, in the 30S ribosomal subunit the U–U base pair in the 5′-GU1406C-3′/5′-GU1495C-3′ sequence of the ribosomal decoding site (A-site) adopts a U1406(closed)–U1495(open) conformation (Wimberly et al. 2000). The U–U base pair in the 5′-GU281A-3′/5′-CU300C-3′ sequence found in the MoMuLV packaging signal adopts a U281(closed)–U300(open) configuration (D'Souza and Summers 2004). The Yuu molecule described here with a 5′-CUC-3′/5′-GUG-3′ sequence contains a U6(closed)–U17(open) base pair. The pseudo-U–U base pairs in the 18 bp RNA helix with six 5′-CUxG-3′/5′-CUyG-3′ repeats, implicated in myotonic dystrophy (Mooers et al. 2005), adopt predominantly Ux(closed)–Uy(open) configurations.
The specific shape of a U–U base pair with its strong electronegative major groove (containing two O4 atoms) and short C1′–C1′ distance creates a unique landscape for ligand binding (Vicens and Westhof 2003). This capacity is used, for instance, in the complexes of the aminoglycoside antibiotics paromomycin, tobramycin, and genetecin with the 16S A-site, where the O4 of U1495 makes a direct hydrogen bond with the N1 of the aminoglycoside ring II (Vicens and Westhof 2001, 2002, 2003). It might also be important for binding of the poliovirus protein 3Cpro to the stem–loop D of the 5′-UTR where a stretch of three consecutive pyrimidine base pairs in a 5′-UCU-3′/5′-UUU-3′ sequence provides for a local narrowed helical element (Du et al. 2004; Ohlenschlager et al. 2004). Interestingly, in contrast to the ribosomal crystal structure the solution structure of a 16S rRNA fragment containing the ribosomal A-site reveals a U1406(open)–U1495(closed) configuration (Lynch and Puglisi 2001). This configuration is swapped to the closed–open configuration, with a widened minor groove and a water-mediated U1406N3H–U1495O2 hydrogen bond, in the aminoglycoside–RNA complexes to fit ligand binding. This provides yet another example of the extreme polymorphic nature of RNA. Furthermore, in a self-complementary sequence like 5′-GUC-3′/5′-GUC-3′ it is not obvious why one out of two possible configurations is occupied. Using the nearest-neighbor model, one might expect both conformations to be equally present. Based on the available structural data it seems that besides ligand interaction also next-neighbor interactions might play a role in determining the specific shape of the of C–U and U–U mismatches.
Thus, despite the substantial amount of structural data it appears to be very difficult to derive simple sequence-dependent rules from the RNA structural database that can be used to predict the specific conformations of single C–U or U–U base pairs in helical regions. Clearly, more research is needed to further understand the stereochemistry and the subtle sequence dependence that determines the choice of a specific form.
Biological implications
We previously suggested that the observed regularities of the local helix geometry in the poliovirus Y-domain could be important for protein interaction involved in virus RNA replication (Lescrinier et al. 2003). Therefore, it was not unexpected that the wild-type Ycu variant with a single C–U pair also shows wild-type-like structural properties and replication efficiencies. However, we did not anticipate finding that, given the significant structural changes of the nonnatural variant with the single U–U mismatch, replication of its viral RNA would be similar to all natural 3′-UTR Y-domain variants. Thus, it would appear that the distinct structural differences of the 3′-UTR Y-domain do not affect RNA replication using an in vitro assay. This does not necessarily mean that there is no effect at all, but rather shows the limited use of an in vitro assay to evaluate functional importance. This is not unprecedented, and has been shown before, for instance, in our study of the structural and functional properties of the 3′-UTR domain of coxsackievirus B3, an enterovirus closely related to poliovirus (Merkle et al. 2002). Poliovirus and coxsackievirus B 3′-UTRs share a high similarity in secondary structure, but differ in the presence of an extra, conserved Z-domain in the coxsackievirus B 3′-UTR (Rueckert 1996). Deletion of the entire Z-domain from coxsackievirus B3 resulted in a virus with wild-type-like growth characteristics in vitro. However, infection of mice showed a severely reduced virulence of the mutant virus (Merkle et al. 2002). In a comparable study Semler and co-workers deleted the entire 3′-UTR of poliovirus and found only a minor defect in viral RNA replication in HeLa cells, but a major replication defect in neuroblastoma cells and a major neurovirulence attenuation in transgenic mice (Todd et al. 1995, 1997; Brown et al. 2004). A cell-type-specific effect of 3′-UTR mutations on virus propagation was also reported for HEK 293 cells by Gromeier and collaborators (Campbell et al. 2005). A coxsackievirus mutant containing the rhinovirus IRES and a deletion of the Z-domain had wild-type growth characteristics in HeLA cells but was severely impaired in growth in neuroblastoma cells (Campbell et al. 2005). We did not detect reduced growth of any mutant in neuroblastoma cells as well. Thus, although an in vitro versus in vivo effect cannot be excluded, the results so far argue for a minor role of the structure dependency of the pseudo-twofold symmetry in the Y-domain of all poliovirus-like enteroviruses.
In summary, we have elucidated the noncanonical pyrimidine base-pair motifs present in the majority of polioviruses and Human Enterovirus Type C group by NMR. Besides the existence of C–U or U–U base pairs, the structural details also revealed that the pseudo-twofold symmetry, typical for a regular A-form RNA helix, is retained for the natural variants. However, despite our efforts, so far we could not find evidence for a specific biological function of these distinctive structural features using an in vitro read-out system. The results we describe here again show that only the tight coupling between virology and structural biology can reveal the true significance of particular RNA entities. The results also open new directions for further research on the question of why some RNA structures without an obvious biological active or essential function nevertheless have evolved and are highly conserved and widespread throughout nature.
MATERIALS AND METHODS
Sample preparation
Unlabeled and uniformly (13C/15N) and U-specific 13C/15N-labeled 22-mer RNAs were prepared by in vitro transcription of partially duplex DNA templates by T7 RNA polymerase as described previously (Milligan et al. 1987). RNA molecules were purified by preparative denaturing polyacrylamide gel electrophoresis and subsequent electroelution. Purified RNA was concentrated by ethanol precipitation and extensively dialyzed against water using Centricon microconcentrators. The final concentration of the RNA samples was 1–2 mM. The pH of the samples was adjusted to 6.0–6.8 (meter reading). Weakly aligned NMR samples were prepared by adding 27 mg/mL filamentous phage Pf1 (Hansen et al. 1998) purchased from ASLA Ltd (ASLA).
NMR spectroscopy
NMR spectra were acquired on Varian Inova 600 MHz and 800 MHz spectrometers. For the assignment of exchangeable imino protons 2D NOESY spectra were recorded in 90% H2O, 10% D2O at 15°C, and 800 MHz using a jump return pulse for water suppression (Plateau and Guéron 1982). Uridine imino protons of the sample with the 5′-CU-3′/5′-UG-3′ motif were unambiguously assigned using a 2D back-and-forth H(N3C5)H5 experiment recorded at 5°C with a U-specific 13C/15N-labeled sample. Using the same sample and conditions a 2D H(N)CO experiment was recorded to assess the hydrogen-bonding patterns of uridines by comparing C2/C4 chemical shifts (Furtig et al. 2003; Theimer et al. 2003; Du et al. 2004; Ohlenschlager et al. 2004). For samples dissolved in D2O, spectra were recorded at 600 or 800 MHz at 25°C. Nonexchangeable protons were assigned using 2D NOESY (50, 100, and 300-msec mixing times), DQF-COSY (Shaka and Freeman 1983), and TOCSY (Griesinger et al. 1988), and for the sample with the 5′-CU-3′/5′-AU-3′motif a 31P-decoupled HCCH-TOCSY (Kolk et al. 1998). Residual dipolar couplings (RDCs) were collected by measuring heteronuclear 1H-13C one-bond couplings in the absence or presence of filamentous phage Pf1 using an f1-proton coupled 2D-HSQC or the IPAP approach (Ottiger et al. 1998) at 600 MHz. For the sample with the 5′-CU-3′/5′-AU-3′motif, RDCs were collected using the uniformly 13C/15N-labeled sample, while for the sample with the 5′-CU-3′/5′-UG-3′ motif RDCs were measured at natural abundance at 600 MHz proton frequency using cryoprobe technology. Typical acquisition times for the IPAP spectra at natural abundance were ∼24 h with 48 scans per FID, 1024 t2 data points, and 320 complex t1 data points. Different data sets were collected involving the aromatic H8, H6, and H2, and the H1′ and H5 with the INEPT-delay set to 2.5 and 2.9 msec, respectively. All data were analyzed with Sparky (T.D. Goddard and D.G. Kneller, University of California, San Francisco).
Structural restraints
Distances involving the nonexchangeable protons were calculated from NOE cross-peak intensities of the 2D NOESYs with 50, 100, and 300 msec mixing times using the modified isolated spin pair approximation approach (Barsukov and Lian 1993). The calculated distances were used with ±20% error bounds. Distance information involving exchangeable protons was obtained from 2D NOESY spectra recorded in H2O. Hydrogen bonding restraints were treated as NOE distance restraints and imposed only on stem residues involved in Watson–Crick base-pairing, evidenced by the imino proton spectra and observed NOEs and chemical shifts.
Torsion angle restraints for the sugar moieties were obtained from DQF-COSY and TOCSY experiments and interpreted as N- or S-type sugar puckers when they were small (<3 Hz) or large (>8 Hz), respectively. For the sample with the 5′-CU-3′/5′-AU-3′motif the β and ε dihedral torsion angles were calculated from a novel spin-echo difference CT-HCCH-TOCSY experiment (E.H.M.P. Lescrinier, G.W. Vuister, C.W. Hilbers, H.A. Heus, and M. Tessari, unpubl.) and restrained to their calculated values with error bounds of ±20° and ±30°, respectively. For the stem residues of the sample with the 5′-CU-3′/5′-UG-3′ motif the ε dihedral angles were restrained to 225 (±60°). Additional dihedral torsion restraints were applied as previously described (Lescrinier et al. 2003). The dihedral angles α and ζ of the residues of the stem were restrained from phosphorous chemical shift considerations to 0 ± 120° (Lescrinier et al. 2003). RDCs were used with 1.5 Hz error bounds.
Structure calculations
Structure calculations were performed with the torsion angle dynamics protocol implemented within XPLOR-NIH (Stein et al. 1997; Schwieters et al. 2003) and included simulated annealing starting from extended strands. For the 5′-UA-3′/5′-CU-3′ and the 5′-UU-3′/5′-CG-3′ samples, out of starting sets of 100 randomly extended structures, 33 and 31 acceptable structures were obtained, respectively, with no dihedral angle violations larger than 5° and no NOE violations larger than 0.5 Å (Table 1), and subjected to refinement using RDCs. Initial values for the axial and rhombic components of the alignment tensor were estimated from the distribution of the RDC values (Clore et al. 1998; Bax et al. 2001). The subsequent grid search yielded optimal values of Da = −50 Hz and R = 0.3 Hz and for the 5′-UA-3′/5′-CU-3′ molecule and Da = −48 Hz and R = 0.3 Hz for the 5′-UU-3′/5′-CG-3′ molecule. In the grid search Da was varied from –35 Hz to –55 Hz and R was varied from 0 Hz to 0.6 Hz in steps of 1 Hz and 0.1 Hz, respectively.
Color figures were generated using MOLMOL (Koradi et al. 1996). Helical parameters were calculated using the programs 3DNA (Lu and Olson 2003) or freehelix98 (Dickerson and Chiu 1997; Dickerson 1998).
Atomic coordinates
Coordinates of ensembles of lowest-energy structures of both hairpins have been deposited in the RCBS Protein Data Bank (accession codes 2GV4 and 2GRW).
UV melting curves
UV absorbance melting curves were recorded at both 260 nm and 280 nm simultaneously on a Cary-300 spectrometer. Samples (A260 = 0.6–0.9 cm−1; corresponding to ∼3.6–5.5 μM) were prepared in 10 mM sodium phosphate, 1 M NaCl, and 0.1 mM Na2 EDTA. The samples were heated a few minutes to 90°C and quickly cooled to 1°C. The melting curves were measured at a heating rate of 0.5°C/min from 20 to 98°C with data collected every 0.25°C increment. Melting temperatures (Tm) were derived assuming a two-state model using the Cary-WIN software.
Plasmids
Mutations of the pyrimidine-rich internal loop were introduced in the poliovirus replicon pRLuc31 in which the region coding for the viral capsid proteins is replaced by the firefly luciferase gene (Andino et al. 1993). Noncanonical base pairs were replaced by Watson–Crick base pairs by substitution of single nucleotides. Mutations were introduced by PCR amplification of a 286-bp fragment containing the 3′-UTR. The fragment included a unique BlnI site at position 6266 and a unique MluI restriction endonuclease site at position 6612, which were used to replace the original BlnI–MluI fragment in the parent plasmid. Desired mutations were confirmed by sequence analysis.
Luciferase replication assay
Upon transfection of cells with in vitro transcribed RNA of the replicon pRLuc31, translation of the viral RNA gives rise to luciferase activity in the cytoplasm. Replication-competent wild-type and mutant viral RNA will produce progeny plus strand RNA and consequently display an enhanced luciferase activity. Discrimination between luciferase production from the transfected RNA molecules and the enhanced production of luciferase from progeny RNAs is achieved by performing the replication assay in the absence and presence of 2 mM guanidine hydrochloride, an inhibitor of the poliovirus protein 2C, necessary for synthesis of minus strand RNA (Barton and Flanegan 1997).
HeLa, mouse neuroblastoma CCL31, and human embryonal kidney (HEK 293) cells, grown in six-well plates to a confluence of 80%, were transfected as previously described (Zoll et al. 1996), with 1 μg of T7 RNA polymerase-generated cRNA derived from MluI linearized replicon plasmid, containing the firefly luciferase gene. Cells were harvested at various times post-transfection, washed two times with phosphate-buffer saline (PBS), and subsequently lysed in 200 μL lysisbuffer (Promega). Luciferase activity was assayed using the Luciferase Assay (Promega) system and quantified using a BioOrbit 1251 Luminometer.
ACKNOWLEDGMENTS
This work was supported by NWO-CW (grant 700.50.021).
Footnotes
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.375607.
REFERENCES
- Andino, R., Rieckhof, G.E., Achacoso, P.L., Baltimore, D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeyens, K.J., De Bondt, H.L., Holbrook, S.R. Structure of an RNA double helix including uracil–uracil base pairs in an internal loop. Nat. Struct. Biol. 1995;2:56–62. doi: 10.1038/nsb0195-56. [DOI] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., Steitz, T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Barsukov, I.L., Lian, L.Y. Structure determinations from NMR data I. Analysis of NMR data. In: Roberts G.C.K., editor. NMR of macromolecules, a practical approach. Oxford University Press; New York: 1993. pp. 315–357. [Google Scholar]
- Barton, D.J., Flanegan, J.B. Synchronous replication of poliovirus RNA: Initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J. Virol. 1997;71:8482–8489. doi: 10.1128/jvi.71.11.8482-8489.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax, A., Kontaxis, G., Tjandra, N. Dipolar couplings in macromolecular structure determination. Methods Enzymol. 2001;339:127–174. doi: 10.1016/s0076-6879(01)39313-8. [DOI] [PubMed] [Google Scholar]
- Bourdelat-Parks, B.N., Wartell, R.M. Thermodynamics of RNA duplexes with tandem mismatches containing a uracil–uracil pair flanked by CG/GC or GC/AU closing base pairs. Biochemistry. 2005;44:16710–16717. doi: 10.1021/bi051659q. [DOI] [PubMed] [Google Scholar]
- Brown, D.M., Kauder, S.E., Cornell, C.T., Jang, G.M., Racaniello, V.R., Semler, B.L. Cell-dependent role for the poliovirus 3′-noncoding region in positive-strand RNA synthesis. J. Virol. 2004;78:1344–1351. doi: 10.1128/JVI.78.3.1344-1351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, S.A., Lin, J., Dobrikova, E., Gromeier, M. Genetic determinants of cell type-specific poliovirus propagation in HEK 293 cells. J. Virol. 2005;79:6281–6290. doi: 10.1128/JVI.79.10.6281-6290.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate, J.H., Gooding, A.R., Podell, E., Zhou, K., Golden, B.L., Kundrot, C.E., Cech, T.E., Doudna, J.A. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- Clore, G.M., Gronenborn, A.M., Tjandra, N. Direct structure refinement against residual dipolar couplings in the presence of rhombicity of unknown magnitude. J. Magn. Reson. 1998;131:159–162. doi: 10.1006/jmre.1997.1345. [DOI] [PubMed] [Google Scholar]
- Cromsigt, J.A.M.T.C., Hilbers, C.W., Wijmenga, S.S. Prediction of proton chemical shifts in RNA. Their use in structure refinement and validation. J. Biomol. NMR. 2001;21:11–29. doi: 10.1023/a:1011914132531. [DOI] [PubMed] [Google Scholar]
- Dickerson, R.E. DNA bending: The prevalence of kinkiness and the virtues of normality. Nucleic Acids Res. 1998;26:1906–1926. doi: 10.1093/nar/26.8.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson, R.E., Chiu, T.K. Helix bending as a factor in protein/DNA recognition. Biopolymers. 1997;44:361–403. doi: 10.1002/(SICI)1097-0282(1997)44:4<361::AID-BIP4>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- D'Souza, V., Summers, M.F. Structural basis for packaging the dimeric genome of Moloney murine leukaemia virus. Nature. 2004;431:586–590. doi: 10.1038/nature02944. [DOI] [PubMed] [Google Scholar]
- Du, Z., Yu, J., Ulyanov, N.B., Andino, R., James, T.L. Solution structure of a consensus stem–loop D RNA domain that plays important roles in regulating translation and replication in enteroviruses and rhinoviruses. Biochemistry. 2004;43:11959–11972. doi: 10.1021/bi048973p. [DOI] [PubMed] [Google Scholar]
- Fourmy, D., Recht, M.I., Blanchard, S.C., Puglisi, J.D. Structure of the A-site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science. 1996;274:1367–1371. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- Furtig, B., Richter, C., Wohnert, J., Schwalbe, H. NMR spectroscopy of RNA. ChemBioChem. 2003;4:936–962. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- Griesinger, C., Otting, G., Wüthrich, K., Ernst, R.R. Clean TOCSY for 1H spin system identification in macromolecules. J. Am. Chem. Soc. 1988;110:7870–7872. [Google Scholar]
- Hansen, M.R., Mueller, L., Pardi, A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat. Struct. Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- Harms, J., Schluenza, F., Zarivach, R., Bashan, A., Gat, S., Agmon, I., Bartels, H., Franceschi, F., Yonath, A. High-resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- Harris, K.S., Xiang, W., Alexander, L., Lane, W.S., Paul, A.V., Wimmer, E. Interaction of poliovirus polypeptide 3CDpro with the 5′- and 3′-termini of the poliovirus genome. J. Biol. Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- Hermann, T., Westhof, E. Non-Watson–Crick base pairs in RNA–protein recognition. Chem. Biol. 1999;12:R335–R343. doi: 10.1016/s1074-5521(00)80003-4. [DOI] [PubMed] [Google Scholar]
- Heus, H.A., Hilbers, C.W. Structures of noncanonical tandem base pairs in RNA helices. Nucleosides Nucleotides Nucleic Acids. 2003;22:559–571. doi: 10.1081/NCN-120021955. [DOI] [PubMed] [Google Scholar]
- Holbrook, S.R., Cheong, C., Tinoco I., Jr, Kim, S.H. Crystal structure of an RNA double helix incorporating a track of non-Watson–Crick base pairs. Nature. 1991;353:579–581. doi: 10.1038/353579a0. [DOI] [PubMed] [Google Scholar]
- Jucker, F.M., Heus, H.A., Yip, P.F., Moors, E.H., Pardi, A. A network of heterogeneous hydrogen bonds in GNRA tetraloops. J. Mol. Biol. 1996;164:968–980. doi: 10.1006/jmbi.1996.0690. [DOI] [PubMed] [Google Scholar]
- Kierzek, R., Burkard, M.E., Turner, D.H. Thermodynamics of single mismatches in RNA duplexes. Biochemistry. 1999;38:14214–14223. doi: 10.1021/bi991186l. [DOI] [PubMed] [Google Scholar]
- Kolk, M.H., Wijmenga, S.S., Heus, H.A., Hilbers, C.W. On the structure determination of a 44n RNA pseudoknot: Assignment strategies and derivation of torsion angle restraints. J. Biomol. NMR. 1998;12:423–433. [Google Scholar]
- Koradi, R., Billeter, M., Wüthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Lescrinier, E.M.H.P., Tessari, M., van Kuppeveld, F.J.M., Melchers, W.J.G., Hilbers, C.W., Heus, H.A. Structure of the pyrimidine-rich internal loop in the poliovirus 3′-UTR: The importance of maintaining pseudo-twofold symmetry in RNA helices containing two adjacent noncanonical base pairs. J. Mol. Biol. 2003;331:759–769. doi: 10.1016/s0022-2836(03)00787-3. [DOI] [PubMed] [Google Scholar]
- Lietzke, S.E., Barnes, C.L., Berglund, J.A., Kundrot, C.E. The structure of an RNA dodecamer shows how tandem U–U base pairs increase the range of stable RNA structures and the diversity of recognition sites. Structure. 1996;4:917–930. doi: 10.1016/s0969-2126(96)00099-8. [DOI] [PubMed] [Google Scholar]
- Lu, X., Olson, W.K. 3DNA: A software package for the analysis, rebuilding, and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003;31:5108–5121. doi: 10.1093/nar/gkg680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky, P.J., Kim, I., Otto, G.A., Puglisi, J.D. Structure of HCV IRES domain II determined by NMR. Nat. Struct. Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- Lynch, S.R., Puglisi, J.D. Structure of a eukaryotic decoding region A-site RNA. J. Mol. Biol. 2001;306:1023–1035. doi: 10.1006/jmbi.2000.4419. [DOI] [PubMed] [Google Scholar]
- McKay, D.B. Structure and function of the hammerhead ribozyme: An unfinished story. RNA. 1996;2:395–403. [PMC free article] [PubMed] [Google Scholar]
- Melchers, W.J., Hoenderop, J.G., Bruins Slot, H.J., Pleij, C.W., Pilipenko, E.V., Agol, V.I., Galama, J.M. Kissing of the two predominant hairpin loops in the coxsackie B virus 3′-untranslated region is the essential structural feature of the origin of replication required for negative-strand RNA synthesis. J. Virol. 1997;71:686–696. doi: 10.1128/jvi.71.1.686-696.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchers, W.J., Bakkers, J.M., Bruins Slot, H.J., Galama, J.M., Agol, V.I., Pilipenko, E.V. Cross-talk between orientation-dependent recognition determinants of a complex control RNA element, the enterovirus oriR. RNA. 2000;6:976–987. doi: 10.1017/s1355838200000480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellits, K.H., Meredith, J.M., Rohll, J.B., Evans, D.J., Almond, J.W. Binding of a cellular factor to the 3′-untranslated region of the RNA genomes of entero- and rhinoviruses plays a role in virus replication. J. Gen. Virol. 1998;79:1715–1723. doi: 10.1099/0022-1317-79-7-1715. [DOI] [PubMed] [Google Scholar]
- Merkle, I., van Ooij, M.J., van Kuppeveld, F.J.M., Glaudemans, D.H., Galama, J.M., Henke, A., Zell, R., Melchers, W.J.G. Biological significance of a human enterovirus B-specific RNA element in the 3′-nontranslated region. J. Virol. 2002;76:9900–9909. doi: 10.1128/JVI.76.19.9900-9909.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan, J.F., Groebe, D.R., Witherell, G.W., Uhlenbeck, O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooers, B.H., Logue, J.S., Berglund, J.A. The structural basis of myotonic dystrophy from the crystal structure of CUG repeats. Proc. Natl. Acad. Sci. 2005;102:16626–16631. doi: 10.1073/pnas.0505873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlenschlager, O., Wohnert, J., Bucci, E., Seitz, S., Hafner, S., Ramachandran, R., Zell, R., Gorlach, M. The structure of the stem–loop D subdomain of coxsackievirus B3 cloverleaf RNA and its interaction with the proteinase 3C. Structure. 2004;12:237–248. doi: 10.1016/j.str.2004.01.014. [DOI] [PubMed] [Google Scholar]
- Ottiger, M., Delaglio, F., Bax, A. Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J. Magn. Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- Pilipenko, E.V., Poperechny, K.V., Maslova, S.V., Melchers, W.J.G., Bruins Slot, H.J., Agol, V.I. Cis-element, oriR, involved in the initiation of (−) strand poliovirus RNA: A quasiglobular multidomain RNA structure maintained by tertiary (“kissing”) interactions. EMBO J. 1996;15:5428–5438. [PMC free article] [PubMed] [Google Scholar]
- Plateau, P., Guéron, M. Exchangeable proton NMR without baseline distortion, using new strong-pulse sequences. J. Am. Chem. Soc. 1982;104:7310–7311. [Google Scholar]
- Rueckert, R. Picornaviridae and their replication. In: Fields B.N., editor. Virology. Raven Press; New York: 1996. pp. 609–654. [Google Scholar]
- Saenger, W. Principles of nucleic acid structure. Springer; New York: 1984. [Google Scholar]
- Schwieters, C.D., Kuszewski, J.J., Tjandra, N., Clore, G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:66–74. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- Shaka, A.J., Freeman, R. Simplification of NMR spectra by filtration through multiple quantum coherence. J. Magn. Reson. 1983;51:169–173. [Google Scholar]
- Stein, E.G., Rice, L.M., Brünger, A.T. Torsion-angle molecular dynamics as a new efficient tool for NMR structure calculation. J. Magn. Reson. 1997;124:154–164. doi: 10.1006/jmre.1996.1027. [DOI] [PubMed] [Google Scholar]
- Tanaka, Y., Fujii, S., Hiroaki, H., Sakata, T., Tanaka, T., Uesugi, S., Tomita, K., Kyogoku, Y. A-form RNA double helix in the single crystal structure of r(UGAGCUUCGGCUC) Nucleic Acids Res. 1999;27:949–955. doi: 10.1093/nar/27.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, Y., Kojima, C., Yamazaki, T., Kodama, T.S., Yasuno, K., Miyashita, S., Ono, A., Ono, A., Kainosho, M., Kyogoku, Y. Solution structure of an RNA duplex including a C–U base pair. Biochemistry. 2000;39:7074–7080. doi: 10.1021/bi000018m. [DOI] [PubMed] [Google Scholar]
- Theimer, C.A., Finger, L.D., Trantirek, L., Feigon, J. Mutations linked to dyskeratosis congenita cause changes in the structural equilibrium in telomerase RNA. Proc. Natl. Acad. Sci. 2003;100:449–454. doi: 10.1073/pnas.242720799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd, S., Nguyen, J.H., Semler, L. RNA–protein interactions directed by the 3′-end of human rhinovirus genomic RNA. J. Virol. 1995;69:3605–3614. doi: 10.1128/jvi.69.6.3605-3614.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd, S., Towner, J.S., Brown, D.M., Semler, B.L. Replication-competent picornaviruses with complete genomic RNA 3′-noncoding region deletions. J. Virol. 1997;71:8868–8874. doi: 10.1128/jvi.71.11.8868-8874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooij, M.J., Glaudemans, D.H., Heus, H.A., van Kuppeveld, F.J., Melchers, W.J. Structural and functional integrity of the coxsackievirus B3 ori/R: Spacing between coaxial RNA helices. J. Gen. Virol. 2006;87:689–695. doi: 10.1099/vir.0.81558-0. [DOI] [PubMed] [Google Scholar]
- Vicens, Q., Westhof, E. Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A-site. Structure. 2001;9:647–658. doi: 10.1016/s0969-2126(01)00629-3. [DOI] [PubMed] [Google Scholar]
- Vicens, Q., Westhof, E. Crystal structure of a complex between the aminoglycoside tobramycin and an oligonucleotide containing the ribosomal decoding a site. Chem. Biol. 2002;9:747–755. doi: 10.1016/s1074-5521(02)00153-9. [DOI] [PubMed] [Google Scholar]
- Vicens, Q., Westhof, E. Crystal structure of geneticin bound to a bacterial 16S ribosomal RNA A-site oligonucleotide. J. Mol. Biol. 2003;326:1175–1188. doi: 10.1016/s0022-2836(02)01435-3. [DOI] [PubMed] [Google Scholar]
- Wang, J., Bakkers, J.M., Galama, J.M., Bruins Slot, H.J., Pilipenko, E.V., Agol, V.I., Melchers, W.J. Structural requirements of the higher order RNA kissing element in the enteroviral 3′-UTR. Nucleic Acids Res. 1999;27:485–490. doi: 10.1093/nar/27.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijmenga, S.S., Mooren, M.W., Hilbers, C.W. NMR of nucleic acids; from spectrum to structure. In: Roberts G.C.K., editor. NMR of macromolecules, a practical approach. Oxford University Press; New York: 1993. pp. 217–288. [Google Scholar]
- Wimberly, B.T., Brodersen, D.E., Clemons W.M., Jr, Morgan-Warren, R.J., Carter, A.P., Vonrhein, C., Hartsch, T., Ramakrishnan, V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Xia, T., McDowell, J.A., Turner, D.H. Thermodynamics of nonsymmetric tandem mismatches adjacent to G.C base pairs in RNA. Biochemistry. 1997;36:12486–12497. doi: 10.1021/bi971069v. [DOI] [PubMed] [Google Scholar]
- Zoll, J., Galama, J.M.D., van Kuppeveld, F.J.M., Melchers, W.J.G. Mengovirus leader is involved in the inhibition of host cell protein synthesis. J. Virol. 1996;70:4948–4952. doi: 10.1128/jvi.70.8.4948-4952.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]