Abstract
In archaeal species, several transfer RNA genes have been reported to contain endogenous introns. Although most of the introns are located at anticodon loop regions between nucleotide positions 37 and 38, a number of introns at noncanonical sites and six cases of tRNA genes containing two introns have also been documented. However, these tRNA genes are often missed by tRNAscan-SE, the software most widely used for the annotation of tRNA genes. We previously developed SPLITS, a computational tool to identify tRNA genes containing one intron at a noncanonical position on the basis of its discriminative splicing motif, but the software was limited in the detection of tRNA genes with multiple introns at noncanonical sites. In this study, we initially updated the system as SPLITSX in order to correctly predict known tRNA genes as well as novel ones with multiple introns. By a comprehensive search for tRNA genes in 29 archaeal genomes using SPLITSX, we listed 43 novel candidates that contain introns at noncanonical sites. As a result, 15 contained two introns and three contained three introns within the respective putative tRNA genes. Moreover, the candidates completely complemented all the codons of two archaeal species of uncultured methanogenic archaeon, RC-I and Thermofilum pendens Hrk 5, with novel candidates that were not detectable by tRNAscan-SE alone.
Keywords: intron-containing tRNA, bulge-helix-bulge (BHB) splicing motif, archaea, SPLITS, tRNAscan-SE, bioinformatics
INTRODUCTION
In archaeal and eukaryal species, transfer RNA (tRNA)-encoding genes (tDNAs), which constitute one of the major noncoding RNA families, have been reported to contain enzyme-dependent spliceable introns. Although most of the introns of eukaryotic tDNAs are located at unique sites in the anticodon loop between nucleotide positions 37 and 38 (referred to as 37/38, or the canonical position), introns of archaeal tDNAs are also located at other positions (noncanonical positions) (Valenzuela et al. 1978; Daniels et al. 1985). In 2003, Marck and Grosjean identified and summarized the predicted locations, RNA structural topologies, and sizes of all introns located on the tDNAs of 18 archaeal chromosomes (Marck and Grosjean 2003). Of the 136 introns in the total of 800 archaeal tDNAs analyzed, 103 are known to be located at position 37/38, and the remaining 33 are located at 14 other sites on tDNAs, such as the anticodon stem, amino acid arm, D- and T-loops, and V-arm, and six of the tDNAs were reported to harbor two introns (Wich et al. 1987; Kjems et al. 1989; Smith et al. 1997; She et al. 2001; Fitz-Gibbon et al. 2002; Marck and Grosjean 2003).
All of these tDNAs have a bulge-helix-bulge (BHB) structural consensus, which is also formed in most archaeal pre-rRNA and pre-mRNA introns (Tang et al. 2002; Watanabe et al. 2002; Yoshinari et al. 2006) around their exon–intron boundaries (Kaine et al. 1983; Daniels et al. 1985; Datta et al. 1989; Thompson et al. 1989; Kleman-Leyer et al. 1997). The canonical BHB is a single-hairpin structure that consists of two bulges (B) of 3 nucleotides (nt) separated by a central helix (H) of 4 base pairs (bp) within consensus motif sequences (hBHBh′). For introns at locations other than 37/38, canonical hBHBh′ motifs are not always formed, but a simplified HBh′ motif consisting of two helices (H and h′) and only one bulge can be discerned (Marck and Grosjean 2003). In either case, the splicing sites are always located two bases downstream of the central helix, and the introns are spliced out by the RNA endonucleases. Three classes of tRNA splicing endonucleases have been described and characterized in 19 archaeal genomes (Tocchini-Valentini et al. 2005): they are a heterotetrameric (α2β2) enzyme in the Crenarchaeota, and homodimeric (α′2) and homotetrameric (α4) enzymes in the Euryarchaeota. Most tDNA introns located at noncanonical positions have been found in Crenarchaeal genomes that encode heterotetrameric enzymes, and a few others have also been observed in the Euryarchaeota whose genomes encode homotetrameric enzymes (Smith et al. 1997; Slesarev et al. 2002). However, no tDNAs that have introns in noncanonical positions in Euryarchaeota whose genome encodes homodimeric enzymes have been reported to date.
Over the last decade, most research on the prediction and annotation of tDNAs has utilized tRNAscan-SE software (Lowe and Eddy 1997), especially in genome sequencing projects. tRNAscan-SE combines three different tRNA search methods: tRNAscan 1.3 (Fichant and Burks 1991), the Pavesi search algorithm (Pavesi et al. 1994), and covariance model analysis (Eddy and Durbin 1994), in order to enable fast and highly sensitive prediction of tDNAs without introns or with one intron at the canonical position. Because of this optimization for canonical tDNAs, using a stochastic model learned from mature tRNA structures consisting of cloverleaf structural constraints and consensus sequences, tRNAscan-SE cannot correctly identify >60% of tDNAs with noncanonical introns (Sugahara et al. 2006). To complement tRNAscan-SE for the identification of tDNAs with noncanonical introns, we previously developed the SPLITS toolkit (Sugahara et al. 2006), which predicts and determines introns with BHB motifs within each putative tDNA sequence predicted by the Virtual Footprint (Munch et al. 2005), and removes the introns before passing the sequence to tRNAscan-SE. SPLITS has contributed to the recent genome sequencing project of Cenarchaeum symbiosum of the Crenarchaea, by the annotation of all tDNAs whose introns are located at noncanonical sites (Hallam et al. 2006). However, SPLITS was unable to detect multiple intron-containing tDNAs whose introns are harbored within the motif regions of tDNAs corresponding to the target sites of Virtual Footprint screening.
In this study, we analyzed 29 archaeal genomes and predicted novel tDNA candidates with multiple introns, and for this purpose we developed SPLITSX, an enhanced, upgraded version of SPLITS. SPLITSX first predicts noncanonical introns from the whole-genome sequence on the basis of the structural prediction of BHB motifs, and the genome sequences after all possible combinational patterns of intron removal are automatically scanned by tRNAscan-SE. We show that the list of comprehensive archaeal tDNAs predicted by SPLITSX contained all documented tDNAs with noncanonical introns reported in the Archaea. Moreover, with the combination of SPLITSX and tRNAscan-SE, we identified a full set of tRNAs corresponding to all 61 sense codons and one initiator codon in the two archaeal genomes of uncultured methanogenic archaeon RC-I (RC-I) and Thermofilum pendens Hrk 5 (T. pendens), where the full set of tRNAs is not detectable by tRNAscan-SE alone. The RC-I genome was further identified to encode the homodimeric type of tRNA-splicing endonuclease, which has previously been suggested not to splice noncanonical introns, and we here present the possibility that the noncanonical introns of tRNAs are actually spliced by the homodimeric enzymes. According to the novel candidates harboring multiple introns, we suggest that many types of tRNA splicing exist in archaeal cells.
RESULTS
Comprehensive screening of novel tDNA candidates in 29 archaeal genomes
For the prediction of novel tDNAs with introns at noncanonical positions by SPLITSX, we analyzed 28 archaeal genomes whose sequences were completely assembled, and the draft assembly genome of T. pendens. In total, 74 tDNA candidates that contained introns at noncanonical positions and one tDNA candidate that contained an intron at 37/38 were screened from 11 archaeal genomes (Table 1); 67 out of the total of 75 candidates were predicted from six Crenarchaeal genomes (Aeropyrum pernix K1, Pyrobaculum aerophilum, Sulfolobus acidocaldarius, Sulfolobus solfataricus, Sulfolobus tokodaii, and T. pendens), seven candidates were from four Euryarchaeal genomes (Methanopyrus kandleri, Methanothermobacter thermautotrophicus, Methanospirillum hungatei, and RC-I), and one candidate was from the Nanoarchaea, Nanoarchaeum equitans. Except for two cases, all novel candidates scored covariance model (COVE) scores (Eddy and Durbin 1994) of >55.00 bits, which was the lowest score of all the documented tDNAs used as positive controls. Furthermore, almost all novel introns were observed to have well-conserved BHB structures whose free energies and position weight matrix (PWM) scores were on a par with those of documented BHB motifs (for details, see Supplemental Materials, Fig. S1, at http://splits.iab.keio.ac.jp/RNA_SupplMat.pdf). The 75 candidates included 32 documented tDNAs, and the remaining 43 were novel candidates. The 32 candidates covered all of the previously documented tDNAs with one or two introns at noncanonical positions. The 43 novel candidates were predicted in the P. aerophilum, S. acidocaldarius, T. pendens, M. hungatei, and RC-I genomes. Twenty-four out of the 43 candidates contained a single intron at noncanonical sites, 15 contained two introns, and three contained three introns within a single tDNA gene. Furthermore, 33 out of 43 overlapped with documented tDNA regions predicted by tRNAscan-SE, and these candidates reassigned the locations of introns or anticodons of the tRNAs. Three candidates from the genome sequence of S. acidocaldarius (SA01 and SA02) and P. aerophilum (PA01) were in agreement with our previous work (Sugahara et al. 2006).
TABLE 1.
All 75 tDNA candidates containing noncanonical introns predicted by SPLITSX
Predicted set of tRNA genes fulfills all 61 codons in T. pendens
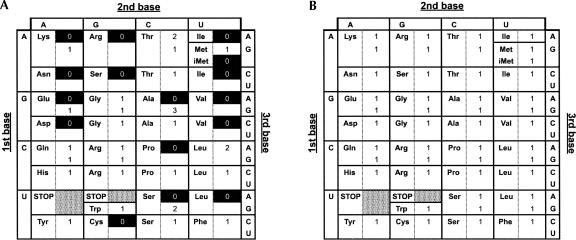
By the analysis of the genome sequence of T. pendens by tRNAscan-SE, a total of 39 tDNAs were predicted (Table 2). The candidates predicted by tRNAscan-SE alone missed a number of tDNAs corresponding to 15 sense codons and one initiator Met tDNA (tDNA-iMet) (Fig. 1A). On the other hand, SPLITSX revealed a total of 35 tDNA genes that contained single or multiple introns at noncanonical sites. Seven out of 35 were predicted from novel genomic regions, and all of the remaining 28 candidates overlapped with tDNA regions predicted by tRNAscan-SE with higher COVE scores. Moreover, the annotations of the corresponding amino acids of 20 tDNAs predicted by tRNAscan-SE alone were reassigned with reliable COVE scores. Combining the 11 candidates predicted only by tRNAscan-SE and the novel 35 candidates by SPLITSX, we identified a total of 46 functional tDNA candidates that were able to read 59 sense codons and one initiator codon AUG, considering the hybridization of dG and rU at a wobble position.
TABLE 2.
The tDNA candidates in T. pendens predicted by tRNAscan-SE alone and by SPLITSX
FIGURE 1.
Codon tables of T. pendens showing the numbers of tRNAs of respective codons. The leftmost column, the top row, and the rightmost column indicate the first, second, and third bases of sense codons, and their coding amino acids are shown in respective fields. (A) Codon table including the 39 tDNA candidates predicted by tRNAscan-SE alone. (B) Codon table including the 46 tDNA candidates complemented and reassigned by SPLITSX, with 10 candidates identified only by tRNAscan-SE.
However, the candidates generated by the merger of the results of SPLITSX and tRNAscan-SE missed only one tDNA encoding tRNASer (GCU), which reads sense codons of AGC and AGU. Therefore, we further analyzed the T. pendens genome with SPLITSX with more relaxed parameters, and the putative tRNASer (TP36) encoded at a genomic location between 200101 and 200210 of Contig 10 (NZ_AASJ01000002) on the complementary strand was identified. The anticodon of TP36 was GCU, the COVE score was 50.35, and the intron was located at 21/22 (for detailed structure, see Supplemental Fig. S1 at http://splits.iab.keio.ac.jp/RNA_SupplMat.pdf). TP36 overlapped with a tRNAAla-encoding gene (SE17) containing an intron located at 37/38 that was predicted by tRNAscan-SE with a COVE score of 64.18. However, the intron (37/38) of SE17 predicted by tRNAscan-SE could not form any type of BHB motifs. In contrast, the 20-nt intron of TP36 predicted by SPLITSX formed a canonical hBHBh′ motif with a stable free energy of −3.20 kcal/mol and high PWM scores of 0.83, and was located at position 21/22, which is reported to harbor a 17-nt intron. Therefore, we suggest that TP36 actually exists in the T. pendens cells to encode tRNASer (GCU), rather than as SE17 encoding tRNAAla. Accordingly, the above-mentioned set of tDNAs fulfills all 61 sense codons and the initiator codon AUG (Fig. 1B).
Structures and isotypes of predicted tDNAs
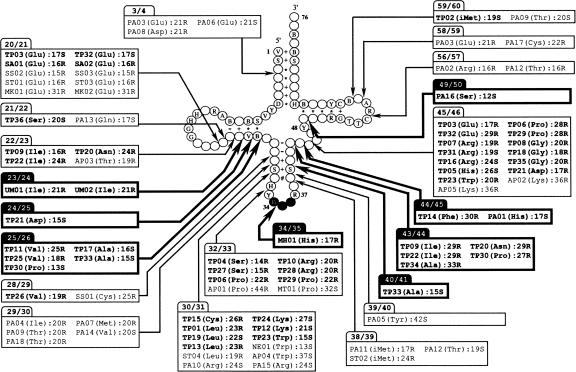
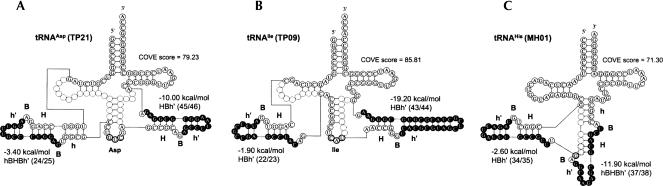
The locations and lengths of the introns, the structures of their BHB motifs, and the isotypes of the predicted tDNAs are summarized in Figure 2. SPLITSX predicted 54 novel introns that were located at noncanonical sites of tDNAs, and ∼65% (36 of 54) of the introns were located at previously reported tDNA sites, whereas the remaining ∼35% (18 of 54) of introns were distributed at eight unreported sites, such as in nucleotide positions 23/24, 24/25, 25/26, 34/35, 40/41, 43/44, 44/45, and 49/50. The schematic diagrams of three typical tDNAs that contain introns at unreported sites are displayed in Figure 3A–C. The gene encoding tRNAAsp (GUC) in T. pendens had two introns at noncanonical positions, 24/25 and 45/46 (Fig. 3A). The intron located at the unreported region 24/25 formed a typical hBHBh′ motif, whereas the other intron located at 45/46 formed an HBh′ motif. The gene encoding tRNAIle (with anticodon CAU reading AUA) also had two introns at 22/23 and 43/44, and both introns formed HBh′ motifs (Fig. 3B). In addition, both of those candidates were significant tDNAs that complement gaps of missing tRNAs in T. pendens. The gene encoding tRNAHis in M. hungatei had a 17-nt intron at the unreported site of 34/35, forming an HBh′ structure, and a 15-nt intron at the canonical site of 37/38, forming an hBHBh′ (Fig. 3C). The intron at 37/38 was found to form the hBHBh′ motif after removal of the intron at 34/35. Moreover, UM01 and UM02 in RC-I were analogous genes encoding tRNAIle (GAU); both had 21-nt introns at unreported sites of 23/24, and UM03 encoding tRNATrp (CCA) harbored the longest 175-nt intron at 37/38. All of these species of tDNA candidates had not been previously annotated by tRNAscan-SE, and our candidates fulfill all codons in RC-I.
FIGURE 2.
Locations of introns of predicted tDNA genes in 11 Archaea. The schematic cloverleaf structure corresponding to the consensus sequence of archaebacterial tRNA sequences has been modified in accordance with the results of a previous work (Marck and Grosjean 2003). The conventional IUB/UPAC degenerate DNA alphabet is used in this figure: R (purine), A or G; Y (pyrimidine), C or T; S (strong), G or C; B (not A), C, G, or T; D (not C), A, G, or T; H (not G), A, C or T; V (not T), A, C, or G; N (any), A, C, G, or T. Base-pairing consensus is denoted by: (+) Watson–Crick base pairing only; (*) Watson–Crick or G-T/T-G pairings; (#) Watson–Crick pairing or mismatch; (−), Watson–Crick pairing or G-T/T-G pairings or mismatches. Intron positions of documented or novel candidates are shown by thin and bold arrows, and by clear and solid tabs over the text boxes, respectively. Each candidate is listed in the text boxes along with the isotype of the amino acid charge in parentheses. The intron length and type of bulge-helix-bulge (BHB) structure are indicated to the right of the colons. The number denotes the nucleotide length, and the capital letter denotes the type of BHB structure: (S) strict hBHBh′ motif, (R) relaxed HBh′ motif. Novel candidates are indicated by bold text.
FIGURE 3.
Schematic diagrams of novel tRNA-encoding genes including multiple introns predicted by SPLITSX, accompanied by COVE scores and free energies of BHB motif. Intron and exon sequences are represented by solid and clear circles, respectively. (Arrows) Location where introns are located. (A) TP21, a putative tRNAAsp with two introns located between nucleotide positions 24 and 25 (24/25) and 45/46 encoded in the T. pendens genomic DNA. The respective position weight matrix (PWM) scores of the BHB motifs were 0.86 and 0.88. (B) TP09, a tRNAIle with two introns at 22/23 and 43/44 synthesized according to the BHB motif with PWM scores 0.85 and 0.63, in T. pendens. (C) MH01, tRNAHis with double introns at 34/35 and 37/38 synthesized according to the BHB motif with PWM scores of 0.67 and 0.90, in the Methanospirillum hungatei.
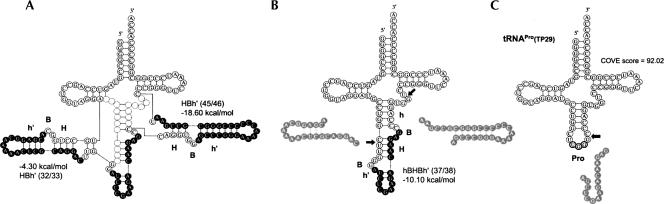
Surprisingly, every proline-charged tDNA of TP06, TP29, and TP30 in the T. pendens genome harbored three introns. TP06 and TP29 were analogous tRNAPro genes that presumably contained three endogenous introns located at 32/33, 37/38, and 45/46. The intron located at the noncanonical site of 32/33 formed an hBHBh′ motif, and another noncanonical intron located at 45/46 formed an HBh′ motif. The intron located at canonical position 37/38 was found to form an hBHBh′ motif after the removal of the outer two noncanonical introns. A schematic representation of the predicted synthetic procedure of the TP29 pre-tRNA is displayed in Figure 4.
FIGURE 4.
Schematic representation of the synthesis procedure in the maturation of tRNA candidate TP29, with three introns located at 32/33, 37/38, and 45/46, along with the COVE score and the free energies of the BHB motifs. (Arrows) Locations where introns were inserted. The position weight matrix (PWM) scores of the respective BHB motifs were 0.74, 0.87, and 0.88. Intron and exon sequences are represented by solid and clear circles, respectively. Two introns that have the potential to form BHB motifs might initially be spliced out at positions 32/33 and 45/46 (A), then another BHB motif is predicted to occur and to be spliced out at position 37/38 (B). Finally, the putative tDNA region of TP29 may form tRNAPro (C).
Identification of tRNA-splicing endonuclease subunits
In order to discuss the novel candidates having noncanonical introns from the viewpoint of the types (homodimer, homotetramer, or heterotetramer) of tRNA-splicing endonuclease subunits, we identified tRNA-splicing endonuclease subunits by using BLASTP (Altschul et al. 1990) searches from the respective genomes. Each type of subunit has been described in the Introduction. The types of tRNA-splicing endonuclease subunits and related genes in seven of the listed 11 genomes have already been identified by Tocchini-Valentini et al. (2005), whereas the types of the subunits in the remaining four genomes (T. pendens, S. acidocaldarius, M. hungatei, and RC-I) have not been identified. We therefore additionally searched for genes homologous to those encoding the tRNA-splicing endonuclease subunits of Methanocaldococcus jannaschii. A set of two homologs was revealed in each of the genomes of T. pendens and S. acidocaldarius. In T. pendens, one was observed in the genomic region between positions 18524 and 19153 in the direct strand of genomic contig 10, with an expectation value (E-value) of 1.3e-24, and the other was in the region between positions 316950 and 317531 in the direct strand, with an E-value of 1.6e-16. On the genome sequence of S. acidocaldarius, one was conserved in the region between positions 687428 and 687973 of the complementary strand, with an E-value of 2.4e-19, and the other was in the region between positions 526525 and 526800 of the complementary strand, with an E-value of 7.5e-05. This suggested that the enzyme structures of the tRNA endonuclease in T. pendens and S. acidocaldarius were heterotetrameric (α2β2). On the other hand, two closely located homologous sequences of lengths of ∼150–200 residues were observed in the RC-I genomic DNA between positions 317008 and 318069 of the direct strand with E-values 6.8e-25 and 4.6e-21, and in M. hungatei genomic DNA between positions 1954717 and 1955724 of the complementary strand with E-values 6.1e-17 and 5.3e-13, which possibly function as one gene, translating to be the homodimer (α′2) with ∼350 amino acid residues. In the results of multiple alignments with documented endonuclease sequences, the novel sequences have been observed with consensus sequences and strong similarities (see Supplemental Fig. S2 at http://splits.iab.keio.ac.jp/RNA_SupplMat.pdf). Except for UM01 and UM02 of RC-I and MH01 of M. hungatei, whose host genomes were predicted to encode homodimeric enzymes, all of the novel tDNA candidates were found in the genomes encoding heterotetrameric or homotetrameric enzymes, which have been reported to recognize introns located at noncanonical positions in tDNAs. We summarize each type of subunit in Table 1.
DISCUSSION
We have described the comprehensive screening of intron-containing tDNAs, including all of the documented tDNAs harboring noncanonical introns with a reasonable number of candidates within the archaeal species analyzed in this work, using enhanced software designated SPLITSX to detect tRNA-encoding genes with multiple introns. Although SPLITSX functions by simply removing BHB motifs from the genome sequence before the execution of tRNAscan-SE, in the absence of any information on the tRNA sequences or their structural specifications and their genomic loci, ∼65% of the predicted introns were located at the sites of previously documented tDNAs. Moreover, ∼74% (32 of 43) of novel tDNA candidates were predicted with their analogous candidates that encode tRNAs of the same isotypes with similar intron lengths and locations. For example, in addition to the known tDNAs that charge glutamic acids containing introns of 15–16-nt conformations located at 20/21, the novel candidates SA01, SA03, TP03, and TP32 were also suggested to charge glutamic acids, containing introns at 20/21 with lengths of 16–17 nt. Additionally, some candidates that presumably contain introns at undocumented sites were also identified with their paralogous or orthologous genes. For example, UM01 and UM02 both encoded tRNAIle (GAU), whose introns were 21-nt long and formed HBh′ structures located at 23/24. According to the results, we therefore suggest that SPLITSX is able to screen reliable tDNA candidates containing introns. Moreover, SPLITSX also detected UM03, which contains a 175-nt intron at canonical position 37/38, from the RC-I genome; this is longer than the 121-nt intron previously reported in the Aeropyrum pernix genome as the longest intron in archaeal tDNAs.
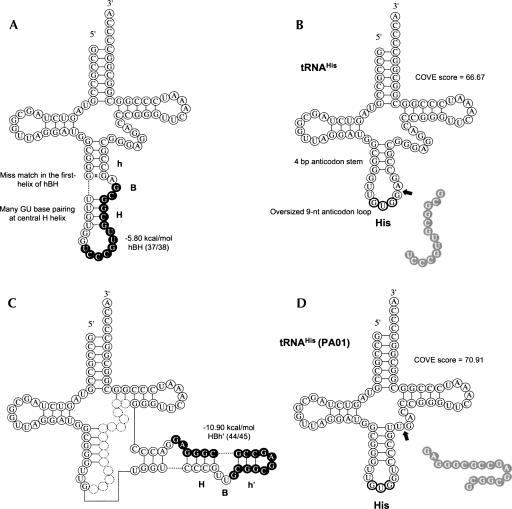
Our novel 43 candidates reassigned 33 tRNA genes, which had been previously documented mainly by utilizing tRNAscan-SE. Here, we suggest that the reassigned tDNAs are more reliable than the previously annotated ones. For example, a tRNAHis-encoding gene had been annotated to contain an intron at 37/38 by Marck and Grosjean (2003), whereas SPLITSX reassigned the tRNAHis-encoding gene as PA01, also encoding tRNAHis in the same genomic locus, containing an intron at the site of 44/45. Although the previous tRNAHis contained a relaxed hBH motif and was located at 37/38, the first helix (h) had one mismatch and the central 4-bp H helix had many rG and rU hybridizations (UUGU/GCGG). On the other hand, the intron located at 44/45 of PA01 predicted by SPLITSX forms a strict hBHBh′ motif with no mismatch, and the central H helix is more convincing (CCCG/CGGG instead of UUGU/GCGG). Moreover, the cloverleaf structure of the reassigned candidate is more mature, with a general tRNA structure consisting of a 5-bp stem at the anticodon arm and a 7-nt anticodon loop. The previous tRNAHis contained the intron at 37/38, and its mature tRNA consisted of a 4-bp anticodon arm and an oversized 9-nt anticodon loop. Therefore, the COVE score was also lower in the previous structure compared with our reassigned candidate (Fig. 5). We thus suggest that PA01 (tRNAHis) containing the intron at 44/45 is actually transcribed, rather than the previous tRNAHis-encoding gene containing the intron at 37/38. Similarly, we claim that other tDNA candidates screened by SPLITSX are more convincing than the previously annotated tDNAs.
FIGURE 5.
Comparison of the P. aerophilum tRNAHis (GUG) structure previously annotated by Marck and Grosjean (2003) with that reassigned by SPLITSX, along with their COVE scores and free energies of the BHB motif. Intron and exon sequences are represented by solid and clear circles, respectively. (A) The previous pre-tRNAHis (GUG) harboring the intron at 37/38 predicted by tRNAscan-SE, (C) the reassigned pre-tRNAHis (GUG) harboring the intron at 44/45 predicted by SPLITSX. Mature tRNA structures are displayed in B and D, respectively. (Arrows) Locations where introns were inserted, (x) a mismatch of base paring in the BHB motif.
We computationally screened a total of 17 tDNAs with multiple introns, including three tDNAs with three introns, encoded in the T. pendens genome. However, only six tDNAs throughout all the archaeal species had been reported to contain two introns within a single gene (Marck and Grosjean 2003), and thus T. pendens is suggested to preferentially encode many tRNA genes with multiple introns compared with other Archaea. In the process of maturation of the pre-tRNAPro of M. thermautotrophicus—the first documented case containing two introns within a single gene (included in our list as MT01)—the first intron, with an hBHBh′ motif of 32 nt located at positions 32/33, is initially spliced out, and then the second intron, with an hBHBh′ motif of 16 nt, is revealed and processed at the canonical location, position 37/38 (Smith et al. 1997). We speculate that the novel candidate tRNAPro in T. pendens (TP06, TP29, and TP30), with the third intron at position 43/44 or 45/46, is spliced in a similar manner (for details, see Fig. 4). In our model, the intron forming the 16-nt hBHBh′ at canonical position 37/38 is spliced out after processing the two introns forming the 22-nt hBHBh′ motif and the 26-nt HBh′ motif located at 32/33 and 45/46, respectively. We propose these genes as a new type of archaeal tDNA containing three introns.
Most of the previously documented introns located at noncanonical positions of tDNAs have been found in Crenarchaeal genomes that encode heterotetrameric endonuclease subunits, and >90% of the novel tDNA candidates predicted in this study contained noncanonical introns (39 of out 42) and are encoded in three Crenarchaeal genomes (P. aerophilum, T. pendens, and S. acidocaldarius) with their heterotetrameric enzymes. On the other hand, the remaining three candidates (UM01, UM02, and MH01) were screened from the Euryarchaeal genomes of RC-I and M. hungatei, which are predicted to encode homodimeric enzymes (Table 1). Although no tDNAs containing introns at noncanonical positions have been found in genomes encoding homodimeric enzymes, and although it is still unclear whether the introns located at noncanonical sites in tDNAs are recognized by homodimeric enzymes, we suggest the existence of such tDNAs processed by homodimeric enzymes in Euryachaea.
In summary, we developed a new program, SPLITSX, for detecting tDNAs containing one or more introns at noncanonical positions on the basis of BHB motif prediction. SPLITSX was able to identify all documented tRNA genes as well as several novel candidates. We further analyzed novel tDNA candidates complementing missing tRNAs in T. pendens and RC-I, and we suggested the existence of a new type of intron-containing tDNA with three introns within a single gene. Our list and the SPLITSX software will be useful for the elucidation of tRNA splicing mechanisms in the Archaea.
MATERIALS AND METHODS
Preparation of genome sequences
The genome sequences of 22 Euryarchaea, five Crenarchaea, and one Nanoarchaea were obtained from GenBank via the National Center for Biotechnology Information (NCBI) ftp server (ftp://ftp. ncbi.nlm.nih.gov). A draft assembly genome of T. pendens (belonging to the Crenarchaeota kingdom) was also used; the genome sequence was obtained from the United States Department of Energy (DOE) Joint Genome Institute (JGI) http server (http://genome.ornl.gov/microbial/tpen/). See Table S1 of the Supplemental Materials (http://splits.iab.keio.ac.jp/RNA_SupplMat.pdf) for a comprehensive listing. The list of the 32 previously identified tDNAs containing introns at noncanonical positions was obtained from the literature (Marck and Grosjean 2003; Randau et al. 2005). Documented tDNAs were used as positive controls for our computational predictions.
SPLITSX
Computational approaches for searching BHB motifs were based on a sequence homology search and structural prediction. For the sequence homology search of BHB motifs, position weight matrices (PWMs) for the sequences of the two outer helices (h and h′), the central helix (H), and the bulge (B) within the 5′ side of the BHB motif sequence (11 nt), and those within the 3′-side sequence (11 nt), were defined by a machine-learning approach from documented BHB motifs (Marck and Grosjean 2003; Randau et al. 2005). To calculate PWMs, the number of occurrences of each base at a given position was compiled. SPLITSX detects pairs of those 5′- and 3′-consensus sequences having lengths between 11 and 200 nt, and determines this region as the first BHB motif candidate. The first BHB motif candidates are further screened for the minimal BHB secondary structure model. The minimal structure of the BHB motif (relaxed HBh′) consists of a 4-bp central helix (H) allowing a 2 bp mismatch, the 3-nt 5′-side bulge (B), and the outer helix (h′) of >1 bp. The free energies of the predicted BHB structure are calculated by using RNAeval (Schuster et al. 1994) implemented within the Vienna-RNA package (http://www.tbi.univie.ac.at/RNA/). A cutoff score of free energy of 3 kcal/mol, which could detect all of the documented BHB motifs in tDNAs, was employed. Finally, the sequences between two bases downstream of the central helix (H) within each BHB were defined as introns.
All possible patterns of genome sequences generated with all combinations of removal of predicted tRNA introns were automatically searched using tRNAscan-SE inside SPLITSX. Because SPLITSX detects false positives within documented tDNA regions that do not contain endogenous introns, the original genome sequence without the removal of the introns was also queried to tRNAscan-SE in order to predict high-integrity candidates. If candidates from the intron-removed sequence overlapped with others, including genes annotated only by the tRNAscan-SE process, we eliminated those candidates whose COVE scores were lower than the COVE scores of their overlapping ones. tRNAscan-SE was invoked using the −A switch to load the specific covariance model for archaeal tDNAs. SPLITSX was performed with the following parameters: −d 2, −p 0.51, −H 2, and −F 3. The SPLITSX source code and the software package are freely available at our Web site (http://splits.iab.keio.ac.jp).
Comparative genomics analysis of tRNA-splicing endonuclease subunits
Using whole-genome sequences whose tRNA-splicing endonuclease subunits were still unclear and unclassified into the types of structural features, we conducted BLASTP searches, using the DNA sequences encoding the tRNA endonuclease subunits of M. jannaschii as the query. According to the analysis of Tocchini-Valentini et al. (2005), a species was defined to have a homotetrameric enzyme if only one homologous gene was found in the genome. Likewise, a heterotetrameric enzyme was defined to be present when two homologous genes were observed in separate positions, and a homodimeric enzyme was defined when two homologous sequences were within the DNA region that has a translated amino acid sequence length of <350 residues and that was encoded by one gene. Finally, using MAFFT software (Katoh et al. 2005), predicted amino acid sequences were aligned with known tRNA endonucleases. Both BLASTP and MAFFT were conducted by default parameters.
ACKNOWLEDGMENTS
We thank the members of MGSP at the Institute for Advanced Biosciences, Keio University, for their critical suggestions. We also thank the United States Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/) for permission to use the sequence data of Thermofilum pendens Hrk 5. This research was supported in part by the Japan Society for the Promotion of Science (JSPS); a grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan (The 21st Century COE Program, entitled Understanding and Control of Life's Function via Systems Biology).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.309507.
REFERENCES
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Daniels, C.J., Gupta, R., Doolittle, W.F. Transcription and excision of a large intron in the tRNATrp gene of an archaebacterium, Halobacterium volcanii. J. Biol. Chem. 1985;260:3132–3134. [PubMed] [Google Scholar]
- Datta, P.K., Hawkins, L.K., Gupta, R. Presence of an intron in elongator methionine-tRNA of Halobacterium volcanii. Can. J. Microbiol. 1989;35:189–194. doi: 10.1139/m89-029. [DOI] [PubMed] [Google Scholar]
- Eddy, S.R., Durbin, R. RNA sequence analysis using covariance models. Nucleic Acids Res. 1994;22:2079–2088. doi: 10.1093/nar/22.11.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichant, G.A., Burks, C. Identifying potential tRNA genes in genomic DNA sequences. J. Mol. Biol. 1991;220:659–671. doi: 10.1016/0022-2836(91)90108-i. [DOI] [PubMed] [Google Scholar]
- Fitz-Gibbon, S.T., Ladner, H., Kim, U.J., Stetter, K.O., Simon, M.I., Miller, J.H. Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc. Natl. Acad. Sci. 2002;99:984–989. doi: 10.1073/pnas.241636498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam, S.J., Konstantinidis, K.T., Putnam, N., Schleper, C., Watanabe, Y., Sugahara, J., Preston, C., de la Torre, J., Richardson, P.M., Delong, E.F. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc. Natl. Acad. Sci. 2006;103:18296–18301. doi: 10.1073/pnas.0608549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaine, B.P., Gupta, R., Woese, C.R. Putative introns in tRNA genes of prokaryotes. Proc. Natl. Acad. Sci. 1983;80:3309–3312. doi: 10.1073/pnas.80.11.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K., Kuma, K., Toh, H., Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjems, J., Leffers, H., Olesen, T., Garrett, R.A. A unique tRNA intron in the variable loop of the extreme thermophile Thermofilum pendens and its possible evolutionary implications. J. Biol. Chem. 1989;264:17834–17837. [PubMed] [Google Scholar]
- Kleman-Leyer, K., Armbruster, D.W., Daniels, C.J. Properties of H. volcanii tRNA intron endonuclease reveal a relationship between the archaeal and eucaryal tRNA intron processing systems. Cell. 1997;89:839–847. doi: 10.1016/s0092-8674(00)80269-x. [DOI] [PubMed] [Google Scholar]
- Lowe, T.M., Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck, C., Grosjean, H. Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: Evolutionary implications. RNA. 2003;9:1516–1531. doi: 10.1261/rna.5132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch, R., Hiller, K., Grote, A., Scheer, M., Klein, J., Schobert, M., Jahn, D. Virtual Footprint and PRODORIC: An integrative framework for regulon prediction in prokaryotes. Bioinformatics. 2005;21:4187–4189. doi: 10.1093/bioinformatics/bti635. [DOI] [PubMed] [Google Scholar]
- Pavesi, A., Conterio, F., Bolchi, A., Dieci, G., Ottonello, S. Identification of new eukaryotic tRNA genes in genomic DNA databases by a multistep weight matrix analysis of transcriptional control regions. Nucleic Acids Res. 1994;22:1247–1256. doi: 10.1093/nar/22.7.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randau, L., Pearson, M., Soll, D. The complete set of tRNA species in Nanoarchaeum equitans. FEBS Lett. 2005;579:2945–2947. doi: 10.1016/j.febslet.2005.04.051. [DOI] [PubMed] [Google Scholar]
- Schuster, P., Fontana, W., Stadler, P.F., Hofacker, I.L. From sequences to shapes and back: A case study in RNA secondary structures. Proc. Biol. Sci. 1994;255:279–284. doi: 10.1098/rspb.1994.0040. [DOI] [PubMed] [Google Scholar]
- She, Q., Singh, R.K., Confalonieri, F., Zivanovic, Y., Allard, G., Awayez, M.J., Chan-Weiher, C.C., Clausen, I.G., Curtis, B.A., De Moors, A., et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slesarev, A.I., Mezhevaya, K.V., Makarova, K.S., Polushin, N.N., Shcherbinina, O.V., Shakhova, V.V., Belova, G.I., Aravind, L., Natale, D.A., Rogozin, I.B., et al. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc. Natl. Acad. Sci. 2002;99:4644–4649. doi: 10.1073/pnas.032671499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D.R., Doucette-Stamm, L.A., Deloughery, C., Lee, H., Dubois, J., Aldredge, T., Bashirzadeh, R., Blakely, D., Cook, R., Gilbert, K., et al. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: Functional analysis and comparative genomics. J. Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara, J., Yachie, N., Sekine, Y., Soma, A., Matsui, M., Tomita, M., Kanai, A. SPLITS: A new program for predicting split and intron-containing tRNA genes at the genome level. In Silico Biol. 2006;6:411–418. [PubMed] [Google Scholar]
- Tang, T.H., Rozhdestvensky, T.S., d'Orval, B.C., Bortolin, M.L., Huber, H., Charpentier, B., Branlant, C., Bachellerie, J.P., Brosius, J., Huttenhofer, A. RNomics in Archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res. 2002;30:921–930. doi: 10.1093/nar/30.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, L.D., Brandon, L.D., Nieuwlandt, D.T., Daniels, C.J. Transfer RNA intron processing in the halophilic archaebacteria. Can. J. Microbiol. 1989;35:36–42. doi: 10.1139/m89-006. [DOI] [PubMed] [Google Scholar]
- Tocchini-Valentini, G.D., Fruscoloni, P., Tocchini-Valentini, G.P. Structure, function, and evolution of the tRNA endonucleases of Archaea: An example of subfunctionalization. Proc. Natl. Acad. Sci. 2005;102:8933–8938. doi: 10.1073/pnas.0502350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela, P., Venegas, A., Weinberg, F., Bishop, R., Rutter, W.J. Structure of yeast phenylalanine-tRNA genes: An intervening DNA segment within the region coding for the tRNA. Proc. Natl. Acad. Sci. 1978;75:190–194. doi: 10.1073/pnas.75.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, Y., Yokobori, S., Inaba, T., Yamagishi, A., Oshima, T., Kawarabayasi, Y., Kikuchi, H., Kita, K. Introns in protein-coding genes in Archaea. FEBS Lett. 2002;510:27–30. doi: 10.1016/s0014-5793(01)03219-7. [DOI] [PubMed] [Google Scholar]
- Wich, G., Leinfelder, W., Bock, A. Genes for stable RNA in the extreme thermophile Thermoproteus tenax: Introns and transcription signals. EMBO J. 1987;6:523–528. doi: 10.1002/j.1460-2075.1987.tb04784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinari, S., Itoh, T., Hallam, S.J., DeLong, E.F., Yokobori, S., Yamagishi, A., Oshima, T., Kita, K., Watanabe, Y. Archaeal pre-mRNA splicing: A connection to hetero-oligomeric splicing endonuclease. Biochem. Biophys. Res. Commun. 2006;346:1024–1032. doi: 10.1016/j.bbrc.2006.06.011. [DOI] [PubMed] [Google Scholar]