Abstract
Adenosine to inosine (A-to-I) pre-mRNA editing by the ADAR enzyme family has the potential to increase the variety of the proteome. This editing by adenosine deamination is essential in mammals for a functional brain. To detect novel substrates for A-to-I editing we have used an experimental method to find selectively edited sites and combined it with bioinformatic techniques that find stem–loop structures suitable for editing. We present here the first verified editing candidate detected by this screening procedure. We show that Gabra-3, which codes for the α3 subunit of the GABAA receptor, is a substrate for editing by both ADAR1 and ADAR2. Editing of the Gabra-3 mRNA recodes an isoleucine to a methionine. The extent of editing is low at birth but increases with age, reaching close to 100% in the adult brain. We therefore propose that editing of the Gabra-3 mRNA is important for normal brain development.
Keywords: RNA editing, adenosine deaminase, ADAR, GABA receptor, Gabra-3
INTRODUCTION
Several gene products involved in neurotransmission undergo site-selective adenosine to inosine (A-to-I) RNA editing in the mammalian brain. This selective editing takes place most notably in the mRNA that codes for several subunits of ionotropic glutamate receptors (Seeburg and Hartner 2003). The editing event is a hydrolytic deamination of adenosine to inosine, catalyzed by the adenosine deaminase that acts on RNA (ADAR) enzymes. Two A-to-I editing enzymes have been shown to be enzymatically active in mammals, ADAR1 and ADAR2. These enzymes differ somewhat in their specificity, but both are essential (Higuchi et al. 2000; Hartner et al. 2004; Wang et al. 2004). However, editing occurs with little sequence specificity in largely double-stranded RNA (Bass 2002). Site-selective editing has been shown to alter amino acid codons and/or splicing patterns in transcripts. This form of RNA processing may therefore contribute to an increased number of protein isoforms. One example is the editing of the α-amino-3-hydroxy-5-methyl-4-isoxazole glutamate receptor subunit B (GluR-B), which alters the properties of the receptor. The Q/R site of GluR-B is nearly fully edited. At this site a CAG, coding for glutamine, is modified to CIG, which is read as an arginine codon (CGG). This change of a single amino acid, situated in the second transmembrane helix of the GluR-B subunit, makes receptors assembled with an edited GluR-B subunit nearly impermeable to calcium (Hume et al. 1991; Burnashev et al. 1992). As in several other cases of site-selective A-to-I editing, an inverted repeat located in the downstream intron is required for Q/R editing (Higuchi et al. 1993). Editing at the Q/R site is essential for a normal brain development. Mice that lack the sequence complementary to the editing site in the downstream intron cannot carry out the Q/R editing, and these mice develop epileptic seizures and die by 3 wk of age (Brusa et al. 1995). Another receptor that is modified by A-to-I editing is the serotonin receptor subtype 5-HT2c, which is a member of the G-protein-coupled receptor family. The pre-mRNA is edited at five sites, creating several changes in codon usage. The edited isoforms code the second intracellular loop differently, and the efficiency of the interaction between the receptor isoforms and the G protein is thus different (Burns et al. 1997).
Bioinformatic techniques have been used to demonstrate abundant hyperediting in Alu sequences within introns and untranslated regions of the human transcriptome (Athanasiadis et al. 2004; Blow et al. 2004; Levanon et al. 2004). The results of these methods suggest that between 13,000 and 30,000 sites in 1600–2600 different genes are A-to-I edited. Many of these sites have been verified experimentally. The purpose of Alu editing is not known, but it is clear that there is an abundant amount of active enzyme that can perform A-to-I editing in the cell.
We have combined bioinformatic analyzes detecting stem–loop structures suitable for A-to-I editing (Pedersen et al. 2006) with an experimental method developed to find novel ADAR substrates that have been edited in a site-selective manner (Ohlson et al. 2005). Several potential editing substrates were discovered using this approach. One of the principal candidate genes is coding for gamma-aminobutyric acid (GABA) type A (GABAA) receptor subunit α3 (Gabra-3). The potential edited site has previously been identified as a site of single nucleotide polymorphism (SNP), entered into the SNP database (Sherry et al. 2001). However, the identification as an SNP is based on two brain-derived expressed sequence tags (ESTs) showing somatic mRNA modifications. The GABA receptor is a member of the Cys-loop ligand-gated ion channel superfamily. These post-synaptic receptors are composed of five subunits with an extracellular ligand-binding domain and ion-channel domains that are integral to the membrane. Ligand binding causes an allosteric activation of the ion channel. The selectivity of the ion channel and the magnitudes of the electrochemical gradients that form across the membrane determine the nature of the electrical signal (Cromer et al. 2002). When the neurotransmitter GABA binds to the GABAA receptor, the chloride channel is activated. This leads to a rapid increase in chloride ion (Cl−) conductance, which reduces the excitatory depolarization (Moss and Smart 2001). However, until post-natal day (P) 8–12, GABA mediates the depolarization rather than hyperpolarization through the GABAA receptors (Ben-Ari 2002).
It has previously been shown that the glutamate receptors (which are responsible for most excitatory post-synaptic signals in the mammalian brain) are regulating the calcium influx by A-to-I editing. In the adult brain an equilibrium between excitation and inhibition is essential. Here we show that the GABAA receptor (which is responsible for most inhibitory post-synaptic signals) also is edited at a single nucleotide that recodes an isoleucine to a methionine with a possibility to function in the regulation of chloride permeability of the channel.
RESULTS AND DISCUSSION
Detecting novel sites of A-to-I editing
We have developed a method by which it is possible to find novel ADAR substrates that have been edited in a site-selective manner (Ohlson et al. 2005). The method is based on extracting intrinsic ADAR2–RNA substrate complexes by coimmunoprecipitation using an anti-ADAR2 antibody. We have used a mouse genome microarray to detect enriched ADAR2–RNA targets. In parallel, we detected stem–loop structures suitable for A-to-I editing in encoded sequences by using EvoFold, a general comparative genomics program for identifying conserved RNA structures (Pedersen et al. 2006). One of the most promising predicted structures suitable for A-to-I editing was found in the Gabra-3 transcript coding for the α3 subunit of the GABAA receptor. The Gabra-3 RNA was enriched on average sevenfold in the microarray after immunoprecipitation using the anti-ADAR2 antibody compared to nonspecific preimmune serum.
Verification of an edited site in the Gabra-3 transcript
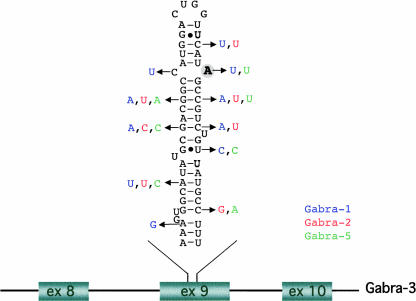
The potential editing site in Gabra-3 is located within exon 9. The stem–loop RNA structure that we believe is required for editing consists of a stem of 22 base pairs (bp), interrupted by three bulges and an A:C mismatch at the edited site (Fig. 1). It is notable that the entire double-stranded RNA structure that creates the editing substrate is located in exon 9. This is rare since most known site-selectively edited substrates consist of an exon sequence and a complementary intron sequence. The Kv1.1 mRNA is the only previously reported example in mammals where the editing site and the editing complementary sequence both are situated in coding sequences (Bhalla et al. 2004). The corresponding RNA sequences of α-subunits 1, 2, and 5 predict a structure that is less favorable for editing than that of α3 (Fig. 1). Further, the adenosine that is edited in Gabra-3 is a uridine in Gabra-1 and -5 (Fig. 1). The amino acid sequence in the vicinity of the isoleucine is conserved across all subunits. However, the nucleotide at the third position of the amino acid codons is frequently different in the transcripts of Gabra-1, -2, and -5 from that in Gabra-3.
FIGURE 1.
Subunit-specific sequences of the gabra genes. The predicted structure of the stem–loop within exon 9 of Gabra-3. The edited A is printed in bold and circled in gray. The differences between Gabra-3 and other Gabra sequences are shown in blue for Gabra-1, red for Gabra-2, and green for Gabra-5.
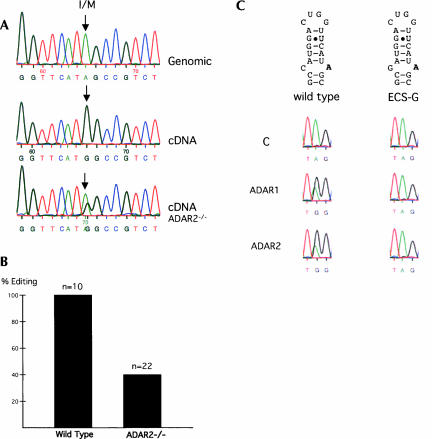
To verify editing of Gabra-3, total RNA from adult mouse brain was isolated and the sequence determined after RT-PCR was compared with the genomic sequence. An adenosine was present in the genomic sequence of exon 9 at the position at which a guanosine was present in the cDNA sequence (Fig. 2A). At this position an isoleucine (I) AUA codon is changed to an AUI read as a methionine (M) codon (AUG), since inosine is read as a guanosine by the cellular machinery. This edited site will now be referred to as the I/M site. We also investigated if the Gabra-2 transcript is edited. Gabra-2 has an adenosine at the I/M site and is therefore a potential substrate for editing. However, no editing at this site could be detected in the Gabra-2 transcript in adult mouse brain (data not shown). This is not surprising since several base pairs in the predicted stem–loop structure are disrupted, making it into a less favorable substrate for editing.
FIGURE 2.
Editing of the Gabra-3 transcript. (A) The editing of Gabra-3 was demonstrated by DNA sequencing. The chromatogram of the genomic DNA sequence shows an adenosine at the I/M-site, indicated by an arrow. Total brain RNA from the same NMRI adult mouse was reverse transcribed (cDNA) and sequenced after PCR amplification. A guanosine was present at the I/M site in the cDNA. The cDNA from an ADAR2−/− adult mouse was amplified by PCR and sequenced. A dual A and G peak appeared with the majority of the transcripts showing an A at the I/M site. (B) PCR products from wild-type and ADAR2−/− cDNA were cloned and sequenced. The sequences of 10 wild-type and 22 ADAR2−/− clones were determined. (C) Wild-type Gabra-3 and Gabra-3 with a mutated ECS (ECS-G) were transfected into HEK 293 cells with an ADAR1 or ADAR2 expression vector. An empty expression vector (labeled “C” for control) was used as a control for endogenous editing. In wild-type Gabra-3 (left), no editing was observed in the control since a single A peak is seen at the I/M site. During cotransfection with ADAR1 or ADAR2 dual G/A peaks were seen with the majority in the G peak. Adenosine was present at the editing site in the ECS-G mutant in all transfection experiments (right).
In summary these results indicate that both sequence and structure are important for editing at the I/M site of Gabra-3 and that neither of these are conserved in the transcripts of Gabra-1, -2, or -5.
Gabra-3 is edited by both ADAR1 and ADAR2
To determine if ADAR2 is catalyzing the editing at this site, cDNA from the brain of an adult ADAR2−/− mouse was analyzed. Editing at the I/M site of Gabra-3 was dramatically lower in the ADAR2−/− mouse (41%) than it was in the wild-type adult mouse (100%) (Fig. 2A,B). We conclude that most Gabra-3 transcripts are edited at the I/M site and that ADAR2 contributes substantially to this editing.
We wanted to investigate if ADAR1 and ADAR2 can edit a Gabra-3 mini-gene containing exon 9, including the I/M site. The Gabra-3 mini-gene was cotransfected into HEK 293 cells, together with an ADAR1 or ADAR2 expression vector. Total RNA was extracted from the cells, and editing was determined by sequencing, after RT-PCR. ADAR1 was able to efficiently edit the I/M site since the major peak in the chromatogram was G (Fig. 2C). In transient transfections using ADAR2 this enzyme was able to edit the reporter construct to about the same extent as ADAR1. As a control the contribution from endogeneous editing in HEK 293 cells was investigated using an empty expression vector cotransfected with the Gabra-3 mini-gene. Since a single A peak was seen in the chromatogram (Fig. 2C, indicated as C) no endogenous ADAR activity in HEK 293 contributed to editing of the mini-gene. In summary, these results show that both ADAR1 and ADAR2 have the potential to efficiently edit the I/M site of the Gabra-3 transcript.
An A-C mismatch at the editing site of Gabra-3
To investigate the importance of a C opposing the I/M editing in the predicted exon 9 stem–loop structure of Gabra-3, we created a point mutation in which the C was changed to a G (ECS-G) (Fig. 2C). It has previously been shown that changing a nucleotide from a C to a G at the site opposing the editing site can have a dramatic negative effect on editing (Wong et al. 2001; Källman et al. 2003). Neither ADAR1 nor ADAR2 produced editing at the I/M site in the ECS-G mutant (Fig. 2C). This result confirms that it is important that the base, 15 nucleotides (nt) from the I/M site, is not a guanosine for the editing to occur. Although the predicted stem–loop structure in Figure 1 is not confirmed by this result, it indicates that the C, 15 nt upstream of the I/M site, is most likely the opposing base to the editing site.
Editing in the α3 subunit of the GABAA receptor causes an amino acid change
The edited site in the α3 subunit of the GABAA receptor is located in the transmembrane (TM) domain 3 (Fig. 3A). The four TM domains of this receptor subunit interact with TM domains from four other subunits to form the channel of the receptor. It is possible that the isoleucine to methionine amino acid change in TM3 of α3 changes the environment in the channel, since TM3 and the large intracellular loop are important for gating and inactivation of the channel (Fisher 2004). A change from the branched isoleucine to methionine will alter the side chain to be longer with a more bulky sulphur atom. Interestingly, frog and pufferfish have a genomically encoded methionine at the equivalent position (Fig. 3B). Noteworthy is that the phylogenetic relationship between the organisms indicates that the more closely related species share the same amino acid at this site (Fig. 3C). The situation is similar for the Q/R editing site of GluR-B, where hagfish, agnathan fish, and teleost fish have an R codon at this position, while species that appeared after the cartilaginous fishes have a Q codon in the same position (Wu et al. 1996; Kung et al. 2001).
FIGURE 3.
The position of the edited site in the α3 subunit and evolutionary conservation. (A) The protein sequence of Gabra-3 consists of four transmembrane domains (TM1–TM4). The edited site was located within TM3 at amino acid 5 (of 22). (B) The I/M sites of Gabra-3 in frog (Xenopus tropicalis) and pufferfish (Tetraodon nigroviridis) have a genomically encoded methionine where all other organisms have an isoleucine (Hinrichs et al. 2006). (C) An evolutionary tree showing the phylogenetic relationship between the species in B (Meyer and Zardoya 2003; Kriegs et al. 2006).
Editing of the I/M site is developmentally regulated
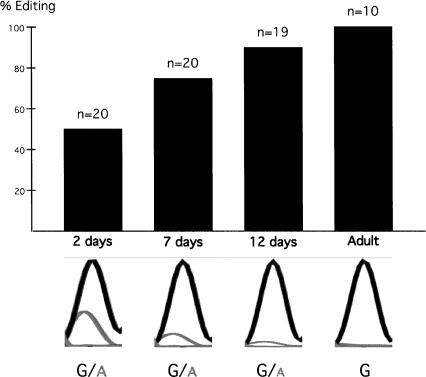
A switch in the GABA response from excitatory to inhibitory post-synaptic potentials occurs during early development where an efflux of chloride ions takes place in immature neurons, while there is an influx of chloride ions in mature neurons (Ben-Ari 2002). GABA switches from being excitatory to inhibitory by an up-regulation of the cotransporter KCC2 that decreases the chloride concentration in the cell. However, if GABA itself promotes the expression of KCC2 is still under debate (Ganguly et al. 2001; Ludwig et al. 2003; Titz et al. 2003). Further, the α subunits are critical elements in determining the nature of the GABAA receptor response to GABA (Böhme et al. 2004). The α3 mRNA (Gabra-3) is present at high levels in several forebrain regions at birth with a major decline after post-natal day 12 (P12), when the expression of α1 is going up (Laurie et al. 1992). The change from α3 to α1 may cause the switch in GABA behavior from excitatory to inhibitory post-synaptic potentials during development. To investigate if editing at the I/M-site of Gabra-3 changes during development, RNA from mouse brain at different developmental stages were extracted. The extent of editing was determined by sequencing of individual clones derived from the RT-PCR products. In newborn mice at day 2 (P2) 50% of the Gabra-3 transcripts were edited while those in adult mouse brain were edited close to 100% (Fig. 4). According to a chi-square statistic analysis this difference is significant (p*<0.01, χ2=16.59, degree of freedom: df=4). The high level of editing in adult mouse was seen in two different mouse strains, NMRI and FVB/N, at 3 wk and 19 wk of age. This result indicates that the extent of editing is low in newborn mice and increases with age. Furthermore, there is a trend of gradual increase in the extent of editing at P7 and P12 (Fig. 4). Although we do not know the effect of this editing event on the nature of the receptor, we do know that a change in editing occurs concurrent with a dramatic change in the function of the GABAA receptor. The α subunits are critical elements in determining the nature of the GABAA receptor response to GABA (Böhme et al. 2004). It is therefore possible that editing at the I/M site of Gabra-3 contributes to this change of function.
FIGURE 4.
Editing of the α3 subunit at different stages during development. Editing of the Gabra-3 transcript extracted from mouse brain at different developmental stages. The RT-PCR products from mice at day 2, day 7, day 12, and adult mouse were cloned and editing at the I/M site was analyzed by sequence determination. The number (n) of clones analyzed at each developmental stage is indicated. Below, the chromatogram from the sequence at the I/M site is based on the population derived from the RT-PCR product.
GABAA receptors respond to anxiolytic drugs such as benzodiazepines and are thus important drug targets. The benzodiazepine binding site is located at the interface of the α and γ2 subunits (Cromer et al. 2002). Antagonists that bind to this site enhance the effect of GABA by increasing the frequency of GABA-induced channel opening events. Post-transcriptional modifications of the α3 subunit, such as the I/M editing described here, could be important in determining the mechanistic features that are responsible for the diversity of GABAA receptors and the variability in sensitivity to drugs.
MATERIALS AND METHODS
RT-PCR and sequencing
RNA was isolated from triplicate FVB/N mouse brains at postnatal days 2, 7, and 12 and adult mouse (19 wk) using TRIzol (Invitrogen). Random primers were used for first-strand cDNA synthesis. cDNA from adult ADAR2−/− mouse was kindly provided by P.H. Seeburg (Max-Planck-Institut) via M.F. Jantsch (University of Vienna). DNA was isolated from mouse tails using the Genomic DNA Purification Kit (Fermenta). PCR was then carried out with primers specific for GABAA receptor subunit α3: exon–exon primers for the cDNA sequences and exon–intron primers for the genomic sequences. Superscript III RT (Invitrogen) was used in all reverse transcription reactions, and platinum Taq (Invitrogen) was used in all PCR reactions. Amplified PCR products were gel purified and sequenced. The PCR products from cDNA wild-type (FVB/N) mice and from cDNA ADAR2−/− mouse were cloned into pGEM-T easy vectors (Promega) and sequenced. Twenty clones from the 2 d mouse, 20 clones from the 7 d mouse, 19 clones from the 12 d mouse, 10 clones from the 3 wk mouse (NMRI wild-type mouse), 10 clones from the 19 wk mouse, and 22 clones from the ADAR2−/− mouse were analyzed. The sequences of primers used in PCR amplification and sequence determination are available from the author on request.
Reporter constructs and substrate mutagenesis
The ADAR1 expression vector pCS DRADA-FLIS6 (Desterro et al. 2003) was a kind gift from M. O'Connell (Medical Research Council, University of Edinburgh). The ADAR2 expression vector pcDNA3 FLAG/rADAR2 has been previously described (Bratt and Öhman 2003). The Gabra-3 editing reporter construct was generated by cloning a fragment of the genomic gabra3 gene (including exon 9 and 319 nt of the downstream intron) into the mammalian expression vector pcDNA3 FLAG. This construct was named “pGARα3-I/M.” The reporter construct named “pGARα3-ECS G” was generated by mutating the cytidine that is located 15 nt upstream of the edited site in the pGARα3-I/M to a guanosine using QuickChange site-directed mutagenesis (Stratagene).
Transfection and editing analysis
HEK 293 cells were transfected with a total of 4 μg of various combinations of the expression vectors, using LIPOFECTAMINE 2000 (Invitrogen). RNA was isolated 24 h after transfection using a GenElute mammalian total RNA isolation kit (Sigma), treated with DNase (TURBO DNA-free; Ambion), and then RT-PCR amplified using gene-specific primers. PCR products were gel purified and sequenced.
Structure analysis
For the protein sequence of Gabra-3, Swiss-Prot (Bairoch et al. 2004), entry P26049, was used. The Modeller 6v2 (Sali and Blundell 1993) program was used to model transmembrane helix 3 (amino acid 338–359 of Gabra-3, I342M) using the entry 1vry in the protein database (Berman et al. 2000) as a template. Figure 3A was drawn using PyMol (http://www.pymol.org).
ACKNOWLEDGMENTS
We thank Britt-Marie Sjöberg and Lars Wieslander for discussions, Peter Seeburg and Michael Jantsch for sharing cDNA, Tomas Ohlson for protein structure predictions, and Anthony Poole for phylogenetic analyses. We are also grateful to the department of Zoophysiology, Stockholm University, for technical assistance. This work was funded by the Swedish Research Council, the U.S. National Cancer Institute, and the Danish Research Council (J.S.P.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.349107.
REFERENCES
- Athanasiadis, A., Rich, A., Maas, S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch, A., Boeckmann, B., Ferro, S., Gasteiger, E. Swiss-Prot: Juggling between evolution and stability. Brief. Bioinform. 2004;5:39–55. doi: 10.1093/bib/5.1.39. [DOI] [PubMed] [Google Scholar]
- Bass, B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari, Y. Excitatory actions of GABA during development: The nature of the nurture. Nat. Rev. Neurosci. 2002;3:728–739. doi: 10.1038/nrn920. [DOI] [PubMed] [Google Scholar]
- Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I.N., Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla, T., Rosenthal, J.J., Holmgren, M., Reenan, R. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nat. Struct. Mol. Biol. 2004;11:950–956. doi: 10.1038/nsmb825. [DOI] [PubMed] [Google Scholar]
- Blow, M., Futreal, P.A., Wooster, R., Stratton, M.R. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhme, I., Rabe, H., Lüddens, H. Four amino acids in the α subunits determine the γ-aminobutyric acid sensitivities of GABAA receptor subtypes. J. Biol. Chem. 2004;279:35193–35200. doi: 10.1074/jbc.M405653200. [DOI] [PubMed] [Google Scholar]
- Bratt, E., Öhman, M. Coordination of editing and splicing of glutamate receptor pre-mRNA. RNA. 2003;9:309–318. doi: 10.1261/rna.2750803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusa, R., Zimmermann, F., Koh, D.S., Feldmeyer, D., Gass, P., Seeburg, P.H., Sprengel, R. Early-onset epilepsy and postnatal lethality associated with an editing- deficient GluR-B allele in mice. Science. 1995;270:1677–1680. doi: 10.1126/science.270.5242.1677. [DOI] [PubMed] [Google Scholar]
- Burnashev, N., Monyer, H., Seeburg, P.H., Sakmann, B. Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron. 1992;8:189–198. doi: 10.1016/0896-6273(92)90120-3. [DOI] [PubMed] [Google Scholar]
- Burns, C.M., Chu, H., Rueter, S.M., Hutchinson, L.K., Canton, H., Sanders-Bush, E., Emeson, R.B. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- Cromer, B.A., Morton, C.J., Parker, M.W. Anxiety over GABAA receptor structure relieved by AChBP. Trends Biochem. Sci. 2002;27:280–287. doi: 10.1016/s0968-0004(02)02092-3. [DOI] [PubMed] [Google Scholar]
- Desterro, J.M., Keegan, L.P., Lafarga, M., Berciano, M.T., O'Connell, M., Carmo-Fonseca, M. Dynamic association of RNA-editing enzymes with the nucleolus. J. Cell Sci. 2003;116:1805–1818. doi: 10.1242/jcs.00371. [DOI] [PubMed] [Google Scholar]
- Fisher, J.L. A mutation in the GABAA receptor α 1 subunit linked to human epilepsy affects channel gating properties. Neuropharmacology. 2004;46:629–637. doi: 10.1016/j.neuropharm.2003.11.015. [DOI] [PubMed] [Google Scholar]
- Ganguly, K., Schinder, A.F., Wong, S.T., Poo, M. GABA itself promotes the developmental switch of neuronal GABAergic responses from excitation to inhibition. Cell. 2001;105:521–532. doi: 10.1016/s0092-8674(01)00341-5. [DOI] [PubMed] [Google Scholar]
- Hartner, J.C., Schmittwolf, C., Kispert, A., Muller, A.M., Higuchi, M., Seeburg, P.H. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- Higuchi, M., Single, F.N., Kohler, M., Sommer, B., Sprengel, R., Seeburg, P.H. RNA editing of AMPA receptor subunit GluR-B: A base-paired intron–exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- Higuchi, M., Maas, S., Single, F.N., Hartner, J., Rozov, A., Burnashev, N., Feldmeyer, D., Sprengel, R., Seeburg, P.H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- Hinrichs, A.S., Karolchik, D., Baertsch, R., Barber, G.P., Bejerano, G., Clawson, H., Diekhans, M., Furey, T.S., Harte, R.A., Hsu, F., et al. The UCSC Genome Browser Database: Update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume, R.I., Dingledine, R., Heinemann, S.F. Identification of a site in glutamate receptor subunits that controls calcium permeability. Science. 1991;253:1028–1031. doi: 10.1126/science.1653450. [DOI] [PubMed] [Google Scholar]
- Källman, A.M., Sahlin, M., Öhman, M. ADAR2 A→I editing: Site selectivity and editing efficiency are separate events. Nucleic Acids Res. 2003;31:4874–4881. doi: 10.1093/nar/gkg681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegs, J.O., Churakov, G., Kiefmann, M., Jordan, U., Brosius, J., Schmitz, J. Retroposed elements as archives for the evolutionary history of placental mammals. PLoS Biol. 2006;4:e91. doi: 10.1371/journal.pbio.0040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung, S.S., Chen, Y.C., Lin, W.H., Chen, C.C., Chow, W.Y. Q/R RNA editing of the AMPA receptor subunit 2 (GRIA2) transcript evolves no later than the appearance of cartilaginous fishes. FEBS Lett. 2001;509:277–281. doi: 10.1016/s0014-5793(01)03183-0. [DOI] [PubMed] [Google Scholar]
- Laurie, D.J., Wisden, W., Seeburg, P.H. The distribution of thirteen GABAA receptor subunit mRNAs in the rat brain. III. Embryonic and postnatal development. J. Neurosci. 1992;12:4151–4172. doi: 10.1523/JNEUROSCI.12-11-04151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon, E.Y., Eisenberg, E., Yelin, R., Nemzer, S., Hallegger, M., Shemesh, R., Fligelman, Z.Y., Shoshan, A., Pollock, S.R., Sztybel, D., et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- Ludwig, A., Li, H., Saarma, M., Kaila, K., Rivera, C. Developmental up-regulation of KCC2 in the absence of GABAergic and glutamatergic transmission. Eur. J. Neurosci. 2003;18:3199–3206. doi: 10.1111/j.1460-9568.2003.03069.x. [DOI] [PubMed] [Google Scholar]
- Meyer, A., Zardoya, R. Recent advances in the molecular phylogeny of vertebrates. Annu. Rev. Ecol. Evol. Syst. 2003;34:311–338. [Google Scholar]
- Moss, S.J., Smart, T.G. Constructing inhibitory synapses. Nat. Rev. Neurosci. 2001;2:240–250. doi: 10.1038/35067500. [DOI] [PubMed] [Google Scholar]
- Ohlson, J., Ensterö, M., Sjöberg, B.M., Öhman, M. A method to find tissue-specific novel sites of selective adenosine deamination. Nucleic Acids Res. 2005;33:e167. doi: 10.1093/nar/gni169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, J.S., Bejerano, G., Siepel, A., Rosenbloom, K., Lindblad-Toh, K., Lander, E.S., Kent, J., Miller, W., Haussler, D. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput. Biol. 2006;2:e33. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali, A., Blundell, T.L. Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Seeburg, P.H., Hartner, J. Regulation of ion channel/neurotransmitter receptor function by RNA editing. Curr. Opin. Neurobiol. 2003;13:279–283. doi: 10.1016/s0959-4388(03)00062-x. [DOI] [PubMed] [Google Scholar]
- Sherry, S.T., Ward, M.H., Kholodov, M., Baker, J., Phan, L., Smigielski, E.M., Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titz, S., Hans, M., Kelsch, W., Lewen, A., Swandulla, D., Misgeld, U. Hyperpolarizing inhibition develops without trophic support by GABA in cultured rat midbrain neurons. J. Physiol. 2003;550:719–730. doi: 10.1113/jphysiol.2003.041863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Miyakoda, M., Yang, W., Khillan, J., Stachura, D.L., Weiss, M.J., Nishikura, K. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- Wong, S.K., Sato, S., Lazinski, D.W. Substrate recognition by ADAR1 and ADAR2. RNA. 2001;7:846–858. doi: 10.1017/s135583820101007x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y.M., Kung, S.S., Chen, J., Chow, W.Y. Molecular analysis of cDNA molecules encoding glutamate receptor subunits, fGluR1 α and fGluR1 β, of Oreochromis sp. DNA Cell Biol. 1996;15:717–725. doi: 10.1089/dna.1996.15.717. [DOI] [PubMed] [Google Scholar]