Abstract
A major function of the p53 tumor suppressor is the induction of a pleiotropic apoptotic program in response to stress through transcription-dependent and -independent mechanisms. In particular, this includes a direct apoptotic role of p53 at the mitochondria. Stress-induced p53 translocation to the mitochondria with subsequent outer membrane permeabilization is a common early component in p53-mediated apoptosis in normal and transformed cells. However, the mechanism of p53 delivery to the mitochondria remains unknown. Here, we show that the cytoplasm contains a separate and distinct p53 pool that is the major source for p53 translocation to the mitochondria upon its stress-induced stabilization. Using various manipulations that enhance or diminish p53 ubiquitylation, our data provide evidence that Mdm2-mediated monoubiquitylation of p53 greatly promotes its mitochondrial translocation and thus its direct mitochondrial apoptosis. On the other hand, p53 does not require Mdm2 as a shuttler. Upon arrival at the mitochondria, our data suggest that p53 undergoes rapid deubiquitylation by mitochondrial HAUSP via a stress-induced mitochondrial p53–HAUSP complex. This generates the apoptotically active non-ubiquitylated p53. Taken together, we propose a novel model for mitochondrial p53 targeting, whereby a distinct cytoplasmic pool of stabilized monoubiquitylated p53, generated in resting cells by basal levels of Mdm2-type ligases, is subject to a binary switch from a fate of inactivation via subsequent polyubiquitylation and degradation in unstressed cells, to a fate of activation via mitochondrial trafficking.
Keywords: apoptosis, mitochondria, p53, ubiquitylation
Introduction
The p53 tumor suppressor exerts its potent antineoplastic function mainly by inducing a pleiotropic apoptotic program after cell damage via transcription-dependent and -independent mechanisms. Recent studies revealed a direct apoptotic role of the p53 protein at the mitochondria in primary and transformed cells and in normal tissues upon genotoxic and hypoxic stress in vivo (Marchenko et al, 2000; Sansome et al, 2001; Dumont et al, 2003; Gilman et al, 2003; Mihara et al, 2003; Bonini et al, 2004; Dagher, 2004; Erster et al, 2004; Leu et al, 2004; Nemajerova et al, 2005b; Endo et al, 2006; Strom et al, 2006). Upon exposure to a broad spectrum of apoptotic stimuli, total cellular p53 levels quickly stabilize and a fraction accumulates at the mitochondria, where it controls a direct apoptotic program. Induced p53 rapidly translocates to the outer membrane of mitochondria, where it engages in inhibitory and activating complexes with the anti- and proapoptotic members of the Bcl family of mitochondrial permeability regulators (BclXL/Bcl2 and BAK, respectively). In sum, this induces outer membrane permeabilization and the release of apoptotic activators (Marchenko et al, 2000; Sansome et al, 2001; Mihara et al, 2003; Nemajerova et al, 2005b).
In unstressed cells, rapid p53 turnover via polyubiquitylation and proteasomal degradation that cause very low levels of p53 is largely owing to basal levels of Mdm2, a p53 ubiquitin E3/E4 ligase and p53's pivotal negative regulator (Kubbutat MH, 1998; Grossman SR, 2003). Mitochondrially-targeted p53 proteins are sufficient to launch apoptosis in all tested p53-null cells in culture (Marchenko et al, 2000; Mihara et al, 2003) and also has tumor suppressor activities in vivo against primary lymphomas that are either p53- or ARF-deficient or harbor high levels of endogenous mutant p53 (Talos et al, 2005). Further support for a direct mitochondrial role of p53 in apoptosis came from an analysis of two common polymorphic variants of human p53, Arg72 and Pro72, that differed greatly in their apoptotic ability. Whereas all the other properties examined failed to explain the difference, the better killer (Arg72) localized better to the mitochondria (Dumont et al, 2003). However, the origin and mechanism of mitochondrial p53 translocation remained unclear. A translocation motif was not found and phosphorylation/acetylation modifications of p53 are not the determining factors for the mitochondrial targeting of p53 (Nemajerova et al, 2005a). Previous work on conditional temperature-sensitive Arg/Pro72 p53 mutants had proposed that polyubiquitylated p53 traffics from the nucleus to the mitochondria in an Mdm2/CRM1-dependent manner (Dumont et al, 2003). However, it is a generally accepted paradigm that Lys48 (and possibly Lys29-linked) polyubiquitylated proteins are destined for proteasomal destruction. Conversely, (multi-lysine)-monoubiquitylation of many cellular proteins determines a fate other than degradation and has been widely implicated as a signal for intracellular trafficking among compartments and organelles (for reviews, see Di Fiore et al, 2003; Sigismund et al, 2004). Importantly, (multi)-monoubiquitylated proteins are stable, as efficient proteasomal degradation minimally requires an at least four-subunit-long multiubiquitin chain per individual lysine residue (Thrower et al, 2000). Of note, ubiquitination was earlier shown to regulate the delivery of specific proteins and phospholipids, such as monoamine oxidase B and phosphatidylserine, to the mitochondria (Zhaung and McCauley, 1989; Schumacher et al, 2002). Moreover, Li et al (2003) have shown that monoubiquitylation of p53 is a trafficking signal for the nuclear–cytoplasmic redistribution of p53. Here, we propose a novel regulation of a p53 function in the cytoplasm in response to stress: an E3 ligase-mediated binary switch from polyubiquitylation to monoubiquitylation of p53 provides a trafficking signal and rapidly redirects the now stress-stabilized pool of cytoplasmic p53 from a fate of polyubiquitylation and degradation in unstressed cells to mitochondrial translocation and activation upon stress.
Results
The cytoplasm contains a separate p53 pool that is the major source of translocated mitochondrial p53
Previous work on ectopic temperature-sensitive p53 mutants had proposed that polyubiquitylated p53 traffics from the nucleus to the mitochondria in an Mdm2/CRM1-dependent manner (Dumont et al, 2003). To determine the subcellular compartment from which mitochondrially translocated p53 originates, we tested mitochondrial translocation under conditions of a nuclear export block. To this end, camptopthecin (Camp)-stressed wtp53 cells were treated with potent inhibitors of CRM1-mediated p53 nuclear export (Figure 1) (Shirangi et al, 2002). When ML1 and RKO cells were treated with leptomycin B (LMB), a non-competitive CRM1 inhibitor, p53 was locked inside the nucleus (Figure 1A, right and data not shown), yet mitochondrial translocation upon stress was only minimally impaired (Figure 1A, left, top and bottom). Moreover, in carefully executed cell fractionation experiments with LMB, cytoplasmic and nuclear p53 pools became independently stabilized upon stress (Figure 1B, lanes 5, 8 and 9, 12). This data was further confirmed by co-immunoprecipitating p53 with the CRM1 protein, the essential component of the nuclear export machinery. Notably, the Crm1–p53 complex was readily detectable in unstressed but not in stressed ML1 cells, indicating that no export occurs during stress (Figure 1C). Also, blocking nuclear export by expressing the competitive CRM1 inhibitor Flag–Rex (the HTLV1 Rex protein fused with strong nuclear export (NES) and import (NLS) signals; (Roth et al, 1998; Shirangi et al, 2002) (Supplementary Figure 1A), had no impact on mitochondrial p53 translocation (Figure 1D and data not shown). Finally, a nuclear import-deficient p53 mutant with mutations in all three C-terminal NLS motifs (p53NLS) was as efficient for mitochondrial translocation as wild-type p53 (wtp53) in p53-null H1299 cells, indicating that p53 does not need to traffic through the nucleus for translocation (Figure 1E, left). According to our model, p53NLS is predicted to efficiently induce apoptosis. Indeed, the apoptotic ability of p53NLS was comparable to that of wtp53 (Figure 1E, right). Together, these data indicate that the cytoplasm is the major source of translocated p53, whereas p53 export from the nucleus contributes little. Accordingly, cytoplasmic ubiquitylated p53 is generated locally and does not depend on nuclear export. Consistent with this result, Yu et al (2000) showed earlier that the cytoplasmic MDM2-NLS mutant is efficient in ubiquitylating cytoplasmic p53. In sum, this supports the presence of two largely independent pools of preexisting p53 in unstressed cells, cytoplasmic and nuclear, which simultaneously but independently respond to stress by stabilization.
Figure 1.
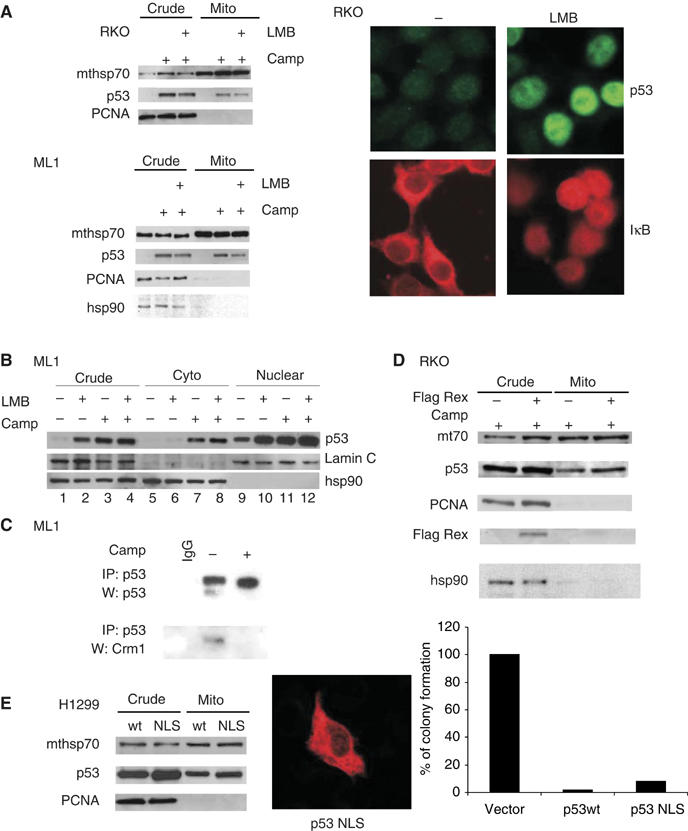
The cytoplasm contains a separate p53 pool that is the major source of translocated mitochondrial p53. (A) Mitochondria were isolated from RKO (top) and ML1 (bottom) cells by sucrose density gradients (both Arg/Pro at codon 72) treated with LMB and/or Camp for 4 h. Purified mitochondrial and crude lysates were immunoblotted and the amount of translocated p53 was detected with DO-1 antibody. Equal mitochondrial loading and purity from nuclear contamination was verified by reblotting with mthsp70 and PCNA and hsp90, respectively in (A), (D) and (E). Nuclear export blockade by LMB was verified in cells treated with LMB and immunostained for endogenous p53 and IκB. (B) Treated ML1 cells were fractionated into nuclear, cytoplasmic and crude fractions (Pierce kit) and immunoblotted as indicated. (C) Crude extract from ML1 cells were treated as indicated, immunoprecipitated with CM1 antibody (p53-specific) and immunoblotted with CRM1 antibody. (D) Immunoblot of crude lysates and mitochondria purified from RKO cells after transfection with Flag-tagged Rex for 24 h, followed by 5 μM Camp for an additional 4 h. (E, left) p53-null H1299 cells were transfected with plasmids encoding wtp53 or NLS-defective mutant p53. Immunoblot as in (A). (E, middle) Immunofluorescence with CM-1 antibody to show cytoplasmic localization of the NLS mutant. (E, right) Colony formation assays. p53-null H1299 cells were transfected with the indicated plasmids. After G418 selection for 18 days, colonies were counted.
Mdm2 is not required to shuttle p53 to the mitochondria
As Mdm2 shuttles in and out of the nucleus, significant Mdm2 levels exist in the cytoplasm (Roth et al, 1998). One possibility is that Mdm2 escorts p53 to the mitochondria, as previously suggested (Dumont et al, 2003). To determine whether physical shuttling by Mdm2 is required to actively transport endogenous p53 to mitochondria, we purified mitochondria from untreated, Camp-treated or proteasome inhibitor ALLN-treated ML1 and RKO cells and tested for the presence of mitochondrial Mdm2. First, although Camp induced p53 translocation, as seen many times before, it did not induce Mdm2 translocation to the mitochondria (Figure 2A, lane 6 and data not shown). This is in complete agreement with the fact that Camp disrupts the p53–Mdm2 complex by inducing phosphorylation and acetylation modifications on p53 and Mdm2 (Figure 2B, lane 11; Appella and Anderson, 2001). In addition, stress induces rapid autodegradation of Mdm2 (Supplementary Figure 1E) (Ashcroft et al, 2000; Stommel and Wahl, 2004). Second, in crude lysates, ALLN greatly stabilized p53 and Mdm2 without post-translational modification, while preserving the p53–Mdm2 complex (Figure 2 and lanes 2 and 8). However, while ALLN induced p53 translocation to the mitochondria, only trace amounts of Mdm2 were detectable at that site (Figure 2A and lane 5); again indicating that Mdm2 is not acting as a mitochondrial p53 shuttler. In further support, in ALLN-treated cells mitochondrial p53 translocation occurred despite a lack of detectable mitochondrial p53–Mdm2 complexes (Figure 2B and lane 10), although abundant amounts of this complex were detectable in total cellular extracts (Figure 2B and lane 8). Finally, transfection of wt, but not catalytically inactive, Mdm2 resulted in enhanced mitochondrial targeting of p53 (Figure 3F), indicating that the enzymatic activity of Mdm2, but not the molecule itself, promotes p53 translocation to this site. Together, these observations suggest as simplest explanation that Mdm2's enzymatic activity, rather than Mdm2 itself is required for the mitochondrial translocation of p53.
Figure 2.
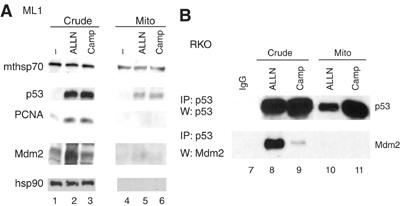
Mdm2 is not required to shuttle p53 to mitochondria. (A) Equal amounts of crude lysates (crude) and purified mitochondria (mito) (4 μg) from ML1 cells treated as indicated were immunoblotted. Note that treatment of cells with proteasome inhibitors like ALLN causes extensive accumulation of mono- and polyubiquitylated p53 (not shown), explaining why ALLN alone mediates mitochondrial p53 translocation (see the text). (B) Mitochondria do not contain p53/Mdm2 complexes despite significantly induced translocated p53. Total cell lysates and purified mitochondria (400 μg each) from untreated or treated RKO cells were immunoprecipitated to detect p53–Mdm2 complexes. Similar results were seen with ML1 cells (data not shown).
Figure 3b.
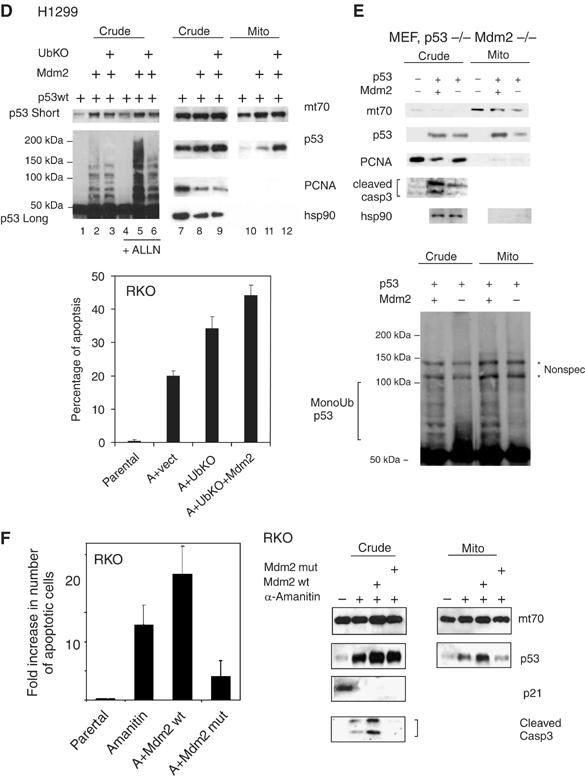
(D) Mdm2-mediated monoubiquitylation of p53 promotes its mitochondrial targeting. Right: immunoblot of crude lysates and purified mitochondria of H1299 cells transfected with wtp53, Mdm2wt and UbKO, left: short and long DO-1 exposures of the same crude lysates +/− proteosome inhibitor ALLN to show preferential Mdm2-mediated p53 monoubiquitylation upon UbKO expression. Sample loading was normalized for the equal amount of nonubiquitilated p53. Lower panel: RKO cells were transfected, as indicated, or left untreated. Twenty-four hours later, α-amanitin was added to the medium, where indicated, for an additional 14 h to block transcription. Apoptosis was determined by TUNEL. (E) Mdm2 deficiency causes defects in mitochondrial p53 translocation and caspase 3 activation. Early passage p53−/− Mdm2−/− double knockout MEFs were retrovirally transduced with p53wt and Mdm2wt as indicated. After 36 h, crude lysates and purified mitochondria were immunoblotted as indicated, including an antibody that is specific for the activated form of caspase 3. Mitochondrial loading was normalized for equal total cellular p53 protein. Lower panel: Mdm2-mediated enhanced mitochondrial p53 translocation is associated with enhanced p53 monoubiquitylation. p53 immunoblot after long exposure. * represents nonspecific bands. (F) α-Amanitin stabilizes wtp53, induces its mitochondrial translocation, activates caspase 3 and induces apoptosis in the absence of transcription, as indicated by, for example, the p53 target gene p21. Expression of wtMdm2 enhances all of these effects, which translates into enhanced apoptosis via the mitochondrial p53 program. In contrast, expression of the catalytically inactive dominant-negative Ring-finger mutant of Mdm2 (C438L) suppresses all of these effects below the level of endogenous Mdm2 (seen with α-amanitin alone). RKO cells were transfected as indicated and 20 h later cells were treated with α-amanitin for an additional 14 h, except for parental cells. Crude and mitochondrial extracts were immunoblotted as indicated. Mitochondrial loading was normalized for equal mitochondrial protein. Apoptosis was assayed by caspase 3 cleavage and bottom by TUNEL staining. Error bars in (C), (E) and (G) are s.d. from three independent experiments.
Mdm2-mediated monoubiquitylation promotes mitochondrial p53 translocation and the mitochondrial p53 apoptotic program
Polyubiquitylation of proteins via Lys-48 including p53 provides a recognition signal for proteosomal degradation, whereas (multi)-monoubiquitylation has been implicated in processes other than proteosomal destruction. In particular, monoubiquitylation can serve as a signal for intracellular trafficking between compartments (Di Fiore et al, 2003; Sigismund et al, 2004). In light of this data and of two pertinent negatives, that is, the absence of a mitochondrial translocation motif within p53 and the fact that phosphorylation/acetylation modifications play no major role in the mitochondrial targeting of p53 (Nemajerova et al, 2005a), we therefore theorized that in response to stress, monoubiquitylation helps to direct cytoplasmic p53 to the mitochondria.
To test this notion directly, p53-null H1299 cells were co-transfected with p53 and wild-type ubiquitin moieties (Ubwt). Interestingly, Ubwt promoted mitochondrial translocation, compared with p53 alone (Figure 3A, lanes 4 and 5). Of note though, polyubiquitylation-defective non-branchable ubiquitin (called UbKO, for ubquitin knockout), which generates mainly monoubiquitylated p53 (Figure 3B, right, Figure 3D top; Supplementary Figure 1B; Li et al, 2003), promoted this process even better than Ubwt (Figure 3A, compare lanes 5 and 6), which generates both poly- and monoubiquitylated p53 (Li et al, 2003). UbKO also promoted mitochondrial translocation of endogenous p53 (Figure 3B, left, lanes 4 and 5) followed by subsequent apoptosis in RKO cells in the absence of any stress and in the absence of transcription (Figure 3B, middle blocked by α-amanitin, see below). Increased apoptosis in RKO cells after expression of Ubwt or UbKO moieties was accompanied by enhanced monoubiquitylation of p53 in these cells (Figure 3B, right). Importantly, enhanced mitochondrial targeting of p53 by Ubwt and even more by UbKO was due to increased p53 modification and not due to p53-independent (H1299 cells in Supplementary Figure 1E) or nonspecific effects. To this end, transfection of UbKO did not upregulate endogenous p53 and thus did not induce nonspecific cellular stress (Figure 3B, left, lanes 1 and 2). Moreover, to rule out that His-tagged ubiquitin per se is toxic to cells, we generated a His-tagged ubiquitin mutant that cannot be attached to other proteins by deleting the last two C-terminal diglycines (called UbΔG) (Haglund et al, 2003). Of note, this mutant neither promoted p53 accumulation and mitochondrial targeting in H1299 and RKO cells, nor did it enhance their apoptosis (Supplementary Figure 1F). To definitively link enhanced mitochondrial targeting of monoubiquitylated p53 with its ability to promote apoptosis via the transcription-independent mitochondrial p53 program, we used α-amanitin, a specific and potent inhibitor of polymerase II-dependent transcription, thereby shutting down the transcription of most protein-coding genes in the cell. Specifically, α-amanitin was shown to efficiently inhibit all p53-dependent transcription, yet to promote apoptosis via the p53 mitochondrial program (Arima et al, 2005). Thus, α-amanitin induces p53-dependent but transcription-independent apoptosis by mediating mitochondrial p53 translocation, without requiring DNA damage and without detectable transcriptional activity of p53, as judged by a panel of sensitive p53 target genes (Arima et al, 2005). Indeed, ML1 and RKO cells, when subjected to transcriptional blockade by α-amanitin, stabilized endogenous cellular p53 and induced p53 mitochondrial translocation, followed by caspase 3 activation and apoptosis (Supplementary Figure 1C and data not shown). α-amanitin synergized in all these effects with camptothecin (Supplementary Figure 1C and data not shown). Moreover, α-amanitin synergized with ubiquitin moieties and Mdm2 in inducing apoptosis of p53-containing RKO cells (Figure 3D, bottom and Supplementary Figure 1D). This was a p53-dependent but transcription-independent effect, as we did not observe apoptosis in p53-null H1299 cells transfected with UbKO, UbΔG or MDM2, irrespective of the presence or absence of α-amanitin (Supplementary Figure 1E).
Figure 3a.
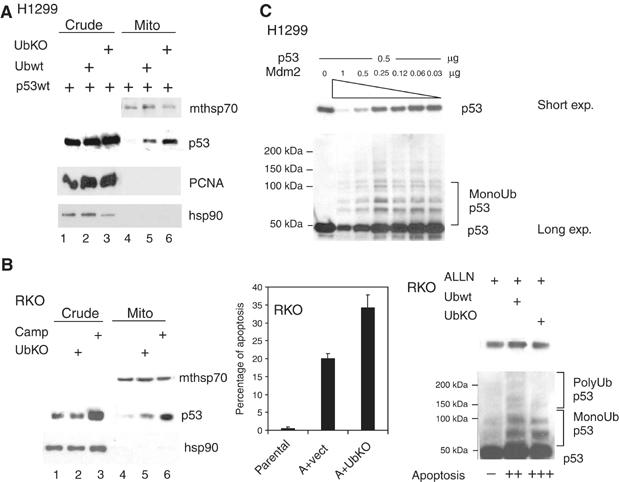
Mdm2-mediated monoubiquitylation promotes mitochondrial p53 translocation and the mitochondrial p53 apoptotic program. (A) Immunoblot of H1299 cells cotransfected with wtp53 and equal amounts of either Ubwt or UbKO. Mitochondrial loading was titrated down to the point where p53wt alone did not produce detectable translocation (lane 4) in order to show increased translocation in lanes 5, 6. (B, left) Immunoblot of RKO cells mock- or UbKO transfected. Camp-treated cells were used as control. (B, middle) RKO cells were transfected, as indicated, or left untreated. Twenty-four hours later α-amanitin was added to the culture medium where, indicated, for an additional 14 h to block transcription. Apoptosis was determined by TUNEL. (B, right) Increased apoptosis in RKO cells is accompanied by enhanced p53 monoubiquitylation after expression of Ubwt or UbKO. RKO cells were treated, as indicated, and normalized amounts of p53 were immunoblotted. (C) The fate of p53 depends on the p53:Mdm2 ratio in the cell. p53-null H1299 cells were cotransfected with 0.5 μg of p53 plasmid and decreasing concentrations of the Mdm2 plasmid. Note that whereas high concentrations of Mdm2 cause polyubiquitylation and degradation of p53, low concentrations of Mdm2 preferentially catalyze monoubiquitylation and stabilization of p53.
Of note, after DNA damage Mdm2 mRNA and protein undergo accelerated auto-degradation, lowering endogenous cellular Mdm2 concentrations (Ashcroft et al, 2000; Stommel and Wahl, 2004), which we confirmed in our systems (Supplementary Figure 1E). As previously shown, Mdm2 can mediate both poly- and monoubiquitylation of p53, depending on the relative stoichiometry: when Mdm2 is more abundant, it promotes polyubiquitylation of p53, while lower amounts of Mdm2 promote monoubiquitylation (Li et al, 2003). We confirmed that the fate of p53 in the cell depends on the p53:Mdm2 ratio. While molar ratios of transfected expression plasmids of 1:2 or 1:1 of p53 to Mdm2 resulted in p53 destabilization and degradation, p53 to Mdm2 plasmid ratios of 2:1 or higher generated p53 monoubiquitylation and stabilization via inefficient degradation (Figure 3C). Therefore, in all subsequent transfections, the minimum amount of Mdm2 sufficient to promote monoubiquitylation was used. Accordingly, Mdm2 promoted p53 translocation in H1299 cells (Figure 3D, top). Moreover, similar to results seen in Figure 3A and B, mitochondrial translocation was greatly enhanced by coexpressing UbKO, which promotes p53 monoubiquitylation by Mdm2 (Figure 3D, top). Finally, UbKO promoted apoptosis in α-amanitin-treated RKO cells, and this was further boosted by co-transfecting 1/10 molar ratios of Mdm2 plasmid (Figure 3D, bottom).
To further strengthen the evidence for a role of Mdm2 in p53 translocation to the mitochondria, we next did reconstitution experiments in null cells. While expression of wtp53 in p53−/− Mdm2-/− double knockout MEFs was sufficient to cause some mitochondrial translocation and caspase 3 activation, co-expression with 1/10 molar plasmid ratios of Mdm2 greatly enhanced both mitochondrial targeting of monoubiquitylated p53 (Figure 3E, top and bottom) and caspase 3 activation (Figure 3E, top). Finally, as seen before, treating RKO cells with α-amanitin stabilizes p53, induces its mitochondrial translocation, activates caspase 3 and induces apoptosis in the absence of transcription, as indicated by, for example, the p53 target gene p21 (Figure 3F). Importantly though, while expression of wt Mdm2 enhances all of these effects, translating into enhanced apoptosis via the mitochondrial p53 program, expression of the dominant-negative Ring-finger mutant of Mdm2 suppresses them below the level of α-amanitin alone (Figure 3F). In order to further evaluate the importance of ubiquitylation for mitochondrial translocation of p53, we wanted to generate a mutant of p53 that can not be ubiquitylated. However, and in agreement with the literature (Krummel et al, 2005), extensive attempts to generate a ubiquitylation-dead mutant of p53 failed. Neither the 5KR nor 6KR mutants which harbor lysine to arginine mutations in all known ubiquitylation sites at the p53 C-terminus, nor a 9KR mutant harboring three additional mutations at putative ubiquitylation sites (lysine residues 319–321) that we generated showed complete loss of ubiquitylation (data not shown). In sum, these data strongly support the idea that Mdm2-mediated monoubiquitylation promotes mitochondrial p53 translocation and the mitochondrial p53 apoptotic program.
Monoubiquitylation promotes mitochondrial p53 translocation in cells, yet the steady-state form of mitop53 is non-ubiquitylated
We next determined the physiological ubiquitylation status of endogenous p53 translocated to the mitochondria upon stress. As p53 deubiquitylases such as HAUSP (USP7) are omnipresent in cells (Cummins et al, 2004; Li et al, 2004), these experiments were performed under deubiquitylase inhibition by ubiquitin aldehyde (UbAL), a specific inhibitor of the ubiquitin-specific protease family of deubiquitylases (DUBs) (Hu et al, 2002). The purpose of adding UbAL throughout the entire experiment was thus to inhibit potential DUBs and therefore be able to ‘freeze' and visualize the ubiquitylation status of p53 as it arrives at the mitochondria. We first ensured that UbAL is also an effective inhibitor of p53 deubiquitylation in cells, as to date it had only been used in cell-free biochemical studies. Growing ML1 and RKO cells under UbAL stabilized ubiquitylated p53 and decreased cellular p53 turnover (Figure 4A and data not shown), indicating that UbAL is a useful albeit not absolute p53 deubiquitylase inhibitor in vivo. To confirm that ubiquitin aldehyde, when added to culture media, enters the cell and interferes with intracellular deubiquitylation in general, we chose Mdm2 as an independent but in this context highly relevant specific HAUSP substrate. As seen in Supplementary Figure 2A, bottom, endogenous Mdm2 is increasingly ubiquitylated in a dose-dependent manner upon adding UbAL to the culture medium. Moreover, compared with Camp and as expected from downregulating HAUSP by siRNA (Cummins et al, 2004; Li et al, 2004), UbAL stabilized non-ubiquitylated p53 only slightly and, importantly, did not induce apoptosis at the concentration used (Supplementray Figure 2A, top). To determine the ubiquitylation status of endogenous stress-translocated mitochondrial p53, ML1 and RKO cells remained untreated or were treated with Camp plus UbAL to block deubiquitylation (Figure 4B and data not shown). As expected, p53 translocated to the mitochondria (Figure 4B, bottom lanes 3 and 4). To compare the ubiquitylation status between total cellular and mitochondrial p53, both fractions were immunoprecipitated for p53 using polyclonal antibody CM1 and their ubiquitin status assessed by immunoblotting with ubiquitin-specific monoclonal antibody P4D1 (Figure 4B, top). Membranes were then reblotted with p53-specific monoclonal DO1 to verify successful p53 immunoprecipitation (middle panel). Of note, while in crude lysates poly- and monoubiquitylated p53 dramatically decreased upon stress, this drop did not occur at the mitochondria (top panel, compare lanes 2 and 4). At mitochondria, it was primarily, although not exclusively, monoubiquitylated p53 that was readily detectable upon stress (top panel, lane 4), while this was not so in stressed crude lysates (lane 2). This is especially the case considering the lower levels of non-ubiquitylated p53 at the stressed mitochondria, compared with the corresponding levels in crude lysate (middle and bottom panels, lanes 2 and 4). Notably, unstressed cytoplasm contains a mixture of mainly unstable polyubiquitylated p53 (Figure 4C, lane 1; see stabilization by ALLN in lane 3) and some stable monoubiquitylated p53. Importantly though, upon stress, a portion of stable endogenous monoubiquitylated p53 (revealed by the fact that it is not stabilized by ALLN, compare lanes 2 and 4) remains in the cytoplasm and serves as the source for p53 translocation to the mitochondria.
Figure 4.
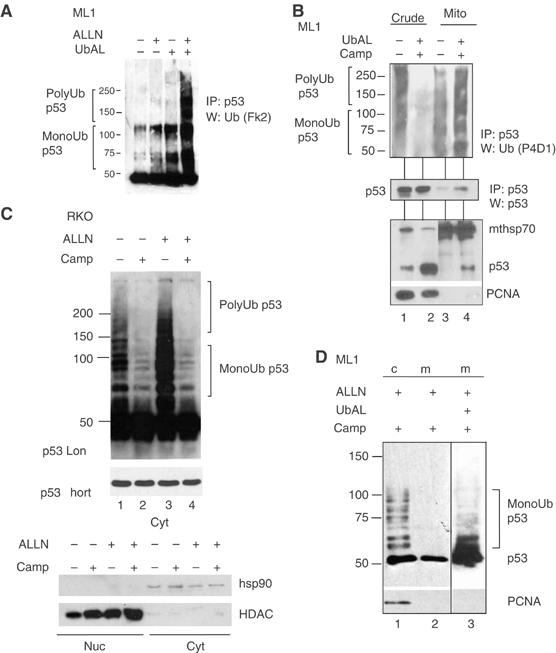
Monoubiquitylation promotes mitochondrial p53 translocation in vivo, yet the steady-state form of mitochondrial p53 is non-ubiquitylated. (A) Ubiquitin aldehyde (UbAL) is a potent deubiquitylase inhibitor in vivo. Growing cells under UbAL prevents p53 degradation and stabilizes cellular ubiquitylated p53. ML1 were cultured in the presence of 50 ng/ml UbAL for 4 h before harvesting. Crude lysates were immunoprecipitated with a mixture of the monoclonal p53 antibodies Pab1801 and DO1 and blotted for ubiquitin with polyclonal FK2 antibody. All buffers contained UbAL throughout the procedures. (B) Monoubiquitylated p53 is preferentially present at stressed mitochondria. Top: purified mitochondria and crude lysates (400 μg) from ML1 cells, treated with 5 μg Camp plus 50 ng/ml UbAL for 4 h or left untreated, were immunoprecipitated for p53 (CM1 antibody) and aliquots containing comparable amounts of non-ubiquitylated p53 in each fraction pair were immunoblotted with a pan-ubiquitin-specific antibody (P4D1). Middle: membrane was reblotted with p53-specific antibody DO1 to verify pairwise loading. Bottom: corresponding direct immunoblots from cell lysates and mitochondria used for IP. (C) Top; cytoplasmic fractions of RKO cells +/− Camp and +/− ALLN treatment were normalized for equal amounts of p53 and immunoblotted with DO1. ‘p53 short' to show equal p53 loading. Bottom; nuclear and cytoplasmic fractions of RKO cells +/− Camp and +/− ALLN. hsp90 and HDAC were used as purity controls for fractionations. (D) The steady-state form of stress-induced mitochondrial p53 is non-ubiquitylated. ML1 cells pretreated for 3 h with ALLN (50 μM), with or without UbAL (50 ng/ml), were then treated for 90 min with Camp. Crude lysates and purified mitochondria (4 μg each) were immunoblotted as indicated.
However, while monoubiquitylation greatly promoted mitochondrial p53 translocation (Figure 3), the majority of stress-induced mitochondrial p53 was nevertheless non-ubiquitylated (Figure 4d and lane 3). Moreover, when analyzed without deubiquitylation inhibition (but with ALLN stabilization for better visualization), the steady-state form of stress-induced mitochondrial p53 was non-ubiquitylated (Figure 4C and lane 2), in stark contrast to total cellular p53 that was heavily ubiquitylated under the same conditions (lane 1). Taken together with Figure 3A–C, 3D–F, in sum this data indicates that monoubiquitylation greatly promotes mitochondrial p53 translocation in vivo, yet it suggests that the steady-state form of mitochondrial p53 is non-ubiquitylated.
HAUSP is constitutively located at the mitochondria and engages in a stress-induced HAUSP–p53 complex upon p53 translocation to this site
The data presented in Figure 4C suggests a relatively higher p53 deubiquitylation activity at the mitochondria compared with total cell lysate. In genetic and biochemical studies HAUSP has been identified as the major p53 deubiquitylase (Li et al, 2002). We therefore theorized that mitochondria harbor HAUSP and that this HAUSP fraction is responsible for rapid p53 deubiquitylation after its translocation. Total cellular HAUSP was not stress-inducible (Figure 5A and B). Of note, HAUSP was readily detectable at the purified mitochondria of ML1 and RKO cells, where it was constitutively present (Figure 5B and C, second panel). By subcellular fractionation, approximately 10% of total cellular HAUSP was located at the mitochondria (Supplementary Figure 2B). Importantly, endogenous mitochondrial HAUSP formed a specific stress-induced complex with mitochondrial p53 after its stress-mediated translocation to this site (Figure 5C and top panel). Moreover, upon stress much higher amounts of endogenous p53–HAUSP complexes were detected at the mitochondria of RKO and ML1 cells compared with crude lysates, indicating higher HAUSP-mediated p53 deubiquitylation at the mitochondria compared with other cellular compartments (Figure 5C, top panel and data not shown). On the other hand, the interaction of total cellular p53 and HAUSP did not change upon DNA damage, a finding also confirmed by others (Meulmeester et al, 2005). The latter finding, however, does not rule out a role for HAUSP in deubiquitylating non-mitochondrial p53 under stress. HAUSP also deubiquitylates Mdm2, thereby blocking autodegradation (Cummins and Vogelstein, 2004; Li et al, 2004). Accordingly, we detected an Mdm2–HAUSP complex in unstressed, and to a much lower extent in stressed crude lysates, but not in the mitochondria (Supplementary Figure 2C), consistent with the absence of Mdm2 and Mdm2–p53 complexes at the mitochondria. This further emphasizes the physiological significance of the stress-induced mitochondrial p53–HAUSP complex.
Figure 5.
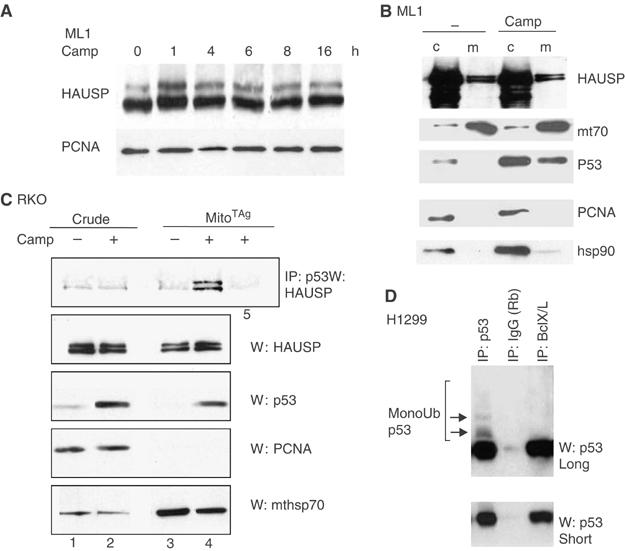
HAUSP is constitutively located at the mitochondria and engages in a stress-induced HAUSP–p53 complex upon p53 translocation. (A, B) HAUSP is constitutively expressed in cells and does not respond to stress. (A) Crude lysates of ML1 cells treated as indicated were immuoblotted with HAUSP antibody. PCNA is used as loading control. (B) HAUSP is also constitutively expressed at the mitochondria. Mitochondrial and crude lysates of ML1 cells treated as indicated were immunoblotted for HAUSP. A 10 μg measure of total protein per lane in (A) and (B). (C) Endogenous HAUSP forms a stress-inducible complex with translocated p53 at the mitochondria. Mitochondrial and crude lysates of RKO cells treated as indicated were immunoprecipitated for p53 (lanes 1–4) and blotted for HAUSP. Lane 5: negative control, the same amount of mitochondria were immunoprecipitated with anti-T antigen antibody. Lower panels: direct immunoblots of aliquots (5 μg each) from the IP input. (D) Mitochondrial p53, when complexed with BclX/L, is selectively non-ubiquitylated. p53–BclX/L complexes contain only non-ubiquitylated p53, whereas the total p53 pool contains detectable ubiquitylated p53 species. H1299 cells cotransfected with p53, MDM2 and BclX/L. p53 immunoprecipitations from parallel aliquots of the same lysate with 1 μg of the indicated antibodies.
A logical consequence of this data is that non-ubiquitylated p53 is the apoptotically active form of mitochondrial p53, and the one that is able to undergo inhibitory complexes with anti-apoptotic BclX/Land Bcl2 (Mihara et al, 2003; Chipuk et al, 2004; Chipuk et al, 2005) and activating complexes with pro-apoptotic Bak (Leu et al, 2004). We therefore examined the ubiquitylation status of p53 in complex with BclX/L. Indeed, in H1299 cells cotransfected with p53, MDM2 and BclX/L, p53–BclX/L complexes contained only non-ubiquitylated p53, while the total p53 pool contained readily detectable ubiquitylated p53 species. This was shown by p53 immunoprecipitations from parallel aliquots with equal amount of coprecipitated p53 (Figure 5D). Collectively, our data supports the notion that active mitochondrial p53 is non-ubiquitylated and that translocated ubiquitylated p53 undergoes rapid deubiquitylation by mitochondrial HAUSP.
Discussion
In this report, we show that a stress-stabilized cytoplasmic pool of p53 is the major source for mitochondrially translocated p53. We further provide evidence that in contrast to polyubiquitylation, monoubiquitylation of p53 provides a trafficking signal that redirects it from a fate of degradation and inactivation in unstressed cells to mitochondrial translocation and activation early during the stress response. Mdm2 appears to not act as a p53 shuttler but promotes mitochondrial p53 translocation by virtue of its enzymatic E3 ligase activity.
Using classical biochemical fractionation as well as competitive and non-competitive export blockade, we found that p53 nuclear export is not required for mitochondrial translocation upon DNA damage. Instead, distinct nuclear and cytoplasmic p53 pools become simultaneously and rapidly stabilized after genotoxic stress (Figure 1B). This is consistent with previous observations that cells can undergo p53-dependent apoptosis in the presence of wheat germ agglutinin, a specific inhibitor of nuclear import (Chipuk et al, 2004). Similarly, we find that an exclusively cytoplasmic version of p53 (NLS-deficient mutant) was as efficient as wtp53 in colony suppression of p53-null cells (Figure 1E). This indicates that mitochondrially translocated p53 arises from a distinct cytoplasmic pool. Our result is further supported by the kinetics of the event, as mitochondrial p53 translocation physiologically coincides with p53 nuclear export block in cells. In unstressed cells, both p53 and Mdm2 constantly shuttle between nucleus and cytoplasm via intrinsic NES and NLS signals (Roth et al, 1998). In contrast, upon stress p53 export is rapidly blocked by tetramerization-dependent masking of its C-terminal NES, and by phosphorylation-mediated inhibition of its N-terminal NES, resulting in rapid nuclear p53 accumulation (Stommel et al, 1999; Zhang and Xiong, 2001). In agreement with that, Crm1–p53 complexes were detectable only in unstressed but not in stressed cells (Figure 1C). Moreover, nuclear p53 export is a slow process requiring 3–8 h (Stommel and Wahl, 2004). In contrast, stress-induced mitochondrial p53 translocation is a fast process detectable within 30 min and peaking at 2 h in, for example, irradiated thymus and spleen and in Camp-treated ML1 and RKO cells (Marchenko et al, 2000; Erster et al, 2004). Together, this indicates the presence of two largely independent pools of preexisting p53 in unstressed cells, cytoplasmic and nuclear, which simultaneously respond to genotoxic stress.
We showed earlier that p53 forcibly targeted to the mitochondria is sufficient to induce p53-dependent apoptosis in cultured cells and mice (Mihara et al, 2003; Talos et al, 2005; Palacios G, 2006). Moreover, Dumont et al (2003) invoked Mdm2 in the proapoptotic mitochondrial activity of p53. Indeed, using various approaches that either enhance or prevent p53 ubiquitylation, we find that Mdm2-mediated p53 monoubiquitylation greatly promotes p53 translocation and direct mitochondrial apoptosis (Figure 3). However, based on the observations that: (i) mitochondria from DNA-damaged or proteasome-inhibited cells are devoid of Mdm2 and of Mdm2–p53 complexes (Figure 2); (ii) that stress-induced mitochondrial translocation of p53 coincides temporally with dissociation of Mdm2–p53 complexes (Appella and Anderson, 2001) and (iii) that a catalytically inactive Mdm2 mutant fails to promote translocation of p53, we conclude that the enzymatic E3 ligase activity, rather than the Mdm2 molecule as a shuttler, is important for trafficking p53 to mitochondrial p53. Notably, monoubiquitylation was previously found to regulate delivery of specific proteins and phospholipids, such as monoamine oxidase B and phosphatidylserine (the latter for use in mitochondrial membrane biogenesis), to the mitochondria (Zhaung and McCauley, 1989; Schumacher et al, 2002). Moreover, monoubiquitylation has previously been shown to act as a trafficking signal for p53 in its nuclear–cytoplasmic redistribution in cells (Li et al, 2003). Importantly, (multi)-monoubiquitylated proteins are stable, as efficient proteasomal degradation minimally requires an at least four-subunits-long multiubiquitin chain per single lysine residue (Thrower et al, 2000). In agreement with this, we show here that monoubiquiytylated p53 is stable, in contrast to polyubiquitylated p53 that is very unstable and subject to immediate proteosomal degradation (Figure 3C). Notably, unstressed cytoplasm contains a mixture of unstable polyubiquitylated and stable monoubiquitylated p53. Importantly though, upon stress, a portion of endogenous monoubiquitylated p53 remains in the cytoplasm and serves as the source for p53 translocation to the mitochondria. (Figure 4C). Furthermore, Dumont et al (2003) found that the better killer Arg72p53, despite higher affinity to Mdm2 than Pro72 p53, is not degraded (which would reduce its apoptotic potential), but exhibits enhanced mitochondrial translocation.
Together, these data led us to propose a novel regulation for mitochondrial translocation of p53, as need arises in the early stages of a cellular stress response (Figure 6). In unstressed conditions, a two-step regulation of p53 enables the creation of a rapid-action binary switch. The product of the first step—monoubiquitylated p53, generated by basal levels of Mdm2-type E3 ligases—can undergo two diametrically opposed fates: (i) either degradation and inactivation by subsequent polyubiquitylation via Mdm2 or E4-type ligases, if no stress occurs. (ii) Alternatively, if stress does occur, this monoubiquiylated intermediate p53 product is rapidly stabilized by stress-induced disruption of the p53–Mdm2 complex and diverted to the mitochondria, hence becoming apoptoticically active. A stress-mediated decrease in Mdm2 levels (Supplementary Figure 1E) (Stommel and Wahl, 2004) might further contribute to the conversion of polyubiquitylated to monoubiquitylated p53. In this way, the pre-existing and now stabilized pool of monoubiquitylated p53 mediates its first apoptotic response wave. Moreover, this first response arises from the cytoplasm, avoiding the extensive investment of time and energy required for nuclear export. This is entirely consistent with the previously published observation that p53 ubiquitylation can occur in two distinct compartments, the nucleus and the cytoplasm, and does not require nuclear import or export (Yu et al, 2000). This model also explains our earlier observation that p53 translocation to the mitochondria precedes the transactivation of p53 target genes. Later in the stress response, stabilization of nuclear p53 reaches a threshold sufficient to activate p53 targets in a second wave, which amplifies the response to a full apoptotic program.
Figure 6.
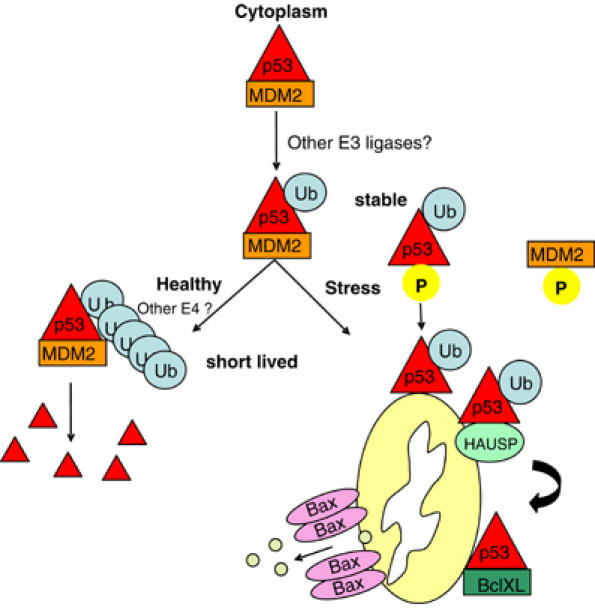
Regulation of mitochondrial translocation of p53 in the cytoplasm. A two-step regulation of p53 enables the creation of a rapid-action binary switch. The product of the first step—monoubiquitylated p53, generated by basal levels of Mdm2-type E3 ligases—can undergo two diametrically opposed fates: (i) either degradation and inactivation by subsequent polyubiquitylation via Mdm2 or E4-type ligases if no stress occurs. (ii) Alternatively, if stress does occur, this monoubiquiylated intermediate p53 product is rapidly stabilized by stress-induced disruption of the p53–Mdm2 complex and diverted to mitochondria, hence becoming apoptoticically active. A stress-mediated decrease in Mdm2 levels might further contribute to the conversion of polyubiquitylated to monoubiquitylated p53 (Stommel and Wahl, 2004). In this way, the pre-existing and now stabilized pool of monoubiquitylated p53 mediates its first apoptotic response wave.
Our data further suggest that upon arrival at the mitochondria, p53 undergoes deubiquitylation by mitochondrial HAUSP. This suggestion is based on: (i) the higher p53 deubiquitylating activity at the mitochondria, compared with other cellular compartments; (ii) the constitutive presence of HAUSP at the mitochondria; (iii) a stress-inducible HAUSP–p53 complex at the mitochondria, in contrast to total cell lysates, which exhibit a constitutive p53–HAUSP complex; (iv) the mitochondrial p53 moieties that are actively engaged in effector complexes with BCL X/L are deubiquitylated. The notion that effector-competent mitochondrial p53 has undergone deubiquitylation after its delivery is supported by structural, mutational and modeling data of inhibitory p53-BclX/L and p53–Bcl2 complexes and activating p53–Bak complexes that are formed at the OMM and involve the central DNA-binding domain of p53 (Mihara et al, 2003; Leu et al, 2004; Petros et al, 2004; Tomita et al, 2006). Moreover, we and others had detected only non-ubiquitylated p53 in endogenous complexes with the Bcl family members and with mitochondrial proteins mtHsp70/60 (Marchenko et al, 2000; Mihara et al, 2003; Chipuk et al, 2004; Leu et al, 2004). Also, non-ubiquitylated recombinant bacterial p53 is robustly capable of all mitochondrial activities in vitro, including BclX/L complex formation, Bak oligomerization and OMM permeabilization (Mihara et al, 2003). Similarly, monoubiquitylation and subsequent HAUSP-mediated deubiquitylation has been shown to be critical for subcellular trafficking and activity of transcription factor FOXO4, while it did not influence the half-life of FOXO4 protein (van der Horst et al, 2006). Interestingly, Ubp16, which shares 30% similarity/identity with human HAUSP, is the only member of the 16 yeast protein deubiquitinases that is exclusively localized to the mitochondria, where it is anchored to the OMM in a Nin–Cout orientation (Kinner and Kolling, 2003). The putative mitochondrial leader sequence of Ubp16 shares 70% homology with a central region of HAUSP. Unfortunately, our extensive attempts at manipulating HAUSP levels by either RNAi-mediated downregulation (almost complete elimination) or ectopic overexpression in stable cell lines in order to further assess the role of HAUSP in mitochondrial p53 function were not informative. This is because HAUSP manipulation simultaneously also affects the total cellular levels of the other two important parameters in the system, that is, Mdm2 and p53 (data not shown). For this reason, HAUSP-null cells (Cummins et al, 2004) will not be useful either. While such a complex and dynamic interplay was already previously reported by others (Li et al, 2004; Meulmeester et al, 2005), it precluded experimental evaluation of p53 deubiquitylation by HAUSP at the mitochondria. Thus, the physiological significance of p53 deubiquitylation by HAUSP at the mitochondria, as well as other cellular compartments during stress requires further evaluation.
In sum, we propose a 2-step regulation of cytoplasmic p53 function involving a binary switch that acts on a monoubiquitylated p53 intermediate in response to stress. This direct pathway of OMM permeabilization might co-transcriptional stages of a p53 stress response, a plethora of additional apoptotic target genes then amplify this p53-initiated apoptosis in a feed-forward loop. Future studies need to address the detailed nature of this ubiquitylation switch and clarify whether alternate E3 ligases also play a role in its regulation.
Materials and methods
Cells
Cells were grown in DMEM plus 10% FBS.
Plasmids
Expression constructs for pcDNA–Flag–Rex, a Flag-tagged HTLV1 Rex fusion protein with strong nuclear export (NES) and import (NLS) signals, and for human wtMdm2 were previously described (Shirangi et al, 2002). pCINeoHdm2 C438L encodes the catalytically inactive Ring-finger mutant of human Mdm2 (gift of A Weissman) and was also described (Fang et al, 2000). pCMV BamNeo-p53NLS expression vector was constructed by deleting the NLS II and III motifs in the extreme C-terminus and mutating lysine residues 319-321 to alanine at NLS I of human p53.
Ubiquitin expression vectors for wild-type ubiquitin (His-Ubwt) and the non-branchable polyubiquitination defective mutant His-UbKO in which all 7 lysines were replaced by arginine, were previously described (Li et al, 2003). To generate UbΔGG, Ubwt was used as PCR template (primers f-gccttctagaatgcagatcttcgtgaag and r-gccccatcctcatctgagacggagga) and inserted into the Xba1–BamH1 sites of expression vector pCGN.
Cells were transfected using lipofectamine (Invitrogen). For Figure 1C, RKO cells were transfected with pcDNA–Flag–Rex for 16 h before treatment with 5 μM Camp for an additional 4 h. For all others, H1299 and RKO cells were transfected for 16 h, followed by mitochondrial isolation.
Treatments
Cells were subjected to Camptothecin (Sigma) treatment (5 μM for 4 h) or mock treated; ML1 and RKO cells were treated with 10 nM leptomycin B (LMB, BioMol International) for 30 min before adding 5 μM Camp for an additional 3 and 4 h, respectively. Where indicated, cells were treated with 25 μM ALLN (Calbiochem). UbAL (BioMol International), a specific inhibitor of the ubiquitin-specific processing protease (UBP) family of deubiquitylases (Hu et al, 2002), was added to cultured cells (50 ng/ml UbAL for 4 h) and included in buffers where indicated. α-Amanitin (Sigma) treatment was at 10 μg/ml for 14 h. Apoptosis was assessed by TUNEL staining.
Mitochondrial purification
Mitochondria were purified by sucrose density gradients (Marchenko et al, 2000). Following transfection, the Pierce Mitochondria Isolation kit was used. In parallel, aliquots of cells were processed for crude lysates. Protease inhibitor cocktail (Roche), UbAL and 2 mM N-ethylmaleimide (Sigma) to prevent deubiquitination were included in all buffers. For Figure 1B, the fractionation kit from Pierce (Rockford, Il) was used.
Immunoblots and co-immunoprecipitation
For immunoblots, equal total protein of mitochondrial and crude cell lysates (typically 2.5–5 μg protein) were loaded. In cases where loadings were normalized for equal p53 loading, a first quantitation immunoblot was run before the definitive second immunoblot. The following antibodies were used: DO1 and Pab 1801 (Calbiochem); CM-1 for human p53; CM-5 for mouse p53 (Vector); Pab 421 (Calbiochem) for human and mouse p53; mt hsp70, hsp90 and histone deacetylase antibody HDAC (Affinity Bioreagents); PCNA, Flag and rabbit IgG (all Sigma); SMP14 for human Mdm2 and P4D1 (Santa Cruz) and Fk2 (Biomol International) for pan-ubiquitin detection and l antibody against human HAUSP (Calbiochem), CRM1 monoclonal (BD Transduction Lab), (for endogenous complexes, mitochondrial and crude lysates from untreated and Camp-treated ML1 or RKO cells were immunoprecipitated with 1 μg antibody for 2 h and beads were washed 3 times with SNNTE plus 2 × RIPA (50 mM Tris, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1% NaDeoxycholate, pH 7.4) before immunoblotting as indicated. For p53 immunoprecipitations, CM-1 was used, except in Figure 5C, where a 1:1 mixture of PAb 1801 and DO1 was used.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Legends to Supplemental Figures
Acknowledgments
This work was supported by National Cancer Institute (CA93853), Philip Morris USA, Inc. and Philip Morris Inc. and New York State Department of Health Research Science Board.
References
- Appella E, Anderson CW (2001) Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem 268: 2764–2772 [DOI] [PubMed] [Google Scholar]
- Arima Y, Nitta M, Kuninaka S, Zhang D, Fujiwara T, Taya Y, Nakao M, Saya H (2005) Transcriptional blockade induces p53-dependent apoptosis associated with translocation of p53 to mitochondria. J Biol Chem 280: 19166–19176 [DOI] [PubMed] [Google Scholar]
- Ashcroft M, Taya Y, Vousden KH (2000) Stress signals utilize multiple pathways to stabilize p53. Mol Cell Biol 20: 3224–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonini P, Cicconi S, Cardinale A, Vitale C, Serafino AL, Ciotti MT, Marlier LN (2004) Oxidative stress induces p53-mediated apoptosis in glia: p53 transcription-independent way to die. J Neurosci Res 75: 83–95 [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Bouchier-Hayes L, Kuwana T, Newmeyer DD, Green DR (2005) PUMA couples the nuclear and cytoplasmic proapoptotic function of p53. Science 309: 1732–1735 [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR (2004) Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 303: 1010–1014 [DOI] [PubMed] [Google Scholar]
- Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B (2004) Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature 428: 486. [DOI] [PubMed] [Google Scholar]
- Cummins JM, Vogelstein B (2004) HAUSP is required for p53 destabilization. Cell Cycle 3: 689–692 [PubMed] [Google Scholar]
- Dagher PC (2004) Apoptosis in ischemic renal injury: roles of GTP depletion and p53. Kidney Int 66: 506–509 [DOI] [PubMed] [Google Scholar]
- Di Fiore PP, Polo S, Hofmann K (2003) When ubiquitin meets ubiquitin receptors: a signalling connection. Nat Rev Mol Cell Biol 4: 491–497 [DOI] [PubMed] [Google Scholar]
- Dumont P, Leu JI, Della Pietra AC III, George DL, Murphy M (2003) The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet 33: 357–365 [DOI] [PubMed] [Google Scholar]
- Endo H, Kamada H, Nito C, Nishi T, Chan PH (2006) Mitochondrial translocation of p53 mediates release of cytochrome c and hippocampal CA1 neuronal death after transient global cerebral ischemia in rats. J Neurosci 26: 7974–7983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erster S, Mihara M, Kim RH, Petrenko O, Moll UM (2004) In vivo mitochondrial p53 translocation triggers a rapid first wave of cell death in response to DNA damage that can precede p53 target gene activation. Mol Cell Biol 24: 6728–6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM (2000) Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem 275: 8945–8951 [DOI] [PubMed] [Google Scholar]
- Gilman CP, Chan SL, Guo Z, Zhu X, Greig N, Mattson MP (2003) p53 is present in synapses where it mediates mitochondrial dysfunction and synaptic degeneration in response to DNA damage, and oxidative and excitotoxic insults. Neuromol Med 3: 159–172 [DOI] [PubMed] [Google Scholar]
- Grossman SR, Deato ME, Brignone C, Chan HM, Kung AL, Tagami H, Nakatani Y, Livingston DM (2003) Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science 300: 342–344 [DOI] [PubMed] [Google Scholar]
- Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I (2003) Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol 5: 461–466 [DOI] [PubMed] [Google Scholar]
- Hu M, Li P, Li M, Li W, Yao T, Wu JW, Gu W, Cohen RE, Shi Y (2002) Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 111: 1041–1054 [DOI] [PubMed] [Google Scholar]
- Kinner A, Kolling R (2003) The yeast deubiquitinating enzyme Ubp16 is anchored to the outer mitochondrial membrane. FEBS Lett 549: 135–140 [DOI] [PubMed] [Google Scholar]
- Krummel KA, Lee CJ, Toledo F, Wahl GM (2005) The C-terminal lysines fine-tune P53 stress responses in a mouse model but are not required for stability control or transactivation. Proc Natl Acad Sci USA 102: 10188–10193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MH, Ludwig RL, Ashcroft M, Vousden KH (1998) Regulation of Mdm2-directed degradation by the C terminus of p53. Mol Cell Biol 10: 5690–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu JI, Dumont P, Hafey M, Murphy ME, George DL (2004) Mitochondrial p53 activates Bak and causes disruption of a Bak–Mcl1 complex. Nat Cell Biol 6: 443–450 [DOI] [PubMed] [Google Scholar]
- Li M, Brooks CL, Kon N, Gu W (2004) A dynamic role of HAUSP in the p53–Mdm2 pathway. Mol Cell 13: 879–886 [DOI] [PubMed] [Google Scholar]
- Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W (2003) Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 302: 1972–1975 [DOI] [PubMed] [Google Scholar]
- Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W (2002) Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416: 648–653 [DOI] [PubMed] [Google Scholar]
- Marchenko ND, Zaika A, Moll UM (2000) Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J Biol Chem 275: 16202–16212 [DOI] [PubMed] [Google Scholar]
- Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, Dirks RW, Jochemsen AG (2005) Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell 18: 565–576 [DOI] [PubMed] [Google Scholar]
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM (2003) p53 has a direct apoptogenic role at the mitochondria. Mol Cell 11: 577–590 [DOI] [PubMed] [Google Scholar]
- Nemajerova A, Erster S, Moll UM (2005a) The post-translational phosphorylation and acetylation modification profile is not the determining factor in targeting endogenous stress-induced p53 to mitochondria. Cell Death Differ 12: 197–200 [DOI] [PubMed] [Google Scholar]
- Nemajerova A, Wolff S, Petrenko O, Moll UM (2005b) Viral and cellular oncogenes induce rapid mitochondrial translocation of p53 in primary epithelial and endothelial cells early in apoptosis. FEBS Lett 579: 6079–6083 [DOI] [PubMed] [Google Scholar]
- Palacios G, Moll U (2006) Mitochondrially targeted wild-type p53 suppresses growth of mutant p53 lymphomas in vivo. Oncogene 25: 6133–6139 [DOI] [PubMed] [Google Scholar]
- Petros AM, Gunasekera A, Xu N, Olejniczak ET, Fesik SW (2004) Defining the p53 DNA-binding domain/Bxl-xL binding interface using NMR. FEBS Lett 28073: 1–4 [DOI] [PubMed] [Google Scholar]
- Roth J, Dobbelstein M, Freedman DA, Shenk T, Levine AJ (1998) Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J 17: 554–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansome C, Zaika A, Marchenko ND, Moll UM (2001) Hypoxia death stimulus induces translocation of p53 protein to mitochondria. Detection by immunofluorescence on whole cells. FEBS Lett 488: 110–115 [DOI] [PubMed] [Google Scholar]
- Schumacher MM, Choi JY, Voelker DR (2002) Phosphatidylserine transport to the mitochondria is regulated by ubiquitination. J Biol Chem 277: 51033–51042 [DOI] [PubMed] [Google Scholar]
- Shirangi TR, Zaika A, Moll UM (2002) Nuclear degradation of p53 occurs during down-regulation of the p53 response after DNA damage. FASEB J 16: 420–422 [DOI] [PubMed] [Google Scholar]
- Sigismund S, Polo S, Di Fiore PP (2004) Signaling through monoubiquitination. Curr Top Microbiol Immunol 286: 149–185 [DOI] [PubMed] [Google Scholar]
- Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM (1999) A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. Embo J 18: 1660–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stommel JM, Wahl GM (2004) Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J 23: 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom E, Sathe S, Komarov PG, Chernova OB, Pavlovska I, Shyshynova I, Bosykh DA, Burdelya LG, Macklis RM, Skaliter R, Komarova EA, Gudkov AV (2006) Small-molecule inhibitor of p53 binding to mitochondria protects mice from gamma radiation. Nat Chem Biol 2: 474–479 [DOI] [PubMed] [Google Scholar]
- Talos F, Petrenko O, Mena P, Moll UM (2005) Mitochondrially targeted p53 has tumor suppressor activities in vivo. Cancer Res 65: 9971–9981 [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J 19: 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita Y, Marchenko N, Erster S, Nemajerova A, Dehner A, Klein C, Pan H, Kessler H, Pancoska P, Moll UM (2006) WT p53, but not tumor-derived mutants, bind to Bcl2 via the DNA binding domain and induce mitochondrial permeabilization. J Biol Chem 281: 8600–8606 [DOI] [PubMed] [Google Scholar]
- van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM (2006) FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol 8: 1064–1073 [DOI] [PubMed] [Google Scholar]
- Yu ZK, Geyer RK, Maki CG (2000) MDM2-dependent ubiquitination of nuclear and cytoplasmic P53. Oncogene 19: 5892–5897 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y (2001) A p53 amino-terminal nuclear export signal inhibited by DNA damage-induced phosphorylation. Science 292: 1910–1915 [DOI] [PubMed] [Google Scholar]
- Zhaung ZP, McCauley R (1989) Ubiquitin is involved in the in vitro insertion of monoamine oxidase B into mitochondrial outer membranes. J Biol Chem 264: 14594–14596 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Legends to Supplemental Figures


