Abstract
Vertebrate cells possess two different condensin complexes, known as condensin I and condensin II, that play a fundamental role in chromosome assembly and segregation during mitosis. Each complex contains a pair of structural maintenance of chromosomes (SMC) ATPases, a kleisin subunit and two HEAT-repeat subunits. Here we use recombinant human condensin subunits to determine their geometry within each complex. We show that both condensin I and condensin II have a pseudo-symmetrical structure, in which the N-terminal half of kleisin links the first HEAT subunit to SMC2, whereas its C-terminal half links the second HEAT subunit to SMC4. No direct interactions are detectable between the SMC dimer and the HEAT subunits, indicating that the kleisin subunit acts as the linchpin in holocomplex assembly. ATP has little, if any, effects on the assembly and integrity of condensin. Cleavage pattern of SMC2 by limited proteolysis is changed upon its binding to ATP or DNA. Our results shed new light on the architecture and dynamics of this highly elaborate machinery designed for chromosome assembly.
Keywords: cohesin, condensin, HEAT repeat, kleisins, SMC proteins
Introduction
Chromosome condensation is an essential prerequisite for the faithful segregation of genetic information, and is therefore crucial in maintaining genome integrity during mitosis and meiosis. Accumulating lines of evidence suggest that a class of multisubunit complexes, known as condensins, plays an important role in this process by collaborating with other chromosomal components (Strunnikov, 2003; Hirano, 2005). The canonical condensin complex (condensin I) is composed of five subunits (Hirano et al, 1997; Sutani et al, 1999). The two core subunits, SMC2/CAP-E and SMC4/CAP-C, belong to a conserved family of chromosomal ATPases, known as structural maintenance of chromosomes (SMC) proteins (Hirano and Mitchison, 1994; Saitoh et al, 1994; Saka et al, 1994; Strunnikov et al, 1995). SMC2 and SMC4 form a heterodimer that adopts a V-shaped structure with two long coiled-coil arms, each containing an ATP-binding cassette (ABC) head domain at the distal end (Melby et al, 1998; Anderson et al, 2002). Among the three non-SMC subunits, two (CAP-D2 and CAP-G) contain multiple HEAT repeats implicated in protein–protein interactions (Neuwald and Hirano, 2000), and the third (CAP-H) belongs to the kleisin family of proteins (Schleiffer et al, 2003). The three non-SMC subunits associate with each other to form an 11S subcomplex (Kimura and Hirano, 2000). As judged by electron microscopy, this subcomplex binds to the head domains of the SMC2–SMC4 dimer, and leads to the formation of a 13S holocomplex that displays a ‘lollipop-like' structure (Anderson et al, 2002).
Recent studies have shown that higher eukaryotes possess a second condensin complex, referred to as condenisn II (Ono et al, 2003; Yeong et al, 2003). This complex shares the same SMC core subunits with condensin I, but has a distinct set of non-SMC subunits (CAP-D3, CAP-G2 and CAP-H2). Although sequence similarities between the non-SMC subunits of condensin I and condensin II are hardly detectable by simple alignment methods, they are related to each other. In fact, it has been found that CAP-D3 and CAP-G2 have HEAT repeats, whereas CAP-H2 belongs to the kleisin family. Condensin II has not yet been visualized by electron microscopy, and it is unknown to what extent its overall architecture is similar to that of condensin I. Equally important, the precise geometry of the five subunits in each complex remains to be determined.
Eukaryotic cells have another SMC protein complex, known as cohesin, that is involved in sister chromatid cohesion during mitosis and meiosis (Losada and Hirano, 2005; Nasmyth and Haering, 2005). The cohesin complex is composed of SMC1, SMC3, a kleisin subunit (Scc1) and a fourth subunit (Scc3) with no obvious structural motifs. Biochemical work has shown that the N-terminal and C-terminal domains of Scc1 associate with the head domains of SMC3 and SMC1, respectively, predicting the formation of a tripartite ‘ring-like' structure (Haering et al, 2002). Consistently, an electron microscopic study showed that cohesin complexes purified from vertebrate cells display a ring-like shape in which the non-SMC subunits apparently bridge the two head domains of the SMC dimer (Anderson et al, 2002). Thus, despite some similarities in their subunit organization, cohesin and condensin I have different, characteristic conformations as judged by electron microscopy. To further understand their actions at a mechanistic level, it is important to compare and contrast the molecular architecture of the two representative SMC complexes conserved in eukaryotic cells.
Functional assays have shown that purified holocomplex of condensin I has the ability to induce positive superhelical tension into DNA in an ATP hydrolysis-dependent manner (Kimura and Hirano, 1997; Kimura et al, 2001). Moreover, condensin I physically compacts DNA by hydrolyzing ATP, as demonstrated by a single-DNA-molecule nanomanipulation assay (Strick et al, 2004). The precise role of ATP in these reactions (or in subunit–subunit interactions) is unknown, although it has been predicted that at least one function of the SMC ATPase cycle is to modulate engagement (association) and disengagement (dissociation) of the SMC head domains (Hirano, 2006).
So far, the biochemical studies on vertebrate condensins have been performed exclusively by using native complexes purified from either Xenopus egg extracts or human tissue culture cells (Kimura and Hirano, 1997; Kimura et al, 2001; Strick et al, 2004). To further dissect the structure and function of condensins, reconstitution of the sub- and holocomplexes from recombinant subunits is essential. In the current study, we have expressed recombinant condensin subunits by using a baculovirus expression system or an in vitro transcription/translation system, and used them to generate a detailed subunit–subunit interaction map. We show that the two HEAT subunits are linked to the SMC dimer through their interactions with separate domains of the kleisin subunit. This pseudo-symmetrical architecture is conserved between condensin I and condensin II. Construction of mutant SMC subunits allows us to test the role of ATP binding and hydrolysis in complex assembly and conformational changes. Our results provide a solid foundation for further structural and functional dissection of this chromosome condensation machinery.
Results
Reconstitution of condensins I and II from their recombinant subunits
We used the baculovirus expression system to produce recombinant subunits of human condensin I and condensin II complexes in Sf9 insect cells. The subunits were expressed individually or coexpressed in different combinations to reconstitute sub- and holocomplexes. Recombinant viruses were titrated carefully so that stoichiometric expression was achieved whenever coexpression was attempted. Nonetheless, the level of expression was somewhat variable among different subunits. In particular, the expression of full-length hSMC4 was substantially lower compared with the other subunits, and was always accompanied with a shorter form that was most likely to be a product translated from an internal methionine. When a lysate of insect cells coexpressing hSMC2 and hSMC4 was subjected to immunoprecipitation with anti-hSMC2, a near-stoichiometric amount of hSMC4 was co-precipitated as judged by Coomassie blue stain of a polyacrylamide gel (Figure 1, lane 1). Successful reconstitution of the SMC dimer in the lysates was further confirmed by sucrose gradient centrifugation. hSMC4 or hSMC2 alone sedimented at ∼4S (Figure 1B, panels 1 and 2). When they were coexpressed, however, most of hSMC4 and a substantial portion of hSMC2 now co-migrated at ∼8S (Figure 1B, panel 3, indicated by bracket), a size consistent with that of the SMC2–SMC4 dimer present in Xenopus egg extracts (Hirano et al, 1997).
Figure 1.
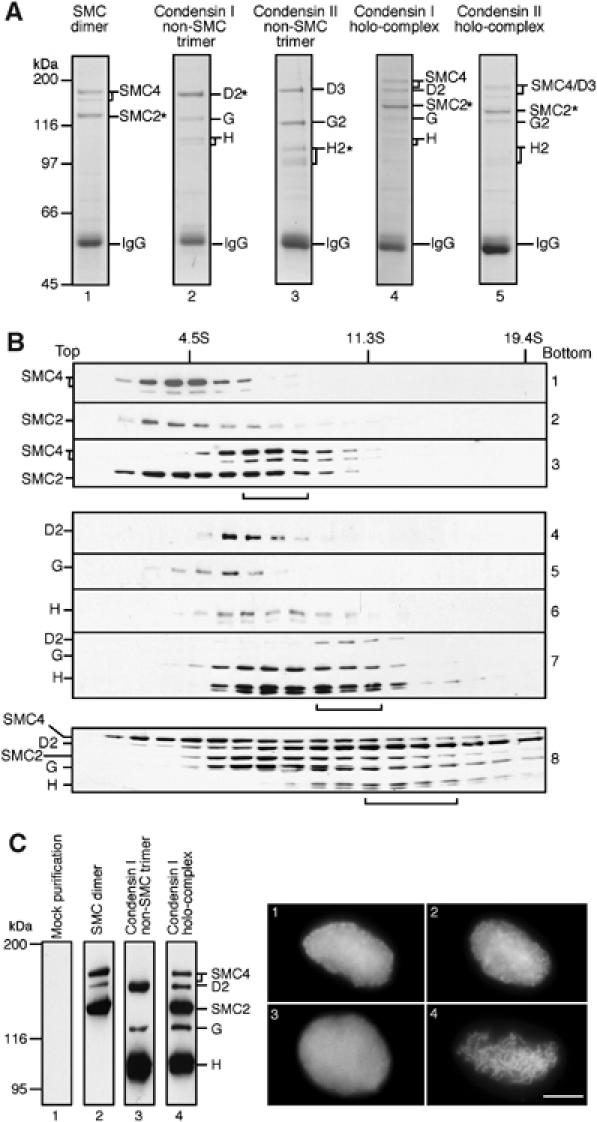
Characterization of recombinant condensin subunits and complexes. (A) Subunits of the condensin complexes were coexpressed in different combinations in Sf9 cells and the cell lysate was subjected to immunoprecipitation using an antibody specific to one of the coexpressed subunits (indicated by asterisks). The sub- and holocomplexes reconstituted were the SMC dimer (hSMC2 and hSMC4; lane 1), the non-SMC trimer of condensin I (hCAP-D2, hCAP-G and hCAP-H; lane 2), the non-SMC trimer of condensin II (hCAP-D3, hCAP-G2 and hCAP-H2; lane 3), the holocomplex of condensin I (lane 4) and the holocomplex of condensin II (lane 5). Proteins were resolved by 7.5% SDS–PAGE and stained with Coomassie brilliant blue (CBB). A very low background was observed as judged by CBB staining or immunoblotting when an untrasnfected Sf9 extract was subjected to immunoprecipitation using these antibodies (data not shown). (B) Lysates of Sf9 cells expressing individual condensin subunits or coexpressing subunits in different combinations were loaded onto 5–20% sucrose gradients and centrifuged at 36 000 r.p.m. for 15 h in an SW50.1 rotor (Beckman). Fractions were precipitated with trichloroacetic acid, resolved by 7.5% SDS–PAGE and analyzed by immunoblotting with the indicated antibodies. The positions of three protein standards (BSA (4.5S), catalase (11.3S) and thyroglobulin (19.4S)) are indicated. The peak fractions of reconstituted complexes are indicated by brackets: the 8S SMC dimer (panel 3); the 11S non-SMC trimer (panel 7); the 13S holocomplex (panel 8). (C) Fractions enriched with the hSMC2–hSMC4 dimer (lane 2), the non-SMC trimer of condensin I (lane 3) and the holocomplex of condensin I (lane 4) were prepared by single-step affinity chromatography and analyzed by immunoblotting. Mock purification was performed using an untransfected cell lysate (lane 1). A Xenopus egg HSS depleted of endogeneous condensin I was supplemented with the mock-purified fraction (panel 1), the SMC dimer fraction (panel 2), the non-SMC trimer fraction (panel 3) or the holocomplex fraction (panel 4), and the ability of each mixture to convert sperm chromatin into mitotic chromosome-like structures was assayed microscopically. Bar, 10 μm.
We had shown previously that the three non-SMC subunits of condensin I could associate with each other independently of the SMC subunits and form an 11S subcomplex in Xenopus egg extracts (Kimura and Hirano, 2000). We were successful in reconstituting a similar ∼11S subcomplex from recombinant hCAP-D2, -G and -H, as judged by co-immunoprecipitation and cosedimentation assays (Figure 1A, lane 2 and B, panels 4–7, non-SMC subcomplex indicated by bracket). Likewise, the non-SMC subunits of condensin II (hCAP-D3, -G2 and -H2) formed a subcomplex of a similar size (Figure 1A, lane 3; data not shown). Finally, the five subunits of condensin I or of condensin II were coexpressed in insect cells, and a cell lysate was subjected to immunoprecipitation with an antibody against hSMC2. All five subunits were detectable by Coomassie blue staining (Figure 1A, lanes 4 and 5) and immunoblotting (data not shown). Sucrose gradient centrifugation of a coexpressing lysate revealed a broad distribution of the five subunits of condensin I, indicating that several different subcomplexes coexisted under the condition tested (Figure 1B, panel 8). Nonetheless, a significant population of the subunits sedimented at a position expected to be the size of a 13S holocomplex (Figure 1B, panel 8, indicated by bracket).
To test the functionality of the reconstituted human condensin I complex, we took advantage of the Xenopus egg cell-free system in which condensin I is primarily responsible for converting sperm chromatin into mitotic chromosome-like structures (Hirano et al, 1997). Fractions enriched with the SMC dimer, the non-SMC trimer of condensin I and the holocomplex of condensin I were prepared by single-step affinity purification from the corresponding lysates (Figure 1C, left), and they were added back into a Xenopus egg mitotic high-speed supernatant (HSS) depleted of the endogenous condensin I. A mock-purified fraction from an untransfected lysate was used as a control. Sperm chromatin was incubated with these mixtures for 2 h and the morphology of the chromatin was observed. We found that only the holocomplex fraction could support the formation of chromosome-like structures (Figure 1C, right), a result consistent with the complementation experiment using native complexes purified from Xenopus egg extracts (Kimura and Hirano, 2000). This result demonstrates that a functional human condensin I complex can be reconstituted from its recombinant subunits, although the fraction used in the current assay was not completely pure.
ATP is not required for holocomplex assembly or SMC–kleisin interactions
The exact role of the ATP binding and hydrolysis cycle of SMC proteins in their action remains unknown. Biochemical studies using the Bacillus subtilis SMC homodimer (BsSMC) had shown previously that ATP binding and hydrolysis regulate engagement and disengagement of SMC head domains, respectively, and thereby modulate dynamic SMC–DNA interactions in vitro (Hirano and Hirano, 1998, 2004; Hirano et al, 2001). A pair of studies using yeast cohesin subunits had suggested another role of SMC's ATP binding in promoting its interaction with a non-SMC subunit (Arumugam et al, 2003; Weitzer et al, 2003). To test the role of ATP binding and hydrolysis in the assembly and functions of the condensin complexes, we introduced three different mutations in each of hSMC2 and hSMC4 that would block the proposed SMC ATPase cycle at three different stages (Figure 2A and B). The Walker A mutations (WA: K38I in hSMC2 and K191I in hSMC4) would prevent ATP binding, whereas the C-motif mutations (CM: S1086R in hSMC2 and S1192R in hSMC4) would allow ATP binding but fail to support head–head engagement (Hirano and Hirano, 1998; Hirano et al, 2001). The third, so-called transition state mutations (TR: E1114Q in hSMC2 and E1220Q in hSMC4) would stabilize head–head engagement by slowing down ATP hydrolysis (Hirano and Hirano, 2004). Whenever we describe mutant dimers in the current work, both subunits contain the same class of mutations.
Figure 2.
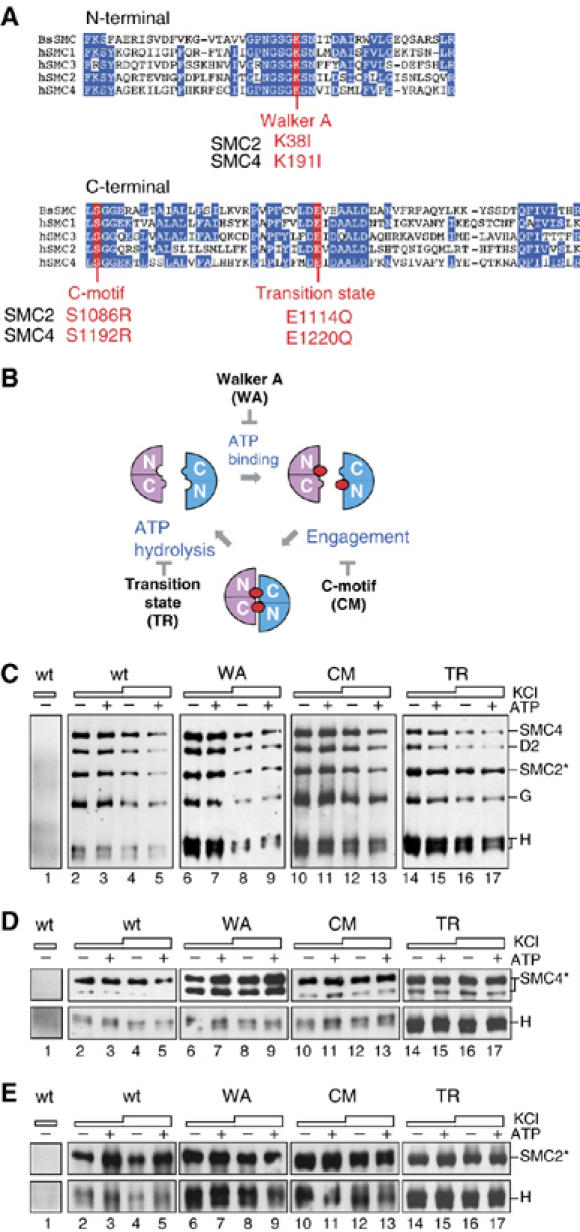
ATP is not required for holocomplex assembly or SMC–kleisin interactions. (A) Sequence alignment of the N- and C-terminal conserved domains among the human SMC proteins (hSMC1–4) and the Bacillus subtilis SMC protein (BsSMC). Also indicated are the mutation sites introduced into hSMC2 and hSMC4 in the current study. (B) The proposed SMC ATPase cycle. Each head domain is composed of the N- and C-terminal domains of an SMC subunit. Binding of ATP (closed circles) to the head domains induces their engagement, and hydrolysis of ATP triggers their disengagement. The Walker A mutation (WA) prevents ATP binding, whereas the C-motif mutation (CM) blocks engagement. The transition-state mutation (TR) stabilizes engagement by slowing down ATP hydrolysis. (C) A pair of wild-type or mutant SMC subunits was coexpressed with the three non-SMC subunits of condensin I in Sf9 cells. The lysates were subjected to immunoprecipitation using a non-immune IgG (lane 1) or anti-hSMC2 (lanes 2–17), and washed with a buffer containing 0.1 M KCl (lanes 1, 2, 3, 6, 7, 10, 11, 14 and 15) or 0.5 M KCl (lanes 4, 5, 8, 9, 12, 13, 16 and 17). No ATP (−) or ATP (+) was added in the lysate and the washing buffers throughout the procedures. The precipitates were analyzed by immunoblotting using the indicated antibodies. When mutant complexes were assayed, both hSMC2 and hSMC4 subunits contained the corresponding mutations. (D) hCAP-H was coexpressed with the wild-type or mutant forms of hSMC4, immunoprecipitated with a non-immune IgG (lane 1) or anti-hSMC4 (lanes 2–17) and analyzed as above. (E) Similarly, hCAP-H was coexpressed with the wild-type or mutant forms of hSMC2 and immunoprecipitated with a non-immune IgG (lane 1) or anti-hSMC2 (lanes 2–17).
We first used these mutant subunits to test the role of the ATPase cycle in the assembly of the condensin I holocomplex. A pair of wild-type or mutant SMC subunits was coexpressed with the three non-SMC subunits in Sf9 cells. Lysates were prepared and supplemented with no ATP or 1 mM ATP. Immunoprecipitations were performed with anti-hSMC2 and the precipitates were washed with either a low-salt buffer (0.1 M KCl) or a high-salt buffer (0.5 M KCl). The concentration of ATP was kept constant throughout the washing steps. For the wild-type complex, virtually no difference was found in the recovery of the five subunits in the presence or absence of ATP (Figure 2C, lanes 2–5). The same was true for the complexes containing the three different pairs of SMC mutant subunits (Figure 2C, lanes 6–17). These results show that neither ATP binding nor hydrolysis is essential for the assembly and structural integrity of condensin I. Furthermore, the comparison of the C-motif and transition-state mutants suggests that the assembly can occur independently of ATP-driven head–head engagement.
The results described above did not exclude the possibility that local interactions between either one of the SMC subunits and a non-SMC subunit(s) might depend on ATP binding. In fact, it has been reported that an ATP-binding mutation in SMC1, but not in SMC3, of yeast cohesin abolishes its interaction with the kleisin subunit Scc1 (Arumugam et al, 2003). By analogy to cohesin, a direct interaction between the kleisin and SMC subunits had been anticipated in condensins, but this notion had never been tested before. By coexpression and co-immunoprecipitation experiments, we found that hCAP-H does indeed interact with either hSMC4 or hSMC2 in the absence of the other subunits of condensin I (Figure 2D and E; also see Figure 3C and D). ATP had little, if any, effect on the interaction between hCAP-H and wild-type or mutant SMC subunits. We therefore conclude that, unlike cohesin, ATP binding is not essential for the SMC–kleisin interactions in condensin I. This conclusion is also most likely to be true for condensin II (see Figure 3C′).
Figure 3.
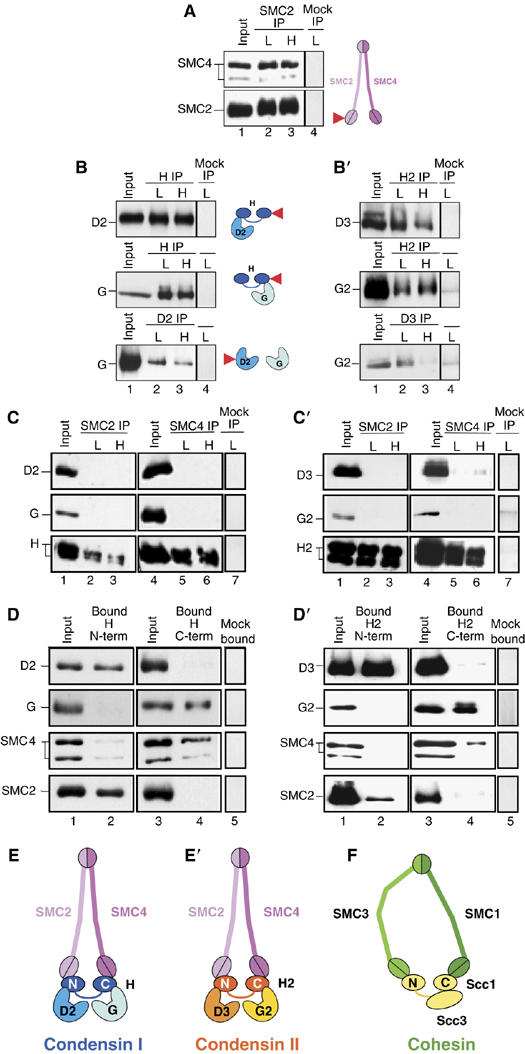
Pseudo-symmetrical structure of condensin I and condensin II as revealed by subunit-subunit interaction assays. (A) A lysate was prepared from Sf9 cells coexpressing hSMC2 and hSMC4 (lane 1) and was subjected to immunoprecipitation with anti-hSMC2 (arrow). The precipitates were recovered on protein A beads, washed with a buffer containing 0.1 M (L, lane 2) or 0.5 M KCl (H, lane 3) and analyzed by immunoblotting with anti-hSMC4 and anti-hSMC2. (B) Two of the three non-SMC subunits of condensin I were coexpressed in different combinations, immunoprecipitated with the indicated antibodies (arrows) and analyzed as above. In (A) and (B), subunit–subunit interactions detected are summarized in the adjacent cartoons. Target subunits used for immunoprecipitations are indicated by the arrows. Mock immunoprecipitation using non-immune rabbit IgG is shown in lane 4 of each panel. (C) hSMC2 was coexpressed with one of the three non-SMC subunits of condensin I, immunoprecipitated with anti-hSMC2 and analyzed as above (left panel, lanes 1–3). The same set of experiments was performed for hSMC4 (right panel, lanes 4–6). A mock immunoprecipitation is shown in lane 7. (D) A hexahistidine-tagged, N-terminal fragment of hCAP-H was coexpressed with either hCAP-D2, hCAP-G, hSMC4 or hSMC2 in Sf9 cells. Lysates were prepared and mixed with Ni-NTA beads. After washing the beads with a buffer containing 0.1 M KCl, bound proteins were analyzed by immunoblotting with the indicated antibodies (left). The same set of experiments was performed for a hexahistidine-tagged, C-terminal fragment of hCAP-H (right). A mock pull-down is shown in lane 5. (E) Deduced subunit geometry in the condensin I complex. (B′–D′) An identical set of experiments was performed as in (B–D) by using condensin II subunits. (E′) Deduced subunit geometry in the condensin II complex. (F) Subunit geometry in the cohesin complex proposed by Haering et al (2002).
Subunit geometry of condensin I and condensin II
To make a subunit–subunit interaction map of condensin I, we coexpressed its subunits in a pairwise manner in insect cells. A cell lysate was prepared from the cells and subjected to immunoprecipitation with a specific antibody against one of the coexpressed subunits. The precipitates were recovered on protein A-beads, washed with a buffer containing either 0.1 or 0.5 M KCl and analyzed by immunoblotting. As expected, hSMC4 co-precipitated with hSMC2 even in a high-salt wash condition (Figure 3A). A pairwise analysis of the non-SMC subunits showed that hCAP-H associated with hCAP-D2 and hCAP-G, whereas hCAP-D2 and hCAP-G interacted only weakly with each other (Figure 3B). A monomeric form of hSMC2 could associate with hCAP-H, but not with hCAP-D2 or hCAP-G (Figure 3C, lanes 1–3). Identical results were obtained for a monomeric form of hSMC4 (Figure 3C, lanes 4–6), indicating that dimerization of hSMC2 and hSMC4 is not a prerequisite for their interaction with hCAP-H. To further narrow down interaction domains, we constructed recombinant viruses that express the N-terminal (amino acids 1–393) or C-terminal half (amino acids 395–730) of hCAP-H. It was found that the N-terminal half of hCAP-H binds to hCAP-D2 and hSMC2, but not to hCAP-G or hSMC4 (Figure 3D, lanes 1 and 2). Conversely, the C-terminal half of hCAP-H interacted with hCAP-G and hSMC4, but not with hCAP-D2 or hSMC2 (Figure 3D, lanes 3 and 4). Previous electron microscopy images suggested that the non-SMC subunits of condensin I associate with the head domains of the hSMC2–hSMC4 dimer (Anderson et al, 2002). Based on these results, we deduced a simple picture of the subunit geometry in condensin I (Figure 3E). In short, the N-terminal domain of hCAP-H links hCAP-D2 to hSMC2, whereas the C-terminal domain of hCAP-H connects hCAP-G to hSMC4. Thus, condensin I has a pseudo-symmetrical structure, each half of which is composed of an SMC subunit, a HEAT-repeat subunit and a half of the kleisin subunit.
An identical set of experiments was then set up for condensin II (Figure 3B′,C′ and D′), which allowed us to conclude that the overall architecture of condensin II is very similar to that of condensin I (Figure 3E′). The position of hCAP-D3 and hCAP-G2 in condensin II is equivalent to that of hCAP-D2 and hCAP-G, respectively, in condensin I. This result is consistent with our previous report that despite their very poor sequence similarities, hCAP-D3 is more related to hCAP-D2 than hCAP-G, whereas hCAP-G2 is more related to hCAP-G than hCAP-D2 (Ono et al, 2003).
Independent binding of two HEAT subunits to a kleisin subunit
The subunit–subunit interaction assay described above involved pairwise expression of two subunits in insect cells, followed by single immunoprecipitations or pull-downs. These results, however, left some ambiguities about the interactions between hCAP-D2 and hCAP-G. It was not very clear whether hCAP-D2 and hCAP-G could make a direct contact with each other, or whether such a mutual interaction between hCAP-D2 and hCAP-G might facilitate their own binding to hCAP-H. We wished to resolve this issue and verify our results with an independent experimental system.
To this end, the three non-SMC subunits of condensin I were individually expressed and labeled with [35S]methionine in an in vitro transcription/translation system. Two or three of the reactions were mixed together and then subjected to immunoprecipitations using specific antibodies. Co-precipitated subunits were visualized by autoradiography. We found that hCAP-D2 interacted with hCAP-H regardless of the presence or absence of hCAP-G (Figure 4A, lanes 5 and 6). Conversely, hCAP-G bound to hCAP-H independently of hCAP-D2 (Figure 4B, lanes 5 and 6). In this assay, hCAP-D2 and hCAP-G barely interacted with each other in the absence of CAP-H (Figure 4A, lane 4 and B, lane 4).
Figure 4.
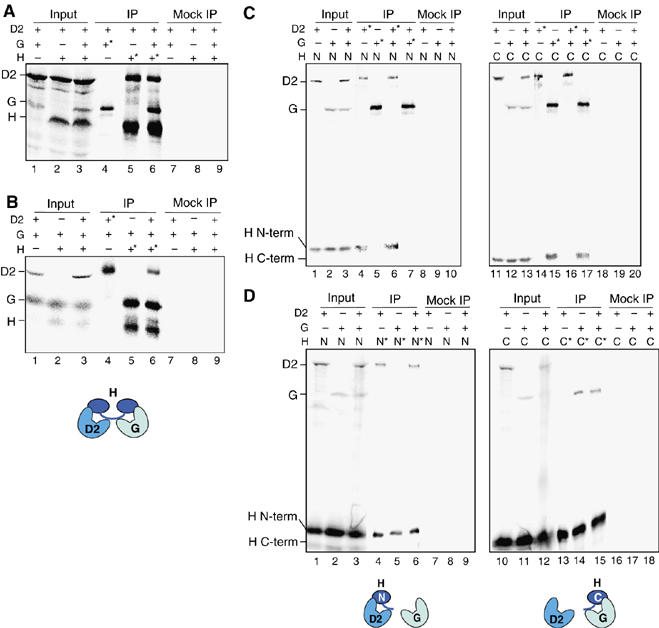
hCAP-D2 and hCAP-G bind independently to separate domains of hCAP-H. (A) Full-length hCAP-D2, hCAP-G and hCAP-H were translated in vitro in the presence of [35S]methionine. The reactions were mixed in different combinations (input; lanes 1–3) and subjected to immunoprecipitation with anti-hCAP-G (lane 4), anti-hCAP-H (lanes 5 and 6) or non-immune rabbit IgG (lanes 7–9). The input and bound fractions were fractionated by SDS–PAGE and analyzed by autoradiography. (B) Full-length hCAP-D2, hCAP-G and hCAP-H were translated in vitro in the presence of [35S]methionine. The reactions were mixed in different combinations (input; lanes 1–3) and subjected to immunoprecipitation with anti-hCAP-D2 (lane 4), anti-hCAP-H (lanes 5 and 6) or non-immune IgG (lanes 7–9). (C) An N-terminal fragment of hCAP-H (1-393), full-length hCAP-D2 and hCAP-G were translated in vitro in the presence of [35S]methionine. The reactions were mixed in different combinations as indicated (input; lanes 1–3), and subjected to immunoprecipitation with anti-hCAP-D2 (lanes 4 and 6), anti-hCAP-G (lanes 5 and 7) or non-immune IgG (lanes 8–10). Alternatively, the N-terminal fragment was replaced with a C-terminal fragment of hCAP-H (395–730), and a similar set of experiments was performed (lanes 11–20). (D) An N-terminal fragment of hCAP-H (1-393), full-length hCAP-D2 and hCAP-G were translated in vitro in the presence of [35S]methionine. The reactions were mixed in different combinations as indicated (input; lanes 1–3) and subjected to immunoprecipitation with the Penta-His antibody (Qiagen) that recognizes the N-terminal fragment of hCAP-H (lanes 4–6) or non-immune rabbit IgG (lanes 7–9). Alternatively, the N-terminal fragment was replaced with a C-terminal fragment of hCAP-H (395–730), and anti-hCAP-H that recognizes its C-terminal fragment or non-immune IgG was used for immunoprecipitation (lanes 10–18). In (A–D), target subunits used for immunoprecipitations are indicated by the asterisks.
We further verified that hCAP-D2 interacted with the N-terminal but not the C-terminal half of hCAP-H (Figure 4C, lanes 4 and 14), whereas hCAP-G interacted with the C-terminal but not the N-terminal half of hCAP-H (Figure 4C, lanes 5 and 15). Again, the interaction of the truncated forms of hCAP-H with one HEAT subunit was not affected in the presence of the other HEAT subunit (Figure 4C, lanes 6, 7, 16 and 17). These observations were further confirmed by reciprocal reactions that pulled down the N-terminal or the C-terminal domain of hCAP-H (Figure 4D). These results provide compelling lines of evidence that hCAP-D2 and hCAP-G bind independently to two separate domains of hCAP-H, and that the interaction between hCAP-D2 and hCAP-G, if any, is very weak in the presence or absence of hCAP-H. The same notion, based on the result presented in Figure 3B′, is most likely applicable to the interactions among the non-SMC subunits of condensin II.
Domain dissection of HEAT repeat subunits
Our original sequence analysis using iterative search and Gibbs sampling alignment procedures identified 12 and 9 HEAT repeats in hCAP-D2 and hCAP-G, respectively (Neuwald and Hirano, 2000). These numbers were by no means definitive because the HEAT repeats are highly degenerated, that is, different algorithms would detect different numbers of the repeat in each subunit. To deduce functionally important domains within the HEAT subunits, the sequences of hCAP-D2 and hCAP-G were compared with their orthologs of the eukaryote parasite Encephalitozoon cuniculi (Katinka et al, 2001). This organism has a very small genome (∼2.9 Mb) whose compaction is reflected by reduced intergenic spacers and the shortness of most encoded proteins relative to their orthologs in other eukaryotes. Our analysis indicated that six out of the 12 HEAT repeats present in hCAP-D2 (HEAT no. 1–3 and no. 9–11) were conserved in EcCAP-D2 (Figure 5A, upper panel). Similarly, the first five HEAT repeats in hCAP-G (HEAT no. 1–5) were readily detectable in EcCAP-G, but the remainders (HEAT no. 6–9) were not (Figure 5B, upper panel).
Figure 5.
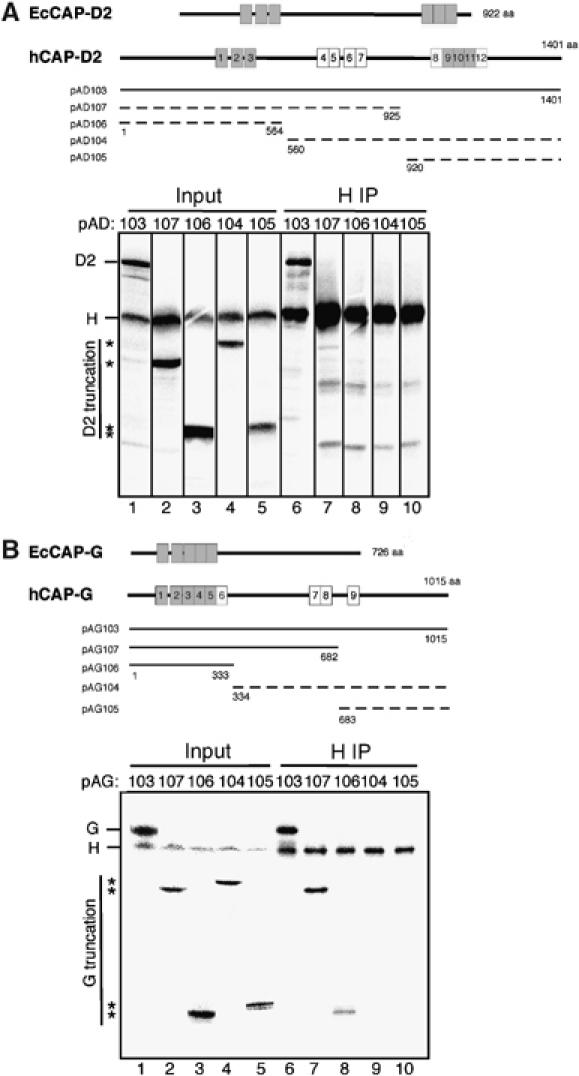
Domain dissection of the HEAT subunits. (A) (upper) Overall structures of hCAP-D2 and its ortholog in Encephalitozoon cuniculi (EcCAP-D2) are shown (upper). Among the 12 HEAT repeats found in hCAP-D2 (shown by boxes), only six are detectable in EcCAP-D2 (gray boxes); (lower) a deletion series of hCAP-D2 (pAD103–107) was translated in vitro and each reaction was mixed with a reaction containing full-length hCAP-H (input; lanes 1–5). The mixtures were subjected to immunoprecipitation with anti-hCAP-H (IP; lanes 6–10). The input and precipitated fractions were fractionated by SDS–PAGE and analyzed by autoradiography. The positions of truncated hCAP-D2 are indicated by asterisks. Only the full-length construct of hCAP-D2 interacted efficiently with hCAP-H (indicated by a solid line in the upper panel), whereas all truncated forms interacted poorly with hCAP-H (indicated by dotted lines). (B) Overall structures of hCAP-G and its ortholog in E. cuniculi (EcCAP-G) are shown. Among the nine HEAT repeats found in hCAP-D2 (shown by boxes), only five are detectable in EcCAP-D2 (gray boxes). A deletion series of hCAP-G (pAG103–107) was translated in vitro and analyzed as described in (A). The positions of truncated hCAP-G are indicated by asterisks. The constructs of hCAP-G that efficiently interact with hCAP-H are indicated by solid lines, whereas those that poorly interact with hCAP-H are indicated by dotted lines in the upper panel.
With this information in our hands, we wished to localize the domain(s) in hCAP-D2 and hCAP-G that is required for their interaction with hCAP-H. To this end, we expressed a panel of 35S-labeled, truncated fragments of hCAP-D2 and hCAP-G in vitro. Each of the reaction mixtures was combined with another mixture that contained an 35S-labeled, full-length hCAP-H and then subjected to immunoprecipitation with anti-hCAP-H antibody. We found that none of the truncated forms of hCAP-D2 were efficiently co-precipitated with hCAP-H, although an interaction between full-length hCAP-D2 and hCAP-H was readily detectable (Figure 5A, lower panel, lanes 6–10). In contrast, the N-terminal 1/3 of hCAP-G appeared to be sufficient for its interaction with hCAP-H (Figure 5B, lower panel, lanes 6–10). Interestingly, the domain(s) found to be important for the subunit–subunit interactions significantly overlapped with the most conserved domains at the primary sequence level. In the case of hCAP-D2, it is conceivable that the two conserved HEAT-repeat clusters, located in the N-terminal and C-terminal 1/3, may cooperate to create an interaction surface for hCAP-H. On the other hand, the N-terminal conserved HEAT cluster of hCAP-G may be sufficient for its proper interaction with hCAP-H.
ATP- and DNA-induced changes in proteolytic cleavage of hSMC2 monomers
How the mechanical cycle of condensins might be coupled to the ATPase cycle of the SMC subunits remains a big question in this field (Hirano, 2006). As an attempt to address this question, we performed limited proteolysis and asked whether ATP binding might induce conformational changes of an SMC subunit. hSMC2 was expressed in the baculovirus system and purified into near-homogeneity by affinity column chromatography. The protein was preincubated in a buffer containing ATP or no ATP and aliquots were taken at time intervals after trypsin was added to the reaction mixtures. The cleaved products were fractionated by SDS–PAGE and analyzed by silver stain (Figure 6A, upper panel; also see Supplementary Figure S1). We observed a set of ATP-induced changes in the cleavage pattern in a molecular mass range of ∼120–130 kDa (Figure 6A, bands a and b). A 60-kDa band accumulated over time, but this occurred independently of ATP (Figure 6A, band f). We then mapped approximate locations of the cleavage sites by means of immunoblotting using two different antibodies that recognize N-terminal and C-terminal epitopes on hSMC2 (Figure 6A, middle and lower panels). Major cleavage occurred in the central hinge domain (Figure 6D, indicated by the arrows), creating two bands derived from the N-terminal half (bands c and d) and a band derived from the C-terminal half (band e). Band f was most likely produced by further cleavage of band e near its C terminus. Prominent ATP-dependent changes in cleavage were mapped in the C-terminal globular domain as well as in the N-terminal domain, as expected (Figure 6D, indicated by the arrowheads). Notably, the addition of ATP produced a subtle yet reproducible difference in the relative ratio of bands c and d (Figure 6A, top and middle panels), raising the possibility that ATP binding at the head domain might alter the conformation of the hinge domain. When the Walker A mutant form of hSMC2 was purified and subjected to the same proteolysis assay, no difference in the cleavage pattern was observed in the presence or absence of ATP (Figure 6B), confirming that the changes observed in wild-type hSMC2 were indeed specific to ATP binding.
Figure 6.
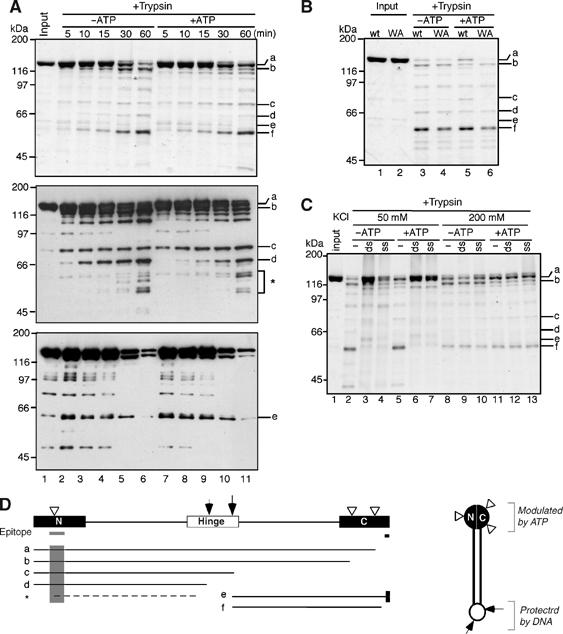
ATP- and DNA-induced changes in proteolytic cleavage of hSMC2 monomers. (A) Purified hSMC2 (lane 1) was preincubated at 37°C in the absence (lanes 2–6) or presence (lanes 7–11) of 1 mM ATP. Trypsin was added at time 0 and the reactions were stopped at the indicated time points (lanes 2–11). The cleaved products were resolved by 7.5% SDS–PAGE and analyzed by silver stain (upper panel) and immunoblotting using antibodies specific to the N-terminal (middle panel) or C-terminal (lower panel) domain of hSMC2. The major cleaved products discussed in the text are labeled as a–f. The asterisk indicates a minor population of N-terminal fragments whose cleavage is altered in the presence of ATP. (B) The wild-type (lanes 1, 3 and 5) and Walker A mutant (lanes 2, 4 and 6) forms of hSMC2 were treated with trypsin at 37°C for 45 min in the absence (lanes 3 and 4) or presence (lanes 5 and 6) of ATP. The cleaved products were analyzed as above. (C) Wild-type hSMC2 was preincubated with no DNA (−), double-stranded DNA (ds) or single-stranded DNA (ss) in a buffer containing either 50 or 200 mM KCl with or without ATP, and then treated with trypsin at 37°C for 45 min. The reaction mixtures were separated by 7.5% SDS–PAGE and analyzed by silver stain (lanes 1–13). (D) Major cleavage sites were mapped on the basis of the results shown in (A). The gray bars and shadows indicate the locations of the N- and C-terminal epitopes recognized by the antibodies used in (A). The white arrowheads indicate the cleavage sites that are modulated by ATP. The arrows represent the major cleavage sites in the hinge domain, whose cleavage is suppressed in the presence of DNA. The model for a folded SMC2 monomer is shown on the right.
We then tested whether the cleavage pattern of hSMC2 might be modulated upon its interaction with DNA (Figure 6C). We found that the cleavage in the hinge domain was greatly suppressed in the presence of double-stranded DNA (dsDNA) or single-stranded DNA (ssDNA), and that such suppression occurred regardless of the presence or absence of ATP (Figure 6C, lanes 2–7). The DNA-dependent changes were no longer observed when the KCl concentration in the reaction mixture was raised from 50 to 200 mM (Figure 6C, lanes 8–13), indicating that hSMC2 interacted with DNA only at a physiological concentration of salt. Naturally, we wanted to extend these analyses to the hSMC2–hSMC4 dimer and to test whether hSMC2 might display a different set of cleavage products in the context of the dimer. However, relatively poor expression of SMC4 hampered us from purifying a large amount of pure dimers with a 1:1 stoichiometry that was required for this type of experiments.
Discussion
Pseudo-symmetrical structure of condensin I and condensin II
The current study is the first to describe the subunit geometry of the condensin complexes in detail. One important conclusion from the current work is that condensin I has a pseudo-symmetrical architecture, in which the N-terminal half of CAP-H links the first HEAT subunit (CAP-D2) to SMC2, whereas its C-terminal half links the second HEAT subunit (CAP-G) to SMC4 (Figure 3E). Recent structural studies have shown that the N- and C-terminal domains of kleisins may contain a common structural fold, known as the winged helix motif, although the two domains share virtually no sequence similarity (Haering et al, 2004; Fennell-Fezzie et al, 2005; Hirano, 2006). Thus, the notion of pseudo-symmetry could be extended further to the structure of CAP-H itself, meaning that each half of the condensin I complex is composed of an SMC subunit, a winged helix domain (from the kleisin subunit) and a HEAT subunit. The kleisin subunit acts as the linchpin in the assembly of the condensin I holocomplex, providing a biochemical explanation for the observation that in vivo depletion of CAP-H/Barren causes destabilization of CAP-D2 in Drosophila cells (Savvidou et al, 2005).
The second conclusion is that condensin II architecture is similar to that of condensin I, in terms of not only its subunit composition, but also its subunit geometry (Figure 3E′). This conclusion should not be underestimated because the sequence similarities between the non-SMC regulatory subunits of the two complexes are very limited. For example, the similarity between CAP-G and CAP-G2 or between CAP-H and CAP-H2 is virtually undetectable by the standard BLAST search. Only biochemical and more advanced bioinformatics approaches have made it possible to identify CAP-G2 and CAP-H2 as the subunits of a second condensin complex (i.e. condensin II) (Ono et al, 2003; Schleiffer et al, 2003). The successful reconstitution of condensin I and condensin II described in the current study provides an excellent starting point toward their detailed functional characterization and comparison in the future.
Similarities and differences between condensins and cohesin
The architecture of condensins reported here displays two notable similarities to that of cohesin (Figure 3F). The first is the geometry of the kleisin subunits. In the cohesin complex, the N-terminal and C-terminal domains of the kleisin subunit Scc1 interact with SMC3 and SMC1, respectively (Haering et al, 2002). We find that in condensin I, the N-terminal and C-terminal halves of CAP-H associate with SMC2 and SMC4, respectively. Because phylogenic analysis of SMC proteins classifies SMC3 and SMC2 into one subfamily and SMC1 and SMC4 into the other (Cobbe and Heck, 2004), the relative orientation of the kleisin subunits to the SMC heterodimers is conserved between condensins and cohesin. The second similarity is that the kleisin subunits bridge the interaction between the SMC dimers and the remaining regulatory subunit(s). The C-terminal domain of Scc1 connects Scc3 to SMC1 in cohesin (Haering et al, 2002). In the case of condensin I, the N-terminal domain of CAP-H mediates the association between CAP-D2 and SMC2, whereas the C-terminal domain of CAP-H links CAP-G to SMC4. Our current assays detect no stable interactions between the HEAT subunits and the SMC dimer in the absence of kleisin. Nevertheless, these observations by no means exclude the possibility that one (or both) of the HEAT subunits may make a direct contact with the SMC subunits in the presence of kleisin.
As judged by the position of Scc3 in cohesin, one could imagine that it might have an analogous function to that of CAP-G in condensin I (or CAP-G2 in condensin II). Nonetheless, no HEAT repeats have been detectable in the sequence of Scc3 by our algorithm (Neuwald and Hirano, 2000). Furthermore, the cohesin complex does not have a fifth subunit and therefore lacks pseudo-symmetry. Thus, the architecture of condensins and cohesin is conserved at the level of the SMC dimer–kleisin interactions, but starts to diverge beyond this point. It should be added, however, that two HEAT-repeat proteins (Scc2 and Pds5) regulate chromatin association of cohesin in vivo, presumably through their transient interactions with cohesin (Neuwald and Hirano, 2000; Arumugam et al, 2003; Losada et al, 2005).
Another difference between cohesin and condensins resides in the requirement for ATP in their complex assembly. Previous studies showed that a mutant form(s) of SMC1 defective in ATP binding expressed in yeast cells failed to interact with Scc1. The corresponding mutant form(s) of SMC3 retained its ability to bind to Scc1, leading to the conclusion that the interaction of Scc1 with SMC1 (but not with SMC3) is ATP-dependent (Arumugam et al, 2003; Weitzer et al, 2003). In contrast, our current results using the three sets of mutant SMC subunits provide strong lines of evidence that neither ATP binding nor its hydrolysis is required for the assembly of the holocomplex of condensin I. The association of CAP-H with SMC2 (or with SMC4) does not require ATP binding, either. We also demonstrated previously that the assembly of a bacterial SMC protein complex is independent of ATP (Hirano and Hirano, 2004). Thus, the requirement for ATP in complex assembly may be a unique property of the yeast cohesin complex rather than a general trait of SMC protein complexes.
ATP- and DNA-dependent conformational changes of SMC subunits
Lammens et al (2004) compared the crystal structure of ATP-bound and nucleotide-free SMC head domains (from Pyrococcus furiosus) and found very little difference in their overall conformations (Lammens et al, 2004). The authors speculated that SMC proteins do not possess a ‘power stroke', and that the role of ATP binding and hydrolysis of SMC proteins may be limited to the regulation of the engagement/disengagement cycle. Our data from the limited proteolysis experiments, however, raise the possibility that full-length SMC proteins may undergo conformational changes upon binding to ATP, and that at least part of such conformational changes may occur independently of head–head engagement. Because only SMC2 monomers were tested in the current study, we were not able to directly compare engagement-independent and engagement-dependent conformational changes, the latter of which would only be observed in the context of SMC2–SMC4 dimers. In the future, the possibility should further be explored that ATP binding in the head domain might alter the conformation of the hinge domain located at the opposite end of the molecule.
The cleavage in the hinge domain of SMC2 is greatly suppressed in the presence of DNA, indicating that the human SMC protein is likely to have an ability to interact with DNA in its monomer form. This observation was somewhat surprising to us because in the case of the bacterial SMC protein, hinge-mediated dimerization was shown to be essential for its basal interaction with DNA (Hirano and Hirano, 2002). Nonetheless, the current result is consistent with the more recent study demonstrating that the hinge domain of SMC proteins may act as an initial sensor for DNA (Hirano and Hirano, 2006). In the future, it will be important to critically compare and contrast the eukaryotic and prokaryotic SMC proteins, which are likely to share a basic mechanism of action, but also have their own unique DNA-binding properties (Sutani and Yanagida, 1997; Hirano and Hirano, 1998, 2004; Hirano et al, 2001; Sakai et al, 2003; Stray and Lindsley, 2003). We anticipate that the limited proteolysis assay described here may offer an alternative and powerful assay to further dissect the very dynamic and complex set of interactions between SMC proteins and DNA.
Materials and methods
Construction of recombinant baculoviruses
Full-length cDNAs encoding human condensin subunits were subcloned into pFASTBac (Invitrogen). The sources of the cDNA clones and the details of plasmid construction are described in Supplementary data. The Bac-to-Bac baculovirus expression system (Invitrogen) was used to produce recombinant viruses according to the manufacturer's instructions. Point mutations were introduced using a QuikChange XL site-directed mutagenesis kit (Stratagene) and confirmed by sequencing. The primer sequences used for mutagenesis are also available in Supplementary data.
Protein expression and purification
To express recombinant proteins, Sf9 cells were infected with the corresponding baculoviruses at a multiplicity of infection (MOI) of 2 and incubated at 27°C for 48 h. For immunoprecipitation experiments, ∼3 × 106 cells were resuspended in 0.5 ml of buffer L (20 mM Hepes (pH 7.7), 100 mM KCl and 2.5 mM MgCl2) and lysed by sonication. Tween 20 (Sigma) was added at a final concentration of 0.05%. The lysate was clarified by centrifugation at 3000 g at 4°C for 10 min, frozen in liquid nitrogen and stored at −80°C. For purification and pull-down experiments using histidine-tagged subunits, buffer L was replaced with buffer N (20 mM Hepes (pH 7.7), 300 mM KCl, 2.5 mM MgCl2, 10% glycerol, 20 mM imidazole and 5 mM 2-mercaptoethanol). The details of protein purification are described in Supplementary data.
Functional complementation of Xenopus egg extracts by human condensin subunits
HSS were prepared from cytostatic factor (CSF)-arrested egg extracts in XBE2 buffer (10 mM K-Hepes (pH 7.7), 100 mM KCl, 2 mM MgCl2, 0.1 mM CaCl2, 5 mM EGTA and 50 mM sucrose) as described previously (Hirano et al, 1997). To deplete endogenous condensin I from the HSS, two rounds of immunodepletion were preformed by mixing 50 μl of HSS and 4 μg of anti-XCAP-G antibody pre-immobilized on 20 μl of protein A–agarose beads for 1 h on ice. Control depletions were performed using beads coated with non-immune rabbit IgG. A 15 μl aliquot of the condensin I-depleted HSS was supplemented with an energy mix (1 mM MgATP, 10 mM creatine phosphate and 50 μg/ml creatine kinase) and the fractions containing human condensin complexes. We roughly estimated the concentration of the condensin subunits present in each fraction by SDS–PAGE and added them back to the HSS at near-physiological levels. Sperm chromatin was then added at a final concentration of 500 nuclei/μl and the reaction mixture was incubated at 22°C for 2 h. Chromatin was stained with DAPI and its morphology was observed by a Zeiss Axiophot microscope (Carl Zeiss Inc.) equipped with a cooled charge-coupled device (CCD) camera (Photometrics Ltd).
Subunit–subunit interaction assays
Frozen aliquots of Sf9 cell lysates prepared in buffer L were thawed and spun at 4°C for 10 min in a microfuge. Affinity-purified antibody or non-immune rabbit IgG (Sigma) was added to the clarified lysates and incubated at 4°C for 1 h. Immunoprecipitates were then recovered on protein A–agarose beads (Invitrogen), washed four times with buffer L (unless other KCl concentrations were indicated) and eluted from the beads by adding 20 μl of SDS buffer (125 mM Tris–HCl (pH 6.8), 2% SDS and 20% glycerol). The proteins were subjected to 7.5% SDS–PAGE and analyzed by Coomassie stain or immunoblotting. When the effect of ATP on subunit–subunit interactions was tested, both the lysates and washing buffers were supplemented with 1 mM ATP. In the experiments using the truncated forms of hCAP-H and hCAP-H2, lysates were prepared in buffer N, mixed with 20 μl of Ni-NTA beads (Qiagen) and incubated under gentle shaking at 4°C for 2 h. The beads were washed four times with buffer N. Proteins were eluted from the beads by adding 20 μl of SDS buffer, subjected to 10% SDS–PAGE and analyzed by immunoblotting, as indicated.
In vitro transcription/translation reactions
DNA fragments encoding different regions of the condensin subunits were amplified by polymerase chain reactions (PCR) and inserted into pTNT vector (Promega). The primers used for PCR are available in Supplementary data. In vitro transcription/translation reactions were performed using TNT Quick Coupled Transcription/Translation Systems (Promega) according to the manufacturer's instructions. Reaction mixtures (50 μl), which contained 1 μg of plasmid DNAs and [35S]methionine (1000 Ci/mmol) at a final concentration of 0.4 mCi/ml, were incubated at 30°C for 90 min. For subunit–subunit interaction assays, two reactions containing different protein fragments were mixed and diluted with nine volumes of XBE2. The mixtures were spun at 10 000 r.p.m. in a microfuge at 4°C for 15 min and the supernatants were subjected to immunoprecipitation with the antibodies indicated. The precipitated polypeptides were resolved by SDS–PAGE and analyzed by an image analyzer (Fuji-film, FLA-5100).
Limited proteolysis
SMC2 monomer (∼70 pmol) was preincubated in 20 μl of buffer (20 mM Hepes (pH 7.7), 50 mM KCl, 10% glycerol and 2.5 mM MgCl2) at 37°C for 30 min. Trypsin (Roche) was then added at a final concentration of 5 μg/ml and the reaction was stopped at various time points by adding 20 μl of SDS sample buffer. Each reaction was divided into aliquots and loaded onto gels and analyzed by silver stain (Bio-Rad) and immunoblotting using antibodies specific to the N-terminal (amino acids 50–100; Bethyl Laboratories, A300-056A-1) and C-terminal domain (amino acids 1184–1197; Kimura et al, 2001) of hSMC2. Whenever indicated, the reaction mixture was supplemented with 1 mM ATP or pBluescript SK dsDNA plasmid or φx174 ssDNA at a molar ratio of one protein per 100 nucleotides of DNA.
Supplementary Material
Supplementary Figure S1
Supplementary data
Acknowledgments
We are grateful to K Yokomori (University of California at Irvine), T Nagase (Kazusa DNA Research Institute) and RZPD German Resource Center for Genome Research for cDNA clones encoding condensin subunits. We thank members of the Hirano Laboratory for critically reading the manuscript. This work was supported by a grant from the National Institutes of Health (to TH) and by a fellowship from the Human Frontier Science Program (to NA).
References
- Anderson DE, Losada A, Erickson HP, Hirano T (2002) Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol 156: 419–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam P, Gruber S, Tanaka K, Haering CH, Mechtler K, Nasmyth K (2003) ATP hydrolysis is required for cohesin's association with chromosomes. Curr Biol 13: 1941–1953 [DOI] [PubMed] [Google Scholar]
- Cobbe N, Heck MMS (2004) The evolution of SMC proteins: phylogenetic analysis and structural implications. Mol Biol Evol 21: 332–347 [DOI] [PubMed] [Google Scholar]
- Fennell-Fezzie R, Gradia SD, Akey D, Berger JM (2005) The MukF subunit of Escherichia coli condensin: architecture and functional relationship to kleisins. EMBO J 24: 1921–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering CH, Lowe J, Hochwagen A, Nasmyth K (2002) Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell 9: 773–788 [DOI] [PubMed] [Google Scholar]
- Haering CH, Schoffegger D, Nishino T, Helmhart W, Nasmyth K, Lowe J (2004) Structure and stability of cohesin's Smc1–kleisin interaction. Mol Cell 15: 951–964 [DOI] [PubMed] [Google Scholar]
- Hirano M, Anderson DE, Erickson HP, Hirano T (2001) Bimodal activation of SMC ATPase by intra- and inter-molecular interactions. EMBO J 20: 3238–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Hirano T (1998) ATP-dependent aggregation of single-stranded DNA by a bacterial SMC homodimer. EMBO J 17: 7139–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Hirano T (2002) Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. EMBO J 21: 5733–5744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Hirano T (2004) Positive and negative regulation of SMC–DNA interactions by ATP and accessory proteins. EMBO J 23: 2664–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Hirano T (2006) Opening closed arms: long-distance activation of SMC ATPase by hinge–DNA interaction. Mol Cell 21: 175–186 [DOI] [PubMed] [Google Scholar]
- Hirano T (2005) Condensins: organizing and segregating the genome. Curr Biol 15: R265–R275 [DOI] [PubMed] [Google Scholar]
- Hirano T (2006) At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol 7: 311–322 [DOI] [PubMed] [Google Scholar]
- Hirano T, Kobayashi R, Hirano M (1997) Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell 89: 511–521 [DOI] [PubMed] [Google Scholar]
- Hirano T, Mitchison TJ (1994) A heterodimeric coiled-coil protein required for mitotic chromosome condensation in vitro. Cell 79: 449–458 [DOI] [PubMed] [Google Scholar]
- Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, Delbac F, El Alaoui H, Peyret P, Saurin W, Gouy M, Weissenbach J, Vivares CP (2001) Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 414: 450–453 [DOI] [PubMed] [Google Scholar]
- Kimura K, Cuvier O, Hirano T (2001) Chromosome condensation by a human condensin complex in Xenopus egg extracts. J Biol Chem 276: 5417–5420 [DOI] [PubMed] [Google Scholar]
- Kimura K, Hirano T (1997) ATP-dependent positive supercoiling of DNA by 13S condensin: a biochemical implication for chromosome condensation. Cell 90: 625–634 [DOI] [PubMed] [Google Scholar]
- Kimura K, Hirano T (2000) Dual roles of the 11S regulatory subcomplex in condensin functions. Proc Natl Acad Sci USA 97: 11972–11977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens A, Schele A, Hopfner K-P (2004) Structural biochemistry of ATP-driven dimerization and DNA-stimulated activation of SMC ATPases. Curr Biol 14: 1778–1782 [DOI] [PubMed] [Google Scholar]
- Losada A, Hirano T (2005) Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev 19: 1269–1287 [DOI] [PubMed] [Google Scholar]
- Losada A, Yokochi T, Hirano T (2005) Functional contribution of Pds5 to cohesin-mediated cohesion in human cells and Xenopus egg extracts. J Cell Sci 118: 2133–2141 [DOI] [PubMed] [Google Scholar]
- Melby TEG, Ciampaglio CN, Briscoe G, Erickson HP (1998) The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J Cell Biol 142: 1595–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH (2005) The structure and function of SMC and kleisin complexes. Annu Rev Biochem 74: 595–648 [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Hirano T (2000) HEAT repeats associated with condensins, cohesins and other chromosome-related complexes. Genome Res 10: 1445–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T (2003) Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 115: 109–121 [DOI] [PubMed] [Google Scholar]
- Saitoh N, Goldberg IG, Wood ER, Earnshaw WC (1994) ScII: an abundant chromosome scaffold protein is a member of a family of putative ATPases with an unusual predicted tertiary structure. J Cell Biol 127: 303–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, Yanagida M (1994) Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J 13: 4938–4952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Hizume K, Sutani T, Takeyasu K, Yanagida M (2003) Condensin but not cohesin SMC heterodimer induces DNA reannealing through protein–protein assembly. EMBO J 22: 2764–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savvidou E, Cobbe N, Steffensen S, Cotterill S, Heck MMS (2005) Drosophila CAP-D2 is required for condensin complex stability and resolution of sister chromatids. J Cell Sci 118: 2529–2543 [DOI] [PubMed] [Google Scholar]
- Schleiffer A, Kaitna S, Maurer-Stroh S, Glotzer M, Nasmyth K, Eisenhaber F (2003) Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol Cell 11: 571–575 [DOI] [PubMed] [Google Scholar]
- Stray JE, Lindsley JE (2003) Biochemical analysis of the yeast condensin Smc2/4 complex: an ATPase that promotes knotting of circular DNA. J Biol Chem 278: 26238–26248 [DOI] [PubMed] [Google Scholar]
- Strick TR, Kawaguchi T, Hirano T (2004) Real-time detection of single-molecule DNA compaction by condensin I. Curr Biol 14: 874–880 [DOI] [PubMed] [Google Scholar]
- Strunnikov AV (2003) Condensin and biological role of chromosome condensation. Prog Cell Cycle Res 5: 361–367 [PubMed] [Google Scholar]
- Strunnikov AV, Hogan E, Koshland D (1995) SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes Dev 9: 587–599 [DOI] [PubMed] [Google Scholar]
- Sutani T, Yanagida M (1997) DNA renaturation activity of the SMC complex implicated in chromosome condensation. Nature 388: 798–801 [DOI] [PubMed] [Google Scholar]
- Sutani T, Yuasa T, Tomonaga T, Dohmae N, Takio K, Yanagida M (1999) Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and cdc2 phosphorylation of Cut3/SMC4. Genes Dev 13: 2271–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzer S, Lehane C, Uhlmann F (2003) A model for ATP hydrolysis-dependent binding of cohesin to DNA. Curr Biol 13: 1930–1940 [DOI] [PubMed] [Google Scholar]
- Yeong FM, Hombauer H, Wendt KS, Hirota T, Mudrak I, Mechtler K, Loregger T, Marchler-Bauer A, Tanaka K, Peters J-M, Ogris E (2003) Identification of a subunit of a novel kleisin-beta/SMC complex as a potential substrate of protein phosphatase 2A. Curr Biol 13: 2058–2064 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary data


