Abstract
XRCC4 and DNA ligase IV form a complex that is essential for the repair of all double-strand DNA breaks by the nonhomologous DNA end joining pathway in eukaryotes. We find here that human XRCC4:DNA ligase IV can ligate two double-strand DNA ends that have fully incompatible short 3′ overhang configurations with no potential for base pairing. Moreover, at DNA ends that share 1–4 annealed base pairs, XRCC4:DNA ligase IV can ligate across gaps of 1 nt. Ku can stimulate the joining, but is not essential when there is some terminal annealing. Polymerase mu can add nucleotides in a template-independent manner under physiological conditions; and the subset of ends that thereby gain some terminal microhomology can then be ligated. Hence, annealing at sites of microhomology is very important, but the flexibility of the ligase complex is paramount in nonhomologous DNA end joining. These observations provide an explanation for several in vivo observations that were difficult to understand previously.
Keywords: DNA repair, DNA recombination, immunoglobulin gene rearrangement, NHEJ, V(D)J recombination
Introduction
Double-strand DNA breaks (DSBs) in mammalian cells are repaired predominantly by either nonhomologous DNA end joining (NHEJ) or homologous recombination (Ma et al, 2005a; Friedberg et al, 2006). NHEJ of DSBs is thought to begin with the binding of the heterodimer Ku to the double-stranded DNA (dsDNA) ends. Like many repair processes, NHEJ utilizes nucleases, polymerases, and ligases; and Ku functions as a ‘toolbelt' protein by recruiting each of these enzymatic components (Ma et al, 2004). The ligase complex is XRCC4:DNA ligase IV in all eukaryotes (Grawunder et al, 1997; Schar et al, 1997; Teo and Jackson, 1997; Wilson et al, 1997; Tomkinson et al, 2006). XLF or Cernunnos in mammalian cells is a homologue of Saccharomyces cerevisiae NEJ1 and forms a complex with XRCC4:DNA ligase IV, the functional consequences of which are yet to be defined (Ahnesorg et al, 2006; Buck et al, 2006; Callebaut et al, 2006). Ku improves the binding of the XRCC4:DNA ligase IV complex to DNA ends (Chen et al, 2000; NickMcElhinny et al, 2000).
Ku also improves the on-rate and slows the off-rate of DNA-PKcs from DNA ends (Gottlieb and Jackson, 1993; West et al, 1998). A significant fraction of Artemis exists in the cell in complex with DNA-PKcs, and becomes an endonuclease after it is phosphorylated by DNA-PKcs (Ma et al, 2002). There are 11 DNA-PKcs phosphorylation sites in the C-terminal half of Artemis (Ma et al, 2005b). Phosphorylation of these sites by DNA-PKcs causes a conformational change in Artemis, allowing it to trim 5′ overhangs to a blunt conformation and trim long 3′ overhangs to much shorter ones, typically about 4 nt in length (Ma et al, 2005b).
DNA polymerases (pol) are thought to be needed for any NHEJ events that require fill-in of gaps or extension of the 3′ end at 5′ overhangs (Ma et al, 2005a). POL4 is responsible for a substantial fraction of the fill-in synthesis of gaps in NHEJ events in S. cerevisiae (Wilson and Lieber, 1999). POL4 is a member of the Pol X family and is most homologous to pol mu and pol lambda in mammalian cells (Tseng and Tomkinson, 2002). The only other two known mammalian proteins in the Pol X family are pol beta and the lymphoid-specific enzyme, terminal deoxynucleotidyl transferase (TdT). Ku recruits pol mu and pol lambda to DNA ends via their BRCT domains (Ma et al, 2004). One group has proposed that pol mu can polymerize across a discontinuous template strand, whereas pol lambda was not reported to do so (NickMcElhinny et al, 2005). This ‘jumping' from one DNA end to another was proposed as a basis for why pol mu deficiency results in an altered V(D)J recombination phenotype, where joining of two DNA ends with incompatible 3′ overhangs is suspected to be common (Schlissel, 1998). However, both the pol lambda null mice and the pol mu null mice turn out to have phenotypes in V(D)J recombination (Bertocci et al, 2003, 2006), raising questions about models based on different abilities of pol mu and pol lambda to synthesize across discontinuous templates.
A fundamental question in NHEJ is the relative contribution of protein factors in configuring the ends for ligation and the role of the intrinsic terminal DNA sequence (Daley et al, 2005b). Here we find that XRCC4:DNA ligase IV plus Ku can ligate several fully incompatible DNA end configurations that do not share even 1 bp of terminal microhomology. Moreover, XRCC4:DNA ligase IV can ligate across short gaps. Terminal annealing of 1–4 bp can obviate the need for Ku, although it is often still stimulatory. When confronted with incompatible DNA ends, pol mu can add nucleotides in a template-independent manner, thereby creating random microhomology in a subset of DNA ends. Pol lambda has only a marginal template-independent polymerase activity. Hence, the template-independent activity of pol mu and the remarkable flexibility of the ligase complex permit a wide range of incompatible DNA ends to be joined via NHEJ.
Results
Pol mu has template-independent polymerase activity under physiological conditions and has a preference for pyrimidine addition
Previous work suggested that human pol mu has template-independent polymerase activity, but nearly all of this work was carried out using Mn2+ as the divalent cation (Dominguez et al, 2000; Garcia-Diaz et al, 2000; Ramadan et al, 2003, 2004; Juarez et al, 2006). Data for Mg2+ conditions were limited to a single-stranded DNA substrate (Dominguez et al, 2000) or to substrates with long (10–20 nt) 5′ overhangs (Covo et al, 2004). We were interested in testing whether pol mu has template-independent polymerase activity under physiological conditions (Mg2+ present) at dsDNA ends of the type subject to NHEJ.
Our substrates for these studies consist of 73 bp dsDNA with an additional 2 nt overhang at each 3′ end (Figure 1A). We used substrates where both ends have an –AG overhang or where both ends have a –TC overhang. The DNA substrates are 5′-radiolabeled with 32P at one end, but the other DNA end has a 5′ OH. We added pol mu to these substrates in free solution, and in selected reactions we also added Ku and XRCC4:DNA ligase IV (Figure 1B). After incubation to permit addition of nucleotides by pol mu, we analyzed the products using denaturing PAGE.
Figure 1.
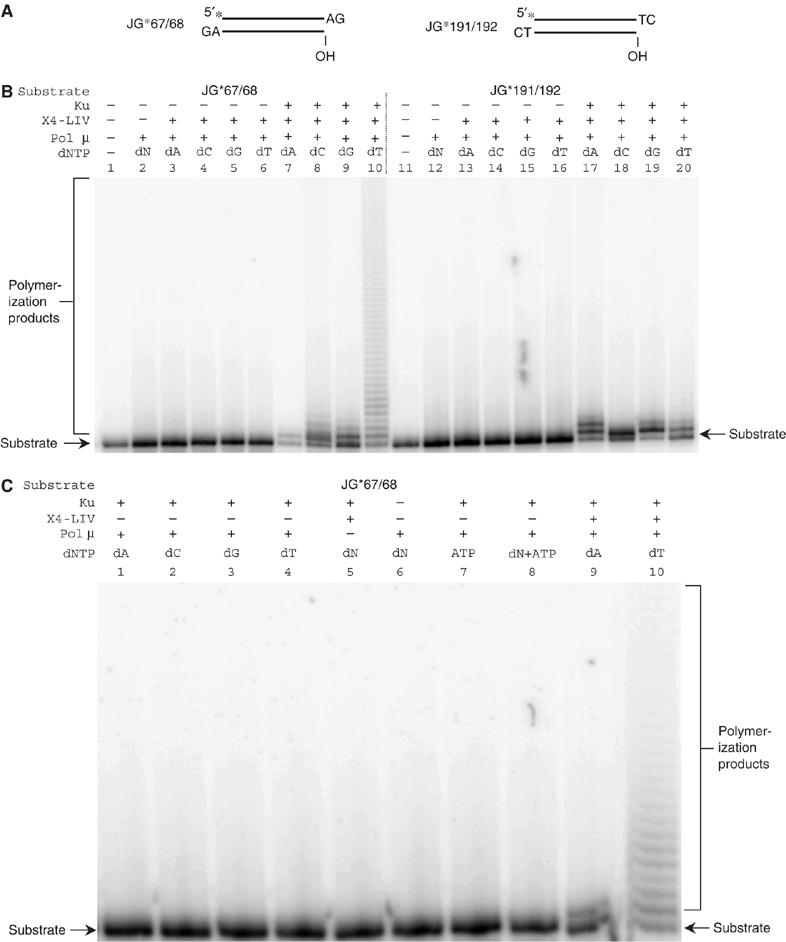
Template-independent polymerase activity of pol mu on substrates in free solution. (A) Two 73 bp substrates with 3′ overhangs were used to test for pol mu template-independent polymerase activity in free solution. An asterisk indicates the position of the radioisotope label. (B, C) In each reaction, 20 nM substrate was incubated with the protein(s) indicated above each lane in a 10 μl reaction for 1 h at 37°C. After incubation, reactions were deproteinized and analyzed using 11% denaturing PAGE. Protein concentrations: Ku, 25 nM; X4-LIV, 50 nM; pol mu, 25 nM, where X4-LIV refers to XRCC4:DNA ligase IV. The specified dNTP was added to 100 μM. ‘dN' means that all the four dNTPs (100 μM each) were included. No ATP was added, unless specified. Template-independent polymerase synthesis results in extension of the radiolabeled strand, and hence, the more slowly moving species located above the substrate band.
We found that pol mu alone had little or no template-independent activity under these conditions (Figure 1B, lanes 2 and 12). But when Ku and XRCC4:DNA ligase IV were present, we noted significant mononucleotide addition to the 3′ overhanging ends (Figure 1B, lanes 7–10 and 17–20). On the basis of single nucleotide iterations, pol mu appears to carry out distributive synthesis on both substrates here. The presence of both Ku and XRCC4:DNA ligase IV were important to achieve maximal pol mu nucleotide addition (Figure 1C, lanes 9 and 10 versus 1–8; also Figure 1B). This is consistent with the fact that Ku can bind to the BRCT domain of pol mu (Ma et al, 2004), and that XRCC4:DNA ligase IV may provide additional stability for the Ku:pol mu interaction (Mahajan et al, 2002).
Interestingly, the two types of DNA ends do not give identical results. Total nucleotide addition to the –AG end is more efficient than to the –TC end. This may reflect better initial binding of the pol mu to this terminal sequence. T is not only the obvious preferred nucleotide for the –AG end (Figure 1B, lane 10), but is seen on longer exposure to be the nucleotide associated with the longest additions at the –TC end also (Supplementary Figure 1, lane 20). However, it is important to note that all four dNTPs could be added to each of the two types of overhangs (Figure 1B, lanes 7–10 and 17–20). Otherwise, one would expect C and T addition only to the –AG end and G and A to the –TC end, and this is clearly not the case. Therefore, pol mu has template-independent activity under physiological conditions.
Polymerization by pol mu at free DNA ends is template-independent and not due to use of another DNA end as the source of a template strand
Pol mu has also well-documented template-dependent activity (Dominguez et al, 2000; Supplementary Figure 4). To definitively rule out any possibility of template-dependent addition by pol mu, we immobilized the DNA at one end to streptavidin–agarose beads using biotin. The DNA was used at a level 100- to 1000-fold below the capacity of the beads to ensure that the DNA ends could not contact one another. Moreover, we showed that the DNA ends of the bead-bound substrates could no longer ligate, verifying the highly dispersed distribution of the DNA substrate on the beads (Supplementary Figure 2B, which is a darker exposure of Figure 2). Polymerase synthesis studies were conducted similar to those described above, using the same types of DNA ends (Figure 2). We still observe template-independent addition that cannot be explained by using another DNA end as a template (NickMcElhinny et al, 2005). Specifically, pol mu could add any of the four nucleotides to the –AG or to the –TC ends (Figure 2B, lanes 2–5 and 9–12). Not surprisingly, the overall efficiency of the reaction on the bead was lower. Therefore, to observe products, we increased the amount of polymerase used. Yet we still observe that any of the four nucleotides can be added. Therefore, pol mu can add nucleotides in a template-independent manner under physiological conditions to 3′ overhangs.
Figure 2.
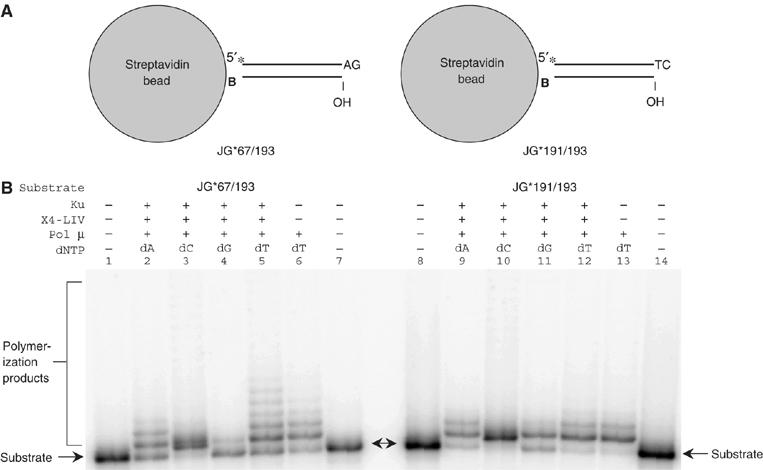
Template-independent synthesis by pol mu on immobilized DNA substrates distributed at low density on agarose beads. (A) Streptavidin agarose beads were used to immobilize two 73 bp DNA substrates with 3′ overhangs. ‘B' designates the 3′-biotin group of the substrate. An asterisk indicates the position of the radioisotope label. (B) In each reaction, 20 nM substrate was incubated with the protein(s) indicated above each lane in a 20 μl reaction for 1 h at 37°C. After incubation, reactions were heated at 100°C for 5 min to disrupt the biotin–streptavidin interaction, and then deproteinized and analyzed using 11% denaturing PAGE. Protein concentrations: Ku, 50 nM; X4-LIV, 100 nM; pol mu, 1.25 μM; dNTP, 5 mM. No ATP was added.
There are some interesting additional points to note. First, C and T are the preferred nucleotide additions at both the –AG and the –TC ends (Supplementary Figure 2, lanes 3 and 5 for the –AG ends and lanes 10 and 12 for the –TC ends). It is clear that over 20 C and T nts can be added to the free –AG ends. For the –TC end, over 20 C nts can be added, and addition of T also clearly occurs, but not as efficiently as C. Hence, pyrimidines seem to be added better than purines, regardless of the sequence of the end. The difference between the number of nucleotides added at –AG and –TC ends may relate to how the polymerase contacts the DNA at the double- to single-strand transition region.
Second, nucleotide addition by pol mu alone without Ku and XRCC4:DNA ligase IV is substantial for the immobilized substrates (Figure 2, lanes 6 and 13). This may mean that immobilization of the DNA makes it easier for pol mu to add nucleotides in a template-independent manner, even without the stabilizing influence of Ku and XRCC4:DNA ligase IV.
We did the same type of study for template-independent addition to the free end of bead-bound DNA substrates, but we replaced the dNTPs with the corresponding ribonucleotides (NTPs). Previous work had shown that pol mu can perform fill-in synthesis using NTPs (NickMcElhinny et al, 2005). However, template-independent synthesis with NTPs had not been reported. Not surprisingly, like dNTPs, the corresponding NTPs can be added in a template-independent manner (Supplementary Figure 3).
In summary, these immobilized substrate experiments demonstrate that pol mu can carry out robust template-independent synthesis under physiological conditions, and the level of this activity suggests that this is the dominant mode of addition at a free DNA end with a 3′ overhang.
We also studied human pol lambda, which previously was shown to have template-independent activity, but only in Mn2+ solutions (Ramadan et al, 2003). At equivalent molar amounts, pol lambda has a similar or even slightly higher template-dependent activity than pol mu (Supplementary Figure 4). However, when tested for template-independent activity, we observed only a very marginal level, regardless of addition of Ku or XRCC4:DNA ligase IV, and regardless of whether the DNA substrate is free in solution or immobilized on beads (data not shown; see below).
The template-independent synthesis by pol mu provides the microhomology for end ligaton by Ku, XRCC4, and DNA ligase IV
Having gained a better understanding of the polymerization properties of pol mu, we were interested in understanding the role of pol mu in the ligation of the same types of DNA ends described above (Figure 3A). For any given ligation reaction, duplex DNA substrates with 2 nt overhangs were used. One 5′ end was radiolabeled with 32P, and the other 5′ end was not ligatable because it had a 5′ OH. To carry out the ligation reactions, we added Ku, XRCC4:DNA ligase IV, dNTPs, and either pol mu or pol lambda. We incubated reactions for 30 min, deproteinized, and analyzed with denaturing PAGE.
Figure 3.
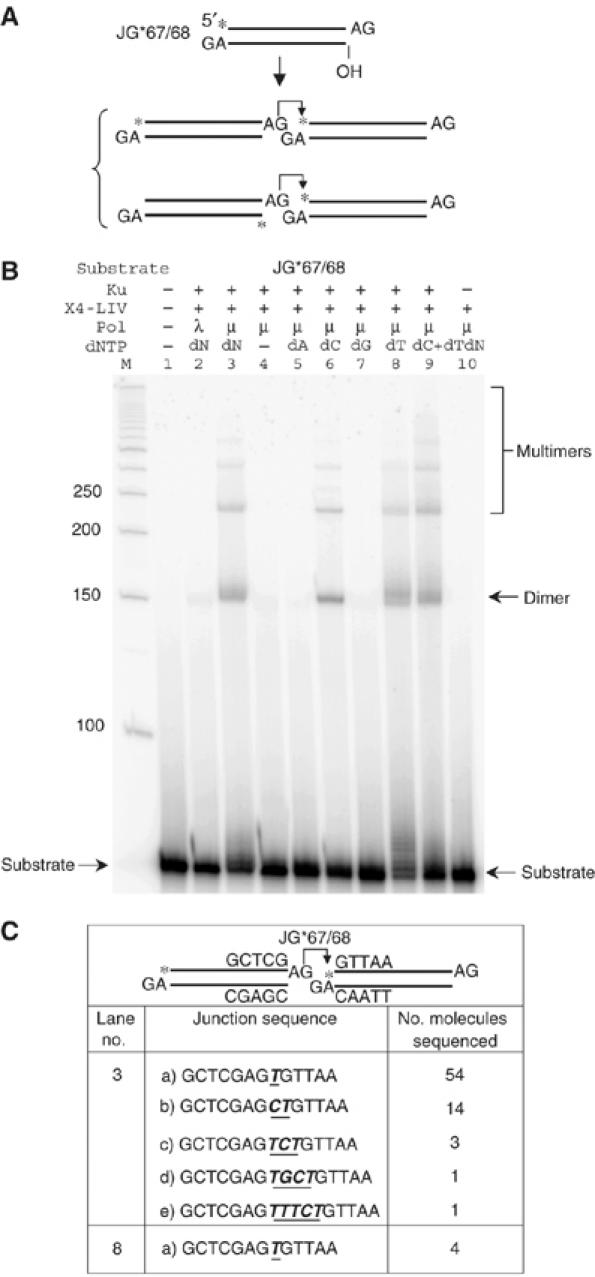
Pol mu template-independent polymerase activity provides terminal microhomology for ligation by XRCC4:DNA ligase IV. (A) The same substrate as in Figure 1A (left side) was tested for ligation. Two alternative joining pathways are proposed below the substrate. From the ligation patterns in (B) and Supplementary Figure 5, we know that the first pathway is favored. An asterisk indicates the position of the radioisotope label. (B) In each reaction, 20 nM substrate was incubated with the protein(s) indicated above each lane in a 10 μl reaction for 30 min at 37°C. After incubation, reactions were deproteinized and analyzed using 8% denaturing PAGE. Protein concentrations: Ku, 25 nM; X4-LIV, 50 nM; pol mu or lambda, 25 nM. dNTP (100 μM) was added to reactions, where indicated. ‘dN' means all the four dNTPs (100 μM each) were included. ATP (100 μM) was also added in indicated reactions. ‘M' indicates 50 bp DNA ladder. The dimer ligation product that results from the joining of two substrate molecules is labeled. Joining of more than two substrates results in trimer and higher-order species labeled as multimers. (C) Dimer products from the selected lanes were cut out of the gel, extracted, and then PCR amplified, TA-cloned, and sequenced. The junction sequences for the ligatable strand were provided. For lane 3, sequencing information was collected and combined from three individual reactions. For lane 8, two bands are apparent in the dimer product, but the longer product was not among the four molecules sequenced.
Even when Ku, XRCC4:DNA ligase IV, and pol mu are all present, we find that the ligation depends on the presence of specific dNTPs that could potentially provide 1 or 2 bp of complementarity (Figure 3B, lanes 6, 8, and 9 versus 4, 5 and 7). To confirm this, we cut out the dimer species from the gel, PCR amplified across the ligation junction, cloned into a TA cloning vector, transformed bacteria, isolated the plasmid, and sequenced the individual ligation junctions (Figure 3C). For reactions in which only dTTP is present, the junctions only contain a T addition, which provides 1 bp of terminal homology. The resulting single A:T bp is adequate to support the ligation. For reactions that contain all four dNTPs (Figure 3B, lane 3), sequencing of the dimer product shows junctions with a spectrum of additions, but predominantly a single T (Figure 3C, upper). As expected based on template-independent synthesis at a DNA end, the number of junctions with CT additions, which would provide 2 bp of terminal homology, was considerably smaller. A few junctions illustrated additional template-independent addition which ended in CT, providing complementarity, but which showed addition of other nucleotides 5′ to the CT. Most of the added nucleotides were pyrimidines, illustrating the pyrimidine preference noted earlier. Hence, in the presence of Ku and XRCC4:DNA ligase IV, pol mu can support template-independent addition at DNA ends, and the resulting subset of ends that acquire complementarity can now be ligated.
Substrates with a –TC overhang can be ligated when dATP or dGTP are provided (Supplementary Figure 1B, lanes 17 and 19). When the dimer ligation products are sequenced, the expected A or G nts are present, consistent with the terminal microhomology for joining being provided by template-independent addition.
The above results indicate that 1 bp of annealing between two DNA ends is sufficient to support ligation, even when the other strand remains in an unligatable configuration. Given this, we reasoned that if we provide that one specific base pair of terminal microhomology in the starting substrate, then the ligation should proceed even without any polymerase present. We found that, indeed, this is the case (Figure 4B, lane 4 on both panels). A substantial amount of dimer and higher ligation multimers can be formed when the one DNA end overhang is –AGT and is ligated to a DNA end with an incompatible 3′ overhang (Figure 4B, right panel, lane 3). The bottom strand in this case would remain unligated for three reasons: because there is no 5′ phosphate at the junction; there is a gap on the bottom strand; and the right bottom strand has a 1 nt 3′ flap.
Figure 4.
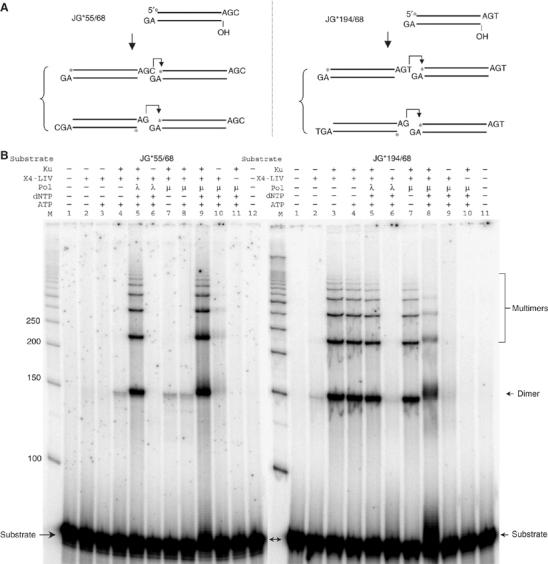
One base pair of terminal microhomology is sufficient for direct ligation by XRCC4:DNA ligase IV. (A) Two substrates with only 1 bp of terminal microhomology for ligation were designed, based on the Figure 3A substrate, to test the direct ligation by XRCC4:DNA ligase IV. Two alternative joining products are proposed below each substrate. From both the ligation patterns in (B) and Supplementary Figure 5, we know that the upper product is favored over the lower product. An asterisk indicates the position of the radioisotope label. (B) In each reaction, 20 nM substrate was incubated with the protein(s) indicated above in a 10 μl reaction for 30 min at 37°C. After incubation, reactions were deproteinized and analyzed by 8% denaturing PAGE. Protein concentrations: Ku, 25 nM; X4-LIV, 50 nM; pol mu or lambda, 25 nM. Twenty-five micromolars of each dNTP were added to the reaction as indicated. ATP (100 μM) was also added in indicated reactions. ‘M' indicates 50 bp DNA ladder.
Surprisingly, even an –AGC 3′ overhang on the top strand can ligate to a GA– 3′ overhang end to form a small, but obvious amount of dimer (Figure 4B, left panel, lane 4). This is unanticipated because the C of the –AGC must ligate across a 1 nt gap to achieve ligation. The amount of ligation was lower than that seen for the –AGT substrate above, perhaps because of the difficulty in ligating across the 1 nt gap (Figure 4B, compare left panel, lane 4 versus right panel, lane 4). The efficiency of the ligation for the –AGT substrate explains the large number of products in which a T is added when the overhang is –AG (Figure 3C, upper, junction sequence a versus b–e). These studies confirm that random addition of a single complementary nucleotide is sufficient to support ligation, and suggests that this occurs even if this ligation must occur across a 1 nt gap.
When we tested a corresponding amount of pol lambda relative to pol mu, as assessed by template-dependent synthesis (Supplementary Figure 4), pol lambda supported only a low level of ligation (Figure 3B, lane 2). We sequenced the small number of junctions formed (from Figure 3B, lane 2, dimer position), and these had T, CT, or GT additions, consistent with a very low level of template-independent addition, some of which provided 1 bp of terminal microhomology.
XRCC4:DNA ligase IV can ligate across gaps
To test formally whether XRCC4:DNA ligase IV could ligate across a gap, we examined substrate ligation by this ligase complex with or without other components (Figure 5). The XRCC4:DNA ligase IV complex alone could ligate a 4 bp terminal microhomology with a 1 nt gap in the ligatable strand (Figure 5B, left panel, lane 2). This ligase complex could also ligate across a 1 nt gap using a 2 bp terminal microhomology (Figure 5B, right panel, lane 2 and Supplementary Figure 6, lane 4). Sequencing of junctions confirmed the ligation across gaps (Figure 5C).
Figure 5.
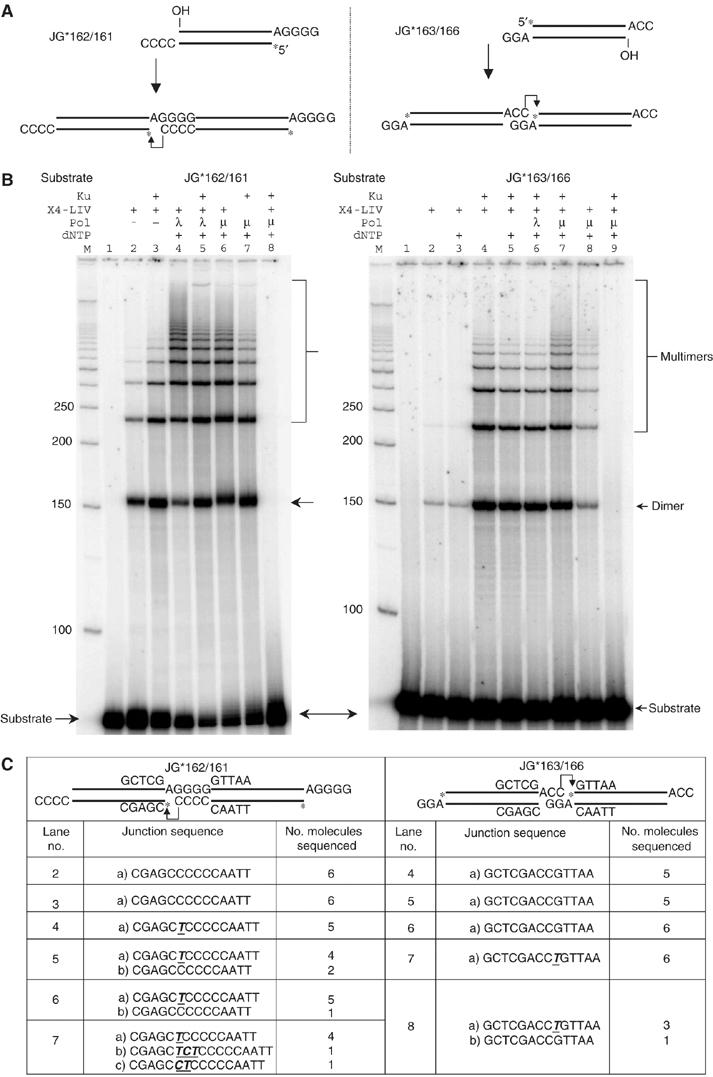
XRCC4:DNA ligase IV can ligate over a gap. (A) Two substrates with different lengths of terminal microhomology for ligation were designed to test the direct ligation over a gap by XRCC4:DNA ligase IV. There is a one-nucleotide gap on the ligatable strand. Only the favored joining product is shown under each substrate. An asterisk indicates the position of the radioisotope label. (B) Reactions were performed as in Figure 4B, except that all the reactions include 100 μM of ATP. (C) Dimer products from the selected lanes were cut out of the gel, extracted, and then PCR amplified, TA-cloned, and sequenced. The junction sequences for the ligatable strand were provided.
Hence, the XRCC4:DNA ligase IV can support ligation across gaps in the absence of other proteins.
Ku stimulated the ligation across a gap (Figure 5B, left panel, lane 3, and right panel, lanes 4 and 5). Addition of pol lambda or pol mu further stimulated the ligation (Figure 5B, left panel, lanes 4–7), but this was because addition of T occurred (sometimes along with additional nucleotides), as documented by sequencing (Figure 5C, left, sequences from lanes 4 to 7, or right, sequences from lanes 7 and 8; Supplementary Figure 6).
The rate of ligation of ends that have gaps adjacent to 2 bp of terminal microhomology is roughly 10 times as slow as the rate of ligation of a nick adjacent to a 4 bp block of terminal microhomology (Supplementary Figure 7).
XRCC4:DNA ligase IV can ligate fully incompatible DNA ends
We were interested in the minimal amount of terminal microhomology needed to support ligation (Figure 6A). We found that, in some cases, Ku and XRCC4:DNA ligase IV were sufficient to complete the ligation of entirely incompatible DNA ends (Figure 6B, lanes 4 and 5 versus 1–3). Sequences of the dimer species confirmed that the ligation occurred with no alteration of either DNA end (see Figure 6C, sequences from the dimer ligation product in lanes 4 and 5).
Figure 6.
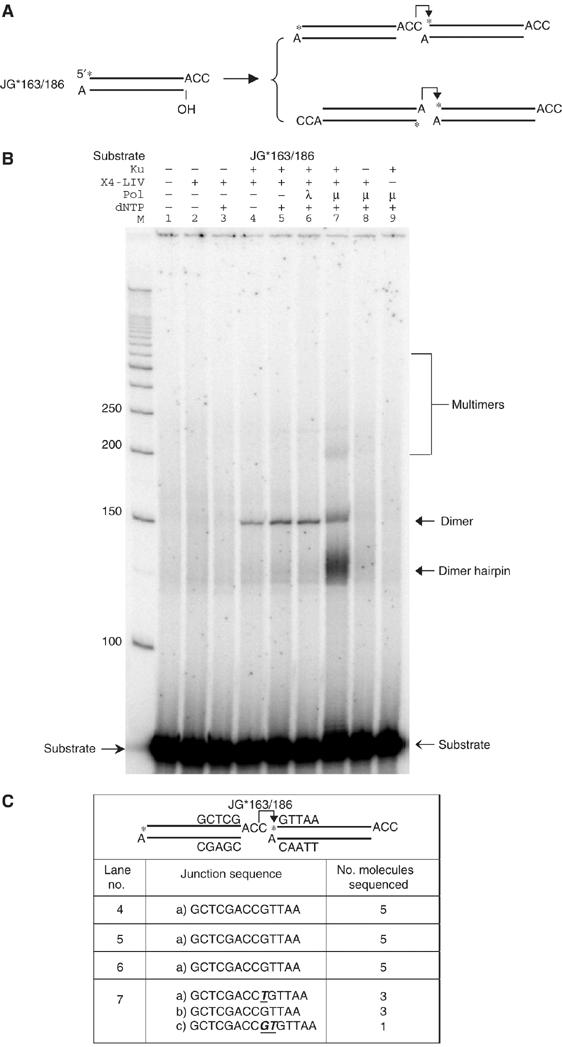
XRCC4:DNA ligase IV and Ku can ligate fully incompatible DNA ends. (A) A substrate without any homology for ligation was designed on the basis of the second substrate in Figure 5A to test the ligation with XRCC4:DNA ligase IV. Two alternative joining pathways are proposed next to the substrate. An asterisk indicates the position of the radioisotope label. (B) Reactions were performed as in Figure 4B, except that all the reactions include 100 μM of ATP. In lane 7, the band below the dimer product is most likely the hairpin structure of the dimer product that is ligated in the manner we proposed in (A), second product. (C) Dimer products from the selected lanes were cut out of the gel, extracted, and then PCR amplified, TA-cloned, and sequenced. The junction sequences for the ligatable strand were provided.
In addition to the ligation of this incompatible DNA end pair, we also observed that a DNA end with a 3′ overhanging A could be ligated to an end with a 3′ overhanging –AGC (data not shown). Another example of the ligation of two incompatible ends is the joining of two DNA ends, each with a 3′ A overhang (Supplementary Figure 8). Hence, Ku and XRCC4:DNA ligase IV can ligate a subset of fully incompatible ends that do not share even 1 bp of terminal microhomology.
The rate of ligation of DNA ends that are incompatible with no terminal microhomology is obviously much slower than for ends that are stabilized by two terminal base pairs or four terminal base pairs plus stacking (Supplementary Figure 7). This is not surprising given the stabilization and alignment contributed by such terminal blocks of microhomology.
How specific is the ability of XRCC4:DNA ligase IV to ligate across gaps at DNA ends and to ligate incompatible DNA ends? We were interested in comparing ligation of various DNA ends by equivalent levels of activity of T4 DNA ligase, human DNA ligase I, DNA ligase III, and DNA ligase IV (plus XRCC4 and Ku). We used a nicked duplex oligonucleotide substrate to establish equal activity levels of each of these four ligases (see Materials and methods; data not shown). We then compared the ligation of four different substrate end configurations (Figure 7). The ligation of a nick adjacent to a 4 bp block of terminal microhomology is similar for all four ligases (Figure 7B, lanes 1–5). However, XRCC4:DNA ligase IV is distinctly better at ligating end pairs that have a 1 nt gap adjacent to a 4 bp block of microhomology (Figure 7B, lanes 6–10) or adjacent to a 2 bp block of microhomology (Figure 7C, lanes 1–5). For incompatible DNA ends with no microhomology, XRCC4:DNA ligase IV ligation was detectable, whereas ligation by the other ligases was undetectable (Figure 7C, lanes 6–10). Ku has no effect on the activity of DNA ligase I or III in these ligations (Figure 7B–D).
Figure 7.
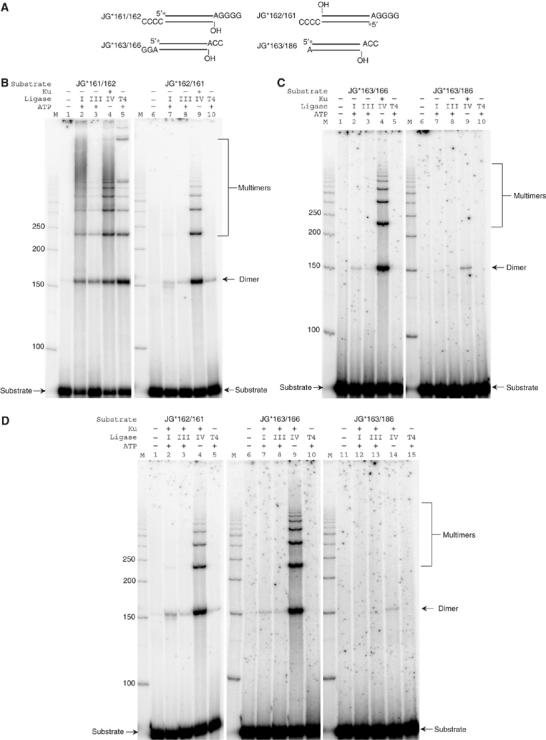
Ligase activity comparison with DNA double-strand break substrates. (A) JG*161/162 is a substrate with a nick on the ligatable strand. JG*162/161 is a substrate with a 1 nt gap on the ligatable strand and has 4 bp of terminal microhomology for ligation. JG*163/166 is a substrate with a 1 nt gap on the ligatable strand and has a 2 bp terminal microhomology for ligation. JG*163/186 is a substrate with fully incompatible ends for ligation. A star indicates the position of the radioisotope label. (B), (C), and (D), in each reaction, 20 nM substrate was incubated with the protein(s) indicated above in a 10 μl reaction for 30 min at 37°C. After incubation, reactions were deproteinized and analyzed by 8% denaturing PAGE. Protein concentrations: Ku, 25 nM; X4-LIV, 50 nM; human ligase I, human ligase III, and T4 DNA ligase were first normalized to the same activity of 50 nM X4-LIV using a single-strand nick substrate, and then used in equivalent amounts of activity for the reactions shown. One millimolar of ATP was also added in specified reactions. ‘M' indicates a 50 bp DNA ladder. Ligase abbreviations: I, human ligase I; III, human ligase III; IV, human XRCC4-ligase IV; T4, T4 DNA ligase.
Although many pairs of ends lacking microhomology can be ligated by Ku and XRCC4:DNA ligase IV, this was not universal. We found that when both ends had 3′ overhangs of two incompatible nucleotides, then joining did not occur (Supplementary Figure 9, lane 5). In summary, pairs of DNA ends that have very short, fully incompatible 3′ overhangs can be ligated by Ku and XRCC4:DNA ligase IV, but longer overhangs cannot be ligated.
Discussion
Ligation of incompatible DNA ends by Ku and XRCC4:DNA ligase IV
Ligation without any terminal microhomology markedly expands the role of Ku and XRCC4:DNA ligase IV in NHEJ. Previously, we and others assumed that nucleases and polymerases were essential to bring two DNA ends to a point where each strand would have a ligatable nick. Although nucleases and polymerases markedly expand the number of ways in which two DNA ends can be ligated, and thereby improve the efficiency of ligation, we now see that many simple overhang combinations may be joined with neither a nuclease nor a polymerase (Figure 8).
Figure 8.
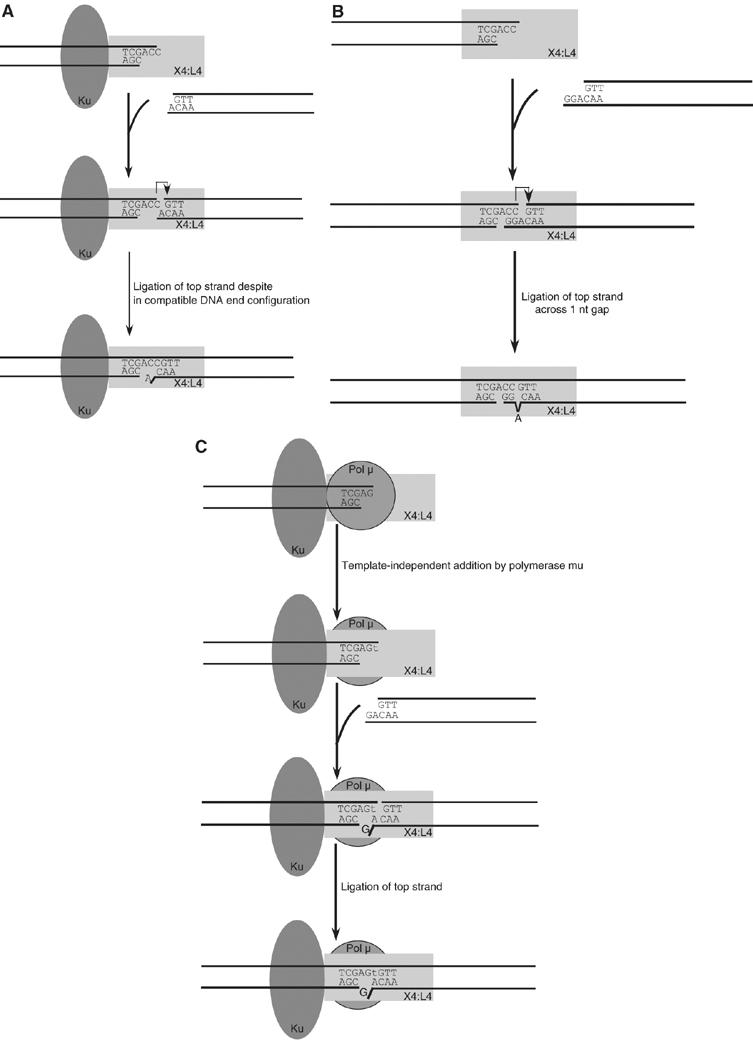
Function of XRCC4:DNA ligase IV in ligating incompatible DNA ends. (A) Ku and XRCC4:DNA ligase IV can ligate fully incompatible DNA ends. XRCC4:DNA ligase IV randomly bind to one DNA end, but this interaction is less stable than if Ku is also bound. Ku may stimulate the interaction by increasing the occupancy time of XRCC4:DNA ligase IV at the DNA end. When another DNA end comes close to this complex, XRCC4:DNA ligase IV binds it and ligates the two ends. (B) XRCC4:DNA ligase IV alone can ligate across a gap. XRCC4:DNA ligase IV randomly binds to one DNA end (although this interaction is not as stable as when Ku is present). The 2 bp of terminal microhomology between the DNA ends increases the chance for XRCC4:DNA ligase IV to ligate. (C) Template-independent polymerase activity of pol mu creates terminal microhomology for ligation by XRCC4:DNA ligase IV. Pol mu can add nucleotides to the DNA end in its template-independent mode. A 1 bp terminal microhomology between the DNA ends (‘t' in this example) permits annealing of the ends and improves the efficiency of ligation.
These biochemical findings fit well with important recent in vivo findings. First, after ionizing radiation, the large majority of DSBs are thought to be incompatible. Recently, it was suggested that only a subset of the DNA ends require the endonucleolytic activity provided by the Artemis:DNA-PKcs complex (Riballo et al, 2004). This observation can now be understood in light of the findings here that Ku and XRCC4:DNA ligase IV may ligate many of the incompatible DNA end configurations generated by the ionizing radiation.
Second, cells from mice fully deficient for both pol mu and pol lambda are not sensitive to ionizing radiation (Bertocci et al, 2006). We and others had previously assumed that this was because of the possible involvement of other polymerases (Wilson and Lieber, 1999). However, the finding that Ku and XRCC4:DNA ligase IV can ligate many incompatible DNA ends without a polymerase provides one clear biochemical basis for why the pol mu/pol lambda double null cells would not be sensitive to ionizing radiation.
Third, the template-independent addition at short 3′ overhangs under physiological conditions finally provides an explanation for a very puzzling aspect of data from V(D)J recombination junctions in TdT−/− mice. Such mice can have template-independent addition at nearly 5% of their V(D)J junctions (Gilfillan et al, 1993; Komori et al, 1993; Bertocci et al, 2006). The source of these template-independent additions has been unclear. The template-independent addition by pol mu (and perhaps a much lower level by pol lambda) can now be seen to explain these in vivo observations.
Incompatible DNA ends with 3′ overhangs are perhaps the most important type of incompatible configuration in the repair of DSBs by NHEJ, because other configurations can be ligated more simply. For example, fill-in synthesis by any of a number of polymerases, including pol mu and pol lambda, can convert 5′ overhangs to blunt configurations, and we previously showed that DNA ligase IV alone can ligate blunt ends (Grawunder et al, 1997; Schar et al, 1997; Teo and Jackson, 1997; Wilson et al, 1997; Tomkinson et al, 2006). Moreover, 5′ overhangs can be endonucleolytically cleaved to a blunt configuration by Artemis:DNA-PKcs or exonucleolytically by Artemis alone (Ma et al, 2002). In contrast, 3′ overhangs (or hairpins in V(D)J recombination) are cleaved by Artemis:DNA-PKcs in a manner that typically leaves a short residual 3′ overhang, which cannot be readily filled in by a polymerase (Ma et al, 2002). Hence, direct ligation of a subset of short incompatible 3′ overhangs by Ku and the ligase complex is important. The only alternative manner of joining is to carry out template-independent addition, which we have demonstrated here for pol mu (and has been previously demonstrated for TdT, in the case of V(D)J recombination (Ramadan et al, 2004)).
Prokaryotic NHEJ systems appear to rely primarily on Ku and a ligase activity with nuclease or polymerases playing roles for only specific types of DNA end configurations (Della et al, 2004; Gong et al, 2005). In yeast NHEJ, Ku and the Lif1:DNA ligase IV complex play major roles in NHEJ, whereas POL4 plays a discernable, but nonessential role (Wilson and Lieber, 1999). As is the case for prokaryotes, a nuclease may be required for only a subset of yeast NHEJ events (Downs and Jackson, 2004; Daley et al, 2005b). Therefore, the ability to handle a diverse set of overhangs using Ku and a ligase complex may have its origins early in evolution, with the polymerases and nucleases contributing to subsets of DNA end configurations to improve joining in selected cases.
Ligation across gaps by XRCC4:DNA ligase IV
XRCC4:DNA ligase IV is distinctive in its ability to ligate across gaps at DNA ends. Studies of ligation by T4 DNA ligase across 1 nt gaps within duplex DNA did not show any data on incompatible DNA end ligation, which is the type shown here (Goffin et al, 1987). The manner in which XRCC4:DNA ligase IV binds the substrate must be quite flexible to tolerate not only the normal nick ligation, but also ligation across 1 nt gaps. The flexibility at the junction could arise from flexibility in the position of the 3′OH and the 5′P at the nick. Alternatively, the nucleotides across from the gap on the anti-parallel strand could be forced into an extrahelical position. Obviously, there are many variations between these two extremes, which are all equally plausible, given that the junctional DNA is unlikely to be in standard B-DNA conformation. The recent cryo-EM studies using Ku and DNA-PKcs suggested that the two DNA ends might be off-set (Spagnolo et al, 2006). Some degree of off-set or alternative nonlinear alignment of the ends (or at least tolerance for such angles) could also facilitate ligation across gaps.
Pol mu template-independent and -dependent addition
In this study, we have shown that pol mu can add nucleotides in a template-independent manner under physiological conditions. The duplex substrates were immobilized on beads at a density that is 100- to 1000-fold lower than the bead capacity, ensuring that synapsis of DNA ends is driven to a negligible level (Yu and Lieber, 2000), and we have shown that ligation is no longer detectable (Supplementary Figure 2). The T, A, G , and C are all added at similar efficiencies to –TC ends (Figure 2, right). A darker exposure shows that more than 15 C or T nts can be added (Supplementary Figure 2).
Interestingly, the immobilized substrate with a –AG overhang also shows addition of all four dNTPs at the DNA terminus, consistent with template-independent addition (Figure 2, left). As described above, a darker exposure shows that runs of either C or T can exceed 15 nts in length (Supplementary Figure 2). The similarity between the –TC and –AG ends in the propensity for C and T addition indicates that there is a bias in favor of pyrimidines in the template-independent addition. Pol mu synthesis in Mn2+ also favors pyrimidine addition at the 3′ end of single-stranded DNA substrates (Dominguez et al, 2000).
When we compare pol mu polymerization at the DNA termini of immobilized DNA fragments with that found for DNA substrates in free solution, the results are indistinguishable for the ends terminating with a –TC 3′ overhang.
There is substantially more pol mu addition of T for the –AG substrate in free solution than for the immobilized −AG substrate, even though multiple rounds of T addition are clearly occurring even at the ends of the immobilized –AG substrate as well. We do not yet know the basis for the extremely efficient T addition, exceeding 40 nt, but it might reflect a combination of several factors: (a) the pyrimidine bias of pol mu template-independent activity; (b) some difference in the ability of pol mu to bind to some overhang sequences versus others; and (c) the frequency of productive encounters between pol mu and the DNA substrate. This last factor may contribute to the difference between the immobilized and free –AG substrates, whereas more than one factor may contribute to the difference between the free –AG and free –TC substrates.
Km measurements for pol mu have been carried out using fill-in synthesis substrates and show tighter binding of dATP and dGTP relative to dTTP (NickMcElhinny and Ramsden, 2003). These measurements may not apply to the template-independent additions by pol mu, given its preference for use of dCTP and dTTP.
The synapsis of DNA ends for ligation by XRCC4:DNA ligase IV
We find here that some pairs of incompatible DNA ends can be ligated with only XRCC4:DNA ligase IV, and others either require Ku to improve the efficiency of joining or are stimulated by Ku. Ku is known to improve the binding of XRCC4:DNA ligase IV to DNA ends (Chen et al, 2000; NickMcElhinny et al, 2000), and the stimulatory effect of Ku for the joining of some pairs of ends may simply reflect this. One could wonder if Ku might actually be the synapsis factor, as was suggested by previous work using purified Ku and DNA (Ramsden and Gellert, 1998). However, synapsis activity of purified Ku has not been seen by others (DeFazio et al, 2002)(Y Ma and MRL, unpublished), nor has a significant ligation stimulation of ligase I and III by Ku (Figure 7 and data not shown).
Moreover, it is quite clear that Ku becomes dispensable when there is terminal annealing of partially compatible DNA ends. The more the annealing occurs, the more negligible the effect of Ku (Figures 5A, B and 6 show the progression from 0, 2, and 4 bp of microhomology). Neither 2 nor 4 bp of terminal annealing would be sufficient to provide a stable synapsis during the 37°C incubations here. Rather, XRCC4:DNA ligase IV appears adequate to provide synapsis without Ku, and terminal microhomology may contribute to the overall stability of the two ends within the XRCC4:DNA ligase IV complex. When there is no terminal microhomology, the addition of Ku may stimulate joining by increasing the occupancy time of XRCC4:DNA ligase IV on either DNA end. Longer occupancy time by XRCC4:DNA ligase IV at either end would then give more opportunity for the partner DNA end to be encountered, bound, and the two ends ligated. However, formal equilibrium and kinetic studies will be needed to discern the precise contribution of each protein and the extent of end annealing to the overall stability of the junctional complex.
We have tested purified DNA-PKcs in our system for its effect on incompatible DNA end joining by Ku plus XRCC4:DNA ligase IV in the presence of pol mu (Supplementary Figure 10). There is only a two-fold increase in the end joining efficiency upon addition of DNA-PKcs. Nevertheless, further study of additional proteins on incompatible DNA end joining is clearly warranted.
Differences in template-dependent addition by pol mu versus pol lambda
Pol mu can improve the joining efficiency of some pairs of ends by XRCC4:DNA ligase IV or Ku:XRCC4:DNA ligase IV. Sequencing shows that a subset of such junctions show direct joining without nucleotide addition; but most show some nucleotide addition, and this addition provides the additional microhomology, which is the basis for the increased joining efficiency.
For some pairs of DNA ends, pol lambda does not fill gaps as efficiently as pol mu. For example, in Figure 6, lane 6, we see no addition by pol lambda (no T added). But pol mu does sometimes add a T or a GT. Part of this difference may be compensated by an increase in the concentration of the polymerase. This is illustrated in Figure 5B, right panel, where pol lambda does not fill in at the concentration of pol lambda shown, but when we increase the concentration of pol lambda five-fold, we do see some ends with addition (Figure 5). This may account for in vivo observations of human pol mu and pol lambda in S. cerevisiae deficient for POL4 (Daley et al, 2005a). The stronger ability of pol mu versus pol lambda could account for joining events where pol mu fills in gaps (after removal of unpaired regions), whereas pol lambda fails to do so (Daley et al, 2005a).
Differences in template-independent addition among pol mu, pol lambda, and TdT
We observe only weak template-independent addition by pol lambda, whereas we observe that pol mu has substantial template-independent addition. Some structural differences between pol mu and pol lambda have been suggested to be due, in part, to variation in a region called loop 1 of these Pol X polymerases (Delarue et al, 2002; NickMcElhinny et al, 2005; Juarez et al, 2006). Loop 1 may fill part of the polymerase catalytic region occupied by a template strand, thereby causing variation in the balance of the template-dependent and template-independent modes.
Although variation in loop 1 may account for differences among pol lambda, pol mu, and TdT, our results differ markedly from previous work regarding the role of pol mu in NHEJ (NickMcElhinny et al, 2005). Previous work suggested that pol mu could polymerize across a discontinuous template strand, effectively jumping from one DNA end to another (see Figure 6A, parts d and e of NickMcElhinny et al, 2005). The data supporting such a mechanism were based on ddNTP utilization by pol mu. However, we would interpret more recent data of this type to indicate that pol mu can add any of the four ddNTPs in a template-independent manner (Figure 8 of Moon et al, 2007). More importantly, a mutation of H329 decreases template-independent synthesis (without an effect on template-dependent synthesis) and simultaneously decreases NHEJ (Moon et al, 2007). This supports the view, based on our data here, that pol mu adds nucleotides template-independently at 3′ overhangs (at junctions lacking any terminal microhomology), thereby permitting annealing between the two DNA ends. Hence, our data indicate a primary role for end annealing (provided by the template-independent activity of pol mu when there is no microhomology) rather than the ability of pol mu to jump or polymerize across a gap in the template strand.
In vivo, pro-B cells derived from mice lacking both pol mu and TdT expression, and still expressing pol lambda, have a very low but detectable level of template-independent addition at their Ig heavy chain junctions (see bottom portion of Supplementary Figure S1 of Bertocci et al, 2006), consisting mostly of pyrimidine addition, particularly T. The low level of template-independent addition by pol lambda that we see may correspond to this.
It is interesting that the Ig heavy chain rearrangement involves pol lambda and TdT, but not pol mu, whereas Ig light chain rearrangement involves pol mu but neither pol lambda nor TdT (Bertocci et al, 2006). Pol mu has robust template-dependent and -independent activity, whereas pol lambda has markedly greater template-dependent synthesis than template-independent synthesis. Hence, the heavy chain rearrangement involves two separate proteins that provide template-dependent (pol lambda) and template-independent activity (TdT), whereas light chain rearrangement involves pol mu, which has both activities in the same polymerase.
Materials and methods
Oligonucleotides
See Supplementary data.
Protein purification
See Supplementary data.
Template-independent polymerization on substrates in free solution
See Supplementary data.
Template-independent polymerization on immobilized DNA substrates
See Supplementary data.
DNA ligation assay
The DNA ligation assay was performed in a 10 μl reaction. DNA substrates (20 nM) were first incubated with or without protein Ku in 1 × ligation reaction buffer supplemented with 10% PEG (MW>8000 kDa), 50 μg/ml BSA, and 5% glycerol at room temperature for 15 min. Ligation was initiated by adding 100 μM of each dNTP and 10 mM MgCl2 with different combinations of proteins as indicated. Reactions were then incubated at 37°C for 30 min. See Supplementary data for more details.
Sequencing
See Supplementary data.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Dr Chih-Lin Hsieh, Dr SC Raghavan, Dr Kefei Yu, Dr Yunmei Ma, Dr S Sasaki, Feng-Ting Huang, Deepankar Roy, and Albert Tsai for advice. This work was supported by NIH grants to MRL.
References
- Ahnesorg P, Smith P, Jackson SP (2006) XLF interacts with the XRCC4-DNA ligase IV complex to promote nonhomologous end-joining. Cell 124: 301–313 [DOI] [PubMed] [Google Scholar]
- Bertocci B, DeSmet A, Weill J-C, Reynaud CA (2006) Non-overlapping functions of polX family DNA polymerases, pol μ, pol λ, and TdT, during immunoglobulin V(D)J recombination in vivo. Immunity 25: 31–41 [DOI] [PubMed] [Google Scholar]
- Bertocci B, Smet AD, Berek C, Weill J-C, Reynaud C-A (2003) Immunoglobulin kappa light chain gene rearrangement is impaired in mice deficient for DNA polymerase mu. Immunity 19: 203–211 [DOI] [PubMed] [Google Scholar]
- Buck D, Malivert L, deChasseval R, Barraud A, Fondaneche M-C, Xanal O, Plebani A, Stephan J-L, Hufnagel M, LeDiest F, Fischer A, Durrandy A, Villartay J-Pd, Revy P (2006) Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 124: 287–299 [DOI] [PubMed] [Google Scholar]
- Callebaut I, Malivert L, Fischer A, Mornon JP, Revy P, Villartay JPd (2006) Cernunnos interacts with the XRCC4/DNA-ligase IV complex and is homologous to the yeast nonhomologous end-joining factor NEJ1. J Biol Chem 281: 13857–13860 [DOI] [PubMed] [Google Scholar]
- Chen L, Trujillo K, Sung P, Tomkinson AE (2000) Interactions of the DNA ligase IV-XRCC4 complex with DNA ends and the DNA-dependent protein kinase. J Biol Chem 275: 26196–26205 [DOI] [PubMed] [Google Scholar]
- Covo S, Blanco L, Livneh Z (2004) Lesion bypass by human DNA polymerase mu reveals a template-dependent, sequence-independent nucleotidyl transferase activity. J Biol Chem 279: 859–865 [DOI] [PubMed] [Google Scholar]
- Daley JM, Laan RLV, Suresh A, Wilson TE (2005a) DNA joint dependence of pol X family polymerase action in nonhomologous end joining. J Biol Chem 280: 29030–29037 [DOI] [PubMed] [Google Scholar]
- Daley JM, Palmbos PL, Wu D, Wilson TE (2005b) Nonhomologous end joining in yeast. Ann Rev Genet 39: 431–451 [DOI] [PubMed] [Google Scholar]
- DeFazio LG, Stansel RM, Griffith JD, Chu G (2002) Synapsis of DNA ends by DNA-dependent protein kinase. EMBO J 21: 3192–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M, Boule JB, Lescar J, Expert-Bezancon N, Jourdan N, Sukumar N, Rougeon F, Papanicolaou C (2002) Crystal structures of a template-independent DNA polymerase: murine terminal deoxynucleotidyltransferase. EMBO J 21: 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Della M, Palmbos PL, Tseng HM, Tonkin LM, Daley JM, Topper LM, Pitcher RS, Tomkinson AE, Wilson TE, Doherty AJ (2004) Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science 306: 683–685 [DOI] [PubMed] [Google Scholar]
- Dominguez O, Ruiz JF, Lera TLd, Garcia-Diaz M, Gonzalez MA, Kirchhoff T, Martinez C, Bernad A, Blanco L (2000) DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J 19: 1731–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JA, Jackson SP (2004) A means to a DNA end: the many roles of Ku. Nat Rev Mol Cell Biol 5: 367–378 [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T (2006) DNA Repair and Mutagenesis. Washington DC: ASM Press [Google Scholar]
- Garcia-Diaz M, Dominguez O, Lopez-Fernandez LA, Lera LTd, Saniger ML, Ruiz JF, Parraga M, Garcia-Ortiz MJ, Kirchoff T, Mazo Jd, Bernad A, Blanco L (2000) DNA polymerase lambda, a novel eukaryotic DNA polymerase with potential role in meiosis. J Mol Biol 301: 851–867 [DOI] [PubMed] [Google Scholar]
- Gilfillan S, Dierich A, Lemeur M, Benoist C, Mathis D (1993) Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science 261: 1755–1759 [DOI] [PubMed] [Google Scholar]
- Goffin C, Bailly V, Verly WG (1987) Nicks 3′ or 5′ to AP sites or to mispaired bases, and one-nucleotide gaps can be sealed by T4 DNA ligase. Nucl Acids Res 15: 8755–8771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Bongiorno P, Martins A, Stephanou N, Zhu H, Shuman S, Glickman MS (2005) Mechanism of nonhomologous end-joining in mycobacteria: a low-fidelity repair system driven by Ku, ligase D, and ligase C. Nat Struct Mol Biol 12: 304–312 [DOI] [PubMed] [Google Scholar]
- Gottlieb T, Jackson SP (1993) The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell 72: 131–142 [DOI] [PubMed] [Google Scholar]
- Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR (1997) Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature 388: 492–495 [DOI] [PubMed] [Google Scholar]
- Juarez R, Ruiz JF, NickMcElhinny SA, Ramsden D, Blanco L (2006) A specific loop in human DNA polymerase mu allows switching between creative and DNA-instructed synthesis. Nucleic Acids Res 34: 4572–4582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T, Okada A, Stewart V, Alt F (1993) Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science 261: 1171–1175 [DOI] [PubMed] [Google Scholar]
- Ma Y, Lu H, Schwarz K, Lieber MR (2005a) Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: the iterative processing model. Cell Cycle 4: 1193–2000 [DOI] [PubMed] [Google Scholar]
- Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh C-L, Schwarz K, Lieber MR (2004) A biochemically defined system for mammalian nonhomologous DNA end joining. Mol Cell 16: 701–713 [DOI] [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Lu H, Niewolik D, Schwarz K, Lieber MR (2005b) The DNA-PKcs phosphorylation sites of human artemis. J Biol Chem 280: 33839–33846 [DOI] [PubMed] [Google Scholar]
- Ma Y, Pannicke U, Schwarz K, Lieber MR (2002) Hairpin opening and overhang processing by an Artemis:DNA-PKcs complex in V(D)J recombination and in nonhomologous end joining. Cell 108: 781–794 [DOI] [PubMed] [Google Scholar]
- Mahajan KN, NickMcElhinny SA, Mitchell BS, Ramsden DA (2002) Association of DNA polymerase mu with Ku and DNA ligase IV: role of in end-joining double-strand break repair. Mol Cell Biol 22: 5194–5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon AF, Garcia-Diaz M, Bebenek K, Davis BJ, Zhong X, Ramsden DA, Kunkel TA, Pedersen LC (2007) Structural insight into the substrate specificity of DNA polymerase mu. Nat Struct Mol Biol 14: 45–53 [DOI] [PubMed] [Google Scholar]
- NickMcElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA (2005) A gradient of template dependence defines distinct biological roles for family x polymerases in nonhomologous end joining. Mol Cell 19: 357–366 [DOI] [PubMed] [Google Scholar]
- NickMcElhinny SA, Ramsden DA (2003) Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol Cell Biol 23: 2309–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NickMcElhinny SA, Snowden CM, McCarville J, Ramsden DA (2000) Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol Cell Biol 20: 2996–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan K, Maga G, Shevelev IV, Villani G, Blanco L, Hubscher U (2003) Human DNA polymerase lambda possesses terminal deoxyribonucleotidyl transferase activity and can elongate RNA primers: implications for novel functions. J Mol Biol 328: 63–72 [DOI] [PubMed] [Google Scholar]
- Ramadan K, Shevelev IV, Maga G, Hubscher U (2004) De novo DNA synthesis by human DNA polymerase lambda, DNA polymerase mu, and terminal deoxynucleotidyl transferase. J Mol Biol 339: 395–404 [DOI] [PubMed] [Google Scholar]
- Ramsden DA, Gellert M (1998) Ku protein stimulates DNA end joining by mammalian DNA ligases: a direct role for Ku in repair of DNA double-strand breaks. EMBO J 17: 609–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riballo E, Kuhne M, Rief N, Doherty A, Smith GCM, Recio M-J, Reis C, Dahm K, Fricke A, Kempler A, Parker AR, Jackson SP, Gennery A, Jeggo PA, Lobrich M (2004) A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell 16: 715–724 [DOI] [PubMed] [Google Scholar]
- Schar P, Herrmann G, Daly G, Lindahl T (1997) A newly identified DNA ligase of S. cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev 11: 1912–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel MS (1998) Structure of nonhairpin coding-end DNA breaks in cells undergoing V(D)J recombination. Mol Cell Biol 18: 2029–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O (2006) Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol Cell 22: 511–519 [DOI] [PubMed] [Google Scholar]
- Teo SH, Jackson SP (1997) Identification of S. cerevisiae DNA ligase IV: involvement in DNA double-strand break repair. EMBO J 16: 4788–4795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkinson AE, Vijayakumar S, Pascal JM, Ellenberger T (2006) DNA ligases: structure, reaction mechanism, and function. Chem Rev 106: 687–699 [DOI] [PubMed] [Google Scholar]
- Tseng HM, Tomkinson AE (2002) A physical and functional interaction between yeast Pol4 and Dnl4-Lif1 links DNA synthesis and ligation in nonhomologous end joining. J Biol Chem 277: 45630–45637 [DOI] [PubMed] [Google Scholar]
- West RB, Yaneva M, Lieber MR (1998) Productive and nonproductive complexes of Ku and DNA-PK at DNA termini. Mol Cell Biol 18: 5908–5920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TE, Grawunder U, Lieber MR (1997) Yeast DNA ligase IV mediates non-homologous DNA end joining. Nature 388: 495–498 [DOI] [PubMed] [Google Scholar]
- Wilson TE, Lieber MR (1999) Efficient processing of DNA ends during yeast nonhomologous end joining: evidence for a DNA polymerase beta (POL4)-dependent pathway. J Biol Chem 274: 23599–23609 [DOI] [PubMed] [Google Scholar]
- Yu K, Lieber MR (2000) The nicking step of V(D)J recombination is independent of synapsis: implications for the immune repertoire. Mol Cell Biol 20: 7914–7921 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


