Abstract
The C/EBPα transcription factor regulates hepatic nitrogen, glucose, lipid and iron metabolism. However, how it is able to independently control these processes is not known. Here, we use mouse knock-in mutagenesis to identify C/EBPα domains that specifically regulate hepatic gluconeogenesis and lipogenesis. In vivo deletion of a proline–histidine rich domain (PHR), dephosphorylated at S193 by insulin signaling, dysregulated genes involved in the generation of acetyl-CoA and NADPH for triglyceride synthesis and led to increased hepatic lipogenesis. These promoters bound SREBP-1 as well as C/EBPα, and the PHR was required for C/EBPα-SREBP transcriptional synergy. In contrast, the highly conserved C/EBPα CR4 domain was found to undergo liver-specific dephosphorylation of residues T222 and T226 upon fasting, and alanine mutation of these residues upregulated the hepatic expression of the gluconeogenic G6Pase and PEPCK mRNAs, but not PGC-1α, leading to glucose intolerance. Our results show that pathway-specific metabolic regulation can be achieved through a single transcription factor containing context-sensitive regulatory domains, and indicate C/EBPα phosphorylation as a PGC-1α-independent mechanism for regulating hepatic gluconeogenesis.
Keywords: C/EBP, gluconeogenesis, lipogenesis, metabolism, transcription
Introduction
The liver is central to metabolic control, and processes, stores and releases glucose, nitrogen metabolites and lipids. It is also central to detoxification and participates in iron metabolism. Adjusting the production and consumption of metabolites to the influx provided by nutrition requires coordinated regulation of groups of genes (regulons) involved in a given metabolic pathway. Thus, sugar intake leads to the suppression of hepatic glucose production and initiation of energy storage in glycogen and lipids. This process is controlled by pancreatic insulin release, and involves the transcriptional suppression of hepatic enzymes rate-limiting for gluconeogenesis (phosphoenol pyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6Pc)) through insulin response elements (IREs) in their promoters (O'Brien et al, 2001). This pathway can be activated during fasting by cAMP/CREB-mediated induction of PGC-1α, which acts as a coactivator for Foxo1 to upregulate a gluconeogenic regulon consisting of PEPCK, G6Pc and fructose-1,6-bisphosphatase (FBP-1) (Yoon et al, 2001). Similarly, lipogenesis is regulated by dedicated transcription factors, the sterol regulatory element binding proteins (SREBPs), which coordinately regulate a large group of genes required for lipid biosynthesis through their cognate binding sites (SREs). SREBP-2 primarily activates cholesterol biosynthetic genes, whereas SREBP-1 preferentially upregulates genes involved in fatty acid production (Horton et al, 2003). SREBPs are activated by proteolysis in the endoplasmatic reticulum, a process that is inhibited by sterols, providing negative feedback regulation. In addition, SREBP-1 is transcriptionally induced by insulin signaling, and repressed by the glucagon/cAMP pathway, providing a means to control energy storage in triglycerides according to food supply (reviewed by Horton et al, 2002).
C/EBPα is of particular interest to the understanding of how the repertoire of hepatic transcription factors can provide independent control of multiple regulons participating in distinct processes, as it has been genetically demonstrated to regulate enzymes important for several metabolic pathways. Loss-of-function studies have shown that at birth C/EBPα is essential for gluconeogenesis (Wang et al, 1995). In adult mice conditional knockout experiments have shown that C/EBPα plays an important role in hepatic glucose, nitrogen, bile acid and iron metabolism. The expression of carbamoyl-phosphate synthase-1 (CPS-1), a rate-limiting enzyme in urea synthesis, and of glucokinase (GcK), rate limiting for hepatic glucose uptake, is strongly decreased in the absence of C/EBPα (Kimura et al, 1998; Inoue et al, 2004). Decreased expression of the bile-metabolizing Br-UGT and UGT-1 (Lee et al, 1997), and of the iron metabolic regulator hepcidin (Courselaud et al, 2002), has also been observed in liver-specific C/EBPα knock-out models. However, how C/EBPα co-operates with other hepatic transcriptional regulators to endow its target genes with distinct regulatory properties, and how it responds to metabolic signaling that requires divergent regulation of target gene expression remain unclear.
C/EBPα contains four highly conserved regions (CR1–4; Ross et al, 1999) in its transactivation domain. CR1–3 interact with the basal transcriptional apparatus (Nerlov and Ziff, 1995), the p300/CBP histone acetyl transferases (Erickson et al, 2001; Kovacs et al, 2003) and the SWI/SNF chromatin-remodeling complex (Pedersen et al, 2001). In contrast, little is known about the function of CR4, which is the most highly conserved among vertebrates. CR4 contains a GSK3 consensus consisting of residues T222, T226 and S230, phosphorylation of which has been proposed to regulate adipogenesis in response to insulin signaling (Ross et al, 1999). A serine residue (S193 in the mouse) is present adjacent to CR4. Phosphorylation of S193 has been reported to control the ability of a proline–histidine rich (PHR) Cdk2/4-binding motif (encompassing residues 180–194) to regulate cell cycle progression of hepatoma cells (Wang et al, 2004). Phosphorylation of S193 was required for Cdk2/4–C/EBPα interaction, which was inhibited by dephosphorylation by PP2A through its activation via the insulin–PI3K-Akt pathway. Insulin signaling therefore controls multiple C/EBPα phosphorylation sites, suggesting a role for these residues in regulating distinct subsets of C/EBPα target genes.
In order to address the in vivo role of phosphorylated C/EBPα domains in the control of cell proliferation and metabolic processes, we generated two mouse strains: one in which S193 and the associated PHR Cdk2/4 interaction motif have been deleted (ΔPHR mice), and one in which the GSK3 phosphorylation sites have been eliminated by changing T222, T226 and S230 to alanines (TTS mice). In vivo, these two mutations affected expression of specific subsets of hepatic genes: in the ΔPHR mice enzymes controlling the production of metabolites (acetyl-CoA and NADPH) required for fatty acid synthesis and β-oxidation were dysregulated in the liver, but not in adipose tissue, leading to increased hepatic triglyceride production. Dysregulated promoters bound both C/EBPα and SREBP-1 and these two factors were found to interact and to cooperate in promoter activation in a PHR-dependent manner. The C/EBPα GSK3 consensus was phosphorylated in all C/EBPα-expressing tissues examined in the fed state, and upon fasting, specifically dephosphorylated in the liver. Simulating the fasted state through alanine mutation of the GSK3 consensus resulted in increased C/EBPα activity of the IRE-controlled G6Pc promoter. In vivo, this mutation led to hepatic upregulation of IRE-controlled genes, including G6Pc and PEPCK, resulting in glucose intolerance. These results show that in vivo C/EBPα uses distinct regulatory motifs to control transcription of genes involved in glucose and lipid metabolic pathways in the liver. Such mechanisms may provide a general means for transcription factors to independently regulate multiple metabolic pathways and other biological processes.
Results
Generation of mice lacking the proline–histidine rich C/EBPα Cdk2/4 interaction motif
CR4 is almost invariant in vertebrate C/EBPα proteins (Figure 1A). In addition to a GSK3 consensus contained within the CR4 a serine (corresponding to S193 in the mouse) is consistently present N-terminally to CR4, embedded in a proline–histidine-rich motif (PHR motif). An in-frame deletion of C/EBPα amino acids 180–194 was introduced into the Cebpa open reading frame in E14.1 embryonic stem cells using the targeting strategy previously described (Porse et al, 2001). Mice heterozygous for the mutant allele (designated ΔPHR) were generated by standard techniques. Intercrosses between CebpaΔPHR/+ mice yielded homozygous mutant mice (ΔPHR mice) at the expected Mendelian ratio (Figure 1B). ΔPHR mice were viable, fertile and displayed no obvious physiological abnormalities. Western blot analysis of liver nuclear extracts confirmed that the ΔPHR allele gave rise to C/EBPα proteins slightly smaller than, but equal in abundance to, the p42 and p30 translation isoforms produced from the wild-type (WT) allele (Figure 1C, a). However, no increase in expression of the PCNA proliferation marker was seen in ΔPHR livers compared with WT controls, indicating that hepatocyte proliferation was not increased (Figure 1C, b). In addition, no differences in the amount and histological appearance of white adipose tissue (Figure 1D and E) were observed. A more detailed analysis of the cell cycle regulatory and granulocyte differentiation properties of the ΔPHR mutant showed no differences compared with WT C/EBPα (Porse et al, 2006). The PHR motif encoded by C/EBPα amino acids 180–194, thus, did not appear to be required for the normal development or proliferation control of the major C/EBPα-expressing tissues.
Figure 1.
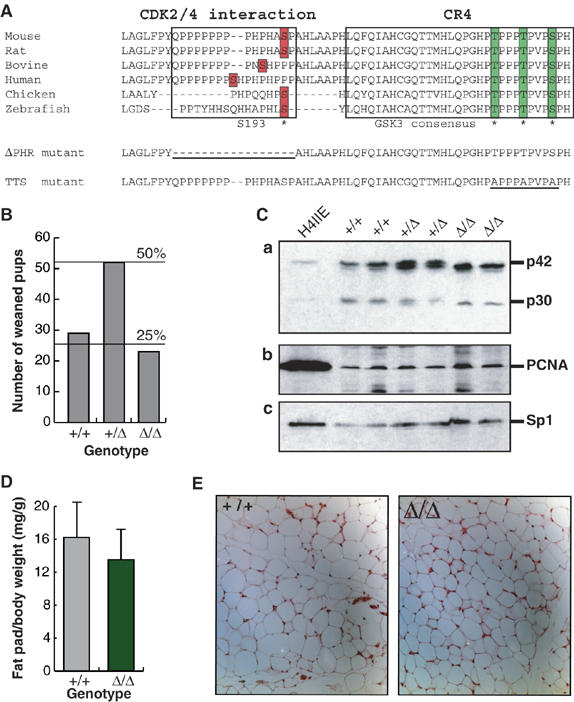
C/EBPα PHR motif is dispensable for WAT development and control of hepatocyte proliferation. (A) Alignment of C/EBPα CR4 fragments from vertebrates. The region required for CDK2/4 interaction (PHR motif) and the CR4 are indicated. The bars and * denote residues shown to be dephosphorylated in response to insulin signaling. The sequences of the ΔPHR and TTS alleles are shown, with the altered area underlined. The position of the first amino acid shown is indicated. (B) Ratios of weaned pups from 15 litters (104 pups) from intercrosses between CebpaΔPHR/+ mice. (C) Levels of C/EBPα (panel a) and PCNA (panel b) in rat H4IIE hepatoma cells and mouse liver were determined by Western blotting. Protein from approximately 3 × 105 H4IIE cells or from nuclei of 2 mg liver was loaded. The Western blot was normalized by probing with Sp1 antibody (panel c). The bands corresponding to C/EBPα p42 and p30 are indicated. (D) Ratio of epididymal fat pad (mg) and total body (g) weight of fed wild-type control (+/+; N=9) or ΔPHR (Δ/Δ; N=8) mice. (E) Sections of epididymal fat pads from control (+/+) and ΔPHR (Δ/Δ) mice stained with hematoxylin–eosin.
Liver-specific deregulation of lipogenic gene expression in ΔPHR mice
Given the absence of a detectable cell-cycle or differentiation defect in ΔPHR mice, and the observation that dephosphorylation of S193 is induced by insulin signaling in hepatocytes (Wang et al, 2004), we analyzed whether expression of liver genes regulated by the metabolic state was affected in mice lacking the PHR motif. Affymetrix DNA microarray analysis was performed on total liver RNA. A greater than two-fold change in expression was consistently seen for 30 entries (Table I). The upregulated genes (23) were generally expressed at low levels, several were of unknown function and no correlation with any known function of C/EBPα was evident. In contrast, of the seven downregulated entries, six represented four enzymes involved in the biosynthesis of fatty acids: ATP citrate lyase (ACL), malic enzyme (ME), acetyl-CoA synthase 2 (ACAS2) and glutamate-oxaloacetate transaminase (GOT1). Of these ACL, ACAS2 and ME are involved in generating cytoplasmic acetyl-CoA and NAPDH for fatty acid biosynthesis, and ME and GOT1 in the regeneration of mitochondrial TCA cycle metabolites to replace the citrate consumed by ACL. ACL, ACAS2, ME and GOT1 thus constitute a coherent functional unit (designated the PHR regulon) that generates metabolites essential for fatty acid biosynthesis.
Table 1.
Genes regulated by PHR domain in vivo
| Affy ID | Gene description | Ratio |
|---|---|---|
| Downregulated in ΔPHR liver | ||
| 1416632_at | Malic enzyme, supernatant | 5.20 |
| 1430307_a_at | Malic enzyme, supernatant | 3.72 |
| 1422479_at | Acetyl-coenzyme A synthetase 2 (ADP forming) | 3.26 |
| 1425326_at | ATP citrate lyase | 2.80 |
| 1450970_at | Glutamate oxaloacetate transaminase 1, soluble | 2.78 |
| 1422478_a_at | Acetyl-coenzyme A synthetase 2 (ADP forming) | 2.72 |
| 1434216_a_at | DNA segment, Chr 7, Roswell Park 2 complex, expressed | 2.35 |
| Upregulated in ΔPHR liver | ||
| 1435716_x_at | Small nuclear ribonucleoprotein N | 0.44 |
| 1449109_at | Suppressor of cytokine signaling 2 | 0.41 |
| 1438676_at | Expressed sequence AI595338 | 0.35 |
| 1436058_at | Mus musculus transcribed sequences | 0.35 |
| 1424126_at | Aminolevulinic acid synthase 1 | 0.35 |
| 1449009_at | T-cell specific GTPase | 0.34 |
| 1416443_a_at | Ubiquitin-like 1 (sentrin) activating enzyme E1A | 0.34 |
| 1418837_at | RIKEN cDNA 2410027J01 gene | 0.30 |
| 1449793_at | AV329014 RIKEN | 0.29 |
| 1417185_at | Lymphocyte antigen 6 complex, locus A | 0.29 |
| 1420835_at | RIKEN cDNA 4933433D23 gene | 0.29 |
| 1425127_at | Hydroxysteroid dehydrogenase-2, delta<5>-3-beta | 0.26 |
| 1448724_at | Cytokine inducible SH2-containing protein | 0.25 |
| 1425120_x_at | RIKEN cDNA 2310061N23 gene | 0.24 |
| 1428022_at | cDNA sequence BC027556 | 0.21 |
| 1456212_x_at | BB831725 RIKEN | 0.19 |
| 1450018_s_at | RIKEN cDNA 4933433D23 gene | 0.19 |
| 1418835_at | Pleckstrin homology-like domain, family A, member 1 | 0.17 |
| 1420836_at | RIKEN cDNA 4933433D23 gene | 0.17 |
| 1423696_a_at | RIKEN cDNA 2400006A19 gene | 0.10 |
| 1434496_at | Cytokine inducible kinase | 0.08 |
| 1425394_at | cDNA sequence BC023105 | 0.07 |
| 1418086_at | Protein phosphatase 1, regulatory (inhibitor) subunit 14A | 0.06 |
| Affymetrix M430A arrays were used to analyze total liver RNA from two wild-type and two ΔPHR mice (fed state). Genes consistently up- or downregulated in the ΔPHR (i.e. two-fold change in all +/+ versus ΔPHR pairwise comparisons), and which were scored as ‘present' or ‘marginally present' in both of the samples with the highest expression are shown. Ratio indicates the ratio of normalized values (+/+/ΔPHR). | ||
The downregulation of mRNAs encoding these four biosynthetic enzymes was confirmed by real-time RT–PCR analysis (Figure 2A). There was no general deregulation of fatty acid synthetic genes, as SREBP-1 and fatty acid synthase (FAS) mRNA expression was unaffected by ΔPHR mutation, as was expression and processing of SREBP-1 (Figure 2B). Likewise, no change in the expression of CPS-1 was seen. However, using real-time PCR, we found that expression of mRNA encoding GcK was significantly upregulated in the ΔPHR mice. As C/EBPα S193 dephosphorylation was observed in adipose cells upon insulin stimulation (Wang et al, 2004), the expression of ACL and ME was analyzed in adipose tissue from WT and ΔPHR mice. This showed no difference (Figure 2C), nor was expression of GLUT4, FAS, or the differentiation markers PPARγ, aP2 and adipsin altered, indicating a hepatocyte-specific requirement for this C/EBPα motif. Analysis of a number of gluconeogenic and IRE-regulated genes (PEPCK, G6Pc, FBP-1, glucose transporter-2 (GLUT2), glycogen synthase (GS), tyrosine aminotransferase (TAT) and insulin-like growth factor binding protein-1 (IGFBP-1)) (Figure 2D) did not identify any that were significantly altered in ΔPHR liver, indicating that the mutation specifically affected fatty acid metabolic genes.
Figure 2.
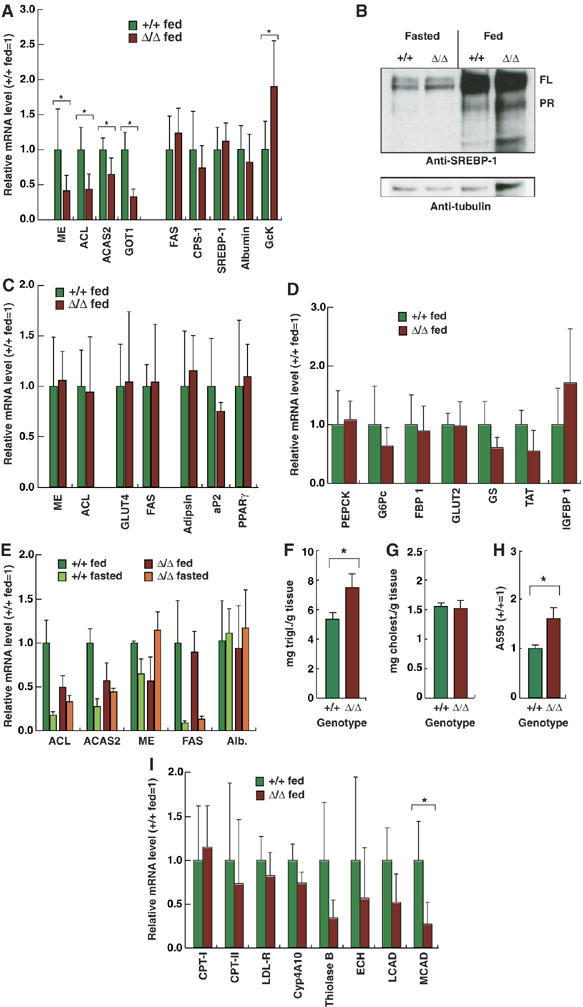
Deregulation of hepatic lipogenic gene expression in ΔPHR mice. (A) Relative mRNA expression of the indicated genes determined by RT–PCR was determined as described in Materials and methods using cDNA prepared from livers of wild-type control (+/+) or ΔPHR mice (N⩾5). The expression level of mRNA in control mice was set to 1. (B) Western blot of liver nuclear extract (100 μg/lane) from fed and overnight fasted +/+ and ΔPHR mice using anti-SREBP-1 (upper panel) and anti-tubulin (lower panel) antibodies. The positions of full-length (FL) and proteolytically cleaved (PR) SREBP-1c are indicated. (C) Relative mRNA expression of the indicated genes determined as in (A) using cDNA prepared from epididymal fat pads (N⩾5). (D) Relative mRNA expression of the indicated genes determined as in A (N⩾5). (E) Relative mRNA expression of the indicated genes determined by RT–PCR as in (A) using cDNA prepared from livers of at least four fed and four overnight fasted (16 h) control (+/+) and ΔPHR mice. (F) Triglyceride content was determined in livers of fed control (+/+; N=3) or ΔPHR (N=3) mice. (G) Cholesterol content in livers of fed control (+/+; N=4) or ΔPHR (N=4) mice. (H) Total triglyceride content of primary hepatocytes isolated from fed control (+/+) or ΔPHR mice. Triplicate cultures of each genotype were analyzed. (I) Relative mRNA expression of the indicated genes involved in lipid metabolism was determined by RT–PCR using cDNA prepared from livers of fed wild-type control (+/+) or ΔPHR mice as in (A) (N⩾5). *Indicated a significant difference (P<0.05, Student's t-test) from the wildtype value.
When hepatic metabolic regulation of ME, ACL and ACAS2 was analyzed, all were found to be downregulated upon fasting in the liver of WT mice. However, in ΔPHR mice, metabolic regulation of ACL and ACAS2 was severely compromised, and in the case of ME, inverted; FAS regulation was preserved (Figure 2E). The PHR motif thus appeared to be specifically required in the liver for coordinated metabolic regulation of genes encoding enzymes generating acetyl-CoA and NADPH for fatty acid biosynthesis. The consequence of this deregulation was evaluated by measuring the hepatic content of triglycerides and cholesterol. It was found that hepatic triglyceride, but not cholesterol, was significantly increased (Figure 2F and G). In addition, primary hepatocytes in ΔPHR mice displayed an increase in lipid content similar to that observed in vivo, when cultured under serum free conditions (Figure 2H). Analysis of genes involved in fatty acid transport and degradation showed that, whereas LDL receptor (LDL-R), Cyp4A10, carnitine-palmitoyl transferase (CPT)-I and -II were unaffected, a clear trend toward lower levels of the β-oxidation enzymes long-chain acyl-CoA dehydrogenase (LCAD), medium-chain acyl-CoA dehydrogenase (MCAD), enoyl-CoA hydratase (ECH) and thiolase B was observed (decrease only significant for MCAD; Figure 2I), indicating that C/EBPα PHR deletion affects rate-limiting steps in hepatic triglyceride production and degradation, resulting in a net increase in hepatic triglyceride levels. Consistent with the lack of effect on gluconeogenic gene expression, blood glucose levels and glucose tolerance were normal in ΔPHR mice (data not shown), indicating intact glucose homeostasis.
C/EBPα and SREBP-1 coregulate PHR-dependent genes
Of the genes identified as dysregulated in ΔPHR mice ACL, ME and ACAS2 have been previously described as SREBP target genes (Horton et al, 2003). Chromatin immunoprecipitation (ChIP) of mouse liver showed association of both C/EBPα and SREBP-1 with the ACL and ACAS2 promoters. In contrast, the G6Pc, IGFBP-1 and GcK promoters showed strong C/EBPα association and no or little SREBP-1 binding (Figure 3A). No binding to the β-globin promoter was detected. Re-ChIP experiments confirmed that SREBP-1 and C/EBPα co-occupied the ACAS2 promoter (Figure 3B); consistent with the ChIP analysis, only weak C/EBPα-SREBP-1 co-occupancy of the GcK promoter was detected. These results suggested a functional interaction between C/EBPα and SREBP on PHR-dependent promoters. Consistent with this, SREBP-1c co-immunoprecipitated with C/EBPα in transiently transfected S293 cells (Figure 3C). This interaction was not PHR-dependent. We next used a knock-in mouse strain in which a tandem affinity purification (TAP) tag (Rigaut et al, 1999) has been fused to the C terminus of C/EBPα to determine whether it associates with SREBP-1 in the liver in vivo. Nuclear extracts from WT (+/+) and tagged (T/+) mice were subjected to pull-down with a protein A matrix in the presence or absence of rabbit immunoglobulin (Ig), which induces the association of the protein A moiety of the TAP tag with the matrix through its Fc domain. This led to the selective pull down of TAP-tagged C/EBPα from nuclear lysates, as assayed by Western blotting with an anti-C/EBPα antibody (Figure 3D, lower panel; weak pull-down in the absence of added Ig is likely due to endogenous mouse Ig). Western blotting using an anti-SREBP-1 antibody showed similar levels of processed nuclear SREBP-1 in both lysates, but SREBP-1 was detected only in the pull-down from T/+ mice (Figure 3D, upper panel), providing evidence that SREBP-1 and C/EBPα form a complex in normal liver.
Figure 3.
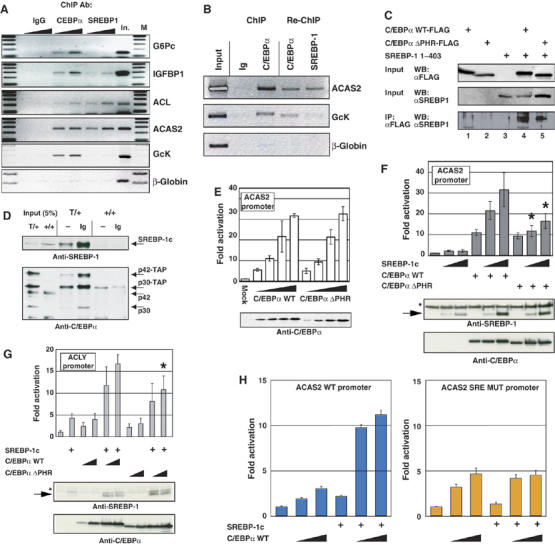
Co-localization and cooperation of C/EBPα and SREBP-1 on PHR-dependent promoters. (A) Chromatin immunoprecipitation on increasing amounts of X-linked liver extracts from fed mice was performed as described in the Materials and methods section using 4 μg anti-C/EBPα, anti-SREBP1 or unspecific rabbit IgG. Input material corresponding to 8 mg liver (In.) was loaded as control. M: 100-bp DNA marker. Each experiment was performed at least twice and representative results are shown. (B) ChIP and re-ChIP were performed as described in the Supplementary data section. Precipitated material from five ChIP reactions using anti-C/EBPα as described in (A), was pooled to perform the second round of ChIP. (C) S293 cells were transfected with expression vectors encoding the N-terminal 403 amino acids of SREBP1c and Flag-tagged full-length C/EBPα or C/EBPαΔPHR, as indicated. Input and anti-FLAG immunoprecipitates were analyzed by Western blotting using the indicated antibodies. (D) Liver nuclear extract (2 mg) from mice with the indicated genotypes were subjected to pull-down in the absence (–) or presence (Ig) of 1 mg rabbit IgG. Bound protein, as well as 5% of the Input (100 μg) was subjected to Western blotting with anti-C/EBPα and anti-SREBP-1 (2A4) antibodies. (E) S293 cells were cotransfected with a pACAS2-LUC and increasing amounts of expression vectors encoding wild-type C/EBPα (WT) or C/EBPαΔPHR, as indicated. Luciferase activity was measured 40 h post transfection, and are expressed as fold activation relative to the basal level of the reporter. C/EBPα expression levels were determined by Western blotting. Western blots where normalized by probing with anti-tubulin antibody (not shown). (F) S293 cells were cotransfected with pACAS2-LUC, expression vectors encoding WT C/EBPα or C/EBPαΔPHR and increasing amount of vector encoding (SREBP1c 1–403) as indicated (N=5). Luciferase activity and protein expression were determined as in (E). (G) S293 cells were cotransfected with pACL-LUC, expression vectors encoding SREBP1c (1–403) and increasing amounts of vector encoding WT C/EBPα or C/EBPαΔPHR as indicated. Luciferase activity and protein expression was determined as in E (N=4). (H) S293 cells were cotransfected with pACAS2-LUC (left panel) or pACAS2mSRE-LUC and expression vectors encoding SREBP1c (1–403) and increasing amounts of vector encoding C/EBPα as indicated (N=3). Luciferase activity was determined as in (E). *Indicated a significant difference (P<0.05, Student's t-test) from the wildtype value.
WT C/EBPα and C/EBPα ΔPHR did not differ in their capacity to directly activate the ACAS2 promoter (Figure 3E). However, whereas WT C/EBPα was found to synergize efficiently with SREBP-1c in ACAS2 promoter activation, the ΔPHR allele was impaired in this function (Figure 3F). Similar results were obtained using the ACL promoter (Figure 3G). The ACAS2 promoter contains numerous SREs. We mutated those previously found to be most important for promoter activity in hepatoma cells (Ikeda et al, 2001), which are found in the vicinity of the C/EBP consensus binding site (see Supplementary Figure S1), and found that this ablated C/EBPα–SREBP synergy (Figure 3H). The PHR motif was therefore not required for C/EBPα and SREBP-1 to interact, but rather to functionally cooperate in promoter activation when associated with adjacent binding sites.
Liver-specific metabolic regulation of C/EBPα GSK3 site phosphorylation
In parallel to generation of the ΔPHR knock-in mice, a triple alanine mutation (T222A, T226A and S230A, designated TTS allele) was introduced into the mouse germ line. Also in this case, homozygous mice (TTS mice) were obtained from intercrosses between CebpaTTS/+ mice at the expected Mendelian frequency (Figure 4A), and TTS mice had no apparent physiological abnormalities. White adipose tissue, lung tissue and granulopoiesis were normal in both amount and morphology, compared with control mice containing an equivalent knockin of WT C/EBPα (WT mice; Porse et al, 2001) (Figure 4B and C and data not shown). In these experiments the WT mice were preferred owing to possible confounding effects of minor amino-acid differences, due to the presence of rat sequences in the knock-in construct, on the phosphorylation state analysis shown below; +/+ controls behaved identically in all cases tested.
Figure 4.
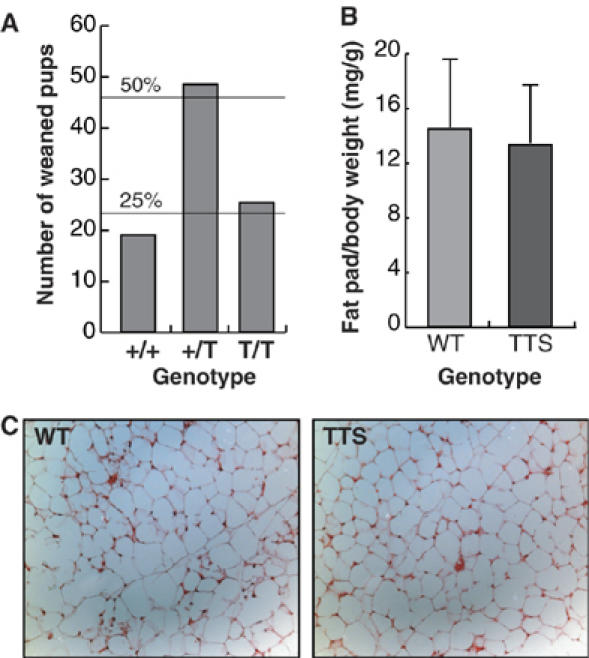
TTS mice are viable and show normal adipocyte development. (A) Ratios of weaned pups from 11 litters (93 pups) from intercrosses between cebpaTTS/+ mice. (B) Ratio of epididymal fat pad (mg) and total body (g) weight of fed WT (N=7) or TTS (N=7) mice. (C) Sections of epididymal fat pads from WT and TTS mice stained with hematoxylin–eosin.
To assess the metabolic regulation of C/EBPα phosphorylation at the GSK3 consensus in vivo, we analyzed three C/EBPα-expressing tissues (liver, adipose tissue and lung) from normally fed TTS mice, as well as from fed and fasted WT mice by Anderson PAGE, which allows resolution of phosphorylated states. We found that C/EBPα from livers of fasted WT mice co-migrated with C/EBPα from TTS mice (Figure 5A, Liver panel), indicating that in this metabolic state hepatic C/EBPα is dephosphorylated. Instead, in livers of fed WT, mice slower migrating, phosphorylated C/EBPα was detectable. This metabolic regulation of C/EBPα phosphorylation on the GSK3 consensus sites was seen only in the liver. In adipose and lung tissue, C/EBPα phosphorylation was constitutive, demonstrating liver-specific metabolic regulation of C/EBPα GSK3 consensus phosphorylation. To confirm that the observed differences reflected phosphorylation of the GSK3 consensus we generated monoclonal antibodies (mAbs) against a peptide representing C/EBPα doubly phosphorylated on T222 and T226, which mutation analysis indicated as the major phosphorylated residues (Ross et al, 1999; data not shown). Two mAbs that recognized this phosphopeptide, but not the corresponding non-phospho peptide, were isolated. Both of these reacted with WT C/EBPα derived from virally transduced NIH3T3 cells, but not a T222A, T226A mutant, confirming that they specifically recognized the phospho-form of full-length C/EBPα (Figure 5B; data not shown). These anti-phospho-TT mAbs detected high levels of phospho-C/EBPα in livers from fed, but not fasted, mice, and in adipose tissue in both conditions (Figure 5C), confirming the phosphorylation pattern inferred above. Injection of glucose into fasted WT and TTS mice showed that the GSK3 consensus was rephosphorylated within 60 min of elevation of blood glucose levels, as detected by both Anderson PAGE and anti-phospho-C/EBPα antibodies (Figure 5D and E). In contrast, we did not observe rephosphorylation by direct insulin challenge (data not shown), indicating that glucose induces phosphorylation of these sites through an insulin-independent mechanism.
Figure 5.
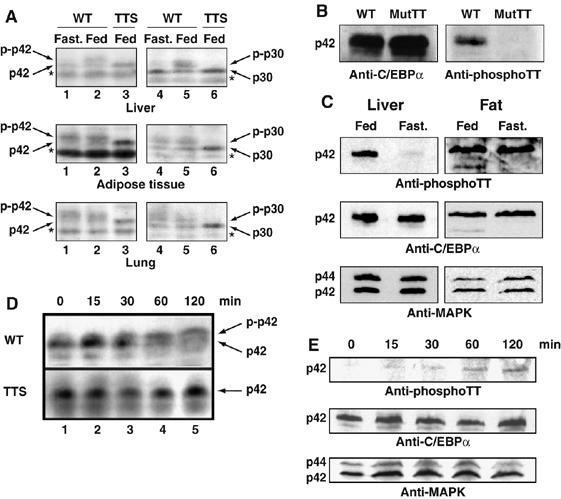
Phosphorylation of the TTS site is metabolically regulated. (A) Protein extracts from approximately 10 mg liver, 20 mg lung and 200 mg epididymal fat pad isolated from fed and fasted WT and fed TTS mice were analyzed by Anderson PAGE and Western blotting. Western blots were developed using anti C/EBPα antibody (14AA). The phosphorylated and non-phosphorylated C/EBPα p42 and p30 forms are indicated. * indicates a nonspecific cross-reactive band. (B) NIH3T3 cells were infected with virus encoding wild-type C/EBPα or mutant C/EBPα T222A,T226A. Cells were analyzed for C/EBPα expression and Thr 222,226 phosphorylation by Western blotting using polyclonal anti-C/EBPα antibody (14AA) or monoclonal anti-C/EBPα phospho-Thr 222,226 (2C6), respectively. (C) Nuclear lysates prepared from 2 mg liver or 40 mg epididymal fat pads from fed or fasted WT mice were analyzed for C/EBPα content and threonine 222, 226 phosphorylation as in (B). Only the C/EBPα p42 form is shown for simplicity. Western blotting was normalized using anti-MAPK p42/44 antibody (MAPK). (D) Liver protein lysates was prepared from WT and TTS mice during a glucose tolerance test (Figure 7E). Mice were killed at the indicated time points and analyzed as in (A). Protein lysate from approximately 10 mg liver was loaded in each lane. (E) Nuclear lysates prepared from 2 mg liver from WT mice during a glucose tolerance test (as in D) were analyzed for C/EBPα content and T222,T226 phosphorylation by Western blotting as in (C).
Deregulation of IRE controlled gene expression in TTS mice
The above results indicated that the C/EBPα GSK3 consensus functions as a liver-specific nutritional sensor in vivo. As the TTS allele mimicks the fasted state of C/EBPα in fed mice, we focused our analysis of the downstream effects of the mutation on mRNAs that are regulated in response to a feeding to fasting transition, as well as known C/EBPα target genes, using quantitative RT–PCR analysis of mRNA from livers of fed TTS and WT mice. We found that a group of four genes consisting of G6Pc, IGFBP-1, PEPCK and TAT were significantly upregulated in the TTS mutants (Figure 6A). These genes all contain IRE sequences in their promoters and are all known to be repressed by insulin signaling through these IREs (O'Brien et al, 2001). In contrast, a number of other genes, including known or putative hepatic C/EBPα target genes involved in glucose (GS, GcK) and nitrogen metabolism (CPS-1), were unaffected by the TTS mutation (Figure 6A). Both PEPCK and G6Pc are part of the gluconeogenic regulon induced by forced hepatic expression of the PGC-1α coactivator (Yoon et al, 2001). We analyzed the expression of PGC-1α, as well the PGC-1α targets FBP-1 and CPT-1 (Louet et al, 2002), which were found not to coregulate with the IRE-controlled genes (Figure 6B). ACL and ME, both dysregulated in ΔPHR mice, were also expressed normally in TTS mice (Figure 6B). To determine if the phosphorylation state of the GSK3 consensus directly affected the activity of C/EBPα on an IRE-containing promoter, we used the proximal G6Pc promoter, which contains three IREs (O'Brien et al, 2001). In transiently transfected HepG2 hepatoma cells, as well as S293 cells, the TTS allele displayed increased transactivation on this promoter compared with WT C/EBPα (Figure 6C and E). In contrast, no significant difference was observed between the WT C/EBPα and the TTS mutant on the ACAS2 promoter in S293 cells (Figure 6F), and a slight, but significant, decrease was observed in HepG2 cells (Figure 6D). Importantly, C/EBPα was phosphorylated on the GSK3 consensus in S293 cells (Figure 6G). ChIP analysis showed similar association of C/EBPα with the G6Pc and ACAS2 promoters in the liver in fed and fasted mice (Figure 6H), whereas SREBP-1 occupancy of the ACAS2 promoter was restricted to the fed state. Together, these results support the notion that phosphorylation of the GSK3 consensus selectively decreases C/EBPα activity on IRE-controlled promoters, and show that the genes regulated by phosphorylation of the C/EBPα GSK3 consensus are distinct from those whose regulation depends on the PHR motif.
Figure 6.
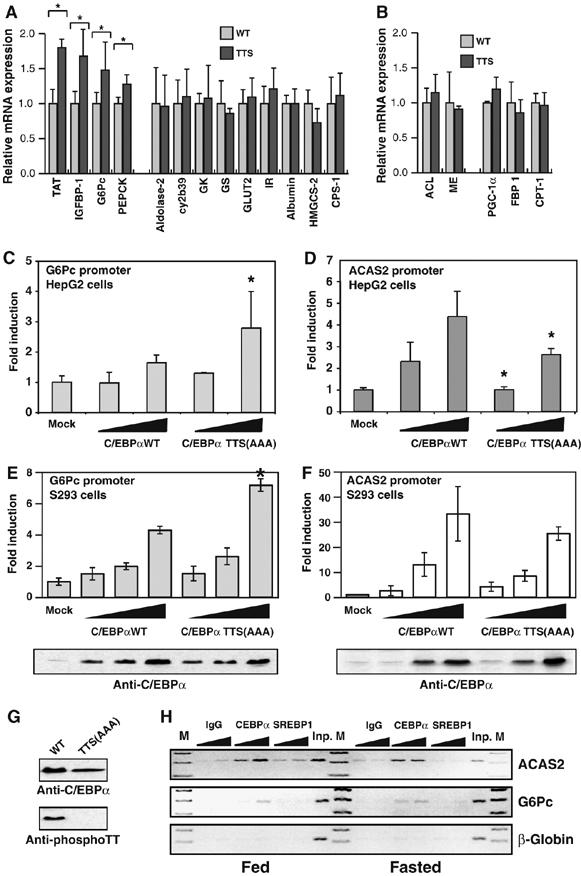
TTS mice show increased hepatic IRE controlled gene expression. (A) mRNA expression was analyzed by RT–PCR as in Figure 2A using cDNA prepared from livers from at least four fed WT or TTS mice. cy2b39: cytochrome 2b39; IR: insulin receptor; HMGCS-2: hydroxymethylglutaryl-CoA synthetase-2 (mitochondrial). (B) mRNA expression as in (A). (C) HepG2 cells were cotransfected with pG6Pc-LUC and increasing amounts (300 ng and 1 μg) of expression vectors encoding wild-type (WT) C/EBPα or C/EBPα TTS(AAA), as indicated. Luciferase activity was measured 40 h post transfection (N=5). (D) As in (C), except pACAS2-LUC reporter was used (N=4). (E) S293 cells were transfected as in C. The amounts of C/EBPα expression vector was 100 ng, 300 ng and 1 μg (N=4). C/EBPα expression levels in transfected cells were determined by Western blotting. (F) As in (E), except pACAS2-LUC reporter was used (N=4) (G) S293 cells were transfected with expression vectors encoding WT C/EBPα or C/EBPα TTS(AAA) as indicated. C/EBPα expression levels and phosphorylation on threonines 222 and 226 were determined by Western blotting as in Figure 5B. (H) ChIP experiments was performed as in Figure 3A using liver from fed or overnight fasted mice, as indicated. *Indicated a significant difference (P<0.05, Student's t-test) from the wildtype value.
TTS motif is required for glucose tolerance
The elevated expression of IRE-controlled gluconeogenic genes (G6Pc and PEPCK) in fed TTS mice suggested that phosphorylation of the GSK3 consensus contributes to limitation of hepatic glucose output in response to a high metabolic state, a regulation lost if these sites are mutated. Consistent with this, hepatic glycogen was decreased in fed TTS mice (Figure 7A) and small, but significant, increase in blood glucose and insulin levels was seen in TTS mice, relative to WT controls (Figure 7B and C), whereas serum-free fatty acids were not elevated (Figure 7D). TTS and WT mice were therefore subjected to a glucose tolerance test. True wildtype (+/+) mice, derived from TTS/+ and WT/+ intercrosses, were tested in parallel to ensure that background differences played no role in the observed effects. TTS mice displayed significant glucose intolerance, compared with both WT and +/+ mice (Figure 7E), and the glucose uptake in skeletal muscle was significantly lowered, with a similar trend in white adipose tissue (Figure 7F), indicating that the deregulation hepatic glucose metabolism caused by C/EBPα GSK3 consensus mutation leads to disturbed systemic glucose tolerance.
Figure 7.
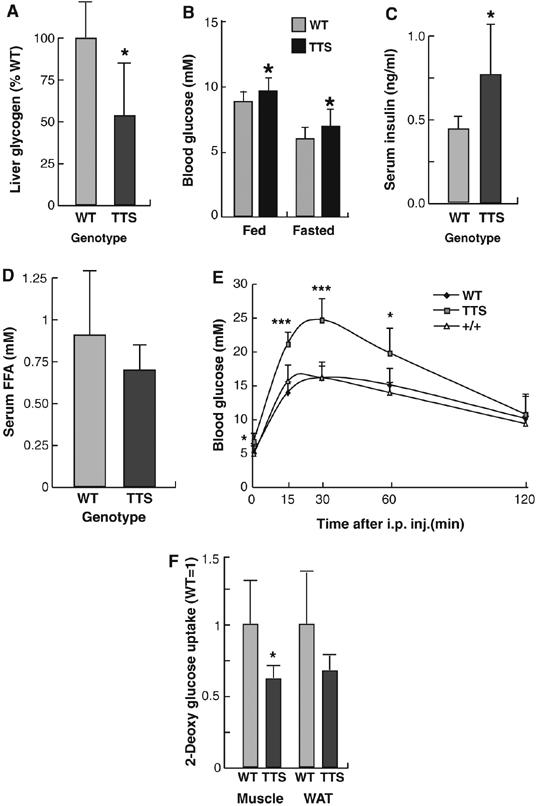
Glucose intolerance in TTS mice. (A) Relative glycogen levels in livers of WT (N=9) and TTS (N=12) mice. (B) Glucose concentration of tail vein blood from fed (N=14) and fasted (N=8) WT or fed (N=34) and fasted (N=19) TTS mice. (C) Insulin levels of serum from WT (N=6) and TTS (N=7) determined by ELISA. (D) Serum-free fatty acid levels from WT (N=6) and TTS (N=7) determined as described in Materials and methods. (E) Glucose tolerance test was performed on overnight fasted WT (N=6), WT (N=8) and TTS (N=10) mice as described in Materials and methods section. Blood glucose was measured immediately before and 15, 30, 60 and 120 min after glucose injection. *** P<10−5. (F) Relative 2-deoxy-glucose uptake in muscle and fat of overnight (16 h) fasted WT (N=6) and TTS (N=4) mice determined 1 h after glucose injection, as described in Materials and methods. *Indicated a significant difference (P<0.05, Student's t-test) from the wildtype value.
Discussion
The results presented here demonstrate that the C/EBPα PHR motif and GSK3 consensus are required for regulation of distinct gene subsets in the liver, showing in vivo target gene specificity of these two regulatory motifs. Second, these gene subsets controlled distinct metabolic processes, and define two regulons involved in lipogenesis (ΔPHR mutant) and gluconeogenesis (TTS mutant), respectively. The PHR regulon is characterized by coregulation by C/EBPα and SREBP-1, whereas the TTS regulon is composed of genes containing IREs in their promoters, providing genetic in vivo evidence that promoter context determines the responsiveness of genes to regulation through the two C/EBPα amino-acid motifs. Finally, the tissue-specific glucose regulation of C/EBPα GSK3 consensus phosphorylation shows that C/EBPα is a hepatic sensor of the metabolic state, and its ability to control hepatic gluconeogenic gene expression in the absence of PGC-1α regulation identifies C/EBPα phosphorylation as a PGC-1α-independent mechanism for controlling hepatic glucose efflux. Together, these observations suggest a model in which independent regulation of metabolic pathways is achieved by context-dependent sensitivity to metabolically regulated signaling via discrete domains within transcriptional regulators.
Regulation and function of the C/EBPα GSK3 consensus
Mutation of the GSK3 consensus leads to increased C/EBPα activity on the IRE-containing G6Pc promoter and specific upregulation in the fed state of a regulon consisting of IRE-controlled genes, including G6Pc and PEPCK. The factors associated with IREs have been extensively investigated, mainly in hepatoma cell lines, and the main candidates identified include Foxo1 and C/EBPβ (Hall et al, 2000; Ghosh et al, 2001). For Foxo1, this notion is supported by the observation that its ectopic expression in kidney cells confers negative insulin regulation upon G6Pc (Nakae et al, 2001). C/EBPα expression, while high in normal liver, is generally low or absent in hepatoma cell lines (see e.g. Figure 1C), and the role of C/EBPα may have been underestimated for this reason. However, in one study, IRE regulation has been shown to be sensitive to antisense C/EBPα downregulation in H4IIE cells (Crosson and Roesler, 2000). Here we show that C/EBPα binds to IRE promoter regions in vivo, both in the fed and fasted states, and our results thus indicate that mutation of the TTS motif specifically affects C/EBPα activity on IRE-containing promoters. This provides direct genetic evidence that C/EBPα participates in IRE regulation in vivo.
In vivo, we find dephosphorylation of the GSK3 consensus to be restricted to fasting hepatocytes, whereas adipose and lung tissue did not show regulation. Since insulin signaling is lower in the fasted state, and GSK3 activity is inhibited by insulin signaling (Cross et al, 1995), this indicates that under physiological conditions C/EBPα GSK3 consensus phosphorylation does not correlate with GSK3 activity. Significantly, hepatic PGC-1α-regulated genes are not systematically affected in TTS mice, and its expression is not changed, and GSK3 consensus phosphorylation was not insulin sensitive in vivo. PGC-1α expression is induced in the liver via the cAMP–CREB axis (Herzig et al, 2001), and regulates gluconeogenic gene expression by acting as a co–factor for Foxo1, an interaction disrupted by insulin-mediated Foxo1 phosphorylation (Puigserver et al, 2003). More recently, the metabolic regulation of gluconeogenic genes, including those containing IREs, was found to be blunted but not absent in mice lacking PGC-1α in the liver (Handschin et al, 2005), indicating the existence of parallel regulatory mechanisms. Our data show that C/EBPα regulates IRE-containing promoters, including those of key gluconeogenic genes, through a mechanism distinct from that defined by the PGC-1α–Foxo1 axis, and C/EBPα GSK3 consensus phosphorylation may therefore constitute a PGC-1α- and insulin-independent mechanism for controlling hepatic glucose efflux.
The physiological consequences of promoter deregulation in TTS mice are decreased liver glycogen, subtle increases in serum insulin and glucose, and glucose intolerance. This phenotype is very similar to that induced by hepatic overexpression of G6Pc in rats (Trinh et al., 1998), and is therefore consistent with the observed changes in gene expression. Nevertheless, the possibility has to be considered that the effects of the TTS mutation on gene expression are caused by metabolic changes in other tissues, and not vice versa. This is unlikely to be the case, because in transient transfection assays in both S293 and HepG2 cells the intrinsic activity of C/EBPα on the G6Pc promoter was increased by mutation of the GSK3 consensus to an extent similar to the increase of G6Pc expression seen in mutant mice. Also, increased blood glucose and insulin would normally downregulate IRE-controlled gene expression, whereas the opposite is in fact observed. We therefore propose that phosphorylation of the C/EBPα GSK3 consensus, induced by elevated blood glucose levels, serves to limit hepatic glucose output through negative regulation of IRE-containing promoters, thereby providing negative feedback regulation, and that in the absence of such control homeostasis occurs at a slightly elevated blood glucose concentration, resulting in systemic glucose intolerance.
Contol of lipogenic gene expression by the C/EBPα PHR motif
The genes in the PHR regulon are involved in the generation of cytoplasmic acetyl-CoA and NADPH, the two main metabolites required for synthesis of fatty acids. ACL, ME and ACAS2 showed the expected metabolic regulation, being repressed in the fasted state, and this regulation was lost when the PHR motif was deleted. In addition, lowered expression of several β-oxidation enzymes was observed. The net result of this was increased triglyceride levels in livers of ΔPHR mice, and hepatocytes isolated from ΔPHR mice had increased triglyceride accumulation. Although more work is required to pinpoint the precise mechanism underlying the increased triglyceride levels observed in the ΔPHR mice, this suggests that the downregulation of the PHR regulon during periods of low food intake may be critical for limiting hepatic triglyceride production. Elevated levels of GcK mRNA and lower β-oxidation may contribute to this effect by increasing hepatic glucose influx and lowering fatty acid degradation, respectively.
At the molecular level, we find that PHR-dependent promoters bind both SREBP-1 and C/EBPα, that the two factors interact in vivo and in cotransfection assays, and synergize in ACAS2 and ACL promoter activation, and that this synergy requires the PHR motif and promoter SREs. The specificity in gene regulation by the PHR motif is therefore most likely provided simply by the SREBP-1 containing promoter context. It is interesting to note that chicken strains selected for high and low levels of abdominal adipose tissue show divergent hepatic expression of ME and ACL: both were upregulated in fat compared with lean animals, whereas FAS and SREBP-1 expression were not significantly altered (Daval et al, 2000; Assaf et al, 2004). This is consistent with ME and ACL being controlled by signaling that does not affect FAS/SREBP-1 expression, and further indicates that specific deregulation of genes from the PHR regulon is sufficient to increase hepatic lipid production. The concept of coregulation of genes constituting functional modules is central to our understanding of genome function, as such modules are predicted to provide regulatory and evolutionary flexibility by allowing pathways, rather than individual molecules, to be used as building blocks when modifying the phenotype of cells, and ultimately organisms (Aderem, 2005). The fat versus lean chickens mentioned above may be an example of how a regulatory mutation, yet to be identified, by changing the expression of the PHR regulon affects the physiology of the whole organism. These studies did not correlate the fat phenotype with increased C/EBPα or SREBP-1 expression. Our results suggest that controlling the functional interaction between these molecules, for example, by phosphorylation/dephosphorylation of C/EBPα, may instead be critical.
In summary, the data presented provide genetic evidence that in vivo the coordinated expression of hepatic enzymes participating in specific metabolic pathways (acetyl-CoA/NADPH generation and gluconeogenesis) is controlled by distinct motifs within the C/EBPα transactivation domain. Neither of the mutations studied had any effect on expression of CPS-I, a key nitrogen metabolic enzyme critically dependent on C/EBPα for its expression (Inoue et al, 2004; Kimura et al, 1998) or other C/EBPα target genes (Albumin, GS). ChIP analysis showed the constitutive presence of C/EBPα on both PHR-(ACAS2) and TTS (G6Pase)-regulated genes. The function of the two motifs is therefore to sensitize target promoters to specific signals, in this case SREBP-1 upregulation and glucose-regulated TTS dephosphorylation, respectively. Regulation via the TTS and PHR motifs also showed cell type specificity: dephosphorylation of the GSK3 consensus in response to fasting was observed only in the liver, and although the lipogenic enzymes constituting the PHR regulon are highly expressed in adipocytes, no effect of the ΔPHR mutation was seen in adipose tissue. The identified C/EBPα motifs and their context-specific gene regulatory properties thus provide a mechanism for how gene and tissue specificity of transcriptional regulation of C/EBPα target genes is achieved.
Materials and methods
DNA constructs and generation of knock-in mice
The C/EBPαΔPHR and TTS mutants were generated by PCR or by using the Quickchange mutagenesis kit (Stratagene) and confirmed by sequencing. Cloning of targeting constructs, electroporation and selection of 14.1 ES cells were performed as described (Porse et al, 2001). The C/EBPα-TAP knock-in mice, containing a tandem affinity tag consisting of a calmodulin binding peptide, a TEV protease cleavage site and a protein A Ig binding domain (Rigaut et al, 1999) fused to the C terminus of C/EBPα, will be described elsewhere (Lopez et al, in preparation). Mouse strains were maintained on a 75% C57BL/6: 25% 129/Ola background. Mice were kept in a 12-hour light/dark cycle on a standard pellet chow diet (Harlan) and water ad libitum, except when otherwise stated. Only male mice between 7 and 8 weeks were used for analysis.
Histology
Analysis of white adipose tissue was performed as described (Porse et al, 2001).
Generation of monoclonal antibodies
Monoclonal antibodies were raised against the peptide QPGHPT[P]PPPT[P]PVPSPH (Sigma Genosys). mAb generation was performed as described (De Masi et al, 2006) by the EMBL Monoclonal Antibody Core Facility. mAbs were screened against both the phosphopeptide and the corresponding non-phospho peptide. Two hybridomas (2C6 and 9G11) were identified that selectively recognized the phospho-form. The 2C6 mAb was used for all experiments, except Figure 6G.
Protein extraction, co-IP, TAP pull-down and Western blotting
Details can be found in Supplementary data.
RNA preparation, affymetrix and real time PCR analysis
Total liver RNA was prepared as described (Chomczynski and Sacchi, 1987). Total RNA was prepared from fat using Trizol (Sigma) as specified by the manufacturer. For microarray analysis, total RNA from liver was analyzed at the RH Microarray Center (Copenhagen University Hospital, Denmark) using Affymetrix Mouse Genome 430 2.0 GeneChips™. For Real-time PCR, cDNA was prepared using the Ready-To-Go T-primed first strand kit (Amersham Bioscience). Genespring software (Silicon Genetics) was used for data mining (see Supplementary data). Real-time PCR protocols, primer sequences and cycling conditions are available as Supplementary data.
Transient transfections
Mouse promoters for ACAS2 (−2059 to +1), ACL (−850 to +50) and G6Pc (−250 to +65) were amplified by PCR from genomic DNA and cloned into pGL3 (Invitrogen) to generate pACAS2-LUC, pACL-LUC and pG6Pc-LUC, respectively. For mutation of three ACAS2 SREs (Ikeda et al, 2001) in pACAS2mSRE-LUC, two successive rounds of PCR mutagenesis were carried out using the the Quickchange mutagenesis kit (Stratagene) and the following primers:
Acas SRE-397* s: CTAACTTGGGAATTCCACAGCCTT;
Acas SRE-397* as: AAGGCTGTGGAATTCCCAAGTTAG;
Acas SRE-352/47* s: AGCCGGGCTAAAGCTTGAATTCACGGG CCCGCCT;
Acas SRE-352/47* as: AGGCGGGGTGAATTCAAGCTTTAGCCC GGCT.
S293 and HepG2 cells were cotransfected with expression vectors encoding SREBP-1c (aa 1–403), WT or mutant rC/EBPα, and, for luciferase assys, reporter construct. Forty hours post transfection, cells were washed with PBS and prepared for use in luciferase assays as described (Porse et al, 2001) or for co-IP.
Preparation of primary hepatocytes and measure of triglyceride content
Hepatocytes were isolated from fed mice as described (Berry and Friend, 1969) and kept in M199 with Earles salts (Sigma) supplemented with 2% ultroser (Biosepra), 0.1% BSA and 1% penicillin/streptomycin (Gibco) before triglyceride content was determined as described (Pedersen et al, 2001).
ChIP
ChIP experiments were performed on chromatin prepared from livers of fed or overnight fasted mice using anti-SREBP-1 (C18), anti-C/EBPα (14AA) or unspecific rabbit IgG (Santa Cruz Biotechnology). For a detailed protocol and PCR primer sequences, see Supplementary data.
Mouse physiology
Details can be found in Supplementary data.
Statistics
Differences between two data sets were tested by calculating P-values using two-tailed Student's t-test. Statistical significance (P<0.05) is indicated by *. Error bars indicate standard deviations.
Supplementary Material
Supplemental data
Acknowledgments
We thank Dr D Tosh for critical reading of the manuscript. OB was supported by a fellowship from the Human Frontiers Science Program. This work was supported by the Association for International Cancer Research.
References
- Aderem A (2005) Systems biology: its practice and challenges. Cell 121: 511–513 [DOI] [PubMed] [Google Scholar]
- Assaf S, Lagarrigue S, Daval S, Sansom M, Leclercq B, Michel J, Pitel F, Alizadeh M, Vignal A, Douaire M (2004) Genetic linkage and expression analysis of SREBP and lipogenic genes in fat and lean chicken. Comp Biochem Physiol B Biochem Mol Biol 137: 433–441 [DOI] [PubMed] [Google Scholar]
- Berry MN, Friend DS (1969) High-yield preparation of isolated rat liver parenchymal cells: a biochemical and fine structural study. J Cell Biol 43: 506–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Courselaud B, Pigeon C, Inoue Y, Inoue J, Gonzalez FJ, Leroyer P, Gilot D, Boudjema K, Guguen-Guillouzo C, Brissot P, Loreal O, Ilyin G (2002) C/EBPalpha regulates hepatic transcription of hepcidin, an antimicrobial peptide and regulator of iron metabolism. Cross-talk between C/EBP pathway and iron metabolism. J Biol Chem 277: 41163–41170 [DOI] [PubMed] [Google Scholar]
- Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA (1995) Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378: 785–789 [DOI] [PubMed] [Google Scholar]
- Crosson SM, Roesler WJ. (2000) Hormonal regulation of the phosphoenolpyruvate carboxykinase gene. Role of specific CCAAT/enhancer-binding protein isoforms. J Biol Chem 275: 5804–5809 [DOI] [PubMed] [Google Scholar]
- Daval S, Lagarrigue S, Douaire M (2000) Messenger RNA levels and transcription rates of hepatic lipogenesis genes in genetically lean and fat chickens. Genet Sel Evol 32: 521–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Masi F, Chiarella P, Wilhelm H, Massimi M, Bullard B, Ansorge W, Sawyer A (2006) High throughput production of mouse monoclonal antibodies using antigen microarrays. Proteomics 5: 4070–4081 [DOI] [PubMed] [Google Scholar]
- Erickson RL, Hemati N, Ross SE, MacDougald OA (2001) p300 coactivates the adipogenic transcription factor CCAAT/enhancer-binding protein alpha. J Biol Chem 276: 16348–16355 [DOI] [PubMed] [Google Scholar]
- Ghosh AK, Lacson R, Liu P, Cichy SB, Danilkovich A, Guo S, Unterman TG (2001) A nucleoprotein complex containing CCAAT/enhancer-binding protein beta interacts with an insulin response sequence in the insulin-like growth factor-binding protein-1 gene and contributes to insulin-regulated gene expression. J Biol Chem 276: 8507–8515 [DOI] [PubMed] [Google Scholar]
- Hall RK, Yamasaki T, Kucera T, Waltner-Law M, O'Brien R, Granner DK (2000) Regulation of phosphoenolpyruvate carboxykinase and insulin-like growth factor-binding protein-1 gene expression by insulin. The role of winged helix/forkhead proteins. J Biol Chem 275: 30169–30175 [DOI] [PubMed] [Google Scholar]
- Handschin C, Lin J, Rhee J, Peyer AK, Chin S, Wu PH, Meyer UA, Spiegelman BM (2005) Nutritional regulation of hepatic heme biosynthesis and porphyria through PGC-1alpha. Cell 122: 505–515 [DOI] [PubMed] [Google Scholar]
- Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, Spiegelman B, Montminy M (2001) CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413: 179–183 [DOI] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS (2002) SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest 109: 1125–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL (2003) Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA 100: 12027–12032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Yamamoto J, Okamura M, Fujino T, Takahashi S, Takeuchi K, Osborne TF, Yamamoto TT, Ito S, Sakai J (2001) Transcriptional regulation of the murine acetyl-CoA synthetase 1 gene through multiple clustered binding sites for sterol regulatory element-binding proteins and a single neighboring site for Sp1. J Biol Chem 276: 34259–34269 [DOI] [PubMed] [Google Scholar]
- Inoue Y, Inoue J, Lambert G, Yim SH, Gonzalez FJ (2004) Disruption of hepatic C/EBPalpha results in impaired glucose tolerance and age-dependent hepatosteatosis. J Biol Chem 279: 44740–44748 [DOI] [PubMed] [Google Scholar]
- Kimura T, Christoffels VM, Chowdhury S, Iwase K, Matsuzaki H, Mori M, Lamers WH, Darlington GJ, Takiguchi M (1998) Hypoglycemia-associated hyperammonemia caused by impaired expression of ornithine cycle enzyme genes in C/EBPalpha knockout mice. J Biol Chem 273: 27505–27510 [DOI] [PubMed] [Google Scholar]
- Kovacs KA, Steinmann M, Magistretti PJ, Halfon O, Cardinaux JR (2003) CCAAT/enhancer-binding protein family members recruit the coactivator CREB-binding protein and trigger its phosphorylation. J Biol Chem 278: 36959–36965 [DOI] [PubMed] [Google Scholar]
- Lee YH, Sauer B, Johnson PF, Gonzalez FJ (1997) Disruption of the c/ebp alpha gene in adult mouse liver. Mol Cell Biol 17: 6014–6022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louet JF, Hayhurst G, Gonzalez FJ, Girard J, Decaux JF (2002) The coactivator PGC-1 is involved in the regulation of the liver carnitine palmitoyltransferase I gene expression by cAMP in combination with HNF4 alpha and cAMP-response element-binding protein (CREB). J Biol Chem 277: 37991–38000 [DOI] [PubMed] [Google Scholar]
- Nakae J, Kitamura T, Silver DL, Accili D (2001) The forkhead transcription factor Foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J Clin Invest 108: 1359–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerlov C, Ziff EB (1995) CCAAT/enhancer binding protein-alpha amino acid motifs with dual TBP and TFIIB binding ability co-operate to activate transcription in both yeast and mammalian cells. EMBO J 14: 4318–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien RM, Streeper RS, Ayala JE, Stadelmaier BT, Hornbuckle LA (2001) Insulin-regulated gene expression. Biochem Soc Trans 29: 552–558 [DOI] [PubMed] [Google Scholar]
- Pedersen TA, Kowenz-Leutz E, Leutz A, Nerlov C (2001) Cooperation between C/EBPalpha TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev 15: 3208–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porse BT, Pedersen TA, Hasemann MS, Schuster MB, Kirstetter P, Luedde T, Damgaard I, Kurz E, Schjerling CK, Nerlov C (2006) The proline–histidine-rich CDK2/CDK4 interaction region of C/EBPalpha is dispensable for C/EBPalpha-mediated growth regulation in vivo. Mol Cell Biol 26: 1028–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porse BT, Pedersen TA, Xu X, Lindberg B, Wewer UM, Friis-Hansen L, Nerlov C (2001) E2F repression by C/EBPalpha is required for adipogenesis and granulopoiesis in vivo. Cell 107: 247–258 [DOI] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, Spiegelman BM (2003) Insulin-regulated hepatic gluconeogenesis through FOXO1–PGC-1alpha interaction. Nature 423: 550–555 [DOI] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol 17: 1030–1032 [DOI] [PubMed] [Google Scholar]
- Ross SE, Erickson RL, Hemati N, MacDougald OA (1999) Glycogen synthase kinase 3 is an insulin-regulated C/EBPalpha kinase. Mol Cell Biol 19: 8433–8441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh KY, O'Doherty RM, Anderson P, Lange AJ, Newgard CB (1998) Perturbation of fuel homeostasis caused by overexpression of the glucose-6-phosphatase catalytic subunit in liver of normal rats. J Biol Chem 273: 31615–31620 [DOI] [PubMed] [Google Scholar]
- Wang GL, Iakova P, Wilde M, Awad S, Timchenko NA (2004) Liver tumors escape negative control of proliferation via PI3K/Akt-mediated block of C/EBP alpha growth inhibitory activity. Genes Dev 18: 912–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ND, Finegold MJ, Bradley A, Ou CN, Abdelsayed SV, Wilde MD, Taylor LR, Wilson DR, Darlington GJ (1995) Impaired energy homeostasis in C/EBP alpha knockout mice. Science 269: 1108–1112 [DOI] [PubMed] [Google Scholar]
- Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, Newgard CB, Spiegelman BM (2001) Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413: 131–138 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental data


