Abstract
ATM and ATR are two related kinases essential for signalling DNA damage. Although ATM is thought to be the principle kinase responsible for signalling ionising radiation (IR)-induced DNA damage, ATR also contributes to signalling this form of genotoxic stress. However, the molecular basis of differential ATM and ATR activation in response to IR remains unclear. Here, we report that ATR is recruited to sites of IR-induced DNA damage significantly later than activation of ATM. We show that ATR is recruited to IR-induced nuclear foci in G1 and S phase of the cell cycle, supporting a role for ATR in detecting DNA damage outside of S phase. In addition, we report that recruitment of ATR to sites of IR-induced DNA damage is concomitant with appearance of large tracts of single-stranded DNA (ssDNA) and that this event is dependent on ATM and components of the Mre11/Rad50/Nbs1 (MRN) protein complex.
Keywords: ataxia telangiectasia mutated and rad3-related (ATR), cell cycle checkpoints, DNA damage and repair
Introduction
The detection and signalling of DNA damage is critical for maintaining genome integrity. Recently, several groups of proteins have been implicated in detecting and signalling DNA damage, including the ATM, ATR, Rad9/Rad1/Hus1 (9-1-1), Rad17 and Mre11/Rad50/Nbs1 (MRN) sensor complexes, adaptor proteins 53BP1, Mdc1 and BRCA1 and the transducer kinases Chk1 and Chk2 (D'Amours and Jackson, 2002; Melo and Toczyski, 2002; Bartek and Lukas, 2003). However, although some of the proteins required for the efficient detection and signalling of DNA damage have been identified, the precise mechanisms by which these different proteins function remain unclear.
The phosphatidylinosotol (PI)–3 kinase-related kinases (PIKKs) ATM and ATR are essential for the activation of cell cycle checkpoints in response to genotoxic stress (Abraham, 2001). ATM is mutated in patients with the clinical disorder ataxia telangiectasia (A-T), a disease characterised by a number of debilitating symptoms, including chromosomal instability and cancer predisposition (Rotman and Shiloh, 1998). Cells obtained from A-T patients are hypersensitive to DNA double-strand breaks (DSBs) induced by agents such as ionising radiation (IR), but not to other forms of genomic insult such as base damage induced by UV irradiation (Rotman and Shiloh, 1998). ATM is rapidly activated in response to IR through autophosphorylation of serine 1981 (S1981), and mediates the phosphorylation of a number of different proteins that regulate various aspects of the DNA damage response (Bakkenist and Kastan, 2003). However, although loss of ATM results in defective phosphorylation of a wide variety of proteins in response to IR, these events remain intact when A-T cells are exposed to other forms of DNA damage such as UV (Shiloh, 2003). The emerging theme, therefore, is that ATM is the primary kinase responsible for detecting and signalling IR-induced DNA damage.
Although deletion of the atr gene in mice results in early embryonic lethality (Brown and Baltimore, 2000; de Klein et al., 2000), hypomorphic mutations of the ATR gene result in Seckel syndrome, an autosomal recessive disorder that displays clinical features consistent with a defect in the DNA damage response (O'Driscoll et al., 2003). ATR is required for signalling base damage induced by agents such as UV (Wright et al., 1998; Tibbetts et al., 1999; O'Driscoll et al., 2003) and work in a variety of model organisms has illustrated a central role for ATR in signalling stalled replication forks and maintaining genome integrity during S phase. For example, the ATR orthologue in Saccharomyces cerevisiae (Mec1p) is required for replication fork stability and inhibition of late origin firing in response to DNA damage and replication stress (Santocanale and Diffley, 1998; Tercero and Diffley, 2001) and ATR is recruited to chromatin when replication forks are induced to stall (Tibbetts et al., 2000; Hekmat-Nejad et al., 2000; Lupardus et al., 2002; You et al., 2002; Lee et al., 2003; Dart et al., 2004).
However, although ATM has long been implicated in signalling IR-induced DNA lesions, ATR also functions in signalling DNA damage induced by this agent. ATR relocates to discrete nuclear foci after administration of IR, an observation that is thought to reflect recruitment of this kinase to sites of DNA damage (Zou and Elledge, 2003). Disruption of ATR function results in sensitivity of cells to agents that induce DNA DSBs such as IR (Cliby et al., 1998; Nghiem et al., 2002) and defective phosphorylation of a variety of effector molecules that initiate cell cycle arrest such as p53, Chk1 and Chk2 (Tibbetts et al., 1999; Brown and Baltimore, 2003; Helt et al., 2005). The observations that loss of ATR function results in defective phosphorylation of p53 and cell cycle arrest at later time points following administration of IR have lead to a model whereby ATM is required for the initial response to IR-induced DNA damage, whereas ATR is responsible for maintenance of this response (Tibbetts et al., 1999; Brown and Baltimore, 2003).
However, although it is apparent that both ATM and ATR are involved in signalling IR-induced DNA damage, the reasons behind the differential activation of these two kinases in response to this form of genotoxic stress remain unclear. Here, we report that although ATM is activated rapidly in response to IR, nuclear retained ATR foci formation is observed at time points following IR-induced ATM activation. ATR is capable of forming IR-induced nuclear retained foci before the onset of S phase, supporting a role for ATR in detecting IR-induced DNA damage outside of S phase. In addition, we present data illustrating recruitment of ATR to sites of IR-induced DNA damage is concomitant with formation of large tracts of single-stranded DNA (ssDNA) and that this event is dependent on ATM and components of the Mre11/Rad50/Nbs1 (MRN) DNA damage sensing complex.
Results
Activation of ATM and recruitment of ATR to sites of IR-induced DNA damage
Given that ATM and ATR have been proposed to signal IR-induced DNA damage at different times following exposure of cells to IR, we assessed the temporal relationship between activation of ATM and recruitment of ATR to sites of DNA damage in response to this form of genotoxic stress. Accordingly, we exposed asynchronous HeLa cell cultures to a sublethal dose of IR (0.5 Gy) and assessed activation of ATM at time points following administration of DNA damage by appearance of nuclear retained ATM phosphorylated at S1981 (Bakkenist and Kastan, 2003). In parallel, we also assessed the recruitment of ATR to sites of DNA damage as judged by the appearance of nuclear retained ATR foci.
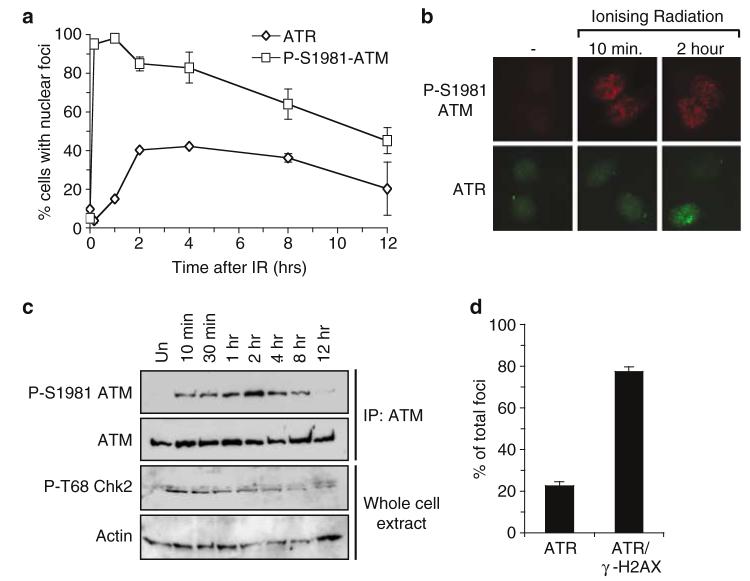
Nuclear retained P-S1981 ATM foci are apparent within 10min following administration of IR (Figure 1a and b). Western blotting using antibodies specific for either P-S1981 ATM or P-T68 Chk2 reveals that appearance of P-S1981 ATM mirrors that of a known ATM substrate (Figure 1c). Furthermore, nuclear retained P-S1981 ATM is not apparent after exposure of A-T cells to IR when compared to wild-type control cells (data not shown). Taken together, these data are consistent with a previous report illustrating that activation of ATM occurs rapidly following administration of IR (Bakkenist and Kastan, 2003).
Figure 1.
IR-induced ATR nuclear retained foci become apparent at times after activation of ATM. (a) HeLa cells were left untreated or treated with 0.5 Gy IR, and subjected to immunofluorescence using P-S1981 ATM or ATR antibodies at time points following irradiation as indicated. The number of cells displaying nuclear foci was scored at each time point from a population of ≥200 cells and error bars represent the s.e.m. (standard error of the mean). (b) Representative pictures of cells described in (a). (c) HeLa cells were left untreated (un.), or exposed to 0.5 Gy of IR and whole-cell extracts prepared at the indicated time points following administration of DNA damage. Immunoprecipitation of ATM was performed before Western blotting with either P-S1981 ATM or ATM antibodies as indicated. In parallel, Western blots on whole-cell extracts were also performed using antibodies specific for either actin or Chk2 phosphorylated on T68 (P-T68 Chk2). (d). HeLa cells were exposed to 0.5 Gy of IR and costained with γ-H2AX and ATR antibodies 2 h after administration of DNA damage. The % of ATR foci that do (ATR/γ-H2AX) or do not (ATR) colocalise with γ-H2AX was scored from ≥200 ATR foci. Error bars represent the s.e.m.
In contrast to ATM, ATR foci only become detectable at between 60 and 120 min following IR treatment (Figure 1a and b). Formation of IR-induced ATR foci is compromised in cells that have been subjected to small interfering RNA (siRNA) mediated repression of ATR expression (data not shown), demonstrating the specificity of the ATR antibody employed in this study. On average, 78% of ATR foci colocalise with γ-H2AX (Figure 1d), suggesting a large proportion of ATR foci occur at sites of DNA DSBs (Downs and Jackson, 2003). Similar kinetics of nuclear retained P-S1981 ATM and ATR foci induction is achieved when 10Gy of IR is administered to HeLa cells, or after administration of 0.5 Gy of IR to Swiss 3T3 cells (data not shown), illustrating this phenomenon is not cell-type specific or limited to exposure of cells to low doses of IR. Taken together, these data support the hypothesis that ATR is recruited to sites of IR-induced DNA damage at time points following activation of ATM.
ATR is recruited to IR-induced DNA damage before and during S phase of the cell cycle
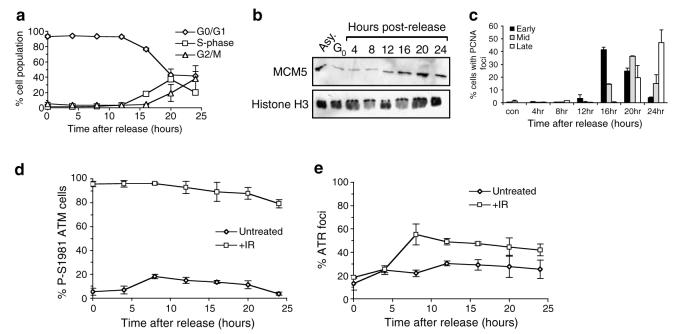
Given the proposed role of ATR in signalling replication stress and maintaining genome integrity during DNA replication, we wished to establish whether formation of nuclear retained ATR foci in response to IR is limited to S phase of the cell cycle. Accordingly, Swiss 3T3 cells were released from quiescence to pass through the cell cycle and left either untreated or exposed to IR at time points that correspond to G0/G1 or S phase of the cell cycle. ATM activation or recruitment of ATR to sites of DNA damage was assessed by staining for P-S1981 ATM and nuclear retained ATR 2 h after administration of DNA damage.
FACS analysis is consistent with almost 100% of cells being in G0 or G1 phase of the cell cycle up to 12 h after release from quiescence (Figure 2a). Loading of MCM5 onto chromatin indicates replication origins begin to be licensed at between 8 and 12 h following release from quiescence (Figure 2b), illustrating cells begin to enter G1 at these time points (Tye, 1999). Replication commences at between 12 and 16 h after release from quiescence, as judged by visualisation of S phase specific PCNA nuclear foci (Celis and Celis, 1985) and FACS analysis (Figure 2a and c).
Figure 2.
IR-induced ATR foci are apparent in both G1 and S phase of the cell cycle. (a) Swiss 3T3 cells were released from quiescence and allowed to progress through the cell cycle. Samples were taken at the time points indicated and the percentage of cells in each phase of the cell cycle established by FACS analysis. Error bars represent the s.e.m. (b) Chromatin extracts were prepared from either asynchronous cells (Asy.), quiescent cells (Go), or cells released from quiescence as described in (a). Equivalent amounts of extracts were subjected to Western blotting using antibodies as indicated. (c) Cells described in (a) were subjected to immunofluorescence using an antibody specific for PCNA. The number of cells in early, mid or late S phase was established by scoring nuclear PCNA staining patterns (Celis and Celis, 1985). Error bars represent the s.e.m. (d and e) Swiss 3T3 cells described in (a) were left either untreated or exposed to 0.5 Gy IR (+IR) at the time points indicated after release from quiescence. Cells were allowed 2 h recovery before being subjected to immunofluorescence with antibodies against either P-S1981 ATM (d) or ATR (e). A population of ≥ 200 cells was scored at each time point and error bars represent the s.e.m.
Nuclear P-S1981 ATM is apparent 2 h after IR treatment at all time points tested, illustrating activation of ATM can occur in G0/G1 and S phase of the cell cycle (Figure 2d). Consistent with a previous report illustrating ATR plays a minor role in signalling IR-induced DNA damage during quiescence (Stiff et al., 2004), little induction of ATR foci is observed in quiescent cells 2 h after exposure to IR (Figure 2e). Induction of ATR foci is also absent when IR is administered to cells 4 h after release from quiescence. Given that replication origins are at least 4 h away from being licensed at this time point (Figure 2b), we believe it likely that cells are still in G0 or early G1 at this time point. IR-induced ATR foci are apparent 10h after release from quiescence (8 h postrelease, plus 2 h postirradiation) and persist at all time points tested thereafter. This time point coincides with replication origin licensing (Figure 2b), but precedes onset of DNA replication as judged by FACS analysis and visualisation of PCNA replication foci (Figure 2a and c). ATR nuclear retained foci were also obtained using HeLa cell cultures exposed to IR in G1 phase of the cell cycle after release from a nocodazole-induced mitotic block (data not shown). Therefore, this phenomenon is not specific to either releasing cells from quiescence, or the cell type used.
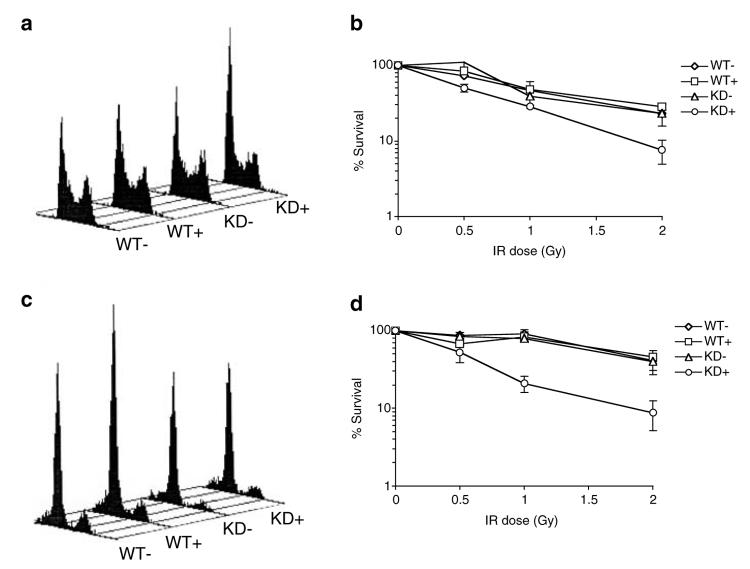
In order to assess the requirement for ATR in signalling IR-induced DNA damage during G1 phase of the cell cycle, U2OS cells were induced to express wild-type ATR (wt-ATR) or dominant-negative kinase-dead ATR (kd-ATR), synchronised in G1 phase of the cell cycle by release from a nocodazole block (Figure 3c), and subjected to cell viability assays after exposure to increasing doses of IR. Consistent with previous reports (Cliby et al., 1998; Nghiem et al., 2002), induction of kd-ATR expression in control asynchronous cell cultures results in mild sensitivity to IR when compared to either uninduced cell cultures, or cells induced to express wt-ATR (Figure 3a and b). However, expression of kd-ATR also results in sensitivity of cell cultures to IR when DNA damage is administered in G1 phase of the cell cycle (Figure 3c and d). These data, taken together with the observations that ATR is capable of forming IR-induced nuclear foci before the onset of S phase, suggest that ATR performs a role in the detection of IR-induced DNA damage outside of DNA replication.
Figure 3.
Overexpression of dominant-negative kinase-dead ATR results in sensitivity to IR administered during G1 phase of the cell cycle. (a and b) U2OS cells were uninduced (−) or induced (+) to express either wild-type (WT) or kinase-dead (KD) ATR for 48 h and the distribution of cells throughout the cell cycle established by FACS (a). Cells were treated with increasing doses of IR and left to recover for 7–10days. Survival of cells was assessed by scoring colonies containing ≥ 50cells. The number of colonies at each IR dose is expressed as a percentage of the untreated control and error bars represent the s.e.m. (b). (c and d) U2OS cells were uninduced (−) orinduced (+) to express either wild-type (WT) or kinase-dead (KD) ATR for 48 h before synchronisation of cells in the G1 by release from a nocodazole block. Synchrony of cell cultures was established by FACS analysis (c). Cell survival following administration of IR 6 h after release from nocodazole (d) was assessed as in b.
IR-induced ATR foci formation coincides with formation of large tracts of ssDNA
Evidence is emerging that processing of different DNA damage architectures into a common intermediate, namely ssDNA, is central to the DNA damage response (D'Amours and Jackson, 2002; Zou and Elledge, 2003). In this regard, although the ssDNA binding protein complex RPA is not required for activation of ATM in response to DNA DSBs (Dodson et al., 2004), this protein complex is critical for the formation of ATR nuclear foci in response to IR (Zou and Elledge, 2003). These data might argue that processing DNA DSBs into ssDNA is required for ATR to recognise IR-induced DNA damage and thus provide an alternative explanation for the differential activation of ATM and ATR in response to IR-induced DNA damage.
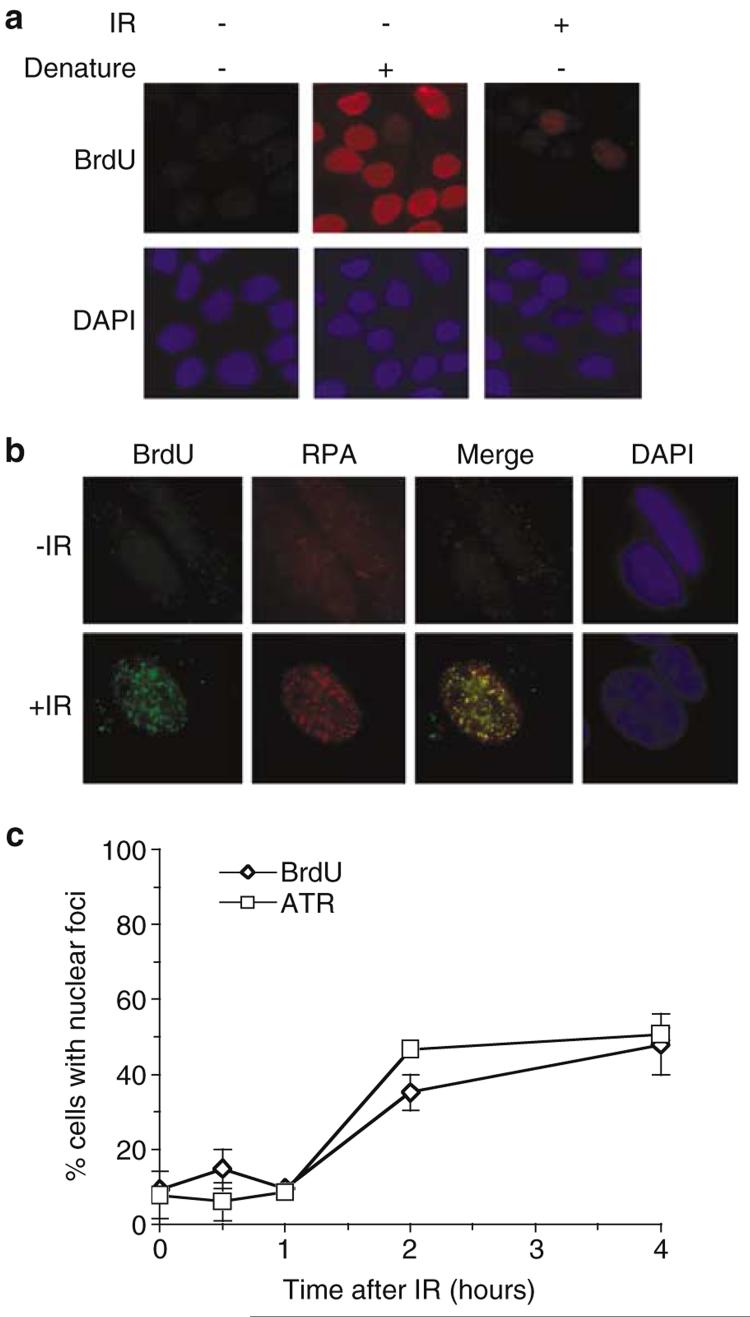
In order to investigate this possibility, we visualised ssDNA formation in situ following exposure of cells to IR by exploiting the fact that BrdU epitopes incorporated into the genome are only revealed as double-stranded DNA (dsDNA) is converted into ssDNA (Raderschall et al., 1999). Accordingly, we incorporated BrdU into the genome of HeLa cells before exposure of cells to IR. As expected, BrdU staining of cells that had not been exposed to IR occurs only after exposure of BrdU epitopes by denaturation of DNA with HCl (Figure 4a). However, exposure of cells to IR results in BrdU foci appearing in 30–40% of cells 2 h postirradiation (Figure 4a and c). Consistent with a previous report (Raderschall et al., 1999), BrdU and RPA34 IR-induced foci co-localise, illustrating that BrdU staining represents visualisation of ssDNA in situ (Figure 4b). Although P-S1981 ATM is detected within 10min of irradiation (Figure 1a), BrdU foci are not apparent at this time point (Figure 4c). However, in contrast to ATM, BrdU and ATR nuclear foci become apparent with almost identical kinetics following administration of IR (Figure 4c), and up to 75% of ATR foci colocalise with BrdU (data not shown). Thus, recruitment of ATR to IR-induced DNA damage is concomitant with processing of DNA lesions into large tracts of ssDNA. Given that a significant proportion of ATR foci colocalise with γ-H2AX 2 h after administration of IR (Figure 1d), one interpretation of these data is that ATR is recruited to sites of DNA DSBs that are converted into ssDNA.
Figure 4.
IR-induced ATR foci formation is concomitant with formation of large tracts of ssDNA in situ. (a) After incorporation of BrdU into their genome (see Materials and methods), asynchronous HeLa cells were exposed to 0.5 Gy IR as indicated. Cells were fixed 2 h after IR and subjected to immunofluorescence with BrdU antibodies. To demonstrate incorporation of BrdU into the genome, a sample was included in which cellular DNA was denatured with 0.2 M HCl before immunofluorescence. (b) Asynchronous HeLa cells were treated as in (a) and subjected to immunofluorescence with RPA34 and BrdU antibodies. (c) Asynchronous HeLa cells were treated as in (a) and subjected to immunofluorescence with the indicated antibodies at time points after administration of IR. Cells were scored for nuclear foci from a population of ≥ 200 cells and error bars represent the s.e.m.
Recognition of IR-induced DNA damage by ATR is dependent on ATM and components of the Mre11 protein complex
We wished to establish the molecular requirements for ATR recruitment to sites of IR-induced DNA damage. One candidate for converting DNA DSBs into ssDNA is the Mre11 protein complex. Mre11 forms a complex with Nbs1 and Rad50 (termed MRN) in vivo and exhibits both exonuclease and endonuclease activities in vitro (D'Amours and Jackson, 2002). Hypomorphic mutations in either NBS1 or MRE11 result in Nijmegen breakage syndrome (NBS) and ataxia-telangiectasia-like disorder (ATLD), respectively (Carney et al., 1998; Stewart et al., 1999), two genome instability syndromes characterised by sensitivity to agents that cause DNA DSBs and defective cell cycle checkpoints in response to IR (D'Amours and Jackson, 2002). In support of a role for the MRN complex in ATR-mediated signalling events, two recent reports have illustrated that dysfunction of MRN complex components can result in defective ATR-mediated signalling in response to UV-induced DNA damage (Stiff et al., 2005; Zhong et al., 2005).
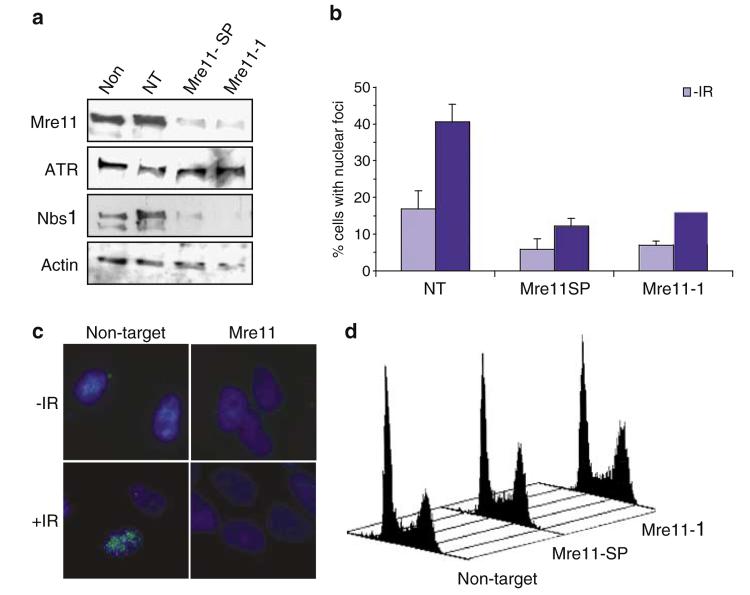
We therefore tested whether components of the MRN complex are required for localisation of ATR to sites of IR-induced DNA damage. In order to achieve this we used two different siRNA oligonucleotides to repress the expression of Mre11 (Figure 5a). In addition to repressing expression of Mre11, these oligonucleotides also result in decreased levels of Nbs1 (Figure 5a), probably through destabilisation of MRN complex components (Stewart et al., 1999). Repression of Mre11 and Nbs1 levels results in a marked decrease in the formation of ATR foci in response to IR (Figure 5b and c). FACS analysis reveals that reduction in Mre11 and Nbs1 levels does not result in a significant degree of apoptosis, as judged by the lack of a sub-G1 peak in these cell cultures (Figure 5d). Although a slight enrichment of cell cultures in G2/M phase of the cell cycle is observed when components of the MRN complex are depleted using siRNA (between 4 and 5%), this is not significant enough to account for the marked reduction in IR-induced ATR foci observed in these cells (Figure 5d). Therefore, it is unlikely that loss of IR-induced ATR foci formation in the absence of MRN complex components is attributable either to extensive apoptosis of cell cultures, or gross enrichment of cells at a stage of the cell cycle in which ATR cannot be recruited to sites of DNA damage.
Figure 5.
Components of the MRN complex are required for ATR foci formation following administration of IR. (a) Asynchronous HeLa cells were left either untreated (Non), or transfected with nontargeting siRNA (NT) or siRNA complementary to two distinct regions of Mre11 (Mre11-SP or Mre11-1). Cells were harvested 72 h after transfection and subjected to Western blotting with the indicated antibodies. (b) Asynchronous HeLa cells were transfected with nontargeting siRNA (NT) or Mre11 siRNA as indicated. At 72 h after transfection, cells were left untreated or treated with 0.5 Gy IR, and after 2 h recovery subjected to immunofluorescence with ATR antibodies. Samples were scored for cells that displayed nuclear retained ATR foci from a population of ≥ 200 cells and error bars represent the s.e.m. (c) Representative pictures of cells described in (b). (d) FACS analysis representing cell cycle distribution of cells 72 h after transfection of cells with either nontargeting or Mre11 (Mre11-SP and Mre11-1) siRNA oligonucleotides.
Given the accumulating evidence that the MRN complex is required for activation of ATM in response to IR (Carson et al., 2003; Uziel et al., 2003), we wished to assess whether ATM is also required for ATR to be recruited to sites of DNA damage induced by this agent. Accordingly, we exposed HeLa cell cultures to 0.5 Gy of IR either in the absence or presence of an ATM-specific inhibitor (KU55933; Hickson et al., 2004). As illustrated in Figure 6a, administration of KU55933 to HeLa cells results in an expected abrogation of ATM phosphorylation on serine 1981. In addition, inhibition of ATM kinase activity using KU55933 results in a decrease in the percentage of cells exhibiting IR-induced ATR nuclear foci to levels approaching those present in untreated cell cultures (Figure 6b). In contrast, repression of ATR expression using siRNA does not result in a gross reduction of P-S1981 ATM accumulation in cells exposed to 0.5 Gy of IR (Figure 6c). Therefore, although IR-induced activation of ATM is not dependent on ATR, ATM kinase activity is required for ATR to be recruited to sites of IR-induced DNA damage.
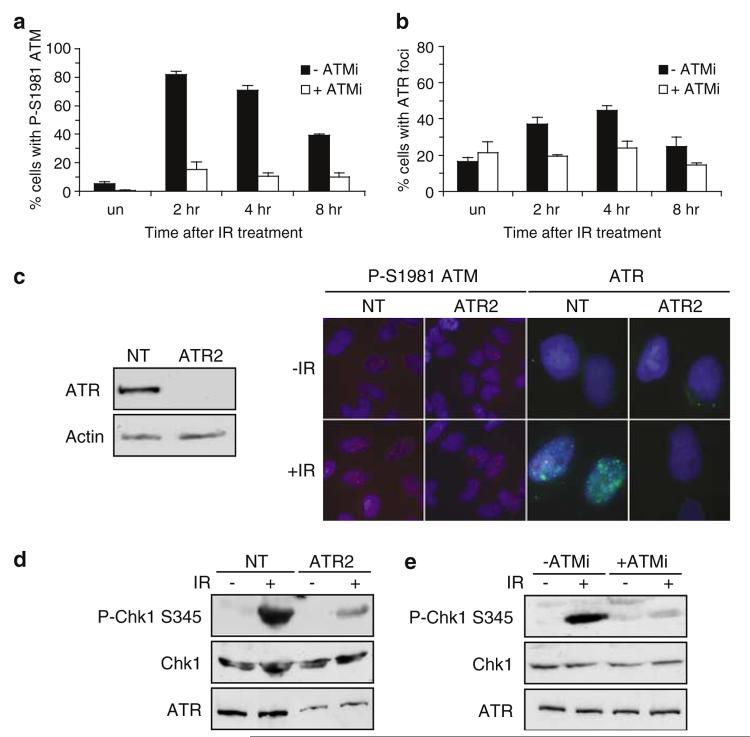
Figure 6.
ATM is required for ATR foci formation and ATR-mediated signalling to Chk1 following administration of IR. (a and b) Asynchronous HeLa cells were either preincubated in 10 μm ATM inhibitor (KU-55933 (+ ATMi)) or left in normal growth media (− ATMi) for 1 h before exposure to 0.5 Gy of IR. Untreated cells (un.), or cells that had been exposed to IR were fixed either 2 h (2 h), 4 h (4 h), or 8 h (8 h) after irradiation and stained for either P-S1981 ATM (A) or ATR (B). Cells with nuclear foci were scored from a population of greater ≥200 cells. Error bars represent the s.e.m. (c) Asynchronous HeLa cells were transfected with either nontargeting siRNA oligonucleotide (NT), or siRNA oligonucleotide complementary to ATR (ATR2) as described in Materials and methods and processed 72 h after the first transfection. Western blots were performed on whole-cell extracts with the indicated antibodies (left hand panel). In parallel, cells were left untreated, or exposed to 0.5 Gy of IR and stained for either P-S1981 ATM or ATR antibodies 2 h after irradiation as indicated. (d) Asynchronous HeLa cells were transfected with either nontargeting siRNA oligonucleotide (NT), or siRNA oligonucleotide complementary to ATR (ATR2) and 72 h after the first transfection left untreated, or exposed to 10Gy IR as indicated. Whole-cell extracts were prepared 2 h after administration of DNA damage and subjected to Western blotting using antibodies as indicated. (e) Asynchronous HeLa cells were either preincubated in 10 μm ATM inhibitor (KU-55933 (+ ATMi)) or left in normal growth media (− ATMi) for 1 h before exposure to 10Gy of IR. Whole-cell extracts were prepared 2 h after administration of DNA damage and subjected to Western blotting using antibodies as indicated.
Consistent with a previous report (Helt et al., 2005), we observe that repression of ATR expression using siRNA results in decreased IR-induced P-S345 Chk1 accumulation at a time point that coincides with ATR foci formation (Figure 6d). Consistent with our data illustrating that ATM kinase activity is required for ATR to be recruited to sites of IR-induced DNA damage, administration of KU55933 to HeLa cells results in abrogation of this ATR-mediated signalling event at this time points following administration of DNA damage (Figure 6e). Taken together, these data support the hypothesis that ATM activation occurs before activation of ATR in response to IR and that ATM kinase activity is required for recruitment of ATR to sites of DNA damage and subsequent signalling to Chk1.
Discussion
ATM and ATR are two kinases that are central to the detection and signalling of DNA damage. We observe that ATM activation occurs within 10min after administration of IR. However, IR-induced ATR foci are observed at time points following activation of ATM. Previously, loss of ATM has been reported to result in a delay in phosphorylation of effector molecules such as p53 (Banin et al., 1998; Canman et al., 1998), whereas disruption of ATR activity abrogates the accumulation of phospho-p53 in response to IR (Tibbetts et al., 1999). These data have led to a model whereby ATM is required for the initial signalling of IR-induced DNA damage, whereas ATR maintains this response. Our data would be consistent with this model, with ATR being recruited to sites of DNA damage at time points after activation of ATM.
One possible explanation for the delayed recruitment of ATR to sites of IR-induced DNA damage is that while ATM becomes activated in response to IR throughout the cell cycle, ATR is only recruited to DNA lesions during S phase. Whether ATR functions by detecting DNA damage exclusively during S phase remains unclear. For example, a recent report has proposed that ATR detects and signals UV-induced base damage specifically as replication forks encounter DNA lesions during S phase (Ward et al., 2004). However, ATR has also been reported to signal UV-induced DNA damage in nonreplicating cells (O'Driscoll et al., 2003) and ATR-mediated signalling of DNA lesions induced by the topoisomerase inhibitor etoposide has been reported to occur independently of DNA replication (Costanzo et al., 2003). As regards IR-induced DNA damage, our observations that ATR foci are capable of forming in response to this form of genotoxic stress before the onset of detectable DNA replication would argue that the role of ATR in signalling this form of DNA damage is not limited to S phase (Figures 2 and 3). In support of this hypothesis, the S. cerevisiae orthologues of RPA (Rfa1p) and the ATR-targeting subunit ATRIP (Ddc2p) are recruited to IR-induced DNA damage either in G1 or S/G2 phases of the cell cycle (Lisby et al., 2004). Indeed, similar to our observations, recruitment of Ddc2p to sites of DNA damage occurs at later time points following administration of IR after initial recognition of DNA lesions by S. cerevisiae ATM (Tel1p) (Lisby et al., 2004).
At present we do not have an explanation for why we do not observe 100% of cells forming ATR foci in either G1 or S phase of the cell cycle 2 h after administration of DNA damage. However, it should be noted that under our assay conditions up to 30% of cells do not release from quiescence (data not shown). Given that we do not observe IR-induced ATR foci when cells are in G0, we believe that the number of cells exhibiting ATR foci in G1 and S phase of the cell cycle illustrated in Figure 2 is an underestimate of the number of cells capable of forming ATR foci. This, taken together with the possibility that formation of ATR foci may occur transiently and without total synchrony between different cells within a given population, may go some way to explaining the lack of complete enrichment of ATR foci at specific stages of the cell cycle.
Our observations that delayed recruitment of ATR to IR-induced sites of DNA damage is concomitant with the appearance of large tracts of ssDNA might be more consistent with a requirement for DNA end processing being responsible for the delayed kinetics of ATR activation. In this regard, we present evidence that components of the Mre11 complex are required for ATR to be recruited to sites of IR-induced DNA damage (Figure 5). Previously, it has been reported that ATR-mediated signalling of UV-induced DNA damage or replication stress remains intact in nbs1-null mice cells (Difilippantonio et al., 2005). However, the observations that hypomorphic mutations in NBS1 and repression of Rad50 using siRNA leads to defective activation of ATR in response to UV provides a link between the MRN complex and ATR-mediated signalling events, at least in the case of base damage induced by this agent (Stiff et al., 2005; Zhong et al., 2005). As regards IR-induced DNA damage, studies in S. cerevisiae would support our data illustrating that the MRN complex is required for ATR to be recruited to sites of DNA damage. For example, deletion of either Mre11 (mre11Δ), or the orthologue of Nbs1 (xrs2Δ), results in diminished Mec1p-dependent signalling to Rad53 (S. cerevisiae Chk2) and recruitment of Mec1p to sites of DNA DSBs has been reported to be partially dependent on Xrs2p (Nakada et al., 2004).
However, although Mre11 is clearly required for the effective signalling of IR-induced DNA damage, whether S. cerevisiae Mre11 is the nuclease responsible for 5′ to 3′ resection of DNA lesions remains controversial. Although S. cerevisiae strains harbouring a nuclease defective allele of Mre11p are sensitive to IR, this is to a lesser extent than mre11 null strains (Moreau et al., 1999). Furthermore, 5′ to 3′ resection of HO endonuclease-induced DNA DSBs is only delayed in xrs2Δ or mre11Δ strains (Ivanov et al., 1994; Tsubouchi and Ogawa, 1998) and no gross defect in 5′ to 3′ DNA end processing is observed when strains containing a nuclease defective allele of Mre11p are exposed to the radiomimetic agent phleomycin (Llorente and Symington, 2004). This apparent paradox has recently been explained, in part, by two reports illustrating that S. cerevisiae exonuclease I (ExoI) is required for processing and signalling DNA DSBs along with components of the MRX complex (Llorente and Symington, 2004; Nakada et al., 2004).
Our observations that ATM kinase activity is required to recruit ATR to sites of IR-induced DNA damage and mediate signalling to Chk1 raises several interesting questions as regards the sequence of events that occur at sites of DNA damage following administration of IR. For example, although ATM activation occurs rapidly in response to IR, we do not observe large tracts of ssDNA until later time points following administration of DNA damage (Figures 1 and 4). Although we cannot discount the possibility that the assay employed in this study is insufficiently sensitive to detect limited MRN-dependent DNA end processing, these data might suggest that extensive DNA end resection is not a requirement for ATM activation. In this regard, recent in vitro studies suggest that Rad50-dependent DNA DSB unwinding is responsible for ATM activation, as opposed to Mre11-dependent end resection (Lee and Paull, 2005). It is interesting to speculate, therefore, that distinct biochemical activities of the MRN complex may be required to activate either ATM or ATR.
Given that both ATM and components of the MRN complex are required for ATR to be recruited to sites of IR-induced DNA damage, one interesting possibility is that ATM phosphorylates components of the MRN complex and modulates its DNA end-processing ability in order to activate ATR. In support of this hypothesis, in addition to being responsible for activation of ATM (Carson et al., 2003; Uziel et al., 2003), components of the MRN complex are also phosphorylated in an ATM-dependent manner in response to IR (Dong et al., 1999; Gatei et al., 2000; Lim et al., 2000; Yuan et al., 2002) and have been proposed to act downstream of ATM in the IR-induced DNA damage signalling pathway (Lavin, 2004). However, an equally plausible explanation of these data is that MRN-dependent activation of ATM may regulate another nuclease, such as ExoI for example, in order to facilitate DNA end processing and activation of ATR. If this is the case, then this might go some way to explaining the functional redundancy between components of the MRX complex and ExoI in yeast (Llorente and Symington, 2004; Nakada et al., 2004).
From our data illustrating that ATM kinase activity is required for ATR-mediated signalling of IR-induced DNA damage, it would be predicted that little if any phosphorylation of effector molecules such as p53 would be observed after exposure of A-T cells to IR. However, although signalling of IR-induced DNA damage is compromised in the absence of ATM, it should be noted that in certain instances phosphorylation of effector molecules such as p53 is still apparent, although with delayed kinetics (Banin et al., 1998; Canman et al., 1998; Brown and Baltimore, 2003). One possible explanation for this apparent discrepancy in data is that although ATR is clearly required for signalling IR-induced DNA damage at later time points following administration of IR to wild-type cells (Tibbetts et al., 1999), in A-T cells a kinase other than ATR is responsible for the delayed kinetics of p53 phosphorylation in response to this form of DNA damage. In this regard, the ATM and ATR-related kinase SMG-1 also contributes towards phosphorylation of p53 in response to IR (Brumbaugh et al., 2004) and the DNA-dependent protein kinase (DNA-PK) has recently been reported to signal IR-induced DNA damage in a manner that is functionally redundant with ATM (Stiff et al., 2004). However, another possible explanation for this disparate set of observations is that loss of ATM function results in a decreased efficiency of DNA end resection and subsequent delay in ATR-mediated signalling events. Clearly, additional studies are required to resolve these issues. Thus, although ATM and components of the MRN complex are required for recruitment of ATR to sites of IR-induced DNA damage, it will be of interest to examine the potential overlapping roles of other kinases such as SMG-1 and DNA-PK and nucleases such as Exo1 and MRN in ATR-mediated DNA damage signalling.
Materials and methods
Cell culture, maintenance and administration of genotoxic stress
Swiss 3T3 and HeLa cells were obtained from the European Collection of Cell Cultures (Health Protection Agency, Porton Down, Wiltshire, UK) and grown using standard procedures. Cells expressing wild-type or kinase-dead ATR were a kind gift from P Nghiem (Massachusetts General Hospital, Charlestown, USA) and grown as described previously (Nghiem et al., 2001). The ATM inhibitor KU-55933 (KuDOS Pharmaceuticals Ltd, Cambridge, UK) was preincubated with cells for 1 h before IR treatment, and used at a concentration of 10 μm. IR was delivered using a Caesium-137 radiation source at a rate of either 0.25 Gy/min for doses <3 Gy, or 2.2 Gy/min for doses >3 Gy.
Cell synchronisation
Swiss 3T3 cells were grown to confluence and forced into quiescence by contact inhibition for 3 days. Cells were released from quiescence by replating at 3.8 × 103 cells/cm2. The percentage of cells in each stage of the cell cycle following release from quiescence was assessed by fluorescence activated cell sorting (FACS). The number of cells in early, mid or late S phase was assessed by PCNA nuclear staining patterns (Celis and Celis, 1985).
Synchronisation of U2OS cell cultures in G1 was achieved by incubation of cells in 40ng/ml nocodazole for 6–10h. Rounded mitotic cells were removed from the plate by extensive washing and replated in fresh media. Cells were left to progress into G1 for 6 h, before exposure to DNA damage.
Cell viability assays
U2OS cells were induced to express either kinase-dead or wild-type ATR for 48 h (Nghiem et al., 2001), before plating cells at a density of 104 cells per cm2. Following IR treatment, cells were grown in media without doxycycline and cell viability assays were performed as described previously (Davies et al., 2004).
Visualisation of ssDNA in situ
Visualisation of ssDNA in situ was performed essentially as described previously (Raderschall et al., 1999). To demonstrate incorporation of BrdU into the genome, a control sample was included in which cellular DNA was denatured by incubation of coverslips in 2 m HCl for 10min, before washing with 0.2 m sodium borate (pH 9). After further washes in PBS, cells were subjected to immunofluorescence.
Preparation of cell extracts
Whole-cell extracts were prepared by washing cultures in PBS before boiling cells in Laemmli buffer for 20min. Between 1 × 104 and 1.5 × 105 cells per sample were subjected to Western blot analysis. Chromatin extracts were prepared as previously described (Dart et al., 2004).
Immunoprecipitation
Cells, 1.5 × 106, were lysed in IP Buffer (50mm Tris (pH 8.0), 200 mm NaCl, 5 mm EDTA, 0.5% NP-40, 1 mm Na3VO4, 50mm NaF, 1 μm microcystine, protease inhibitor cocktail (Roche)). ATM was immunoprecipitated from lysates by addition of 5 μg affinity purified ATM antibody and incubation for 1 h at 4°C. Protein G sepharose beads were then added and lysates incubated at 4°C for an extra hour with rotation. Finally, beads were washed three times in IP buffer before boiling in Laemmli sample buffer and precipitated proteins subjected to Western blotting.
Transfection of siRNA
Transfection of cells with siRNA was performed as previously described (Dart et al., 2004) and samples analysed 72 h after the first transfection. Custom siRNA oligonucleotides, Mre11 siGENOME Smart Pool (Mre11-SP) reagent and controls were obtained from Dharmacon (Lafayette, CO, USA). Sequences of oligonucleotides are as follows:
Mre11-1 5′-GAGCAUAACUCCAUAAGUAdTdT-3′
ATR2 5′-GCCAAGACAAAUUCUGUGUdTdT-3′.
As a negative control we employed siCONTROL nontargeting siRNA#1, an oligonucleotide that contains at least four mismatches to all genes identified in the human genome.
Antibodies
Antibodies obtained from commercial sources were as follows: Actin (C-11; Santa Cruz Biotechnology), ATM (5C2; Abcam), ATR (Santa Cruz Biotechnology (N-19) and ATR-N; Davies et al., 2004), phospho-S1981 ATM (Rockland), mouse anti-BrdU (Caltag Laboratories), sheep anti-BrdU (Biodesign International), Chk1, Phospho-Chk1 S345 (Cell Signaling Technology), Phospho-Chk2 T68 (Cell Signaling Technology), γH2AX (Upstate cell signaling solutions), histone H3 (Abcam), Mcm5 (Serotec), Nbs1 (Abcam), RPA34 (CR-UK). Antibodies that recognise Mre11 and PCNA were kind gifts from S P Jackson (Wellcome/CR-UK Institute, Cambridge) and L Cox (Department of Biochemistry, Oxford University, UK) respectively.
Immunofluorescence
Cells were grown as monolayers on glass coverslips. After specified treatments, pre-extraction of cells and immunofluorescence was performed as previously described (Dart et al., 2004; Mirzoeva and Petrini, 2001).
Acknowledgements
We thank GC Smith (KuDOS Pharmaceuticals Ltd, Cambridge, UK) for providing KU55933 and SP Jackson (Wellcome Trust and Cancer Research UK Gurdon Institute, Cambridge, UK) for communicating unpublished data. We are grateful to P Nghiem and L Cox for reagents in addition to members of the Lakin laboratory for comments during the preparation of this manuscript. This work was supported by Cancer Research UK and the Medical Research Council. While this manuscript was under review, reports by Garcia-Muse and Boulton, in addition to Jazayeri et al., illustrated that Mre11 is required for ATR-mediated signaling in response to IR. Furthermore, Jazayeri et al. reported that this event is also dependent on ATM.
Footnotes
Note added in proof
While this manuscript was under review, reports by Garcia-Muse and Boulton, in addition to Jazayeri et al., illustrated that Mre11 is required for ATR-mediated signalling in response to IR. Furthermore, Jazayeri et al. reported that this event is also dependent on ATM.
References
- Abraham RT. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Banin S, Moyal L, Shieh SY, Taya Y, Anderson CW, Chessa L, et al. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- Bartek J, Lukas J. Cancer Cell. 2003;3:421–429. doi: 10.1016/s1535-6108(03)00110-7. [DOI] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Baltimore D. Genes Dev. 2003;17:615–628. doi: 10.1101/gad.1067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbaugh KM, Otterness DM, Geisen C, Oliveira V, Brognard J, Li X, et al. Mol Cell. 2004;14:585–598. doi: 10.1016/j.molcel.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, et al. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, LeBeau M, Yates JR, et al. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- Carson CT, Schwartz RA, Stracker TH, Lilley CE, Lee DV, Weitzman MD. EMBO J. 2003;22:6610–6620. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celis JE, Celis A. Proc Natl Acad Sci USA. 1985;82:3262–3266. doi: 10.1073/pnas.82.10.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliby WA, Roberts CJ, Cimprich KA, Stringer CM, Lamb JR, Schreiber SL, et al. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo V, Shechter D, Lupardus PJ, Cimprich KA, Gottesman M, Gautier J. Mol Cell. 2003;11:203–213. doi: 10.1016/s1097-2765(02)00799-2. [DOI] [PubMed] [Google Scholar]
- D'Amours D, Jackson SP. Nat Rev Mol Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- Dart DA, Adams KE, Akerman I, Lakin ND. J Biol Chem. 2004;279:16433–16440. doi: 10.1074/jbc.M314212200. [DOI] [PubMed] [Google Scholar]
- Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Mol Cell Biol. 2004;24:1279–1291. doi: 10.1128/MCB.24.3.1279-1291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smit B, Carr AM, et al. Curr Biol. 2000;10:479–482. doi: 10.1016/s0960-9822(00)00447-4. [DOI] [PubMed] [Google Scholar]
- Difilippantonio S, Celeste A, Fernandez-Capetillo O, Chen HT, Reina San Martin B, Van Laethem F, et al. Nat Cell Biol. 2005;7:675–685. doi: 10.1038/ncb1270. [DOI] [PubMed] [Google Scholar]
- Dodson GE, Shi Y, Tibbetts RS. J Biol Chem. 2004;279:34010–34014. doi: 10.1074/jbc.C400242200. [DOI] [PubMed] [Google Scholar]
- Dong ZW, Zhong Q, Chen P-L. J Biol Chem. 1999;274:19513–19516. doi: 10.1074/jbc.274.28.19513. [DOI] [PubMed] [Google Scholar]
- Downs JA, Jackson SP. Nature. 2003;424:732–734. doi: 10.1038/424732a. [DOI] [PubMed] [Google Scholar]
- Garcia-Muse T, Boulton SJ. Embo J. 2005;24:4345–4355. doi: 10.1038/sj.emboj.7600896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatei M, Young D, Cerosaletti KM, Desai-Mehta A, Spring K, Kozlov S, et al. Nature Genet. 2000;25:115–119. doi: 10.1038/75508. [DOI] [PubMed] [Google Scholar]
- Hekmat-Nejad M, You ZS, Yee MC, Newport JW, Cimprich KA. Curr Biol. 2000;10:1565–1573. doi: 10.1016/s0960-9822(00)00855-1. [DOI] [PubMed] [Google Scholar]
- Helt CE, Cliby WA, Keng PC, Bambara RA, O'Reilly MA. J Biol Chem. 2005;280:1186–1192. doi: 10.1074/jbc.M410873200. [DOI] [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, et al. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- Ivanov EL, Sugawara N, White CI, Fabre F, Haber JE. Mol Cell Biol. 1994;14:3414–3425. doi: 10.1128/mcb.14.5.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, et al. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- Lavin MF. DNA Repair (Amsterdam) 2004;3:1515–1520. doi: 10.1016/j.dnarep.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Lee J, Kumagai A, Dunphy WG. Mol Cell. 2003;11:329–340. doi: 10.1016/s1097-2765(03)00045-5. [DOI] [PubMed] [Google Scholar]
- Lee JH, Paull TT. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Lim DS, Kim ST, Xu B, Maser RS, Lin JY, Petrini JHJ, et al. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Llorente B, Symington LS. Mol Cell Biol. 2004;24:9682–9694. doi: 10.1128/MCB.24.21.9682-9694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupardus PJ, Byun T, Yee MC, Hekmat-Nejad M, Cimprich KA. Genes Dev. 2002;16:2327–2332. doi: 10.1101/gad.1013502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo J, Toczyski D. Curr Opin Cell Biol. 2002;14:237–245. doi: 10.1016/s0955-0674(02)00312-5. [DOI] [PubMed] [Google Scholar]
- Mirzoeva OK, Petrini JHJ. Mol Cell Biol. 2001;21:281–288. doi: 10.1128/MCB.21.1.281-288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Ferguson JR, Symington LS. Mol Cell Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakada D, Hirano Y, Sugimoto K. Mol Cell Biol. 2004;24:10016–10025. doi: 10.1128/MCB.24.22.10016-10025.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghiem P, Park PK, Kim YS, Desai BN, Schreiber SL. J Biol Chem. 2002;277:4428–4434. doi: 10.1074/jbc.M106113200. [DOI] [PubMed] [Google Scholar]
- Nghiem P, Park PK, Kim Y-S, Vaziri C, Schreiber SL. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9092–9097. doi: 10.1073/pnas.161281798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. Nat Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- Raderschall E, Golub EI, Haaf T. Proc Natl Acad Sci USA. 1999;96:1921–1926. doi: 10.1073/pnas.96.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotman G, Shiloh Y. Hum Mol Genet. 1998;7:1555–1563. doi: 10.1093/hmg/7.10.1555. [DOI] [PubMed] [Google Scholar]
- Santocanale C, Diffley JFX. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- Stewart GS, Maser RS, Stankovic T, Bressan DA, Kaplan MI, Jaspers NGJ, et al. Cell. 1999;99:577–587. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- Stiff T, O'Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA. Cancer Res. 2004;64:2390–2396. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- Stiff T, Reis C, Alderton GK, Woodbine L, O'Driscoll M, Jeggo PA. EMBO J. 2005;24:199–208. doi: 10.1038/sj.emboj.7600504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh SY, et al. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts RS, Cortez D, Brumbaugh KM, Scully R, Livingston D, Elledge SJ, et al. Genes Dev. 2000;14:2989–3002. doi: 10.1101/gad.851000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Ogawa H. Mol Cell Biol. 1998;18:260–268. doi: 10.1128/mcb.18.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye BK. Annu Rev Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- Uziel T, Lerenthal Y, Moyal L, Andegeko Y, Mittelman L, Shiloh Y. EMBO J. 2003;22:5612–5621. doi: 10.1093/emboj/cdg541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward IM, Minn K, Chen J. J Biol Chem. 2004;279:9677–9680. doi: 10.1074/jbc.C300554200. [DOI] [PubMed] [Google Scholar]
- Wright JA, Keegan KS, Herendeen DR, Bentley NJ, Carr AM, Hoekstra MF, et al. Proc Natl Acad Sci USA. 1998;95:7445–7450. doi: 10.1073/pnas.95.13.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Kong L, Newport J. J Biol Chem. 2002;277:27088–27093. doi: 10.1074/jbc.M204120200. [DOI] [PubMed] [Google Scholar]
- Yuan SS, Chang HL, Hou MF, Chan TF, Kao YH, Wu YC, et al. Toxicology. 2002;177:123–130. doi: 10.1016/s0300-483x(02)00220-2. [DOI] [PubMed] [Google Scholar]
- Zhong H, Bryson A, Eckersdorff M, Ferguson DO. Hum Mol Genet. 2005;14:2685–2693. doi: 10.1093/hmg/ddi302. [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]