Abstract
Divergent relatives of the Hsp70 protein chaperone such as the Hsp110 and Grp170 families have been recognized for some time, yet their biochemical roles remained elusive. Recent work has revealed that these “atypical” Hsp70s exist in stable complexes with classic Hsp70s where they exert a powerful nucleotide-exchange activity that synergizes with Hsp40/DnaJ-type cochaperones to dramatically accelerate Hsp70 nucleotide cycling. This represents a novel evolutionary transition from an independent protein-folding chaperone to what appears to be a dedicated cochaperone. Contributions of the atypical Hsp70s to established cellular roles for Hsp70 now must be deciphered.
INTRODUCTION
Molecular chaperones are an ancient group of proteins dedicated to protein folding and remodeling, and therefore are ubiquitous from bacteria to humans. The Hsp70 class of chaperones are perhaps the most well known and best-studied due to their cellular abundance and high degree of conservation. Hsp70s are implicated in a wide range of cellular roles, ranging from protein translocation across membranes to regulation of apoptosis, and in all cases are likely to operate by similar fundamental mechanisms of substrate binding/sequestration and release (Bukau and Horwich 1998; Mayer et al 2001). Hsp70s are weak adenosine triphosphatases (ATPases) and cycle through low- and high-affinity substrate-binding states driven by nucleotide hydrolysis: adenosine triphosphate– (ATP) bound Hsp70 binds substrates with low affinity and adenosine 5′-diphosphate–(ADP) bound and nucleotide-free Hsp70 exhibits tight binding to substrate (Schmid et al 1994). Additional proteins, many chaperones themselves, modulate Hsp70 function through regulation of the ATPase cycle and/or substrate targeting. The Hsp40 or DnaJ class of chaperones work in this manner, transiently binding and accelerating ATP hydrolysis to enhance Hsp70 function (Cyr et al 1992). In addition, the minimal J-domain, an approximately 70 amino acid α-helical module found at the amino terminus of the Hsp40 chaperones, is present in multiple nonchaperone proteins where it binds Hsp70 to recruit the chaperone to assist in biological processes (Cheetham and Caplan 1998). A number of evolutionarily unrelated proteins, including bacterial GrpE and mammalian BAG-1 and HspBP1, participate in the other half of the cycle by facilitating exchange of ADP for ATP through distinct mechanisms (Hohfeld and Jentsch 1997; Kabani et al 2002a; Kabani et al 2002b; Shomura et al 2005). However, evidence is accumulating that atypical Hsp70 homologs, including the Hsp110 and Grp170 families, stably interact with the typical Hsp70s in multiple cellular compartments to regulate function through acceleration of nucleotide exchange. This review will focus on the theme that a major role of the atypical Hsp70s is to function as modulators of Hsp70 cellular activities.
ARCHITECTURE OF HSP70 CHAPERONES
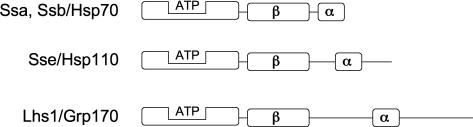
The Hsp70 superfamily of protein chaperones share remarkable sequence and structural similarity, largely based on their highly conserved bipartite domain structure composed of a nucleotide-binding domain (NBD) and a substrate or peptide-binding domain (PBD). The NBD is an independently folding unit of approximately 44 kD that can be observed upon proteolytic cleavage of intact Hsp70 (Buchberger et al 1995). The PBD in the archetypal DnaK-type Hsp70s can be further divided into an 18 kD substrate interaction region, followed by a variable domain of 10 kD. Both of these subdomains, and the variable domain in particular, are divergent in length and serve as the primary characteristic distinguishing the typical DnaK/Hsp70 group from the related Hsp110/Sse and Grp170/Lhs1 groups, that together constitute the Hsp70 superfamily of protein chaperones (Figure 1). For example, the acidic “loop” between the β-sandwich and α-helical lid regions of the PBD is 9 amino acids in length in DnaK, nearly 100 amino acids in the Hsp110s, and greater than 135 in the Grp170 family. A detailed overview of cloning, sequence identities, and phylogenetic relationships between these family members was the subject of a previous review and will not be reiterated here (Easton et al 2000). Although overall sequence identity between the more divergent family members is low, all three groups are clearly derivatives of the ancestral DnaK chaperone. Although eukaryotic Hsp70s generally share about 45% sequence identity, the Hsp110 family is only about 32–35% identical, and identity in the Grp170 family drops to approximately 25% (Boorstein et al 1994; Easton et al 2000). In addition to sequence divergence, the Grp170/Lhs1 family appears restricted to the endoplasmic reticulum (ER) lumen, and the others are cytosolic (Baxter et al 1996; Chen et al 1996; Craven et al 1996; Hamilton and Flynn 1996).
Fig 1.
Domain architecture of the Hsp70 superfamily. The relative arrangement of the nucleotide-binding domain ([NBD]; shown binding adenosine triphosphate [ATP]) and the peptide-binding domain (PBD), composed of the β sandwich and α helical lid subdomains, is indicated. At left are shown the yeast and generic names for the three major Hsp70 subfamilies. The typical Hsp70 also is referred to as the DnaK type, in reference to the sole bacterial Hsp70-type chaperone
HSP110 AND GRP170 ARE NOT PROTEIN-FOLDING CHAPERONES
A hallmark of the Hsp70 chaperone family is their ability to assist in protein folding either of newly synthesized polypeptides or of thermally or chemically denatured substrates by blocking off-pathway reactions such as aggregation. However, the Hsp110 chaperones do not appear to share this function. In contrast, a number of studies have demonstrated that both rodent and yeast Hsp110s are very efficient “holdases” in vitro, binding to and preventing aggregation of denatured proteins (Brodsky et al 1999; Oh et al 1999). In fact, Hsp110 may be a better chaperone in this regard than Hsp70 itself (Oh et al 1997). Mutational analysis of hamster Hsp110 localized the holdase activity to the predicted peptide-binding domain with an additional requirement for C-terminal sequences of unknown function (Oh et al 1999). As with Hsp70, the NBD is not required for substrate binding (Freeman et al 1995). Hsp110-protected substrates are fully competent for subsequent folding by either purified Hsp70/Hsp40 or by cell-free lysates, whereas folding of substrates denatured in the absence of Hsp110 frequently is unproductive, presumably due to the formation of irreversible aggregates (Oh et al 1997; Brodsky et al 1999; Oh et al 1999).
Because protein folding by Hsp70s is mediated by nucleotide cycling within the NBD, the inability of Hsp110 chaperones to fold substrates calls into question the role of this conserved domain. This was addressed biologically through mutant analysis using the yeast Hsp110 homolog Sse1. Site-directed amino acid substitutions of ATPase domain residues previously demonstrated to be crucial for ATP hydrolysis (K69Q) or nucleotide binding (G233D) of the Hsp70 chaperones Ssa1 and BiP reduce the ability of overexpressed SSE1 to suppress the temperature sensitive ydj1-151 allele, but not the ability of Sse1 to promote hormone binding by heterogeneously expressed androgen receptor (Goeckeler et al 2002). However, in a more detailed analysis, a series of additional mutations predicted to impede either nucleotide hydrolysis or nucleotide binding were generated, and the ability of these SSE1 alleles to complement growth and Hsp90 regulatory defects in an sse1Δ strain was assessed. Without exception, only substitutions demonstrated to block or substantially reduce ATP binding abrogated complementation (Shaner et al 2004). Together these data suggest that ATP hydrolysis is unnecessary for the critical functions of Sse1, although nucleotide binding is essential.
Direct biochemical measurements using rodent Hsp110 purified from E. coli initially suggested that the Hsp110 family lacked ATPase activity (Oh et al 1999). Moreover, purified full-length Hsp110 is unable to bind ATP, whereas the isolated NBD does, suggesting the nucleotide-binding pocket may be masked by interaction with the C-terminal region of the protein (Oh et al 1999). This model was further supported by the demonstration that the N- and C-terminal halves of Sse1 interacted with each other when expressed independently in vivo (Shaner et al 2004). However, a recent report successfully described nominal ATPase activities for yeast Sse1 (kcat = 10−2 s−1) and mammalian Apg-2 (kcat = 10−2 s−1) very close to that of Hsp70 (Raviol et al 2006). The Km for ATP binding for both Hsp110 molecules is substantially weaker than Hsp70, making it likely that in vivo Hsp110s likely are poor ATPases (Raviol et al 2006). The Hsp110/170 families therefore must operate by a fundamentally different mechanism than Hsp70s.
HSP110 AND GRP170 BIND HSP70
The first indication that the Hsp110s might interact with other chaperones was provided by studies showing that murine Hsp105 exists in vivo in high molecular-weight protein complexes with Hsp70 and Hsp25 (Hatayama and Yasuda 1998; Wang et al 2000). Although these complexes could be reconstituted in vitro using purified components, the relative abundance of the chaperones was unclear (Wang et al 2000). In addition, thermally denatured luciferase associated with these complexes, raising the possibility that native substrates may have contributed to the broad range of apparent masses from 160– 500 kD observed using size exclusion chromatography (Wang et al 2000). A series of recent papers utilizing immunoprecipitation approaches from yeast extracts firmly has established interaction between Hsp70 and Hsp110/ 170s. A search for binding partners for the Saccharomyces cerevisiae Grp170 homolog Lhs1, an ER-resident chaperone, identified the ER Hsp70, Kar2/BiP (Steel et al 2004). Likewise, two groups discovered that the yeast Sse proteins existed in stable but distinct complexes with the two classes of cytosolic Hsp70, Ssa and Ssb (Shaner et al 2005; Yam et al 2005). Unlike the murine Hsp110·Hsp70 complexes, the yeast complexes observed were strongly suggestive of a binary interaction, based on observed molecular masses and in vitro reconstitution studies (Shaner et al 2005; Yam et al 2005). Furthermore, other Hsp70-modulating proteins, such as Ydj1 in the case of Sse1·Ssa, and Sil1 in the case of Lhs1·Kar2, are not found in native complexes and are not required for interaction in vitro (Steel et al 2004; Shaner et al 2005; Yam et al 2005). Subsequent work has demonstrated that Sse1 binds yeast and mammalian Hsp70s in a 1:1 stoichiometry, with nanomolar affinities (Dragovic et al 2006; Raviol et al 2006; Shaner et al 2006). Moreover, ATP binding by Sse1, but not the Hsp70 partner, appears to be a requirement for heterodimerization, as ascertained through the use of SSE1 ATP-binding mutants (Dragovic et al 2006; Raviol et al 2006; Shaner et al 2006). This also was demonstrated biochemically by significant enhancement of heterodimer formation in vitro with ATP or transition state nucleotide analogs versus ADP (Shaner et al 2006).
The domain arrangements within the Hsp110·Hsp70 or Grp170·Hsp70 heterodimers at present are unknown. Native gel electrophoresis and protease protection experiments support a compact association that ultimately may facilitate future cocrystallization (Shaner et al 2006). Nevertheless, in the absence of high-resolution structural studies, analysis of truncation mutants and expression of individual domains of both proteins already is yielding clues. Both the Sse1 and Sse2 NBDs are capable of interacting with the Ssa and Ssb proteins, although Sse2 interacts less well with the Ssb chaperones for unknown reasons (Shaner et al 2005; Yam et al 2005; Shaner et al 2006). Moreover, Sse1 strongly stimulates the ATPase activity of purified bovine Hsc70 NBD, demonstrating that both binding and nucleotide exchange do not require the Hsp70 PBD (Shaner et al 2006). Interestingly, the Sse1, but not the Sse2, NBD is highly unstable when expressed alone in either yeast or bacteria, but is stabilized in the presence of the Sse PBD (Shaner et al 2004). It is possible, therefore, that the PBD may bind and stabilize the NBD in vivo. In fact, both fragments coimmunoprecipitate when expressed in yeast, suggesting either the presence of a docking site on one domain for the other, or concurrent binding of both fragments to the same Hsp70 monomer (Shaner et al 2004).
HSP110 AND GRP170 ARE HSP70 NUCLEOTIDE EXCHANGE FACTORS
A functional consequence of the interaction between the atypical and typical Hsp70s was provided by the observations that Lhs1 stimulates Kar2 ATPase activity, and Sse1 stimulates ATPase activity of yeast and mammalian cytosolic Hsp70s. Several lines of evidence demonstrate that the stimulatory mechanism involves enhanced nucleotide exchange. First, both Sse1 and Lhs1 increase steady state ([ATP]>[Hsp70]), but not single-turnover ([ATP]<[Hsp70]) ATPase activity, indicative of an increased aggregate cycling rate versus increased ATP hydrolysis alone (Steel et al 2004; Shaner et al 2005). Second, substantial stimulation requires inclusion of a J-protein, which stimulates ATP hydrolysis, making nucleotide exchange the rate-limiting step in the reaction (Steel et al 2004; Shaner et al 2005). This hypothesis was confirmed for Lhs1, Sse1, and mammalian Hsp110, which were demonstrated to be potent nucleotide exchange factors (NEF) for their respective Hsp70s (Steel et al 2004; Dragovic et al 2006; Raviol et al 2006; Shaner et al 2006). In fact, Sse1 and Lhs1 are more potent NEFs than the previously described cognate NEFs (Sil1 for Kar2; Fes1 for Ssa) based on their ability to stimulate steady-state ATPase activity (Tyson and Stirling 2000; Kabani et al 2002a; Steel et al 2004; Raviol et al 2006). This comparison is confounded in the case of Lhs1, however, because binding with Kar2 reciprocally stimulates its own cryptic ATPase (Steel et al 2004). In fact, ATP hydrolysis by Kar2 is required for hydrolysis by Lhs1, suggesting that the ATPases in the heterodimer are coupled functionally (Steel et al 2004).
By what mechanism do the atypical Hsp70 chaperone enhance Hsp70 nucleotide exchange? To date, two distinct mechanisms of stimulating nucleotide release from Hsp70 have been characterized. The Hsp70 NBD is composed of two lobes framing a deep nucleotide-binding cleft (Flaherty et al 1990). The bacterial GrpE and eukaryotic BAG proteins operate through a common mechanism involving conformational change in lobe II with little structural perturbation elsewhere in the NBD (Harrison et al 1997; Sondermann et al 2001). In contrast, the mammalian NEF HspBP1 induces substantial deformation between the two lobes of the NBD upon binding lobe II (Shomura et al 2005). HspBP1 structurally is unrelated to both GrpE and BAG domain-containing proteins, suggesting that Hsp70 nucleotide exchange activity has evolved independently multiple times. It is likely that binding of the Hsp110 or Grp170 NBD to the Hsp70 NBD results in structural deformation of the latter to a state with significantly lower nucleotide-binding affinity. The nature of this conformational change awaits detailed analysis of a cocrystal structure. A role for the Sse/Hsp110 PBD and C-terminus also must be considered, because the Sse2 NBD, though capable of Hsp70 binding, does not complement sse1Δ growth phenotypes (Shaner et al 2006). In addition, C-terminal truncations of Sse1 eliminate both exchange activity and complementation (Dragovic et al 2006; Shaner et al 2006). The Sse/Hsp110 PBD therefore may be required to properly position the paired NBDs. The observation that the Sse1 NBD and PBD functionally complement the sse1Δ mutation when expressed in trans suggests that the two domains need not be linearly coupled to effect nucleotide exchange (Shaner et al 2004).
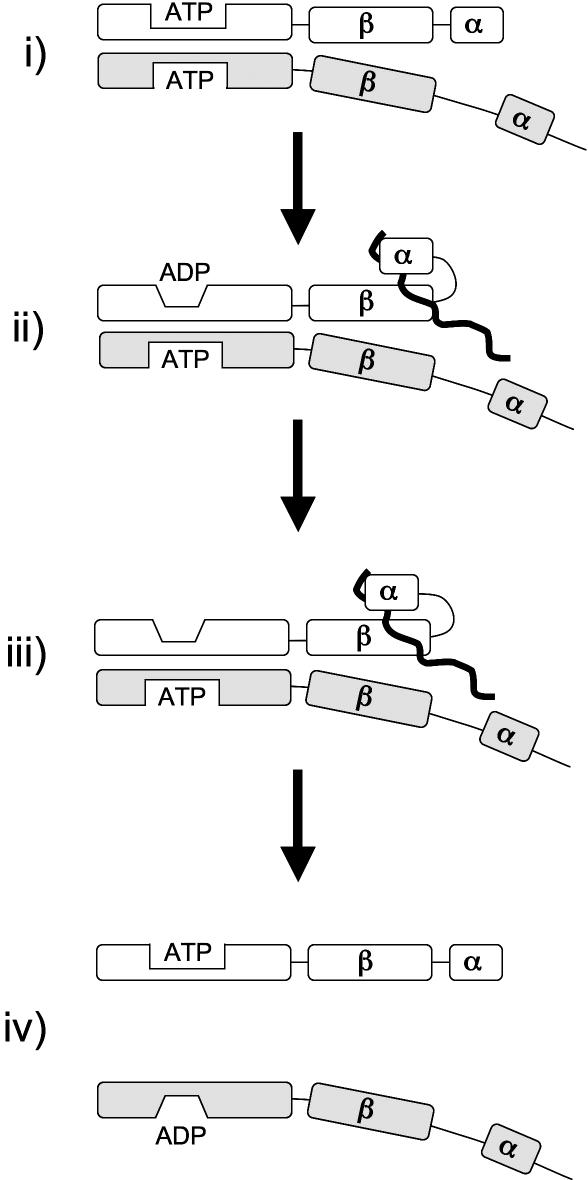
A revised model for the Hsp70 nucleotide and protein-folding cycle incorporating the atypical Hsp70s is presented in Figure 2. In step (i), a heterodimeric Hsp110·Hsp70 or Grp170·Hsp70 complex is formed with both proteins in the ATP bound state. Though the nucleotide status of Hsp70 is not a determinant of dimerization, ATP bound within the Hsp110 (Sse1) NBD is required for complex formation. Substrate recruitment and stimulation of Hsp70 ATP hydrolysis by an Hsp40 J-type chaperone yields step (ii), with the Hsp70 PBD complexed with a substrate in the high-affinity state. Contributions of the atypical Hsp70 PBD to substrate binding at present are unknown. Complex (ii) likely transitions rapidly to step (iii), with Hsp70 in the nucleotide-free state, which resembles the ADP-bound state in terms of Hsp70 conformation and substrate affinity (Raviol et al 2006). We then postulate that the weak intrinsic ATPase activity of the Hsp110 chaperones, and the stimulated ATPase of Lhs1 (and perhaps Grp170), hydrolyzes bound ATP, resulting in dissociation of the heterodimer and subsequent recharging of both NBDs.
Fig 2.
Hsp70 heterodimer nucleotide and protein-folding cycle. The well-established Hsp70 nucleotide cycle is shown taking into account heterodimerization with an atypical Hsp70 and nucleotide status of both proteins. Characteristics of each step are outlined in the main text. The typical Hsp70 is represented with white boxes and the atypical with grey boxes. A peptide substrate is represented as the black squiggle line associated with the typical Hsp70
SSZ1 AS A J-PROTEIN MODULATOR
Not all atypical Hsp70 proteins may operate as heterodimeric partners of a classic Hsp70. The J-protein zuotin (Zuo1) acts as an ATPase activator for the ribosome-associated classic Hsp70s Ssb1/2 and is itself tightly associated with assembled ribosomes (Huang et al 2005). In addition, another protein resembling a classic Hsp70 called Ssz1 is highly enriched with the ribosome (Hallstrom et al 1998; Gautschi et al 2001). Unlike most J-protein–Hsp70 interactions, which are weak and transient, Zuo1, Ssz1, and the Ssbs exist in a stable heterotrimer denoted the ribosome-associated complex (RAC) (Mayer et al 1999; Gautschi et al 2002; Hundley et al 2002). In keeping with the idea that all three proteins operate within the same complex, mutation of each gene independently results in the identical set of phenotypes: slow growth, cold sensitivity, and profound sensitivity to the translation inhibitor paromomycin (Gautschi et al 2002; Hundley et al 2002). Interestingly, Ssz1 lacking its peptide-binding domain fully complements ssz1Δ mutants, and Ssb chaperones require substrate binding to function, suggesting that, despite close sequence homology to other typical cytosolic Hsp70s, Ssz does not function as one (Hundley et al 2002). Instead, Ssz appears to partner with Zuo1 to activate Ssb's ATPase through an unknown mechanism. In fact, Ssb cannot be crosslinked to emerging nascent chains in the absence of Ssz1 (Gautschi et al 2002). Unlike the clear requirement for ATP binding for Sse1 to stimulate Hsp70 nucleotide exchange, mutations in the Ssz1 ATPase domain have little to no effect on RAC formation or Ssb stimulation (Huang et al 2005). Ssz1 is not required absolutely for Zuo1 J-protein activity, as ZUO1 overexpression partially suppresses ssz1Δ phenotypes (Hundley et al 2002). In further support of this observation, purified Mpp11, the human zuotin ortholog, is capable of activating Hsp70 ATPase activity alone (Hundley et al 2005). Together, these findings suggest that Ssz1 has evolved a unique scaffolding role to organize Zuo1 and Ssb into a dedicated ribosomal protein-folding complex.
ATYPICAL HSP70S REGULATE HSP70 FUNCTIONS IN VIVO
Hsp70 chaperones are essential to life, presumably due to the myriad of protein folding and translocation roles they play. However, many Hsp70 modulatory proteins, for example Ydj1 and Fes1 (for Ssa) and Sil1 (for Kar2) in budding yeast, display varying degrees of growth impairment while remaining viable (Caplan and Douglas 1991; Tyson and Stirling 2000; Kabani et al 2002a). Loss of Lhs1 is lethal in combination with deletion of SIL1, suggesting that the nucleotide exchange activities of these two proteins are partially redundant (Tyson and Stirling 2000). Though sse1Δ mutants are slow growing and temperature sensitive, sse2Δ mutants display no detectable phenotypes (Mukai et al 1993). Simultaneous deletion of both genes is synthetically lethal, although contrasting studies have suggested otherwise (Mukai et al 1993; Trott et al 2005; Yam et al 2005; Raviol et al 2006). The primary role for the Sse proteins in the yeast cytosol appears to be nucleotide exchange, because overexpression of the structurally unrelated NEF Fes1 confers viability to sse1Δ sse2Δ mutants (Raviol et al 2006; Shaner et al 2006). Likewise, Sil1 overexpression rescues lethality caused by simultaneous deletion of LHS1 and IRE1, encoding the master regulator of the unfolded protein response (Steel et al 2004). Given that the Sse and Fes1 proteins exhibit some degree of functional redundancy, it is possible that strain-specific perturbations in expression of Fes1 or perhaps the BAG homolog Snl1 may contribute to the differential results of SSE1 and SSE2 deletion (Sondermann et al 2002).
Are atypical Hsp70s involved in all Hsp70-mediated cellular processes, or are Hsp110·Hsp70 and Grp170·Hsp70 heterodimers recruited for specific substrates? To date only a few biological processes have been demonstrated to be dependent on function of an atypical Hsp70. The budding yeast mating pheromone alpha factor (αF) is post-translationally inserted into the ER, followed by extensive glycosyl and proteolytic processing prior to secretion (Brodsky et al 1993). Productive interaction with Hsp70s is required on both the cytosolic (Ssa) and lumenal (Kar2) sides of the ER membrane for maximal αF biosynthesis, because mutations in either chaperone system result in accumulation of precursor forms of the protein (Vogel et al 1990; Brodsky et al 1993). In keeping with the notion that the atypical Hsp70 NEFs promote Hsp70 activities, mutations in LHS1 or SSE1 result in αF precursor accumulation, but do not affect cotranslational, Hsp70-independent import (Shaner et al 2004; Steel et al 2004).
Hsp70s intimately are associated with protein synthesis, hinting at potential roles for the cytosolic Hsp110 in translation. The yeast cytosol contains two classes of Hsp70 required for efficient protein synthesis, the Ssb family that interacts directly with the ribosome and nascent chains, and the Ssa group that binds post-translationally to a subset of newly synthesized polypeptides (James et al 1997). Sse1 forms independent complexes with Ssa and Ssb chaperones, and both Sse·Ssa and Sse·Ssb complexes can be crosslinked to newly translated proteins (Shaner et al 2005; Yam et al 2005). In cells lacking SSE1, translating polypeptides and synthesized proteins exhibit delayed maturation as evidenced by slower release from the Hsp70s (Yam et al 2005). Hsp110 therefore serves to modulate protein translation through regulation of Hsp70 folding kinetics. Based on this relationship, misfolding defects may be expected in sse1Δ mutants, perhaps severe enough to explain the lethality observed in sse1Δ sse2Δ cells. In support of this prediction, fes1Δ cells also exhibit translational defects, functionally implicating both NEFs in protein synthesis (Kabani et al 2002a). Hsp110s also are likely to be involved in suppression of aggregation and protein refolding after proteotoxic stresses, such as thermal shock, as demonstrated for luciferase in yeast and polyglutamine-containing androgen receptor in mammalian cells (Ishihara et al 2003; Dragovic et al 2006; Raviol et al 2006). Sse1 is implicated additionally in folding and regulation of substrates of the Hsp90 chaperone complex, including heterologous steroid receptors and the heat shock transcription factor, Hsf1 (Liu et al 1999; Lee et al 2004). Sse1 physically associates with Hsp90, but detailed protein-protein interaction studies have not been carried out, nor is there yet evidence for Hsp110 interacting with mammalian Hsp90 homologs (Liu et al 1999). Because Hsp70 is a critical component of Hsp90 complex function, it is tempting to speculate that Sse1 partners with the Ssa Hsp70 to accelerate substrate maturation within this multichaperone machine (Picard 2002).
CONCLUSION
The atypical Hsp70 protein chaperones were identified over 25 years ago, yet have received a fraction of the attention paid to their molecular cousins, despite being highly abundant and conserved components of the heat shock response (Subjeck et al 1982). The identification of the Hsp110 and Grp170 chaperones as essential Hsp70 binding partners with potent NEF activity brings these proteins from the periphery of the stress and chaperone field into the limelight. Clearly Hsp70 NEFs are clinically relevant, because mutations in the Kar2 NEF Sil1 are responsible for the rare autosomal recessive neurodegenerative disorder Marinesco-Sjögren Syndrome (Anttonen et al 2005; Senderek et al 2005). To date no human disease loci have been mapped in the vicinity of the Hsp110 or Grp170 genes. However, the demonstrated high expression of Hsp110 in human brain tissue is in keeping with the growing realization that the pathology of many neurodegenerative diseases is based on aberrant protein folding (Hylander et al 2000).
A number of questions remain. Though the function of the Grp170 group is limited to ER lumenal events requiring the sole resident Hsp70 Kar2/BiP, Hsp110 may be recruited to multiple cytosolic processes that involve distinct populations of Hsp70. How is this distribution regulated? The relative abundance of Hsp110 and Hsp70 in yeast (1:9) suggests that the Sse/Ssa and Sse/Ssb heterodimers constitute a small fraction of the total active Hsp70 in the cytosol (Ghaemmaghami et al 2003). However, this subset is also more “active” in the sense that nucleotide cycling is accelerated dramatically versus that of Hsp70 alone. The question of how this interaction arose is also significant. Little evidence exists for homodimerizaton of the typical Hsp70s, yet some reports have provided genetic and biochemical support for this idea (Nicolet and Craig 1991; Tokunaga et al 1992). A tantalizing possibility is that the Hsp70 NBDs possess cryptic binding interfaces that mediate weak and transient homodimerization, and that the atypical Hsp70s have unmasked this association to result in highly stable heterodimers. Simultaneously these chaperones lost the ability to engage in productive protein refolding while retaining substrate-binding capability. Biochemistry and genetics have provided a mechanism for the atypical Hsp70 family members. We will now look to structural biology to explain the architecture, and cell biology to provide a purpose.
Acknowledgments
The authors apologize to workers whose individual contributions could not be cited directly due to space constraints. Work in the authors' laboratory is supported by Research Scholar grant MBC-103134 from the American Cancer Society and grant GM074696 from the National Institutes of Health.
REFERENCES
- Anttonen AK, Mahjneh I, and Hamalainen RH. et al. 2005 The gene disrupted in Marinesco-Sjogren syndrome encodes SIL1, an HspA5 cochaperone. Nat Genet. 37:1309–1311. [DOI] [PubMed] [Google Scholar]
- Baxter BK, James P, Evans T, Craig EA. SSI1 encodes a novel Hsp70 of the Saccharomyces cerevisiae endoplasmic reticulum. Mol Cell Biol. 1996;16:6444–6456. doi: 10.1128/mcb.16.11.6444.1098-5549(1996)016[6444:SEANHO]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorstein WR, Ziegelhoffer T, Craig EA. Molecular evolution of the Hsp70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490.0022-2844(1994)038[0001:MEOTHM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Brodsky JL, Hamamoto S, Feldheim D, Schekman R. Reconstitution of protein translocation from solubilized yeast membranes reveals topologically distinct roles for BiP and cytosolic Hsc70. J Cell Biol. 1993;120:95–102. doi: 10.1083/jcb.120.1.95.0021-9525(1993)120[0095:ROPTFS]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, Werner ED, Dubas ME, Goeckeler JL, Kruse KB, McCracken AA. The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J Biol Chem. 1999;274:3453–3460. doi: 10.1074/jbc.274.6.3453.0021-9258(1999)274[3453:TRFMCD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Buchberger A, Theyssen H, Schroder H, McCarty JS, Virgallita G, Milkereit P, Reinstein J, Bukau B. Nucleotide-induced conformational changes in the ATPase and substrate binding domains of the DnaK chaperone provide evidence for interdomain communication. J Biol Chem. 1995;270:16903–16910. doi: 10.1074/jbc.270.28.16903.0021-9258(1995)270[16903:NCCITA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9.0092-8674(1998)092[0351:THAHCM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Caplan AJ, Douglas MG. Characterization of YDJ1: a yeast homologue of the bacterial DnaJ protein. J Cell Biol. 1991;114:609–621. doi: 10.1083/jcb.114.4.609.0021-9525(1991)114[0609:COYAYH]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham ME, Caplan AJ. Structure, function, and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2.1466-1268(1998)003[0028:SFAEOD]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Easton D, Oh HJ, Lee-Yoon DS, Liu X, Subjeck J. The 170 kDa glucose regulated stress protein is a large Hsp70-, Hsp110-like protein of the endoplasmic reticulum. FEBS Lett. 1996;380:68–72. doi: 10.1016/0014-5793(96)00011-7.0014-5793(1996)380[0068:TKGRSP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Craven RA, Egerton M, Stirling CJ. A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of a number of protein precursors. EMBO J. 1996;15:2640–2650.1460-2075(1996)015[2640:ANHOTY]2.0.CO;2 [PMC free article] [PubMed] [Google Scholar]
- Cyr DM, Lu X, Douglas MG. Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J Biol Chem. 1992;267:20927–20931.0021-9258(1992)267[20927:ROHFBA]2.0.CO;2 [PubMed] [Google Scholar]
- Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25:2519–2528. doi: 10.1038/sj.emboj.7601138.1460-2075(2006)025[2519:MCOTHF]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton DP, Kaneko Y, Subjeck JR. The Hsp110 and Grp170 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaperones. 2000;5:276–290. doi: 10.1379/1466-1268(2000)005<0276:thagsp>2.0.co;2.1466-1268(2000)005[0276:THAGSP]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KM, DeLuca-Flaherty C, McKay DB. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990;346:623–628. doi: 10.1038/346623a0.1476-4687(1990)346[0623:TSOTAF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Freeman BC, Myers MP, Schumacher R, Morimoto RI. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding, and interaction with HDJ-1. EMBO J. 1995;14:2281–2292. doi: 10.1002/j.1460-2075.1995.tb07222.x.1460-2075(1995)014[2281:IOARMI]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi M, Lilie H, Funfschilling U, Mun A, Ross S, Lithgow T, Rucknagel P, Rospert S. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc Natl Acad Sci USA. 2001;98:3762–3767. doi: 10.1073/pnas.071057198.1091-6490(2001)098[3762:RASRCI]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi M, Mun A, Ross S, Rospert S. A functional chaperone triad on the yeast ribosome. Proc Natl Acad Sci USA. 2002;99:4209–4214. doi: 10.1073/pnas.062048599.1091-6490(2002)099[4209:AFCTOT]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046.1476-4687(2003)425[0737:GAOPEI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Goeckeler JL, Stephens A, Lee P, Caplan AJ, Brodsky JL. Overexpression of yeast Hsp110 homolog Sse1p suppresses ydj1-151 thermosensitivity and restores Hsp90-dependent activity. Mol Biol Cell. 2002;13:2760–2770. doi: 10.1091/mbc.02-04-0051.1059-1524(2002)013[2760:OOYHHS]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallstrom TC, Katzmann DJ, Torres RJ, Sharp WJ, Moye-Rowley WS. Regulation of transcription factor Pdr1p function by an Hsp70 protein in. Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:1147–1155. doi: 10.1128/mcb.18.3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton TG, Flynn GC. Cer1p, a novel Hsp70-related protein required for posttranslational endoplasmic reticulum translocation in yeast. J Biol Chem. 1996;271:30610–30613. doi: 10.1074/jbc.271.48.30610.0021-9258(1996)271[30610:CANHPR]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Harrison CJ, Hayer-Hartl M, Di Liberto M, Hartl F, Kuriyan J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science. 1997;276:431–435. doi: 10.1126/science.276.5311.431.0193-4511(1997)276[0431:CSOTNE]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hatayama T, Yasuda K. Association of Hsp105 with Hsc70 in high molecular mass complexes in mouse FM3A cells. Biochem Biophys Res Commun. 1998;248:395–401. doi: 10.1006/bbrc.1998.8979.0006-291X(1998)248[0395:AOHWHI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hohfeld J, Jentsch S. GrpE-like regulation of the hsc70 chaperone by the antiapoptotic protein BAG-1. EMBO J. 1997;16:6209–6216. doi: 10.1093/emboj/16.20.6209.1460-2075(1997)016[6209:GROTHC]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Gautschi M, Walter W, Rospert S, Craig EA. The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat Struct Mol Biol. 2005;12:497–504. doi: 10.1038/nsmb942.1545-9985(2005)012[0497:THSMTF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hundley H, Eisenman H, Walter W, Evans T, Hotokezaka Y, Wiedmann M, Craig E. The in vivo function of the ribosome-associated Hsp70, Ssz1, does not require its putative peptide-binding domain. Proc Natl Acad Sci USA. 2002;99:4203–4208. doi: 10.1073/pnas.062048399.1091-6490(2002)099[4203:TIVFOT]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundley HA, Walter W, Bairstow S, Craig EA. Human Mpp11 J protein: ribosome-tethered molecular chaperones are ubiquitous. Science. 2005;308:1032–1034. doi: 10.1126/science.1109247.0193-4511(2005)308[1032:HMJPRM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hylander BL, Chen X, Graf PC, Subjeck JR. The distribution and localization of Hsp110 in brain. Brain Res. 2000;869:49–55. doi: 10.1016/s0006-8993(00)02346-5.0006-8993(2000)869[0049:TDALOH]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Ishihara K, Yamagishi N, Saito Y, Adachi H, Kobayashi Y, Sobue G, Ohtsuka K, Hatayama T. Hsp105α suppresses the aggregation of truncated androgen receptor with expanded CAG repeats and cell toxicity. J Biol Chem. 2003;278:25143–25150. doi: 10.1074/jbc.M302975200.0021-9258(2003)278[25143:HSTAOT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- James P, Pfund C, Craig EA. Functional specificity among Hsp70 molecular chaperones. Science. 1997;275:387–389. doi: 10.1126/science.275.5298.387.0193-4511(1997)275[0387:FSAHMC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kabani M, Beckerich JM, Brodsky JL. Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol Cell Biol. 2002a;22:4677–4689. doi: 10.1128/MCB.22.13.4677-4689.2002.1098-5549(2002)022[4677:NEFFTY]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M, McLellan C, Raynes DA, Guerriero V, Brodsky JL. HspBP1, a homologue of the yeast Fes1 and Sls1 proteins, is an Hsc70 nucleotide exchange factor. FEBS Lett. 2002b;531:339–342. doi: 10.1016/s0014-5793(02)03570-6.0014-5793(2002)531[0339:HAHOTY]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Lee P, Shabbir A, Cardozo C, Caplan AJ. Sti1 and Cdc37 can stabilize Hsp90 in chaperone complexes with a protein kinase. Mol Biol Cell. 2004;15:1785–1792. doi: 10.1091/mbc.E03-07-0480.1059-1524(2004)015[1785:SACCSH]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XD, Morano KA, Thiele DJ. The yeast Hsp110 family member, Sse1, is an Hsp90 cochaperone. J Biol Chem. 1999;274:26654–26660. doi: 10.1074/jbc.274.38.26654.0021-9258(1999)274[26654:TYHFMS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Mayer MP, Brehmer D, Gassler CS, Bukau B. Hsp70 chaperone machines. Adv Protein Chem. 2001;59:1–44. doi: 10.1016/s0065-3233(01)59001-4.0065-3233(2001)059[0001:HCM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Mayer MP, Laufen T, Paal K, McCarty JS, Bukau B. Investigation of the interaction between DnaK and DnaJ by surface plasmon resonance spectroscopy. J Mol Biol. 1999;289:1131–1144. doi: 10.1006/jmbi.1999.2844.0022-2836(1999)289[1131:IOTIBD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Mukai H, Kuno T, Tanaka H, Hirata D, Miyakawa T, Tanaka C. Isolation and characterization of SSE1 and SSE2, new members of the yeast Hsp70 multigene family. Gene. 1993;132:57–66. doi: 10.1016/0378-1119(93)90514-4.0378-1119(1993)132[0057:IACOSA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Nicolet CM, Craig EA. Functional analysis of a conserved amino-terminal region of Hsp70 by site-directed mutagenesis. Yeast. 1991;7:699–716. doi: 10.1002/yea.320070706.0749-503X(1991)007[0699:FAOACA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Oh HJ, Chen X, Subjeck JR. Hsp110 protects heat-denatured proteins and confers cellular thermoresistance. J Biol Chem. 1997;272:31636–31640. doi: 10.1074/jbc.272.50.31636.0021-9258(1997)272[31636:HPHPAC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Oh HJ, Easton D, Murawski M, Kaneko Y, Subjeck JR. The chaperoning activity of Hsp110. Identification of functional domains by use of targeted deletions. J Biol Chem. 1999;274:15712–15718. doi: 10.1074/jbc.274.22.15712.0021-9258(1999)274[15712:TCAOHI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Picard D. Heat shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci. 2002;59:1640–1648. doi: 10.1007/PL00012491.1420-682X(2002)059[1640:HSPACF]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raviol H, Bukau B, Mayer MP. Human and yeast Hsp110 chaperones exhibit functional differences. FEBS Lett. 2006;580:168–174. doi: 10.1016/j.febslet.2005.11.069.0014-5793(2006)580[0168:HAYHCE]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 2006;25:2510–2518. doi: 10.1038/sj.emboj.7601139.1460-2075(2006)025[2510:CNITYC]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid D, Baici A, Gehring H, Christen P. Kinetics of molecular chaperone action. Science. 1994;263:971–973. doi: 10.1126/science.8310296.0193-4511(1994)263[0971:KOMCA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Senderek J, Krieger M, and Stendel C. et al. 2005 Mutations in SIL1 cause Marinesco-Sjogren syndrome, a cerebellar ataxia with cataract and myopathy. Nat Genet. 37:1312–1314. [DOI] [PubMed] [Google Scholar]
- Shaner L, Sousa R, Morano KA. Characterization of Hsp70 binding and nucleotide exchange by the yeast Hsp110 chaperone Sse1. Biochemistry. 2006;45:15075–15084. doi: 10.1021/bi061279k.0006-2960(2006)045[15075:COHBAN]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner L, Trott A, Goeckeler JL, Brodsky JL, Morano KA. The function of the yeast molecular chaperone Sse1 is mechanistically distinct from the closely related Hsp70 family. J Biol Chem. 2004;279:21992–22001. doi: 10.1074/jbc.M313739200.0021-9258(2004)279[21992:TFOTYM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Shaner L, Wegele H, Buchner J, Morano KA. The yeast Hsp110 Sse1 functionally interacts with the Hsp70 chaperones Ssa and Ssb. J Biol Chem. 2005;280:41262–41269. doi: 10.1074/jbc.M503614200.0021-9258(2005)280[41262:TYHSFI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Shomura Y, Dragovic Z, and Chang HC. et al. 2005 Regulation of Hsp70 function by HspBP1: structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol Cell. 17:367–379. [DOI] [PubMed] [Google Scholar]
- Sondermann H, Ho AK, Listenberger LL, Siegers K, Moarefi I, Wente SR, Hartl FU, Young JC. Prediction of novel Bag-1 homologs based on structure/function analysis identifies Snl1p as an Hsp70 cochaperone in Saccharomyces cerevisiae. J Biol Chem. 2002;277:33220–33227. doi: 10.1074/jbc.M204624200.0021-9258(2002)277[33220:PONBHB]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Sondermann H, Scheufler C, Schneider C, Hohfeld J, Hartl FU, Moarefi I. Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science. 2001;291:1553–1557. doi: 10.1126/science.1057268.0193-4511(2001)291[1553:SOAHCC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Steel GJ, Fullerton DM, Tyson JR, Stirling CJ. Coordinated activation of Hsp70 chaperones. Science. 2004;303:98–101. doi: 10.1126/science.1092287.0193-4511(2004)303[0098:CAOHC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Subjeck JR, Sciandra JJ, Johnson RJ. Heat shock proteins and thermotolerance: a comparison of induction kinetics. Br J Radiol. 1982;55:579–584. doi: 10.1259/0007-1285-55-656-579.0007-1285(1982)055[0579:HSPATA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Tokunaga M, Kawamura A, Kohno K. Purification and characterization of BiP/Kar2 protein from Saccharomyces cerevisiae. J Biol Chem. 1992;267:17553–17559.0021-9258(1992)267[17553:PACOKP]2.0.CO;2 [PubMed] [Google Scholar]
- Trott A, Shaner L, Morano KA. The molecular chaperone Sse1 and the growth control protein kinase Sch9 collaborate to regulate protein kinase A activity in Saccharomyces cerevisiae. Genetics. 2005;170:1009–1021. doi: 10.1534/genetics.105.043109.0016-6731(2005)170[1009:TMCSAT]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson JR, Stirling CJ. LHS1 and SIL1 provide a lumenal function that is essential for protein translocation into the endoplasmic reticulum. EMBO J. 2000;19:6440–6452. doi: 10.1093/emboj/19.23.6440.1460-2075(2000)019[6440:LASPAL]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Misra LM, Rose MD. Loss of BiP/GRP78 function blocks translocation of secretory proteins in yeast. J Cell Biol. 1990;110:1885–1895. doi: 10.1083/jcb.110.6.1885.0021-9525(1990)110[1885:LOGFBT]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XY, Chen X, Oh HJ, Repasky E, Kazim L, Subjeck J. Characterization of native interaction of Hsp110 with Hsp25 and Hsc70. FEBS Lett. 2000;465:98–102. doi: 10.1016/s0014-5793(99)01733-0.0014-5793(2000)465[0098:CONIOH]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Yam AY, Albanese V, Lin HT, Frydman J. Hsp110 cooperates with different cytosolic Hsp70 systems in a pathway for de novo folding. J Biol Chem. 2005;280:41252–41261. doi: 10.1074/jbc.M503615200.0021-9258(2005)280[41252:HCWDCH]2.0.CO;2 [DOI] [PubMed] [Google Scholar]