Abstract
Phage P22 wild-type (WT) coat protein does not require GroEL/S to fold but temperature-sensitive–folding (tsf) coat proteins need the chaperone complex for correct folding. WT coat protein and all variants absolutely require P22 scaffolding protein, an assembly chaperone, to assemble into precursor structures termed procapsids. Previously, we showed that a global suppressor (su) substitution, T166I, which rescues several tsf coat protein variants, functioned by inducing GroEL/S. This led to an increased formation of tsf:T166I coat protein:GroEL complexes compared with the tsf parents. The increased concentration of complexes resulted in more assembly-competent coat proteins because of a shift in the chaperone-driven kinetic partitioning between aggregation-prone intermediates toward correct folding and assembly. We have now investigated the folding and assembly of coat protein variants that carry a different global su substitution, F170L. By monitoring levels of phage production in the presence of a dysfunctional GroEL we found that tsf:F170L proteins demonstrate a less stringent requirement for GroEL. Tsf:F170L proteins also did not cause induction of the chaperones. Circular dichroism and tryptophan fluorescence indicate that the native state of the tsf: F170L coat proteins is restored to WT-like values. In addition, native acrylamide gel electrophoresis shows a stabilized native state for tsf:F170L coat proteins. The F170L su substitution also increases procapsid production compared with their tsf parents. We propose that the F170L su substitution has a decreased requirement for the chaperones GroEL and GroES as a result of restoring the tsf coat proteins to a WT-like state. Our data also suggest that GroEL/S can be induced by increasing the population of unfolding intermediates.
INTRODUCTION
The amino acid sequence determines the final fold for a protein (Anfinsen 1973). Relatively small changes such as single amino acid substitutions can disrupt the folding process and misfolded proteins that aggregate can result (Thomas et al 1995). To alleviate these folding defects, second site amino acid substitutions, termed suppressor substitutions (su), are able to revert the misfolding caused by the original substitution (Mitraki et al 1991; Beißinger et al 1995; Sideraki et al 2001). Sites of both the single amino acid substitutions and second site suppressor substitutions pinpoint areas in the protein that are crucial for the proper folding and stability of the molecule.
The coat protein of bacteriophage P22, a double-stranded (ds) DNA phage that infects Salmonella, was the model system chosen for this study. During phage P22 assembly, 420 coat protein monomers, along with 60–300 scaffolding protein monomers (Casjens and King 1974; Casjens 1979; Eppler et al 1991; Thuman-Commike et al 1996; Parent et al 2004, 2006); 12–20 copies of each of the injection proteins, gp7, gp16, and gp20 (Botstein et al 1973; Israel 1977); and the unique portal complex (Bazinet et al 1988), coassemble into a precursor structure termed the procapsid. On completion of the procapsid, dsDNA is packaged via the portal (Casjens and King 1974), and scaffolding protein exits, likely through holes in the procapsid lattice; concomitantly, a change in overall capsid morphology converts the round-appearing procapsid to the classic icosahedral shape of many viruses (Prasad et al 1993) and is accompanied by an expansion in head volume by ∼10% (Earnshaw et al 1976). Finally, the phage head is stabilized by the addition of plug and tailspike proteins to form the mature, infectious phage (Strauss and King 1984).
P22 coat protein is an established model for protein folding, especially for proteins destined for macromolecular assembly. P22 coat protein comprises 429 amino acids and has a monomeric molecular mass of 47 kDa (Eppler et al 1991). The wild-type (WT) coat protein of phage P22 folds independently of GroEL, and does not significantly aggregate in vivo even at elevated temperatures (Nakonechny and Teschke 1998). Eighteen single amino acid substitutions in coat protein render the molecule temperature sensitive for folding (tsf) (Gordon and King 1993). These coat protein variants require GroEL/S for in vivo folding and are highly prone to aggregate (Gordon et al 1994). In addition, several second site suppressor substitutions (su) revert the tsf phenotype back to WT. Some suppressor substitutions, called global suppressors, are able to revert the phenotype of several tsf substitutions (Aramli and Teschke 1999).
Previously, we found that 1 global suppressor substitution, T166I, functions to alleviate folding defects caused by tsf substitutions by inducing GroEL and GroES expression. In addition, tsf:T166I coat proteins demonstrate enhanced interactions with both the co-chaperone complex, as well as P22 scaffolding protein, to alleviate the folding defects of the tsf parents even at low temperatures (Parent et al 2004). Here, we present evidence that a different global suppressor substitution, F170L, does not utilize the GroEL chaperone to alleviate the tsf folding defects in the same manner as the T166I variants. This leads to the question of why the su substitutions function differently, and how substrate specificity is determined for GroEL. The F170L su substitution, although only 4 residues from the T166I substitution, results in coat proteins that fold in a manner that does not induce GroEL/S. Our data show that the tsf:F170L proteins have a decreased propensity to unfold from the assembly-competent state compared with tsf:T166I proteins. Ultimately, this leads to a decreased population of unfolding intermediates. We propose that the observed difference between variants of P22 coat protein in both induction and requirement of GroEL is largely due to the propensity of the coat proteins to unfold.
MATERIALS AND METHODS
Bacteria
Salmonella strain DB7136 (leuA414 am, hisC525am, su–) and the su+ derivative DB7155 (supE20 gln, leuA414 am, hisC525 am) have been previously described in Winston et al (1979). The Escherichia coli strains DW720 (WT groEL and groES) and DW716 (groEL44) were transformed with the plasmid pPR1347 (Neal et al 1993), which encodes for the rfb gene cluster and rfc gene so that E. coli synthesizes the O antigen needed for P22 infection. The plasmid conferred kanamycin resistance to the cells.
Bacteriophage
The P22 bacteriophage used in these studies either were WT in gene 5, which codes for coat protein, or carried mutations that result in the tsf amino acid substitutions D174N, S223F, and F353L. In addition, phage with the global su substitutions D163G, T166I, and F170L were used in conjunction with the D174N, S223F, and F353L mutations (tsf:su). All P22 strains carried the c1-7 allele to prevent lysogeny. Bacteriophage preparation was performed as previously described (Aramli and Teschke 1999).
Media
Luria Broth (LB, from Invitrogen Life Technologies, Paisley, Scotland, UK) was used to support bacterial growth for plating experiments, and for the preparation of phage stocks. For pulse-chase experiments, minimal medium was used. Minimal medium contains M9 plus 1 mM MgSO4, 1 μM FeCl3 and CaCl2, 1.2% glucose, 0.008% leucine, and 0.003% histidine. M9 medium contains 1.28% NaHPO4, 0.3% KH2PO4, 0.05% NaCl, and 0.1% NH4Cl. For storing phage stocks, M9 with 2 mM MgSO4 was used.
Relative titer determination
Phage were either WT in gene 5, or carried tsf or tsf:su mutations, as described previously (Aramli and Teschke 1999). Phage were plated on E. coli strains at temperatures of 28–39°C. The relative titer was calculated by dividing the resultant titer of each phage at the experimental temperature and experimental strain by the titer of the phage at the permissive condition (28°C on DW720 cells).
Pulse-chase experiments and sucrose gradient centrifugation
Overnight cultures were grown in minimal medium from a single isolated colony of DB7136 at 30°C. The overnight cultures were used to inoculate a culture in minimal medium, grown at 30°C. The cells were grown to a density of 2 × 108 cells/mL and then infected with a multiplicity of infection of 17, which produces maximal infection of the cells. The infected cultures were grown for 45 minutes, followed by a 1-minute pulse of 20 μCi/mL of [35S]-methionine and [35S]-cysteine protein labeling mix (10 mCi/mL, NEN Life Science Products, Boston, MA, USA). After 1 minute, a chase of unlabeled cysteine/methionine at a final concentration of 450 μM each was added. An aliquot was taken 30 seconds after the chase, and transferred immediately into a microfuge tube on ice. The cells were then pelleted at 13 200 rpm at 4°C for 5 minutes and resuspended in the same volume of resuspension buffer (100 μg/mL lysozyme, 0.1% Triton, and 5 mM ethylenediaminetetraacetic acid) and then frozen at –20°C. The radiolabeled samples were thawed on ice and incubated for 1 hour with 20 mM MgSO4 and 100 μg/mL DNase. Samples were then frozen and thawed 2 times to be sure of complete lysis.
After the freeze-thaw lysis, the lysates were centrifuged for 30 seconds in a microcentrifuge to pellet any unlysed cells. A 100-μL aliquot of the supernatant was applied to a 2.2-mL 5–20% linear sucrose gradient and run in an RP55S rotor in a Sorvall (Newtown, CT, USA) RC M120EX centrifuge for 3 hours and 10 minutes at 50 000 rpm at 4°C. The linear sucrose gradients were made with a Gradient Master Model 106 (Biocomp Instruments, Fredericton, New Brunswick, Canada). The gradients were fractionated from the top taking 100-μL aliquots. The samples were run on a 10% sodium dodecyl sulfate (SDS) polyacrylamide gel. The amount of radioactivity in the protein bands was quantified by densitometry of the autoradiographs with a Kodak (Rochester, NY, USA) EDAS system to determine the amount of GroEL induction. The total pixels of radiolabeled GroEL expressed for each mutant was calculated. The ratio of GroEL to total soluble coat protein for the same mutant was calculated. Soluble coat protein was considered to be any coat protein that is not in the last fraction of the sucrose gradient, which contains procapsids. Therefore, soluble coat protein consists of free monomeric coat protein, as well as GroEL bound coat protein. To correct for small differences between samples (such as radioactive decay, infection levels, time of exposure, etc), this ratio was used to normalize the data. All ratios were then normalized to the WT ratio so that the WT levels equal 1.
Purification of coat proteins
WT as well as the tsf and tsf:su proteins used in the following experiments were obtained from empty procapsid shells that had been prepared as previously described (Anderson and Teschke 2003; Parent et al 2005).
In vitro refolding to monitor aggregation
Empty procapsid shells were unfolded at 2 mg/mL in 6.75 M urea and 20 mM sodium phosphate buffer, pH 7.6, for 30 minutes at room temperature. Refolding was initiated by rapid dilution with phosphate buffer at temperatures ranging from 4–33°C to yield a final protein concentration of 0.1 mg/mL, with a 0.34 M residual urea content. The reactions were stopped at various times (range 0.5–15 minutes) by taking aliquots into native sample buffer (30% glycerol, 112 mM Tris, and 120 mM glycine) held on ice. The samples were then run on a native polyacrylamide gel.
Temperature shift from native monomer
Empty procapsid shells were unfolded at 2 mg/mL in 6.75 M urea and 20 mM sodium phosphate buffer, pH 7.6, for 30 minutes at room temperature. Refolding was initiated by overnight dialysis against phosphate buffer at 4°C. Any aggregated structures were removed by centrifugation at 175 000 × g for 20 minutes at 4°C. The protein concentration was determined by absorbance at 280 nm and adjusted to 0.1 mg/mL by dilution with ice cold phosphate buffer. The samples were held on ice for 30 minutes and then shifted to 15°C and 33°C. The reactions were stopped at various times (range 0–30 minutes) by taking aliquots into native sample buffer held on ice. The samples were then run on a native polyacrylamide gel.
Native polyacrylamide gel electrophoresis
Native polyacrylamide gels comprise a 4.3% stacking gel (pH 8.3) and a 7.5% resolving gel (pH 9.5). The amount of protein loaded into each lane was 0.5 μg. The bands on the native polyacrylamide gels were visualized by silver staining as described previously (Rabilloud et al 1988). The resulting bands were quantified by densitometry with a Kodak EDAS system. For the tsf proteins, the monomeric bands were quite diffuse; therefore, all density in the monomeric forms was totaled. The fraction of remaining native forms was determined by dividing the intensity of monomers as a function of time by the values at the initial time.
Fluorescence spectra
Fluorescence experiments were done with an SLM Aminco-Bowman 2 spectrofluorometer (Spectronic Instruments, Urbana, IL, USA). The temperature of the cuvette was maintained at 20°C with a circulating water bath. A 1-cm pathlength cell was used for fluorescence wavelength scans. Solutions of refolded coat protein monomers were at a final concentration of 100 μg/mL in sodium phosphate buffer (20 mM, pH 7.6). Excitation was set to 295 nm, and emission was monitored from 300–410 nm, with bandpasses of 4 nm.
Circular dichroism wavelength spectra
Circular dichroism (CD) was done with an Applied Photophysics (Leatherhead, Surrey, UK) Pi-Star 180 circular dichroism spectrapolarimeter with the cuvette maintained at 20°C with a circulating water bath. A 0.1-cm pathlength cell was used for CD wavelength scans. Solutions of refolded coat protein monomers were at a final concentration of 1 mg/mL in sodium phosphate buffer (20 mM, pH 7.6). Wavelength scans were done over 180– 250 nm and sampled every 1 nm with entrance and exit slit widths of 2 nm and a scan time of 45 minutes.
Assembly of refolded coat proteins
Empty procapsid shells were unfolded at 2 mg/mL in 6.75 M urea and 20 mM sodium phosphate buffer, pH 7.6, for 30 minutes at room temperature. Refolding was initiated by continuous-flow dialysis with phosphate buffer at 4°C at a rate of 0.45 mL/min. Coat protein monomers were collected when urea was no longer detected by refractometry. Aggregates and other structures were removed by centrifugation at 175 000 × g for 20 minutes at 4°C. Refolded coat protein was added to native scaffolding protein for a final concentration of 0.5 mg/ mL each (∼11 μM coat protein and ∼15 μM scaffolding protein) in the presence of 60 mM NaCl. Assembly was monitored by light scattering at 500 nm with 4-nm bandpasses. The samples were then run on a 1.2% agarose gel to confirm procapsid production. Sucrose gradient sedimentation, sodium dodecyl sulfate– polyacrylamide gel electrophoresis (SDS-PAGE), and densitometry were performed as previously described (Parent et al 2005).
RESULTS
Previously, second site su substitutions were isolated for P22 coat protein that alleviate folding defects caused by single amino acid substitutions (Aramli and Teschke 1999). Three su substitutions, D163G, T166I, and F170L, were determined to be “global” suppressors, which can improve folding defects caused by more than 1 tsf substitution (Aramli and Teschke 1999). T166I functions through concerted interactions with the folding chaperones GroEL/S, as well as the assembly chaperone, P22 scaffolding protein, to drive the process of procapsid assembly (Parent et al 2004). Preliminary data showed that tsf:D163G and tsf:T166I coat proteins behaved similarly in vivo (data not shown). For this study, we focused on the effects of the F170L global su substitution because the F170L su substitution appeared to function via a dissimilar mechanism. We chose to study F170L in the context of the 3 tsf parents, D174N, S223F, and F353L because all 3 were rescued by both su substitutions T166I and F170L.
Global suppressors have different requirements for GroEL in vivo
Plating experiments that count the number of phage produced from infected cells are a convenient way to determine whether various types of coat proteins can fold under different conditions because misfolded coat proteins cannot assemble into infectious phage. We characterized the chaperone requirement of the T166I and F170L global su substitutions by comparing the efficiency of plating for phage producing the substituted coat proteins in the presence and absence of functional GroEL at various temperatures. Phage were plated on cells containing GroEL and compared with phage plated on cells containing GroEL44, a temperature-sensitive variant of GroEL that does not support growth of various bacteriophages (Zeilstra-Ryalls et al 1993). Phage strains containing either WT, tsf, or tsf:su coat proteins were plated at a temperature range of 28–39°C. The titer at each temperature was divided by the titer of phage produced for each strain when plated under the permissive condition (28°C with WT GroEL).
Phage production with WT and tsf:su coat proteins was unaffected by temperature when plated on cells with WT GroEL, as expected. However, phage carrying the various tsf:su substitutions in coat protein demonstrate different restrictive temperatures in the presence of GroEL44 (Fig 1). Phage carrying the tsf substitutions D174N or S223F in combination with the T166I su substitution became temperature sensitive (ts) at 35°C in the presence of GroEL44. However, when D174N or S223F were paired with the F170L su substitution, the ts temperature was elevated to 37°C, indicating a less severe phenotype. For the tsf substitution, F353L, when paired with either su substitution T166I or F170L, the ts temperature of the phage is 37°C.
Fig 1.
The F170L suppressor substitution has a less stringent GroEL requirement than T166I. Phage that had tsf:su substitutions or were WT in coat protein were grown on DW720 cells (WT GroEL), or on DW716 cells (dysfunctional GroEL44). The relative titer is the titer of phage produced at each experimental temperature and condition divided by the titer of the phage at the permissive temperature and condition (28°C, DW720 cells). Open squares are phage production with tsf:T166I coat proteins grown on DW716 cells (GroEL44); closed squares are phage production with tsf:T166I coat proteins on DW720 cells (WT GroEL); open circles are phage production with tsf:F170L coat proteins grown on DW716 cells; closed circles are phage production with tsf:F170L coat proteins grown on DW720 cells
Because phage carrying the F170L su substitution demonstrate a ts phenotype when GroEL is dysfunctional, clearly GroEL is important for the proper folding of coat proteins carrying the F170L substitution at elevated temperatures. However, because the restrictive temperature was different for tsf mutants carrying the F170L su substitution than for phage carrying the T166I su substitution, we reasoned that these coat proteins could have different requirements for GroEL.
GroEL is not induced by the F170L su substitution in vivo
Tsf:T166I coat proteins induce GroEL/S because they fold poorly (Doyle et al 2003; Doyle et al 2004; Parent et al 2004). This induction is seen even at 30°C, at which aggregation does not occur. We determined whether induced GroEL expression was responsible for the increase in the ts temperature observed for phage carrying D174N: F170L or S223F:F170L coat proteins in the plating experiments above (eg, if simply more GroEL molecules were produced, the increase in the ts temperature might not be an indication of a change in chaperone requirement). Alternatively, if the F170L su substitution resulted in no change in GroEL expression, coupled with the upward shift in the ts temperature, we could conclude that the molecular chaperones were required less for proper folding of tsf:F170L coat proteins when compared with the tsf:T166I proteins.
To determine whether more GroEL was induced at 30°C when the tsf and F170L substitutions were present in coat protein, phage-infected cells were labeled with 35S-cysteine and 35S-methionine and briefly chased with nonradioactive amino acids, as described previously (Parent et al 2004). The radioactively labeled proteins were separated by sucrose gradient sedimentation, and the fractions were analyzed by SDS-PAGE and autoradiography. The amount of radioactivity incorporated into both coat protein and GroEL protein bands was quantified by densitometry. We calculated the relative GroEL induction, expressed as the ratio of GroEL intensity to soluble coat protein intensity, and normalized the values to induction of GroEL by WT phage (Fig 2; Parent et al 2004). If a variant coat protein induces GroEL expression, we would expect to see a higher ratio that corresponds to more GroEL molecules produced per soluble coat protein molecules. WT coat protein has been shown to induce the lowest levels of GroEL expression, which is reasonable because WT coat protein does not require GroEL for proper folding (Gordon et al 1994; Nakonechny and Teschke 1998). As expected, the 3 tsf mutants shown here induce GroEL expression compared with WT coat protein (Fig 2A; Gordon et al 1994). Consistent with the plating experiments described herein, and in contrast to what was observed for T166I coat proteins, infection with phage carrying the F170L coat su substitution did not induce relative GroEL expression (Fig 2A). Although both tsf:T166I and tsf:F170L coat protein variants have a stronger ts phenotype in the presence of GroEL44 compared with WT coat protein, induction of the chaperones occurs even at permissive temperatures for the tsf:T166I proteins, but not tsf:F170L proteins. This effect is striking because phage production levels are the same for the 2 variants at permissive temperatures (data not shown). GroEL appears to be necessary for proper tsf:F170L folding at elevated temperatures, but induction of GroEL expression is not likely the mechanism by which the F170L suppressor substitution functions to alleviate folding defects in P22 tsf coat proteins.
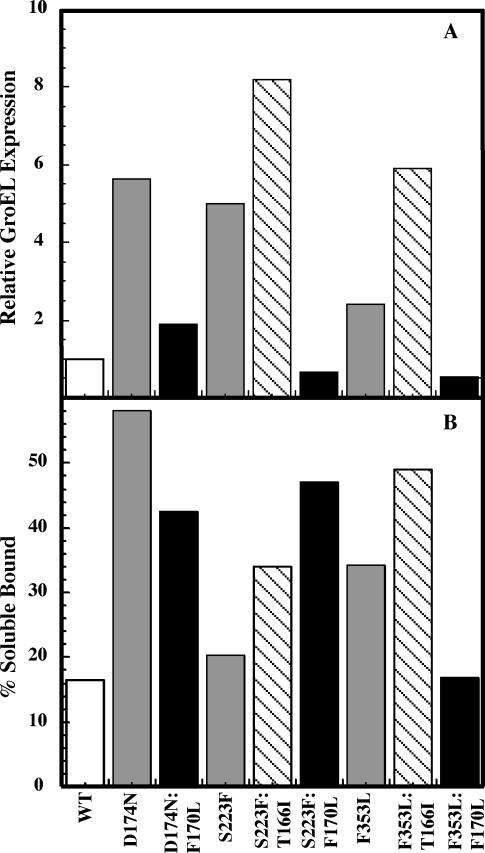
Fig 2.
The F170L global suppressor substitution does not induce GroEL expression in vivo. Pulse-chase experiments were done with cells infected with phage that carried tsf, tsf:su, or WT coat protein. Lysates were applied to 5–20% linear sucrose gradients and fractionated after centrifugation. The fractions were run on 10% SDS-acrylamide gels. Protein bands from the autoradiographs were quantified using densitometry. The total amount of labeled soluble coat protein, as well as the total amount of labeled GroEL for each variant was calculated. (A) The ratio of labeled GroEL per labeled soluble coat protein for each mutant was determined and normalized to the WT ratio. (B) The amount of soluble coat proteins bound to GroEL. In both panels, open bars represent WT data, gray bars represent tsf data, and black bars represent tsf:F170L data. The striped bars represent tsf:T166I data taken from Parent et al (2004). Although only a representative experiment is shown here, errors for these experiments are typically ±5% (Parent et al 2004)
We also determined the amount of soluble coat protein bound to GroEL at 30°C (Fig 2B), as described previously (Parent et al 2004). In brief, the soluble coat protein is equal to the intensity of coat protein that co-migrates with GroEL during sucrose gradient sedimentation divided by the intensity of the total soluble coat protein. For all 3 tsf coat proteins, coat protein bound to GroEL is increased compared with WT coat protein. D174N:F170L and F353L:F170L coat proteins have a decreased level of GroEL binding when compared with their tsf parents. These data are consistent with the notion that tsf:F170L coat proteins have a decreased requirement for GroEL compared with the corresponding T166I variants. Interestingly, the S223F:F170L coat protein variant shows an increased amount of binding compared with its S223F tsf parent or the S223F:T166I variant. So, why does S223F: F170L require GroEL for folding and yet not induce the chaperone?
The F170L su substitution stabilizes the native state
Because GroEL expression was not increased by infection with phage producing tsf:F170L coat proteins and the ts temperature was elevated in the presence of GroEL44, we reasoned that coat proteins containing the F170L su substitution could be less aggregation prone during folding than their tsf parents. To test this, unfolded coat protein monomers were refolded in vitro by rapid dilution to a residual urea content of ∼0.34 M at various temperatures ranging from 4°C to 33°C. The presence of native or aggregated structures at a time range of 0–15 minutes after dilution was visualized by running samples on nondenaturing acrylamide gels. The gels were silver stained, and 2 representative temperatures are shown in Figure 3.
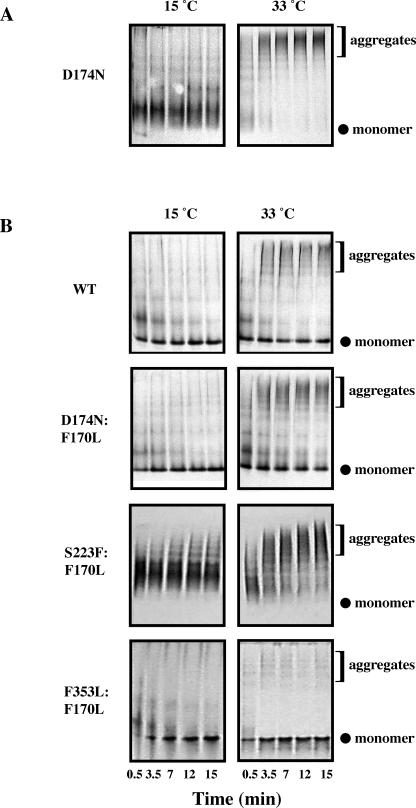
Fig 3.
Aggregation is decreased for some, but not all, tsf coat proteins when the F170L substitution is also present. The propensity for aggregation while folding was determined by refolding urea-denatured coat proteins by rapid dilution with buffer at various temperatures. Aliquots were taken with time after dilution and run on native polyacrylamide gels silver stained to detect the presence of native monomers and aggregates. Native gels of a time course ranging from 0.5–15 minutes at 15°C and 33° C are shown. (A) A representative sample of aggregation of tsf coat protein during folding; D174N is shown because F353L and S223F were previously published (Aramli and Teschke 2001). (B) A representative sample of tsf:F170L coat protein aggregation
For all 3 tsf mutants—D174N, S223F, and F353L—aggregated structures were easily visualized at 33°C, consistent with previously published reports (a representative pair is shown in Fig 3A; Teschke 1999; Aramli and Teschke 2001). In addition, tsf:T166I coat proteins readily aggregated while folding at 33°C (Teschke 1999; Aramli and Teschke 2001) showing no monomeric bands. In contrast, the addition of the F170L su substitution decreased aggregation for D174N and F353L, as determined by a decrease in the amount of aggregated structures, coupled with an increase in the monomeric species at 33°C relative to their respective tsf parents. However, this trend was not observed for the S223F and S223F:F170L pair. These data indicate that decreasing the aggregation propensity during folding is not the mechanism by which F170L suppresses the folding defect.
S223F:F170L and S223F are equally aggregation prone during folding, yet S223F:F170L does not induce GroEL/ S. We next determined whether the tsf:F170L coat proteins were aggregation prone when unfolding from the monomeric state, by doing temperature shift-up experiments from the native state. Unfolded coat protein monomers were refolded by dialysis at 4°C so that there was no residual urea. Monomeric WT, tsf, and tsf:F170L coat proteins were held on ice and then shifted to 33°C. At times ranging from 0 to 30 minutes, samples were transferred to ice and diluted with ice-cold native gel sample buffer. The presence of native or aggregated structures was visualized by silver staining the samples run on nondenaturing acrylamide gels. The amount of native monomer for each protein at each time point was quantified by densitometry and normalized to the amount present initially. The fraction of native protein remaining at 33°C is shown in Figure 4A. The tsf:F170L coat proteins retained much of their monomeric native state at 33°C, whereas the tsf parents showed a decrease in native monomer and an increase in aggregated structures with time. WT coat protein demonstrated a small decrease in native monomer with time and had the slowest rate of monomer loss compared with the other proteins observed. Although the tsf:F170L coat proteins did eventually aggregate with time at 33°C, all 3 were considerably more stable than their tsf parents, retaining at least 45% of their monomer after 30 minutes. The tsf:F170L coat proteins also showed a decrease in rate of monomer loss compared with their tsf parents. Aramli and Teschke (2001) observed that native tsf:T166I coat proteins were not stable in these experiments, and significant aggregation started to occur in as little as 7 minutes at 33°C, indicating a less stable native state than the tsf:F170L proteins. Figure 4B shows the average fraction of native monomer for each protein remaining after 30 minutes at 33°C, as averaged from 5 data sets. These data indicate that monomeric tsf:F170L coat proteins are significantly more stable than their tsf parents.
Fig 4.
The native state is stabilized for tsf:F170L coat proteins. Temperature shift experiments were performed as described in the Materials and Methods. In brief, natively folded coat protein held on ice was transferred to 33°C for 0–30 minutes. Samples were run on native polyacrylamide gels and silver stained to detect the presence of native monomers and aggregated forms. (A) Densitometry data taken from the native gels. The native fraction remaining is the intensity of the native band at the experimental time divided by the intensity at time 0. The lines drawn do not reflect a fit of the data to any model; they are drawn to aid the eye. Closed squares are data for WT coat protein; open circles are tsf coat protein; closed circles are tsf:F170L coat proteins. (B) The average fraction of native protein remaining after 30 minutes at 33°C; error bars are the standard deviation taken from 5 data sets
The difference in native state stabilization between tsf: T166I and tsf:F170L coat proteins might be the cause of the difference in chaperone induction (ie, GroEL might be required to fold the coat proteins to their native state, but once a folded structure is attained, the native state is stabilized, driving the reaction toward assembly of the folded monomers, thereby decreasing the population of folding intermediates).
Overall structure of the tsf:F170L coat proteins is similar to WT
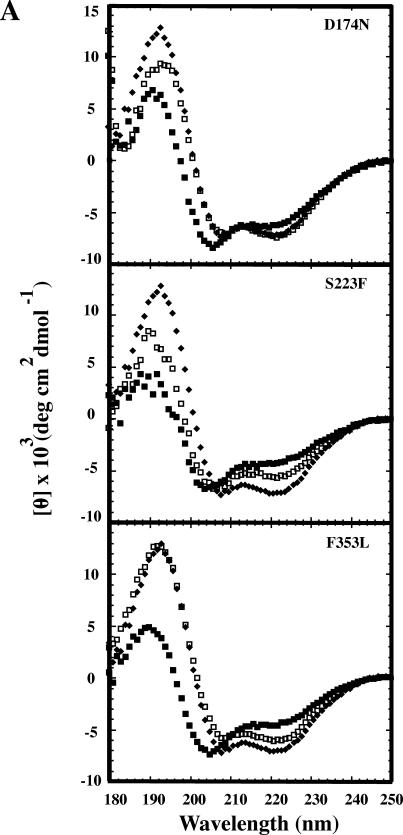
Because the F170L su substitution stabilized the native state when coupled with the tsf substitutions, we determined the effect this substitution had on the secondary and tertiary structures of the coat proteins. CD was used to observe the effects of su substitutions on the secondary structures, compared with the structures of their tsf parents and WT coat protein. The tsf substitution resulted in altered CD spectra compared with WT coat protein. For all cases, the addition of the F170L su substitution resulted in spectra that were restored to the WT coat protein spectrum, most notably in the wavelength range 180– 210 nm, which is indicative of β-strand (Fig 5).
Fig 5.
Tsf:F170L coat proteins have WT-like secondary structures compared with their tsf parents. Circular dichroism spectra of WT, tsf, and tsf:F170L coat proteins were done as described in the Materials and Methods and are shown. Closed diamonds are WT coat protein; closed squares are tsf coat proteins; open squares are tsf: F170L coat proteins
To determine whether the overall tertiary structure was also restored to values similar to WT coat protein, the intrinsic tryptophan fluorescence emission was measured for WT, tsf, and tsf:F170L coat proteins. The emission scans for WT and the tsf parents were in excellent agreement with previous results (Teschke et al 1991). For D174N, S223F, and F353L tsf substitutions, a red shift of about 2–4 nm compared with WT coat protein was observed, indicating that the tryptophans were more solvent exposed. The presence of F170L in combination with the tsf parents D174N and F353L restored the emission maximum to that of WT, ∼339 nm. The emission maximum for S223F:F170L did not return to the exact WT emission maximum but was blue shifted compared with S223F, indicating that the environment of the tryptophans was approaching a WT-like state (data not shown). Together, these data indicate that the native state of the tsf:F170L coat proteins was similar to that of WT coat protein.
tsf:F170L Mutants are more assembly-competent than their tsf parents
Because the tsf:F170L coat proteins demonstrate a monomeric state with a lower propensity to unfold compared with their tsf parents, it would be reasonable for these proteins to also demonstrate enhanced ability to assemble. In vitro procapsid assembly simply requires the mixing of coat protein and scaffolding protein. The resulting particles are similar in size and shape to procapsids formed in vivo. Assembly of procapsids was performed as described in the Materials and Methods and monitored by light scattering; procapsid production was confirmed by agarose gel electrophoresis and quantified by sucrose gradient sedimentation followed by SDS-PAGE as described previously (Parent et al 2005).
The rates of procapsid assembly were monitored by the change in light scattering with time and increased significantly for all tsf coat proteins when the F170L substitution was present (Fig 6A). The yield of procapsid production was determined by calculating the percentage of coat protein found in the sucrose gradient fractions to which procapsids migrate. For all tsf:F170L coat proteins, procapsid formation increased relative to their tsf parents and were similar to WT coat protein levels (Fig 6B). These data indicate that the F170L su substitution has enhanced assembly compared with their tsf parents. These data demonstrate that tsf:F170L coat protein folding results in coat proteins that have similar structure and activity to WT coat protein.
Fig 6.
The F170L substitution results in coat proteins that are more assembly competent. In vitro assembly reactions were performed as described in Materials and Methods. (A) The light scattering data for WT and variant coat proteins at 20°C. Solid lines represent tsf:F170L coat protein assembly, and dashed lines represent tsf coat protein assembly. (B) The percentage of procapsids formed during these experiments was determined by the fraction of coat protein sedimenting in the procapsid position of sucrose gradient sedimentation
DISCUSSION
Previously, tsf:T166I coat proteins were shown to be more aggregation prone than their tsf parents (Aramli and Teschke 2001) and that in vivo GroEL/S were induced even at permissive temperatures (Parent et al 2004). In vitro folding and unfolding experiments showed that the tsf:T166I proteins have a more populated folding intermediate because of changes in rates of folding and unfolding (Doyle et al 2004). These data explain why the tsf: T166I proteins are more aggregation prone than their tsf coat protein parents. These results are surprising because the typical effect of a suppressor substitution is to decrease aggregation (Tsai et al 1991; Schuler and Seckler 1998). We found that the su substitution T166I functions by recruiting GroEL, as well as through enhanced scaffolding protein interactions (Parent et al 2004). In contrast to T166I, the global substitution F170L does not function by inducing or recruiting GroEL. Instead, we provide evidence that F170L alleviates the folding defects of their tsf parents through stabilization of the native state by decreasing the unfolding propensity.
Chaperone roles in protein folding and in capsid assembly
The GroEL and GroES co-chaperone complex has been established as a promiscuous chaperone complex that acts on a wide range of substrates, such as monomeric rhodanese (Langer et al 1992), barnase (Gray and Fersht 1993), staphylococcal nuclease (Tsurupa et al 1998), maltose-binding protein (Horwich et al 1993), dihydrofolate reductase (Gorovits and Horowitz 1997), and lactate dehydrogenase (Badcoe et al 1991). In vivo, the essential GroEL and GroES are responsible for folding ∼10% of all cytoplasmic bacterial proteins (Kerner et al 2005). The chaperones can accommodate folding of a wide array of proteins between the sizes of 20–70 kDa in the traditional cis arrangement of the GroEL/ES molecule (Langer et al 1992).
How GroEL/S distinguishes between substrate and nonsubstrate polypeptides is not understood. GroEL/ES has been observed to bind only misfolding proteins and not proteins in their native state (Fink 1999). Substrate polypeptide interaction with GroEL could be driven by either conformation of folding intermediates or folding kinetics (Frieden and Clark 1997). Both hydrophobic and ionic interactions have been implicated in the binding of substrate polypeptides to GroEL, although hydrophobic interactions are the favored hypothesis (Schmidt and Buchner 1992; Zahn et al 1994; Perrett et al 1997; Persson et al 1997). Slow folding kinetics have also been implicated as crucial to substrate polypeptide recognition (Tieman et al 2001).
The folding and assembly of P22 coat protein variants shed light on GroEL substrate recognition. Here, and in previous experiments, we have shown that the tsf and tsf: su coat proteins require GroEL for proper folding, but WT coat protein folds independently of the chaperone (Gordon et al 1994; Nakonechny and Teschke 1998; Doyle et al 2003, 2004; Parent et al 2004). Both in vivo and in vitro data for P22 coat protein support the idea that altered unfolding kinetics are the cause for interaction with GroEL. Our current model for P22 coat protein is as follows and is illustrated in Figure 7.
Fig 7.
Our current model for P22 coat protein folding and assembly. See Discussion for a description of this model. The dashed boxes show the predominant forms for each folding reaction, and arrows represent the flux of the folding reaction determined by equilibrium and kinetic experiments (Anderson and Teschke 2003; Doyle et al 2003, 2004). The GroEL dependence is based on the ts temperature when GroE44 is present ( Gordon et al 1994; Nakonechny and Teschke 1998; Aramli and Teschke 1999). A minus sign (−) means no GroEL dependence, and the number of plus signs (+) indicates the degree of dependence
WT, tsf, and tsf:su coat proteins fold to their native conformation via a 3-state equilibrium folding model, unfolded ⇔ intermediate ⇔ native (U ⇔ I ⇔ N) (Teschke and King 1993; Anderson and Teschke 2003; Doyle et al 2003, 2004). A late-folding intermediate has been identified to interact with GroEL (de Beus et al 2000). WT coat protein does not significantly aggregate and does not require GroEL (Nakonechny and Teschke 1998). WT coat protein folds rapidly to the native state and has a very slow unfolding rate from N ⇒ I (Anderson and Teschke 2003); therefore, the population of intermediates is small. This is likely the reason why WT coat protein does not require GroEL.
Amino acid substitutions in coat protein affect the folding and unfolding kinetics and change the relative amounts of intermediate when compared with WT coat protein. Tsf coat proteins have been shown to fold rapidly, but then flicker between native and intermediate structures, thereby significantly populating intermediates. The intermediates also rapidly unfold to U (Doyle et al 2003). Relative to the tsf coat proteins, the tsf:T166I proteins have been shown to populate the intermediates via a slower unfolding to U (Doyle et al 2004). Shifting the population of intermediate species explains the GroEL and GroES requirement of the tsf and tsf:T166I coat proteins. Both types of substitutions result in an increased population of intermediates compared with WT. This phenomenon causes the tsf and tsf:T166I proteins to become aggregation prone, and therefore the co-chaperones are induced.
Tsf:F170L coat proteins do not function by decreasing aggregation during folding, which explains why tsf: F170L coat proteins require the GroEL chaperone when compared with WT coat protein. Although 1 tsf:F170L coat protein, S223F:F170L, has an enhanced interaction with the chaperone while not inducing it, once successfully folded, all tsf:F170L coat proteins demonstrate a stabilized native structure compared with their tsf parents and tsf:T166I coat proteins. Therefore, the tsf:F170L coat proteins have a decreased unfolding propensity. The equilibrium is shifted toward the direction of proper folding and assembly by decreasing aggregation caused by the unfolding of N. This explains why the tsf:F170L coat proteins do not induce the chaperones; the intermediate population is much smaller than the intermediate population of tsf and tsf:T166I coat proteins because of the decreased unfolding to I.
P22 scaffolding protein is another example of a molecular chaperone that aids in the folding of coat protein. By favorably interacting with correctly folded coat proteins, the scaffolding protein drives the folding reaction toward native structures by trapping the coat proteins in procapsids (Aramli and Teschke 2001; Parent et al 2004). By increasing native state stabilization, tsf:F170L coat proteins enhance the assembly of P22 coat proteins by creating a greater pool of correctly folded coat proteins that can interact with scaffolding protein, thereby decreasing the presence of intermediate structures even further.
Su substitutions aid in folding defects by original amino acid substitutions
Su substitutions generally highlight areas in a protein that are responsible for proper folding. Su substitutions were originally isolated in P22 tailspike protein (Mitraki et al 1993), and as it is for many suppressor substitutions, the mechanism of suppression is due to an increase in the stability of the native state. Suppressor substitutions that increase stabilization have been isolated for a variety of proteins and usually have a very specific effect on the particular protein involved, as is shown for the tumor suppressor p53 core domain (Joerger et al 2004) and T4 lysozyme mutants (Wray et al 1999). We hypothesize that the su substitutions in coat protein do not lead to more stable coat protein monomers because of the plasticity required for capsid assembly.
Viral capsids are not immobile static structures, but macromolecular assemblies that undergo large morphological changes as part of their normal morphogenesis (Caspar 1980; Steven et al 1997; Steven et al 2005). Bacteriophage P22 has a capsid composed of only 1 type of protein. The 420 coat proteins that are needed to form a P22 phage are all chemically identical, yet according to the theory of quasiequivalence, these proteins must be plastic enough to adopt several different conformations to assemble and mature (Caspar and Klug 1962; Johnson and Speir 1997). It is therefore not surprising that super-stable mutants are not observed for P22 coat proteins. In addition, suppressor substitutions in P22 coat protein likely affect more than 1 region of the protein. Although a high-resolution structure is not available for P22 coat protein, the protein has been proposed to have 2 putative folding domains joined by a hinge region (Lanman et al 1999; Kang et al 2006). The 3 global suppressors lie within the hinge loop. These global suppressors are able to rescue tsf substitutions that are contained in either domain. This indicates that the hinge region of coat protein is sensitive to the folding of the overall structure.
Clearly, the “global effect” of each individual su substitution is universal for P22 coat protein. All tsf:T166I coat proteins studied thus far demonstrate chaperone recruitment and a destabilized native state, whereas the tsf: F170L coat proteins demonstrate a stabilized native state with no GroEL/S induction. Although residing only 4 amino acids apart, these global su substitutions function via disparate mechanisms, yet they are able to rescue the same original tsf substitutions. Both tsf:T166I and tsf: F170L result in protein native states that are not exactly the same as WT coat protein, yet all improve phage production relative to the tsf parents. Amino acid substitutions in this region affect not only protein folding, but procapsid formation as well, indicating that this hinge area of the protein is important for all types of capsomer function.
Acknowledgments
We acknowledge Eric Anderson for his help with protein purification and some preliminary data. This work was supported by PHS grant GM53567 to C.M.T., a PHS fellowship GM073598 to K.N.P., and a University of Connecticut Research Foundation Grant.
REFERENCES
- Anderson E, Teschke CM. Folding of phage P22 coat protein monomers: kinetic and thermodynamic properties. Virology. 2003;313:184–197. doi: 10.1016/s0042-6822(03)00240-x.0042-6822(2003)313[0184:FOPPCP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223.0193-4511(1973)181[0223:PTGTFO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Aramli LA, Teschke CM. Alleviation of a defect in protein folding by increasing the rate of subunit assembly. J Biol Chem. 2001;276:25372–25377. doi: 10.1074/jbc.M101759200.0021-9258(2001)276[25372:AOADIP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Aramli LA, Teschke CM. Single amino acid substitutions globally suppress the folding defects of temperature-sensitive folding mutants of phage P22 coat protein. J Biol Chem. 1999;274:22217–22224. doi: 10.1074/jbc.274.32.22217.0021-9258(1999)274[22217:SAASGS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Badcoe IG, Smith CJ, Wood S, Halsall DJ, Holbrook JJ, Lund P, Clarke AR. Binding of a chaperonin to the folding intermediates of lactate dehydrogenase. Biochemistry. 1991;30:9195–9200. doi: 10.1021/bi00102a010.0006-2960(1991)030[9195:BOACTT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Bazinet C, Benbaset J, King J, Carazo JM, Carrascosa JL. Purification and organization of the gene 1 portal protein required for phage P22 DNA packaging. Biochemistry. 1988;27:1849–1856. doi: 10.1021/bi00406a009.0006-2960(1988)027[1849:PAOOTG]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Beißinger M, Lee SC, Steinbacher S, Reinemer P, Huber R, Yu MH, Seckler R. Mutations that stabilize folding intermediates of phage P22 tailspike protein: folding in vivo and in vitro, stability, and structural context. J Mol Biol. 1995;249:185–194. doi: 10.1006/jmbi.1995.0288.0022-2836(1995)249[0185:MTSFIO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Botstein D, Waddell CH, King J. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22. J Mol Biol. 1973;80:669–695. doi: 10.1016/0022-2836(73)90204-0.0022-2836(1973)080[0669:MOHAAD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Casjens S. Molecular organization of the bacteriophage P22 coat protein shell. J Mol Biol. 1979;131:1–19. doi: 10.1016/0022-2836(79)90298-5.0022-2836(1979)131[0001:MOOTBP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Casjens S, King J. P22 morphogenesis. I: catalytic scaffolding protein in capsid assembly. J Supramol Struct. 1974;2:202–224. doi: 10.1002/jss.400020215.0091-7419(1974)002[0202:PMICSP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Caspar DLD. Movement and self-control in protein assemblies: quasi-equivalence revisited. Biophys J. 1980;32:103–138. doi: 10.1016/S0006-3495(80)84929-0.0006-3495(1980)032[0103:MASIPA]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp Quant Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005.0091-7451(1962)027[0001:PPITCO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- de Beus MD, Doyle SM, Teschke CM. GroEL binds a late folding intermediate of phage P22 coat protein. Cell Stress Chaperones. 2000;5:163–173. doi: 10.1379/1466-1268(2000)005<0163:gbalfi>2.0.co;2.1466-1268(2000)005[0163:GBALFI]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SM, Anderson E, Parent KN, Teschke CM. A concerted mechanism for the suppression of a folding defect through interactions with chaperones. J Biol Chem. 2004;279:17473–17482. doi: 10.1074/jbc.M400467200.0021-9258(2004)279[17473:ACMFTS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Doyle SM, Anderson E, Zhu D, Braswell EH, Teschke CM. Rapid unfolding of a domain populates an aggregation-prone intermediate that can be recognized by GroEL. J Mol Biol. 2003;332:937–951. doi: 10.1016/s0022-2836(03)00955-0.0022-2836(2003)332[0937:RUOADP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Earnshaw W, Casjens S, Harrison SC. Assemby of the head of bacteriophage P22: x-ray diffraction from heads, proheads and related structures. J Mol Biol. 1976;104:387–410. doi: 10.1016/0022-2836(76)90278-3.0022-2836(1976)104[0387:AOTHOB]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Eppler K, Wykoff E, Goates J, Parr R, Casjens S. Nucleotide sequence of the bacteriophage P22 genes required for DNA packaging. Virology. 1991;183:519–538. doi: 10.1016/0042-6822(91)90981-g.0042-6822(1991)183[0519:NSOTBP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Fink A. Chaperone-mediated protein folding. Physiol Rev. 1999;79:425–449. doi: 10.1152/physrev.1999.79.2.425.0031-9333(1999)079[0425:CPF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Frieden C, Clark AC. Protein folding: how the mechanism of GroEL action is defined by kinetics. Proc Natl Acad Sci U S A. 1997;94:5535–5538. doi: 10.1073/pnas.94.11.5535.1091-6490(1997)094[5535:PFHTMO]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon CL, King J. Temperature-sensitive mutations in the phage P22 coat protein which interfere with polypeptide chain folding. J Biol Chem. 1993;268:9358–9368.0021-9258(1993)268[9358:TMITPP]2.0.CO;2 [PubMed] [Google Scholar]
- Gordon CL, Sather SK, Casjens S, King J. Selective in vivo rescue by GroEL/ES of thermolabile folding intermediates to phage P22 structural proteins. J Biol Chem. 1994;269:27941–27951.0021-9258(1994)269[27941:SIVRBE]2.0.CO;2 [PubMed] [Google Scholar]
- Gorovits BM, Horowitz PM. Conditions of forming protein complexes with GroEL can influence the mechanism of chaperonin-assisted refolding. J Biol Chem. 1997;272:32–35. doi: 10.1074/jbc.272.1.32.0021-9258(1997)272[0032:COFPCW]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Gray TE, Fersht AR. Refolding of barnase in the presence of GroE. J Mol Biol. 1993;232:1197–1207. doi: 10.1006/jmbi.1993.1471.0022-2836(1993)232[1197:ROBITP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Horwich AL, Low KB, Fenton WA, Hirshfield IN, Furtak K. Folding in vivo of bacterial cytoplasmic proteins: role of GroEL. Cell. 1993;74:909–917. doi: 10.1016/0092-8674(93)90470-b.0092-8674(1993)074[0909:FIVOBC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Israel V. E proteins of bacteriophage P22. J Virol. 1977;23:91–97. doi: 10.1128/jvi.23.1.91-97.1977.1098-5514(1977)023[0091:EPOBP]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joerger AC, Allen MD, Fersht AR. Crystal structure of a superstable mutant of human p53 core domain. Insights into the mechanism of rescuing oncogenic mutations. J Biol Chem. 2004;279:1291–1296. doi: 10.1074/jbc.M309732200.0021-9258(2004)279[1291:CSOASM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Johnson JE, Speir JA. Quasi-equivalent viruses: a paradigm for protein assemblies. J Mol Biol. 1997;269:665–675. doi: 10.1006/jmbi.1997.1068.0022-2836(1997)269[0665:QVAPFP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kang S, Hawkridge AM, Johnson KL, Muddiman DC, Prevelige PJ. Identification of subunit-subunit interactions in bacteriophage P22 procapsids by chemical cross-linking and mass-spectrometry. J Proteome Res. 2006;5:370–377. doi: 10.1021/pr050356f.1535-3893(2006)005[0370:IOSIIB]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kerner MJ, Naylor DJ, and Ishihama Y. et al. 2005 Proteome-wide analysis of chaperone-dependent protein folding in Escherichia coli. Cell. 122:209–220. [DOI] [PubMed] [Google Scholar]
- Langer T, Pfeifer G, Martin J, Baumeister W, Hartl FU. Chaperone-mediated protein folding: GroES binds to one end of the GroEL cylinder, which accommodates the protein substrate within its central cavity. EMBO J. 1992;11:4757–4765. doi: 10.1002/j.1460-2075.1992.tb05581.x.1460-2075(1992)011[4757:CPFGBT]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanman J, Tuma R, Prevelige PE Jr.. Identification and characterization of the domain structure of bacteriophage P22 coat protein. Biochemistry. 1999;38:14614–14623. doi: 10.1021/bi9915420.0006-2960(1999)038[14614:IACOTD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Mitraki A, Danner M, King J, Seckler R. Temperature-sensitive mutations and second-site suppressor substitutions affect folding of the P22 tailspike protein in vitro. J Biol Chem. 1993;268:20071–20075.0021-9258(1993)268[20071:TMASSS]2.0.CO;2 [PubMed] [Google Scholar]
- Mitraki A, Fane B, Haase-Pettingell C, Sturtevant J, King J. Global suppression of protein folding defects and inclusion body formation. Science. 1991;253:54–58. doi: 10.1126/science.1648264.0193-4511(1991)253[0054:GSOPFD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Nakonechny WS, Teschke CM. GroEL and GroES control of substrate flux in the in vivo folding pathway of phage P22 coat protein. J Biol Chem. 1998;273:27236–27244. doi: 10.1074/jbc.273.42.27236.0021-9258(1998)273[27236:GAGCOS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Neal BL, Brown PK, Reeves PR. Use of Salmonella phage P22 for transduction in Escherichia coli. J Bacteriol. 1993;175:7115–7118. doi: 10.1128/jb.175.21.7115-7118.1993.0021-9193(1993)175[7115:UOSPPF]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Doyle SM, Anderson E, Teschke CM. Electrostatic interactions govern both nucleation and elongation during phage P22 procapsid assembly. Virology. 2005;340:33–45. doi: 10.1016/j.virol.2005.06.018.0042-6822(2005)340[0033:EIGBNA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Parent KN, Ranaghan MJ, Teschke CM. A second site suppressor of a folding defect functions via interactions with a chaperone network to improve folding and assembly in vivo. Mol Microbiol. 2004;54:1036–1054. doi: 10.1111/j.1365-2958.2004.04326.x.0950-382X(2004)054[1036:ASSSOA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Parent KN, Zlotnick A, Teschke CM. Quantitative analysis of multi-component spherical virus assembly: scaffolding protein contributes to the global stability of phage P22 procapsids. J Mol Biol. 2006;359:1097–1106. doi: 10.1016/j.jmb.2006.03.068.0022-2836(2006)359[1097:QAOMSV]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Perrett S, Zahn R, Stenberg G, Fersht AR. Importance of electrostatic interactions in the rapid binding of polypeptides to GroEL. J Mol Biol. 1997;269:892–901. doi: 10.1006/jmbi.1997.1081.0022-2836(1997)269[0892:IOEIIT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Persson M, Carlsson U, Bergenhem N. GroEL provides a folding pathway with lower apparent activation energy compared to spontaneous refolding of human carbonic anhydrase II. FEBS Letters. 1997;411:43–47. doi: 10.1016/s0014-5793(97)00663-7.0014-5793(1997)411[0043:GPAFPW]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Prasad BVV, Prevelige PE, Marieta E, Chen RO, Thomas D, King J, Chiu W. Three-dimensional transformation of capsids associated with genome packaging in a bacterial virus. J Mol Biol. 1993;231:65–74. doi: 10.1006/jmbi.1993.1257.0022-2836(1993)231[0065:TTOCAW]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Rabilloud T, Carpentier G, Tarroux P. Improvement and simplification of low-background silver staining of proteins by using sodium dithionite. Electrophoresis. 1988;9:288–291. doi: 10.1002/elps.1150090608.0173-0835(1988)009[0288:IASOLS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Schmidt M, Buchner J. Interaction of GroE with an all-beta-protein. J Biol Chem. 1992;267:16829–16833.0021-9258(1992)267[16829:IOGWAA]2.0.CO;2 [PubMed] [Google Scholar]
- Schuler B, Seckler R. P22 tailspike folding mutants revisited: effects on the thermodynamic stability of the isolated β-helix domain. J Mol Biol. 1998;281:227–234. doi: 10.1006/jmbi.1998.1944.0022-2836(1998)281[0227:PTFMRE]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Sideraki V, Huang W, Palzkill T, Gilbert HF. A secondary drug resistance mutation of TEM-1 beta-lactamase that suppresses misfolding and aggregation. Proc Natl Acad Sci U S A. 2001;98:283–288. doi: 10.1073/pnas.011454198.1091-6490(2001)098[0283:ASDRMO]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steven AC, Heymann JB, Cheng N, Trus BL, Conway JF. Virus maturation: dynamics and mechanism of a stabilizing structural transition that leads to infectivity. Curr Opin Struct Biol. 2005;15:227–236. doi: 10.1016/j.sbi.2005.03.008.0959-440X(2005)015[0227:VMDAMO]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steven AC, Trus BL, Booy FP, Cheng N, Zlotnick A, Caston JR, Conway JF. The making and breaking of symmetry in the virus capsid assembly: glimpses of capsid biology from cryoelectron microscopy. FASEB J. 1997;11:733–742. doi: 10.1096/fasebj.11.10.9271358.0892-6638(1997)011[0733:TMABOS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Strauss H, King J. Steps in the stabilization of newly packaged DNA during phage P22 morphogenesis. J Mol Biol. 1984;172:523–543. doi: 10.1016/s0022-2836(84)80021-2.0022-2836(1984)172[0523:SITSON]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Teschke C, Kim J, Song T, Park S, Park C, Randall L. Mutations that affect the folding of ribose-binding protein selected as suppressors of a defect in export in Escherichia coli. J Biol Chem. 1991;266:11789–11796.0021-9258(1991)266[11789:MTATFO]2.0.CO;2 [PubMed] [Google Scholar]
- Teschke CM. Aggregation and assembly of phage P22 temperature-sensitive coat protein mutants in vitro mimic the in vivo phenotype. Biochemistry. 1999;38:2873–2881. doi: 10.1021/bi982739f.0006-2960(1999)038[2873:AAAOPP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Teschke CM, King J. Folding of the phage P22 coat protein in vitro. Biochemistry. 1993;32:10839–10847. doi: 10.1021/bi00091a040.0006-2960(1993)032[10839:FOTPPC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Thomas PJ, Qu BH, Pedersen PL. Defective protein folding as a basis of human disease. TIBS. 1995;20:456–459. doi: 10.1016/s0968-0004(00)89100-8.0376-5067(1995)020[0456:DPFAAB]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Thuman-Commike PA, Greene B, Jokana J, Prasad BVV, King J, Prevelige PE, Chiu W. Three-dimensional structure of scaffolding-containing phage P22 procapsids by electron cryo-microscopy. J Mol Biol. 1996;260:85–98. doi: 10.1006/jmbi.1996.0383.0022-2836(1996)260[0085:TSOSPP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Tieman BC, Johnston MF, Fisher MT. A comparison of the GroE chaperonin requirements for sequentially and structurally homologous malate dehydrogenases: the importance of folding kinetics and solution environment. J Biol Chem. 2001;276:44541–44550. doi: 10.1074/jbc.M106693200.0021-9258(2001)276[44541:ACOTGC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Tsai AYM, Toh M, Streuli M, Thai T, Saito H. Isolation and characterization of temperature-sensitive and thermostable mutants of the human receptor-like protein tyrosine phosphatase LAR. J Biol Chem. 1991;266:10534–10543.0021-9258(1991)266[10534:IACOTA]2.0.CO;2 [PubMed] [Google Scholar]
- Tsurupa GP, Ikura T, Makio T, Kuwajima K. Refolding kinetics of staphylococcal nuclease and its mutants in the presence of the chaperonin GroEL. J Mol Biol. 1998;277:733–745. doi: 10.1006/jmbi.1998.1630.0022-2836(1998)277[0733:RKOSNA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Winston R, Botstein D, Miller TH. Characterizations of amber and ochre suppressors in Salmonella typhimurium. J Bacteriol. 1979;137:433–439. doi: 10.1128/jb.137.1.433-439.1979.0021-9193(1979)137[0433:COAAOS]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray JW, Baase WA, Lindstrom JD, Weaver LH, Poteete AR, Matthews BW. Structural analysis of a non-contiguous second-site revertant in T4 lysozyme shows that increasing the rigidity of a protein can enhance its stability. J Mol Biol. 1999;292:1111–1120. doi: 10.1006/jmbi.1999.3102.0022-2836(1999)292[1111:SAOANS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Zahn R, Spitzfaden C, Ottiger M, Wuthrich K, Pluckthun A. Destabilization of the complete protein secondary structure on binding to the chaperone GroEL. Nature. 1994;368:261–265. doi: 10.1038/368261a0.1476-4687(1994)368[0261:DOTCPS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Zeilstra-Ryalls J, Fayet O, Baird L, Georgopoulos C. Sequence analysis and phenotypic characterization of groEL mutations that block λ and T4 bacteriophage growth. J Bacteriol. 1993;175:1134–1143. doi: 10.1128/jb.175.4.1134-1143.1993.0021-9193(1993)175[1134:SAAPCO]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]