Abstract
In Saccharomyces cerevisiae, Sgt2 was thought to be the homologue of vertebrate SGT (small glutamine tetratricopeptide repeat–containing protein). SGT has been known to interact with both Hsp70 and Hsp90. However, it was not clear whether Sgt2 might have a similar capacity. Here, we showed that Ssa1/Ssa2 (yeast heat shock cognate [Hsc]70), Hsc82 (yeast Hsp90), and Hsp104 coprecipitated with Sgt2 from yeast lysates. Another molecular chaperone, Ydj1, known to interact with Ssa1 and Hsc82, also coprecipitated with Sgt2. Synthetic lethality between SGT2 and YDJ1 was observed after the cells were under stress, although Sgt2 might not interact physically with Ydj1. We also found that Mdy2 interacted with the N-terminal region of Sgt2 and that Mdy2 appeared to interact physically with Ydj1. Mdy2 therefore may mediate the association of Ydj1 and Sgt2. In addition, the mating efficiency of mdy2Δ, sgt2Δ, and mdy2Δsgt2Δ strains was reduced to a similar extent. Compared with mdy2Δ and ydj1Δ cells, ydj1Δmdy2Δ cells, however, showed a further suppression in mating efficiency. Moreover, MDY2 interacted genetically with YDJ1. These results suggest that protein complexes containing Sgt2 and Mdy2 bring molecular chaperones together to carry out certain chaperoning functions.
INTRODUCTION
Considerable evidence supports the view that the stress-induced 70-kDa heat shock protein (Hsp70) and its cognate (Hsc70) are ATP-dependent molecular chaperones. It is also clear that Hsc70 cooperates with other cellular proteins to perform many cellular functions, including protein transport into organelles and refolding of denatured proteins (for reviews, see Bukau and Horwich 1998; Hartl and Hayer-Hartl 2002). Arguably, the most well characterized Hsc70-interacting partners are members of the ubiquitous DnaJ protein family. Although DnaJ proteins can be divided into 3 subfamilies, they all contain a conserved J-domain that has the capacity to stimulate ATP-hydrolytic activity of Hsc70 (for reviews, see Cheetham and Caplan 1998; Young et al 2004). Moreover, in mammalian systems, Hsc70 has been shown to work in concert with HDJ2 to assist refolding of denatured luciferase and import of preornithine transcarbamylase into mitochondria (Terada et al 1997).
Characterization of molecular chaperones in Saccharomyces cerevisiae has been documented. For instance, it has been shown that 14 Hsp70 homologues in yeast can be divided into several functionally distinct groups (James et al 1997). Four SSA genes in S. cerevisiae are the equivalent of vertebrate Hsc70/Hsp70, and this subfamily is essential (Werner-Washburne et al 1987). The products of these 4 genes are functionally similar, but the interplay among them is complicated. For instance, under normal growth conditions, only SSA1 and SSA2 are expressed, whereas the expression of SSA3 and SSA4 is induced by stress. Deletion of both SSA1 and SSA2 also induces the high-level expression of SSA3 and SSA4 (Werner-Washburne et al 1987). The predominant function of Ssa1/Ssa2 could be in protein folding and refolding because the refolding activity of the yeast lysates prepared from ssa1Δssa2Δ cells is dramatically reduced (Bush and Meyer 1996). Nevertheless, Ssa1 also plays an important role in transporting a subset of proteins into the endoplasmic reticulum (ER) and mitochondrion (Becker et al 1996).
Several DnaJ-like proteins, including Ydj1, have been characterized in S. cerevisiae (Cheetham and Caplan 1998). Ydj1 was first identified as a gene participating in protein import into the ER and mitochondrion (Atencio and Yaffe 1992; Caplan et al 1992). Just like that of the ssa1Δssa2Δ yeast strain, the ydj1 mutant strains also show a slow growth at permissive temperature but are unable to grow at a higher temperature (Atencio and Yaffe 1992; Caplan et al 1992). Direct interactions between Ydj1 and Ssa1 have also been shown. For instance, the J-domain of Ydj1 is capable of stimulating the ATP-hydrolytic activity of Ssa1 (Cyr et al 1992), and YDJ1 is synthetically lethal to SSA1 (Becker et al 1996). Evidently, Ydj1 works with Ssa1 to carry out certain cellular functions, including protein folding. However, to refold in vitro denatured proteins at high efficiency, besides Ydj1 and Ssa1, the participation of Hsp104 (Glover and Lindquist 1998) or Sse1 (Goeckler et al 2002) is required.
Recently, with the use of yeast 2-hybrid screening, several tetratricopeptide repeat (TPR)–containing proteins were shown to interact with Hsc70 (Liu et al 1999). Previously, the TPR domains found in many proteins were implicated to mediate protein-protein interactions. Each repeat in the TPR domains is composed of 34 amino acids and forms an anti-parallel pair of helices of equal length (for reviews, see Lamb et al 1995; D'Andrea and Regan 2003). The Hsc70-interacting TPR domains contain 3 repeats in tandem that form a structural unit that interacts with the PTIEEVD sequence at the C-terminus of Hsc70 (Scheufler et al 2000; Wu et al 2001). More frequent than not, these Hsp70-interacting TPR domains also have the capacity to interact with Hsp90 (Connell et al 2001; Liou and Wang 2005), which has a C-terminal sequence (MEEVD) similar to that of Hsp70's PTIEEVD. The aspartate at the C-terminus of Hsc70 indeed appears essential for this interaction (Wu et al 2001). Recently, the structure of the complexes formed by the TPR domain of Hop (mammalian homologue of yeast Sti1) with a C-terminal peptide of Hsp70 was determined by X-ray diffraction (Scheufler et al 2000). Thus, the residues in this TPR domain responsible for interacting with the 2 carboxylic groups of the Asp residue were identified. These residues, known to form the “dicarboxylate clamp,” are highly conserved among different TPR domains that interact with Hsp70 (Scheufler et al 2000).
The C-terminal sequences of Ssa1 and Hsc82 (yeast homologue of Hsp90) are virtually identical to those of their vertebrate homologues. Proteins containing TPR domains with the dicarboxylate clamp in S. cerevisiae has also been shown to interact with molecular chaperones. Tom70 has been shown to interact with both Ssa1 and Hsc82 to facilitate protein import into mitochondrion (Young et al 2003). Moreover, Sti1, Cpr6, Cpr7, and Cns1 use their TPR domains to associate with Hsc82/Hsp82 (Abbas-Terki et al 2001; Tesic et al 2003). Sti1 and Cpr7 also interact with Hsp104 (Abbas-Terki et al 2001). More recently, it was demonstrated that Sti1 (Wegele et al 2003) and Cns1 (Hainzl et al 2004) are capable of stimulating the ATP-hydrolytic activity of Ssa1. In this report, we demonstrated that a previously uncharacterized TPR-containing protein, Sgt2 (Kordes et al 1998), in S. cerevisiae has the capacity to interact with members of Ssa molecular chaperones. Moreover, SGT2 interacted genetically with YDJ1 when yeast cells were under stress. On the basis of Saccharomyces genome database (SGD) searches, Mdy2, which plays a role in the yeast mating process (Hu et al 2006), is one of the proteins interacting with Sgt2. We show here that the N-terminal domain of Sgt2 interacted with Mdy2, and that Ydj1 co-immunoprecipitated with Mdy2. Moreover, compared with the mdy2Δ and ydj1Δ strains, the mdy2Δydj1Δ strain not only showed a sever retardation in growth but also showed a pronounced reduction in mating efficiency. Evidently, MDY2 interacted genetically with YDJ1. These results are in agreement with the view that Sgt2 and Mdy2 are components of protein complexes that bring molecular chaperones together to exert chaperoning functions.
MATERIALS AND METHODS
Yeast strains
Strains with an S288C genetic background were used in this study (Table 1), except for TFW 4272 (MATa ade8), which is in the SK1 background. The wild-type strain (BY4741 and BY4742) and strains with 1 additional deletion were purchased from Open Biosystems (Huntsville, AL, USA), two of which (sgt2Δ and mdy2Δ) were used after verification of the specific mutations. However, different isolates of wild-type cells and these 2 mutants obtained through mating and tetrad dissection behaved the same as those originally purchased. On the other hand, ydj1Δ strains were obtained by tetrad dissection after crossing ydj1Δ (MATα) with mdy2Δ (Table 1); and ydj1Δmdy2Δ was also obtained from the same tetrad analysis. Moreover, ssa1Δssa2Δ was made by crossing ssa1Δ (MATa) with ssa2Δ (MATα), and mdy2Δsgt2Δ was obtained after crossing mdy2Δ (MATa) with sgt2Δ (MATα). To generate ssa1Δssa2Δsgt2Δ and ydj1Δsgt2Δ strains, we first amplified the hygromycin-resistant cassette (hphMX4) by polymerase chain reaction (PCR) with plasmid pAG32 (Goldstein and McCusker 1999) as template. One of the primers used was composed of PR29 (Goldstein and McCusker 1999) flanked at its 5′-end by 49 nucleotides upstream of the initiation codon of SGT2, and the other primer was PR32 (Goldstein and McCusker 1999) flanked by the complementary sequence of the 49 nucleotides downstream of the stop codon of SGT2. The PCR products were transformed into ssa1Δssa2Δ and ydj1Δ strains. The transformants were selected by hygromycin, and the deletions were subsequently verified by immunoblotting analysis with specific antiserum.
Table 1.
Saccharomyces cerevisiae strains with S288C background used in this study
Preparation of antibodies
Polypeptides were expressed in bacteria and were purified. The purified proteins were used as antigen to immunized rabbits. The antiserum, with at least 3 boost injections, was collected and used in this study. To construct the expression plasmids, the encoding sequences of full-length Ydj1, Ssa1, Sgt2, and Mdy2 and the DNA fragments corresponding to the C-terminal regions of Hsc82 (amino acids 591–785) and Hsp104 (amino acids 570–908) were generated by PCR. Colonies of BY4741 were used as templates. Specifically, to express Ydj1 in bacteria, its full-length DNA was amplified with primers A and B (Table 2). Here, NdeI and XhoI sites were introduced at the 5′-end of the N- and C-terminal primers, respectively. The PCR products were cloned into the pET-15b vector (EMD Biosciences, San Diego, CA, USA) with NdeI and XhoI sites by following the procedures previously described (Liou and Wang 2005). An identical approach was used to clone the DNA of Ssa1 into the pET-22b vector (EMD Biosciences). The primers used for PCR amplification were C and D (Table 2). To express the C-terminal fragment of Hsc82, the corresponding DNA was amplified with primer E (containing the coding sequence from amino acids 589–594) and the C-terminal primer F (Table 2). The EcoRI-XhoI fragment was cloned into the pGEX-4T-1 vector (GE Healthcare, Buckinghamshire, UK) for protein expression. To generate the plasmid-containing Hsp104 fragment, the DNA was amplified with primers G (amino acids 570–571 of Hsp104) and the C-terminal primer H. The NdeI-XhoI fragment was cloned into pET-15b for expressing the C-terminal fragment of Hsp104. (Construction of expression plasmids for Sgt2 and Mdy2 will be described in the following paragraphs.) Expression and purification of the proteins were as described (Liu et al 1999; Liou and Wang, 2005), and the purified polypeptides were used as antigens to immunize rabbits.
Table 2.
The sequences of the primers used for PCR amplifications
In some cases, the antibodies were affinity purified before use. To purify the antibodies, the antigens were dialyzed against phosphate-buffered saline (PBS) with or without 2 M guanidine hydrochloride, and were coupled to solid support (AminoLink resin; Pierce, Rockford, IL, USA). The antiserum was then incubated with the appropriate resin. The bound antibodies were eluted with 20 mM hydrogen chloride (HCl), and the eluates were immediately neutralized with 1 M Tris (base). They were stored in a refrigerator at 4°C after the addition of sodium azide (NaN3) to a final concentration of 5 mM.
Immunoprecipitation
Yeast cells harvested by centrifugation were homogenized with glass beads in buffer A (75 mM potassium chloride [KCl], 25 mM Tris, 0.5% Triton X-100, pH 7.0) with protease inhibitors (Complete EDTA–free; Roche Diagnostic GmbH, Mannheim, Germany). The homogenates was centrifuged, and the supernatant collected (lysate) was used for immunoprecipitation. The precipitation was carried out with the use of Protein A-Sepharose beads as described by the manufacturer (Pierce) or with some modification. Briefly, the affinity-purified antibodies were first incubated with Protein A-Sepharose, and the unbound antibodies were removed by washing with PBS. The antibodies bound to the resin were then cross-linked to Protein A with dissucinimidyl suberate (Pierce). The uncross-linked antibodies were washed away with 0.2 M glycine (pH 2.4). The resin coupled with the antibodies was quickly neutralized and subsequently used for immunoprecipitation by following conventional protocols.
Plasmids used for the yeast 2-hybrid and glutathione S-transferase pull-down assays
We carried out yeast 2-hybrid analysis and glutathione S-transferase (GST) pull-down assays to investigate the interaction of Sgt2 and Mdy2. A DNA fragment containing the coding sequence of Sgt2 with an additional 250 base pairs at the 3′-end was generated by PCR with primers I and J (Table 2). The resulting NdeI-HindIII fragment was cloned into the pET-15b vector with the use of the same 2 restriction sites to obtain plasmid Sgt2/15b. For yeast 2-hybrid analysis, the NdeI-XbaI and XbaI-EcoRI fragments containing the coding sequence of Sgt2 were separately isolated from Sgt2/15b. A unique XbaI site is located at the middle of the SGT2 (amino acids 174–175), and EcoRI lies in the pET-15b vector downstream of the HindIII site. The 2 fragments were ligated with the pAS2-1 vector (Clontech, Palo Alto, CA, USA), having previously been treated with restriction enzymes NdeI and EcoRI. The plasmid obtained, Sgt2/pAS, was used to screen for proteins interacting with Sgt2 with the use of yeast library pGAD-C(x) (James et al 1996). One of the clones obtained, MDY2/GAD, was further used for 2-hybrid analysis.
Several deletion mutants of Sgt2 were engineered and cloned into the pAS2-1 vector for yeast 2-hybrid analysis and into the pET15b vector for pull-down assays. To generate plasmids containing SGT2 with its TPR domain disrupted, primer K (Table 2, complementary to the coding sequence of Sgt2 from amino acids 136–140) and primer I (Table 2) were used to produce a new NdeI-XbaI fragment for the N-terminal region of Sgt2. The fragment was used to substitute for the original DNA fragment (amino acids 1–173) in Sgt2/pAS. To obtain a plasmid containing the C-terminal truncated form of Sgt2, the corresponding SGT2 DNA fragment (amino acids 1–233) was amplified by PCR with primers I and L (Table 2). The NdeI-PstI fragment was isolated and inserted into the pAS2-1 vector with the same 2 sites. The DNA fragment for Sgt2 with the N-terminal deletion (amino acids 59–346) was also obtained by PCR amplification with primer M (Table 2, containing the coding sequence of SGT2 from amino acids 59–64) and primer N (Table 2, complementary to the coding sequence from amino acids 341–amber codon). The NdeI-XhoI fragment obtained was subsequently inserted into the pET-15b vector with the NdeI and XhoI sites, and into the pAS2-1 vector with the NdeI and SalI sites, respectively. To obtain the DNA fragment for the N-terminal region of Sgt2, primers I and O (Table 2) were used for PCR amplification. The PCR products were then cloned into the pAS2-1 vector with the NdeI and SalI sites and cloned into the pET-15b vector with the NdeI and XhoI sites, respectively.
To engineer the plasmid containing Mdy2 fused with GST, a fragment of the gene was amplified with primers P and Q (Table 2). MDY2/GAD was used as template. The PCR products were cloned into the pGEM-Teasy vector (Promega, Madison, WI, USA). Subsequently, the insert was excised with EcoRI and XhoI digestion. (The EcoRI site was in the vector.) The fragment was then inserted into the pGEX-4T-3 vector (GE Healthcare) with the same 2 restriction sites. In addition, the NdeI-XhoI fragment containing the Mdy2 coding sequence was excised and cloned into pET-15b for expressing the protein in bacteria.
To generate plasmids containing GST fused with Sgt2 and N-terminal truncated Sgt2, we first amplified the coding sequence of Sgt2 with the use of primers N and R (Table 2). The PCR products were cloned into the pGEM-Teasy vector, resulting in plasmid Sgt2/Te. The BamHI-XhoI fragment was excised from Sgt2/Te and ligated with the pGEX-4T-1 vector previously treated with the same 2 restriction enzymes. To obtain the fusion plasmid with the N-terminal–truncated Sgt2, the EcoRI-XhoI fragment was excised from Sgt2/Te and ligated with pGEX-4T-1 with the use of these 2 restriction sites. (An EcoRI site is located at amino acids 59–60 of Sgt2.)
Purification of recombinant Ydj1 from yeast cells
The plasmid Ydj1-15b, containing the cDNA of Ydj1 in the pET-15b vector, was first digested with restriction enzyme NcoI. The digested product was ligated with a primer (5′-CATGGAAGCTTC). The oligonucleotide has the capacity to self-anneal, producing a 4-base overhang that is identical to that resulting from NcoI digestion; once self-annealed, it also contains a HindIII restriction site. Subsequently, the newly generated plasmid was digested with enzymes HindIII and XhoI. The insert, containing the full-length DNA for Ydj1 with a his-tag at the N-terminus, was ligated with the vector pYES2 (Invitrogen, Carlsbad, CA, USA) with the same 2 restriction sites. The resulting plasmid, Ydj1-15b/Yes2, was transformed into yeast strain ydj1Δ. The plasmid has the capacity to complement the slow growth and high temperature lethality of the ydj1Δ strain (data not shown). The transformants were grown in liquid synthetic complete medium containing 2% galactose but lacking uracil. The cells were then harvested and homogenized with glass beads in 2 volumes of buffer B (75 mM KCl, 25 mM Tris, 0.5% CHAPS, and protease inhibitors). The homogenates were subjected to high-speed centrifugation, and the clear supernatant was used to purify his-tagged Ydj1 with the use of Talon resin (Clontech) by the procedures recommended by the manufacturer.
Mating assays
We performed the assays by following the procedures of Leu and Murray (2006) with some modifications. Briefly, overnight cultures were diluted to an optical density at 600 nm (OD600) of 0.15 in YPD medium (1% yeast extract, 2% peptone, 2% dextrose), except that ydj1Δmdy2Δ cells were diluted to OD600 = 0.25. Except for ydj1Δ and ydj1Δmdy2Δ, the cells were allowed to grow at 30°C for 4 hours before use for the mating assays. For ydj1Δ and ydj1Δmdy2Δ, 6-hour cultures were used for the assays. To carry out the mating assays, 0.03 OD unit of log-phase BY4742 and the mutant cells with an α mating type first were mixed with 0.3 OD unit of log-phase a-type cells (TFW4272) and were spread uniformly on YPD medium plus adenine plates with a diameter about 4 cm. Simultaneously, to determine the number of α-type cells used for the assays, identical amounts of cells were subjected to serial dilution and were spread on YPD plates. The mating was allowed to continue at 30°C for an hour. Then, the cells were collected, serially diluted, and plated on synthetic dextrose (SD) plates for selecting diploid cells. The number of diploid cells and the number of cells with an α mating type originally used for the assays were counted, and the ratio was taken as the mating efficiency.
Other methods
Yeast cells were grown in YPD medium. Mutants carrying genes replaced by kanMX4 and hphKM4 cassettes were selected with YPD plates supplemented with G418 (200 μg/ml) and hygromycin (300 μg/ml), respectively. Yeast transformation, mating, and sporulation were essentially carried out by following the procedures of Guthrie and Fink (1991). Tetrad dissection was performed with a TMD 400 tetrad dissecting microscope (MVI, Avon, MA, USA). To generate anti-hexokinase antiserum, commercial hexokinase (Sigma, St Louis, MO, USA) was used as an antigen to immunize rabbits. Yeast 2-hybrid assays and GST pull-down assays were performed as described previously (Liu et al 1999; Liou and Wang 2005).
RESULTS
It has been documented that certain TPR domains interact with the PTIEEVD sequence at the C-terminus of Hsp70 (Liu et al 1999; Wu et al 2001). The structure of the TPR domain of Hop cocrystallized with the C-terminal peptide of Hsp70 has been determined (Scheufler et al 2000), and residues at specific locations within this TPR domain responsible for interacting with the Asp residue of Hsp70 have been unequivocally defined. These residues are conserved among different Hsp70-interacting TPR domains, so using this conservation as a criterion, we searched the genome of S. cerevisiae to identify putative Ssa1-interacting proteins that contain the TPR domain. Among the proteins obtained, only Sgt2 (Yor007c) was not shown previously to interact with Ssa1, Hsc82, or Hsp104. The alignment of the TPR domain of Sgt2 with that of Hop is shown in Figure 1. Therefore, the first question considered in this study was whether Sgt2 might interact with Ssa1. Yeast lysates were prepared and immunoprecipitation was performed with anti-Sgt2 antibodies. The precipitated proteins were resolved by sodium dodecyl sulfate (SDS) gel electrophoresis. Subsequently, antibodies prepared against Ssa1 were used to determine whether Ssa1 could co-immunoprecipitate with Sgt2. The results shown in Figure 2 indicated that Ssa1/Ssa2 indeed was associated with Sgt2, albeit only a small fraction of Ssa proteins coprecipitated with Sgt2. Because SGT (putative vertebrate homologue of Sgt2) is known to interact with both Hsc70 and Hsp90, we then examined whether Hsc82 (yeast Hsp90) might also coprecipitate with Sgt2. In addition, we investigated whether Hsp104 might be associated with Sgt2 because not only is the C-terminal sequence of Hsp104 (DDID) similar to those of Ssa proteins and Hsc82 (EEVD), but also Hsc82 and Hsp104 are known to associate with some other TPR-containing proteins found in our initial database search, including Tom70, Cns1p, Sti1p, and Cpr7 (Abbas-Terki et al 2001; Tesic et al 2003; Young et al 2003). Indeed, these 2 molecular chaperones coprecipitated with Sgt2 (Fig 2). Conceivably, Sgt2 had the capacity to associate with Ssa proteins, Hsc82, and Hsp104.
Fig 1.
Sequence comparison of the tetratricopeptide repeat (TPR) domains. The amino acid sequence of the Hsc70-interacting TPR domain (TPR1) of Hop is aligned with the TPR domain of Sgt2. The locations of the 2 helices in each repeat are shown, and the residues in Hop known to interact with the C-terminal Asp residue of Hsp70 are typed in bold and are underlined
Fig 2.
Co-immunoprecipitation of chaperones with Sgt2. Yeast lysates were incubated with anti-Sgt2 antibodies, and the proteins bound were then resolved by sodium dodecyl sulfate gel electrophoresis for immunoblotting analysis with the antibodies against various proteins indicated. The specificities of antibodies against chaperones and cochaperones were validated by the deletion strains. The apparent molecular masses of these proteins on the blots were similar, if not identical, to those previously reported or predicted except for Mdy2. The apparent molecular mass of Mdy2 (29 kDa) is 20% larger than that predicted from the sequence (24 kDa). Antibodies against hexokinase were used as a negative control. Lane 1 contains 1/10 of the total lysates used for the precipitation; lane 2 is the final wash before eluting the bound proteins with acid; lanes 3 to 5 are 3 consecutive fractions of the acid eluates
We then asked whether SGT2 might interact genetically with members of SSA molecular chaperones. We therefore generated cells with SSA1, SSA2, and SGT2 deleted with several different ssa1Δssa2Δ isolates and examined the growth phenotype of the mutant. In most cases, the growth of the triple mutant (ssa1Δssa2Δsgt2Δ) appeared identical to the double mutant (ssa1Δssa2Δ). Moreover, simultaneous deletions of SGT2 with HSC82 or HSP104 showed no synthetic lethality (data not shown).
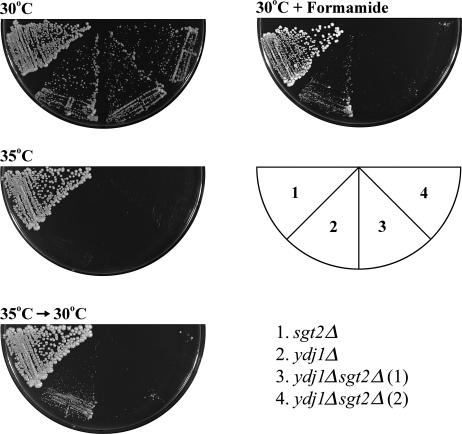
Because SSA1 interact physically and genetically with YDJ1 (Becker et al 1996), we wondered whether SGT2 might interact with YDJ1, and we first investigated whether or not this interaction might be genetic. Deletion of SGT2 alone in yeast cells had no effect on its growth (not shown). We then deleted SGT2 in ydj1Δ cells and examined the phenotype of the double mutant. As shown in Figure 3, under normal growth conditions, deletion of SGT2 did not have a significant effect on the growth of the ydj1Δ mutant either. However, if the mutants were exposed to a nonpermissive temperature (35°C) for 2 days and then shifted back to a permissive temperature, ydj1Δ cells remained viable but the growth of ydj1Δsgt2Δ cells was severely retarded (Fig 3). The double mutant thus appeared more problematic to thermal stress. We therefore investigated whether other type of stress might also have an effect on the growth of ydj1Δsgt2Δ cells. Hence, the cells were allowed to grow on plates containing formamide, and the phenotypes were then examined. Indeed, in the presence of formamide, the growth of the double mutant was much worse than that of ydj1Δ (Fig 3). Thus, a genetic interaction between SGT2 and YDJ1 was observed after the cells were placed under stress. We next asked whether or not Ydj1 associated with Sgt2. Figure 2 clearly demonstrates that a significant fraction of Ydj1 did indeed coprecipitate with Sgt2. To further investigate whether Sgt2 interacts physically with Ydj1, we performed a pull-down assay with purified recombinant GST-Ydj1 and Sgt2, but the result was negative (data not shown).
Fig 3.
Genetic interaction between SGT2 and YDJ1. Liquid cultures (optical density at 600 nm [OD600] ≈ 4 to 6) were diluted to OD600 = 0.1 with sterile water. Three microliters of cell suspension were then spotted on yeast extract/peptone/dextrose (YPD) plates with or without 2% formamide and were streaked. The growth phenotypes were subsequently examined. The mutations in ydj1Δsgt2Δ were verified by Western blot analysis (not shown). Left panel: Different yeast strains as indicated were grown on YPD plates at 30°C and 35°C for 2 days. The cells grown at 35°C were then shifted to 30°C, and they were allowed to grow for 2 more days. Although ydj1Δ cells appeared fully recovered, ydj1Δsgt2Δ cells did not recover as well. Right panel: The cells with the indicated mutations were allowed to grow on YPD plates with 2% formamide at 30°C and were photographed after 2 days. Two different isolates of ydj1Δsgt2Δ obtained through independent tetrad analysis were showed here
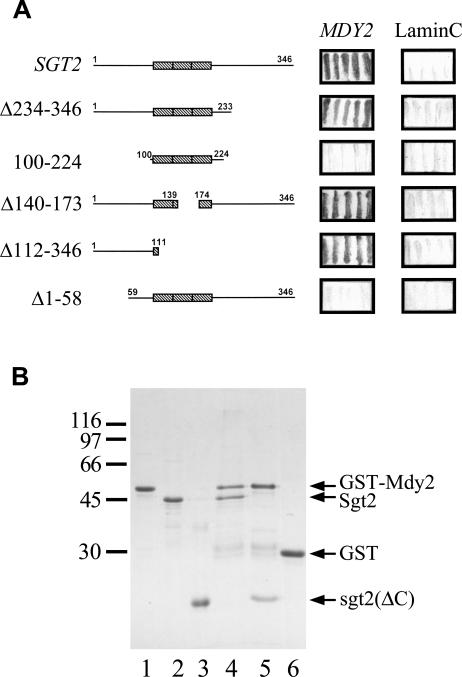
It was intriguing as to why a large fraction of Ydj1 coprecipitated with Sgt2 (Fig 2), even though these 2 proteins did not appear to interact physically with each other (pull-down assays in previous paragraph). One of the possibilities was that the coprecipitation of Ydj1 with Sgt2 was mediated by some other unidentified protein or proteins. Examination of the documented protein-protein interaction in the SGD database revealed that Mdy2 is one of the proteins that interacts with Sgt2. Mdy2 contains a ubiquitin-like domain; and deletion of MDY2 in S. cerevisiae brings about a defect in nuclear migration during the mating process, resulting in a reduction in mating efficiency (Hu et al 2006). Previously, the use of Sgt2 as bait for a yeast 2-hybrid screen, we also discovered Mdy2 as the predominant Sgt2-interacting protein (unpublished observation). Moreover, a substantial amount of Mdy2 was indeed coprecipitated with Sgt2 (Fig 2), further supporting the notion that these 2 proteins interacted with each other. We subsequently identified the region in Sgt2 responsible for its interaction with Mdy2: Deletion mutants of Sgt2 were generated for yeast 2-hybrid assays. As shown in Figure 4A, the TPR domain and the C-terminal region of Sgt2 were not required for its interaction with Mdy2. In fact, the N-terminal fragment of Sgt2 is necessary and sufficient for interacting with Mdy2. This conclusion was in agreement with the results of the pull-down assays shown in Figure 4B. Here, full-length Mdy2 was fused with GST and mixed with recombinant Sgt2 and its fragments, and the polypeptides bound to GST-Mdy2 were isolated with glutathione-Sepharose. Clearly, Sgt2 physically interacted with Mdy2 (Fig 4B, lane 4). Indeed, the N-terminal fragment of Sgt2 was sufficient for the association of Sgt2 with Mdy2 (Fig 4B, lane 5).
Fig 4.
The N-terminal region of Sgt2 interacts with Mdy2. (A) Yeast 2-hybrid analysis. The open reading frame of SGT2 and its deletion mutants were constructed in pAS2-1, and MDY2 was constructed in pGAD-Cx. The plasmids were transformed into yeast strain Y190 for 2-hybrid assays, and β-galactosidase activity was determined by filter assays. Lamin C in pACT2 was used as a negative control, and the resulting blue filters are shown here. (B) Glutathione S-transferase (GST) pull-down assays. GST-Mdy2 and GST were purified and allowed to bind to glutathione (GSH)-Sepharose. The resin was then mixed with purified Sgt2 or its N-terminal fragment, sgt2(ΔC). The bound proteins were eluted with 50 mM GSH and were then analyzed with sodium dodecyl sulfate gel electrophoresis. A Coomassie Brilliant Blue–stained gel is shown. Lanes 1, 2, and 3 contain purified GST-Mdy2, Sgt2, and its N-terminal fragment, respectively. Lanes 4 and 5 contain GST-Mdy2 incubated with Sgt2 and the N-terminal fragment of Sgt2, respectively. Purified GST incubated with Sgt2 (lane 6) or the N-terminal fragment of Sgt2 (not shown) were used as controls. Both Sgt2 and its N-terminal fragment failed to associate with GST. The molecular mass markers are β-galactosidase (116 000), phosphorylase (97 000), bovine serum albumin (66 000), ovalbumin (45 000), and carbonic anhydrase (30 000)
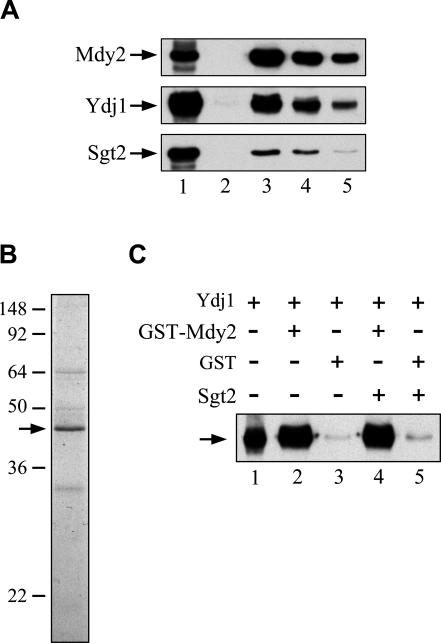
We next determined whether Mdy2 might interact physically with Ydj1. We first determined whether Ydj1 could be precipitated from yeast lysates by antibodies against Mdy2. The results shown in Figure 5A clearly demonstrated that this was the case. We then determined whether Mdy2 physically interacts with Ydj1. Therefore, we prepared GST-Mdy2 from bacteria and Ydj1 from yeast cells (Fig 5B), and we then incubated the proteins. Subsequently, proteins associated with GST-Mdy2 were analyzed with anti-Ydj1 antibodies to determine whether Ydj1 interacted with Mdy2. Immunoblotting was necessary because the electrophoretic mobilities of GST-Mdy2 and Ydj1 are similar, if not identical, on SDS gels. The results shown in Figure 5C (lanes 2 and 3) clearly demonstrated that Ydj1 was associated with Mdy2. Moreover, addition of Sgt2 in the reaction mixtures for the pull-down assay did not affect the association of Ydj1 with Mdy2 (Fig 5C; lanes 4 and 5), implying that the interaction between Mdy2 and Ydj1 was independent of Sgt2.
Fig 5.
Association of Ydj1 with Mdy2. (A) Co-immunoprecipitation of Ydj1 with Mdy2. Yeast lysates were prepared and were incubated with resin coupled with anti-Mdy2 antibodies. The bound proteins were eluted by acid, and the eluates were subjected to immunoblot analysis with antibodies against Mdy2, Ydj1, and Sgt2, respectively. Lane 1 contains 1/10 of the total lysates; lane 2: the final wash before acid elution; lanes 3 to 5: the 3 consecutive fractions of the eluates. (B) Purified Ydj1. To purify Ydj1 from yeast, we first transformed plasmid harboring YDJ1 with an N-terminal his-tag into ydj1Δ yeast strain; Ydj1 was then purified from detergent-extracted yeast lysates. The purified protein was visualized with the use of sodium dodecyl sulfate (SDS) gel electrophoresis, and a Coomassie Blue–stained gel is shown. The location of Ydj1 on the gels is indicated by the arrow. Prestained molecular mass markers (kDa) were purchased from Invitrogen (SeeBlue Plus2) (C) In vitro interaction of Mdy2 with Ydj1. Purified Ydj1 was incubated with GST-Mdy2 or glutathione S-transferase (GST) immobilized on glutathione (GSH)-Sepharose with or without the addition of purified Sgt2. Then, protein bound to the resin was eluted with GSH. Immunoblotting was used to determine whether Ydj1 was specifically associated with GST-Mdy2. Lane 1 contains 10% of the total purified Ydj1 used for each pull-down assay; lanes 2 and 4: Ydj1 incubated with GST-Mdy2 in the absence or presence of Sgt2; lanes 3 and 5: Ydj1 with GST in the absence or presence of Sgt2, respectively
We next verified in vitro whether coprecipitation of Ydj1 with Sgt2 could be mediated by Mdy2. Therefore, recombinant GST-Sgt2, GST-sgt2(ΔN) (GST fusion protein of Sgt2 with its first 58 amino acids deleted), and Mdy2 were purified (Fig 6A). We then performed GST pull-down assays. The results shown in Figure 6B demonstrated that GST-Sgt2 interacted with Mdy2, but that GST-sgt2(ΔN) lost the capacity to form complexes with Mdy2. Subsequently, we incubated the GST-Sgt2 and GST-sgt2(ΔN) with purified Ydj1 in the presence and absence of Mdy2. The Ydj1 pull-down by the fusion proteins was then determined by immunoblotting analysis. As shown in Figure 6C, a small fraction of Ydj1 was associated with GST-Sgt2 in the absence of Mdy2 (lane 2). However, the association of Ydj1 with Sgt2 was diminished once the N-terminal region of Sgt2 was deleted (lane 3). More importantly, the association of Ydj1 with GST-Sgt2 was much greater when Mdy2 was added to the reaction mixtures (lane 5). These results indicated that Mdy2 had the capacity to mediate the association of Ydj1 with Sgt2.
Fig 6.
Mdy2 effects on the interaction of Sgt2 and Ydj1. (A) Purified proteins: GST-Sgt2 (lane 1), GST-sgt2(ΔN) (lane 2), glutathione S-transferase (GST) (lane 3), and Mdy2 (lane 4) were expressed in bacteria and were purified as described in “Materials and Methods.” Five micrograms each were displayed by sodium dodecyl sulfate (SDS) gel electrophoresis, and a Coomassie Brilliant Blue–stained gel is given here. Molecular mass markers (kDa) are phosphorylase b (97 000), bovine serum albumin, ovalbumin (66 000), carbonic anhydrase (30 000), and soybean trypsin inhibitor (22 000). The apparent molecular mass of the recombinant Mdy2 is larger than that predicted, and the lower band (marked by a circle) is a degradation product of his-tagged Mdy2 (lane 4). (B) Interaction between Sgt2 and Mdy2. GST pull-down assays were carried out as described and were used to determine whether the region containing the N-terminal 58 amino acids of Sgt2 was essential for interacting with Mdy2. The proteins pull-down by glutathione (GSH)-Sepharose were analyzed by SDS gel, and a Coomassie Blue–stained gel is shown here. Lanes 1, 2, and 3 are GST-Sgt2, GST-sgt2(ΔN), and GST with Mdy2, respectively. The location of Mdy2 on the gel is indicated with an asterisk. (C) Association of Ydj1 with Sgt2 is elevated by Mdy2. GST-Sgt2, GST-sgt2(ΔN), and GST were incubated with Ydj1 purified from yeast in the presence and absence of Mdy2. The association of Ydj1 with GST fusion proteins was determined by Western blot analysis with anti-Ydj1 antibodies. Lane 1 contains 1/10 of the Ydj1 used for the assays. Lanes 2 and 5 contain GST-Sgt2 without and with Mdy2, respectively. Lanes 3 and 6 contain GST-sgt2(ΔN) without and with Mdy2, respectively. Lanes 4 and 7 contain GST without and with Mdy2, respectively.
Now that we have shown that Mdy2 interacts with Ydj1 and Sgt2, we wanted to determine whether or not Mdy2 might interact with Ssa chaperones. We first prepared yeast lysates and performed immunoprecipitation with anti-Mdy2 antibodies. The precipitated proteins were subjected to immunoblotting with anti-Ssa1 antibodies. Our result demonstrated that little Ssa1/Ssa2 co-immunoprecipitated with Mdy2 (data not shown). Similarly, antibodies against Mdy2 did not bring down any Hsc82 or Hsp104 either (data not shown). We then generated a triple mutant ssa1Δssa2Δmdy2::hphMX4 and examined its growth phenotype. The results demonstrated that the growth of this triple mutant was indistinguishable from that of ssa1Δssa2Δ in the presence or absence of formamide (data not shown).
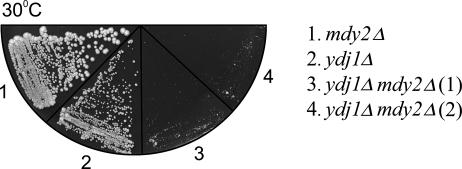
The next question asked was whether or not MDY2 and YDJ1 might interact genetically. We therefore generated a double mutant ydj1Δmdy2Δ yeast strain with both YDJ1 and MDY2 deleted and examined the growth phenotype of the mutants. Although the growth of mdy2Δ was identical to that of the wild type yeast (not shown), the result shown in Figure 7 clearly indicated that the growth of the double mutant was severely retarded compared with that of ydj1Δ. Evidently, MDY2 and YDJ1 interact genetically.
Fig 7.
Genetic interaction of YDJ1 and MDY2. Mutant ydj1Δmdy2Δ was generated and verified by Western blot analysis. Equal units of cells at an optical density of 600 nm (OD600) carrying the indicated mutations were streaked onto YPD plates and were allowed to grow for 54 hours before being photographed. The growth of the mdy2Δ strain is identical to that of the wild-type cells (not shown). Two different isolates of ydj1Δmdy2Δ are shown here
Previously, it was shown that the deletion of MDY2 results in a reduction in the mating efficiency of S. cerevisiae (Hu et al 2006). The last question considered in this study was whether deletion of SGT2 or YDJ1 in mdy2Δ background might further suppress the mating efficiency of the yeast. Therefore, we mixed the mutants (MATα) with a tester strain (MATa) at a ratio of 1:10 for mating. Subsequently, we measured the mating efficiency. Under these conditions, the difference in growth rate of haploid cells on mating efficiency would be minimized, and small variations in mixing and plating of the cells were not likely to affect the measurement. As shown in Figure 8, 70% of the wild-type cells were mated in an hour. The mating efficiency of the mdy2Δ strain was reduced, albeit the level of the reduction was not as dramatic as that previously reported (Hu et al 2006). Interestingly, deletion of SGT2 also resulted in a small reduction in mating efficiency. However, deletion of SGT2 in an mdy2Δ background did not further suppress the mating efficiency compared with the mdy2Δ strain. On the other hand, compared with the wild-type cells, deletion of YDJ1 resulted in a 15-fold reduction in mating efficiency. In addition, deletion of both YDJ1 and MDY2 brought about a further 10-fold reduction in mating efficiency. Therefore, the genetic interaction between YDJ1 and MDY2 as determined by growth (Fig 7) was also revealed in the mating efficiency measurement.
Fig 8.
Mutants show reduction in mating efficiency. BY4742 and various mutants with an α mating type were mixed with an a-type tester strain (TFW4272) at a ratio of 1:10, and they were allowed to mate at 30°C for an hour. The mating efficiency was then quantified. The results shown here are the average of 3 separate determinations, except for the ydj1Δ and ydj1Δmdy2Δ strains. In both cases, the average of 2 determinations is given. The average deviations for the measurements also are shown
DISCUSSION
Herein, use of the conserved dicarboxylate clamp residues in certain TPR domains as one of the criteria to search the genome of S. cerevisiae, we predicted that Sgt2 might interact with members of Ssa proteins. Although the documented yeast databases (including SGD) provide little evidence indicating that Sgt2 interacts with Ssa proteins, we nevertheless demonstrated through immunoprecipitation that Sgt2 can indeed associate with Ssa1/Ssa2. With the use of yeast 2-hybrid assays, it has been demonstrated that the 30-kDa domain of Hsc70 interacts with human SGT (Liu et al 1999). To further verify the interaction between Ssa1 and Sgt2, we performed a similar analysis with the C-terminal 30-kDa domain of Ssa1 in pAS2-1 and Sgt2 in pACT-2, respectively, but failed to demonstrate this predicted interaction (data not shown). The lack of the predicted interaction might reflect that such an interaction in yeast could be more transient than the interaction between their vertebrate counterparts, although a lack of significant interaction between Ssa1 and Sgt2 is not ruled out.
In vertebrates, SGT is thought to participate in several cellular events. For instance, SGT, Hsc70, and cysteine string protein (a protein containing J-domain) form trimeric complexes (Tobaben et al 2001). This complex regulates the nucleotide exchange of heterotrimeric GTP-binding protein, promoting inhibition of the Ca2+ channel (Natochin et al 2005). More recently, it has been demonstrated that depletion of SGT by the RNA interference approach results in an increase in cells with misaligned chromosomes and affects cell proliferation (Winnefeld et al 2006). In contrary to that of SGT, deletion of SGT2 in S. cerevisiae has little effect on the growth of the cells. The reason for this disagreement is not known. However, it is evident that the structural features of Sgt2 and SGT are similar but not identical. Both Sgt2 and SGT possess highly conserved TPR domains for interacting with Hsp70 chaperones, and they could function as cochaperones, but equivalent regions outside the TPR domain of these 2 proteins are less conserved. In particular, Sgt2 lacks the glutamine-rich sequence that is located at the C-terminal region of SGT. It is conceivable that the functional diversity between these 2 homologous proteins originates from variations in the structural domains. Of course, this hypothesis needs to be further verified.
To investigate whether SGT2 might interact genetically with SSA1/SSA2, we have generated ssa1Δssa2Δsgt2Δ strains with the use of several ssa1Δssa2Δ isolates obtained after tetrad analysis. Triple mutants generated from most of the ssa1Δssa2Δ strains show no synthetic lethality between SGT2 and SSA1/SSA2. One of the possible explanations is that induction of Ssa3 and Ssa4 could compensate for the loss of Ssa1 and Ssa2. Consequently, genetic interaction between SSA1/SSA2 and SGT2 cannot be observed. Our studies nevertheless show that SGT2 interacts genetically with YDJ1. An interesting question raised here is why the synthetic lethality of SGT2 with YDJ1 becomes more evident in the presence of formamide. Under our experimental conditions, the growth in log phase of the wild-type and mutant yeast strains, including sgt2Δ and ydj1Δ, was retarded by only 20–30% with the addition of formamide (data not shown). Thus, although the effect of formamide on the growth might be mild, formamide does represent a form of stress to the yeast cells. It is generally believed that, in vitro, formamide has the capacity to disrupt hydrogen bonds, causing denaturation of macromolecules. However, its is not clear whether formamide indeed brings about nonspecific denaturation of proteins in the yeast cells or affects some other as yet undefined process in the cells. In any case, the nature of the cellular stress caused by formamide and the significance of the genetic interaction between SGT2 and YDJ1 remains to be elucidated.
We also examined whether or not Sgt2 and Ydj1 interact physically. Although Ydj1 coprecipitated with Sgt2, our study suggests that the interaction of the 2 proteins might not be a direct one but one mediated by Mdy2 with the N-terminal region of Sgt2 interacting with Mdy2. This hypothesis is supported by the observation that addition of Mdy2 to the reaction mixtures containing purified GST-Sgt2 and Ydj1 for pull-down assays enhanced the association of Ydj1 with Sgt2. Further proof that MDY2 interacts with YDJ1 was demonstrated by the finding that MDY2 and YDJ1 are synthetically lethal and that Ydj1 purified from yeast cells associated with bacterium-expressed GST-Mdy2.
Previously, deletion of MDY2 was shown to cause a 5-fold reduction in mating efficiency (Hu et al 2006). However, under our experimental conditions, deletion of this gene only results in a marginal reduction in mating efficiency. This disagreement could originate either from the difference in the definition of mating efficiency or form the difference in the genetic background of the strains used. Nevertheless, it is evident that MDY2 plays a role in the mating process. More importantly, the level of reduction in mating efficiency for the mdy2Δ, sgt2Δ, and mdy2Δsgt2Δ strains appears similar, if not identical. This observation suggests that SGT2 and MDY2 might act on the same pathway in the yeast mating process. Moreover, deletion in YDJ1 in cells with an α mating type brings about a pronounced reduction in mating efficiency. It has been shown that mutation of YDJ1 in a-type cells results in reduction of mating efficiency because of a severe defect in a-factor production (Meacham et al 1999). However, the defect in α-factor processing in ydj1Δ cells is mild at permissive temperatures (Becker et al 1996). It needs to be clarified whether the reduction in mating efficiency in ydj1Δ cells with an α mating type is solely the result of changes in α-factor production. In any event, the mating efficiency was further reduced in ydj1Δmdy2Δ strain. Although other possibilities cannot be ruled out entirely, the result implies that YDJ1 and MDY2 might act on different pathways for the mating process.
More recently, MDY2 was shown to be synthetically lethal with HSP82 (Zhao et al 2005). We carried out immunoprecipitation to determine whether Hsp82/Hsc82 might be associated with Mdy2. The results were negative. In fact, Ssa proteins were not associated with Mdy2 either (data not shown). This observation was somewhat surprising because Mdy2 formed complexes with Sgt2, and Sgt2 has the capacity to precipitate Ssa proteins and Hsc82. One of the possible interpretations was that only a relatively small fraction of Sgt2 was coprecipitated with Mdy2. Thus, the association of Ssa proteins and Hsc82 with Mdy2 via Sgt2 might be beyond the limit of the detection. During the past few years, large-scale analysis of protein complexes (Ho et al 2002; Gavin et al 2006; Krogan et al 2006) and high-throughput 2-hybrid screening (Uetz et al 2000; Ito et al 2001) have been carried out in S. cerevisiae. From these data, interaction of Sgt2 with Mdy2 is evident. However, interactions of these 2 proteins with molecular chaperones were not suggested. Our results, nevertheless, clearly show that protein complexes containing Sgt2 and Mdy2 interact physically with Ydj1. Because Sgt2 also has the capacity to associate with Ssa proteins, Hsc82, and Hsp104, Sgt2 and Mdy2 therefore might bring different molecular chaperones together to facilitate some specific chaperoning function in yeast cells. If this is the case, one needs to elucidate the functional role of these complexes.
Acknowledgments
We thank Dr Jun-Yi Leu for help in mating assays, Dr Ting-Fang Wang for yeast strain TFW4272, and Tsai-Ching Chang for the SGT2 clones. We are also grateful to Dr Kenrick Deen for comments on the manuscript. This work was supported by grants from Academia Sinica and the National Science Council (NSC92-2311-B-001-096; NSC93-2311-B-001-055) of Taiwan, Republic of China.
REFERENCES
- Abbas-Terki T, Donez O, Briand P-A, Picard D. Hsp104 interacts with Hsp90 cochaperones in respiring yeast. Mol Cell Biol. 2001;21:7569–7573. doi: 10.1128/MCB.21.22.7569-7575.2001.1098-5549(2001)021[7569:HIWHCI]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atencio DP, Yaffe MP. MAS5, a yeast homolog of DnaJ involved in mitochondrial protein import. Mol Cell Biol. 1992;12:283–291. doi: 10.1128/mcb.12.1.283.1098-5549(1992)012[0283:MAYHOD]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Walter W, Yan W, Craig EA. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol Cell Biol. 1996;16:4378–4386. doi: 10.1128/mcb.16.8.4378.1098-5549(1996)016[4378:FIOCHA]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9.0092-8674(1998)092[0351:THAHCM]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Bush GL, Meyer DI. The refolding activity of the yeast heat shock proteins Ssa1 and Ssa2 defines their role in protein translocation. J Cell Biol. 1996;135:1229–1237. doi: 10.1083/jcb.135.5.1229.0021-9525(1996)135[1229:TRAOTY]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan AJ, Cyr DM, Douglas MG. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell. 1992;71:1143–1155. doi: 10.1016/s0092-8674(05)80063-7.0092-8674(1992)071[1143:YFPTAD]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Caplan AJ, Tsai J, Casey PJ, Douglas MG. Farnesylation of YDJ1p is required for function at elevated temperatures in S. cerevisiae. J Biol Chem. 1992;267:18890–18895.0021-9258(1992)267[18890:FOYIRF]2.0.CO;2 [PubMed] [Google Scholar]
- Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2.1466-1268(1998)003[0028:SFAEOD]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hofeld J, Patterson C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618.1465-7392(2001)003[0093:TCCRPT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Cyr DM, Lu X, Douglas MG. Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J Biol Chem. 1992;267:20927–20931.0021-9258(1992)267[20927:ROHFBA]2.0.CO;2 [PubMed] [Google Scholar]
- D'Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007.0376-5067(2003)028[0655:TPTVH]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Gavin A-C, Aloy P, and Grandi P. et al. 2006 Proteome survey reveals modularity of the yeast cell machinery. Nature. 440:631–636. [DOI] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4.0092-8674(1998)094[0073:HHAHAN]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Goeckeler JL, Stephens A, Lee P, Caplan AJ, Brodsky JL. Over-expression of SSE1 (Hsp110) rescues a Ydj1p mutant defect through the restoration of Hsp90 activity. Mol Biol Cell. 2002;13:2760–2770. doi: 10.1091/mbc.02-04-0051.1059-1524(2002)013[2760:OOSHRA]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K.0749-503X(1999)015[1541:TNDDRC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR 1991 Guide to Yeast Genetics and Molecular Biology. Methods in Enzymology series, vol 194. Academic Press, San Diego, CA, USA. [PubMed] [Google Scholar]
- Hainzl O, Wegele H, Reinstein J, Buchner J. Cns1 is an activator of the Ssa1 ATPase activity. J Biol Chem. 2004;279:23267–23273. doi: 10.1074/jbc.M402189200.0021-9258(2004)279[23267:CIAAOT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408.0193-4511(2002)295[1852:MCITCF]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, and Heilbut A. et al. 2002 Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 415:180–183. [DOI] [PubMed] [Google Scholar]
- Hu Z, Potthoff B, Hollenberg CP, Ramezani-Rad M. Mdy2, a ubiquitin-like (UBL)-domain protein, is required for efficient mating in Saccharomyces cerevisiae. J Cell Sci. 2006;119:326–338. doi: 10.1242/jcs.02754.0021-9533(2006)119[0326:MAUUPI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa T, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498.1091-6490(2001)098[4569:ACTATE]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425.0016-6731(1996)144[1425:GLAAHS]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Pfund C, Craig EA. Functional specificity among Hsp70 molecular chaperones. Science. 1997;275:387–389. doi: 10.1126/science.275.5298.387.0193-4511(1997)275[0387:FSAHMC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Kordes E, Savelyeva L, Schwab M, Rommelaere J, Jaunlaux J-C, Cziepluch C. Isolation and characterization of human SGT and identification of homologues in Saccharomyces cerevisiae and Caenorhabditis elegans. Genomics. 1998;52:90–94. doi: 10.1006/geno.1998.5385.0888-7543(1998)052[0090:IACOHS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, and Yu H. et al. 2006 Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 440:637–643. [DOI] [PubMed] [Google Scholar]
- Lamb JR, Tugendreich S, Hieter P. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4.0376-5067(1995)020[0257:TPRITT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Leu J-Y, Murray A. Experimental evolution of mating discrimination in budding yeast. Curr Biol. 2006;16:280–286. doi: 10.1016/j.cub.2005.12.028.0960-9822(2006)016[0280:EEOMDI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Liou S-T, Wang C. Small glutamine-rich tetratricopeptide repeat–containing protein is composed of three structural units with distinct functions. Arch Biochem Biophys. 2005;435:253–263. doi: 10.1016/j.abb.2004.12.020.0003-9861(2005)435[0253:SGTRPI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Liu F-H, Wu S-J, Hu S-M, Hsiao C-D, Wang C. Specific interaction of the 70-kDa heat shock cognate protein with the tetratricopeptide repeats. J Biol Chem. 1999;274:34425–34432. doi: 10.1074/jbc.274.48.34425.0021-9258(1999)274[34425:SIOTKH]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Meacham GC, Browne BL, Zhang W, Kellermayer R, Bedwell DM, Cyr DM. Mutations in the yeast Hsp40 co-chaperone protein Ydj1 cause defects in Axl1 biogenesis and pro-a-factor processing. J Biol Chem. 1999;274:34396–34402. doi: 10.1074/jbc.274.48.34396.0021-9258(1999)274[34396:MITYHC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Natochin M, Campbell TN, Barren B, Miller LC, Hameed S, Artemyev NO, Braun JEA. Characterization of Gαs regulator cysteine string protein. J Biol Chem. 2005;280:30236–30241. doi: 10.1074/jbc.M500722200.0021-9258(2005)280[30236:COGRCS]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Scheufler C, Brinker A, Bourenkov G, Pegorado S, Moroder L, Bartunik H, Hartl FU, Moarefi I. Structure of TPR domain– peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 2000;101:199–210. doi: 10.1016/S0092-8674(00)80830-2.0092-8674(2000)101[0199:SOTDPC]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Terada K, Kanazawa M, Bukau B, Mori M. The human DnaJ homologue dj2 facilitates mitochondrial protein import and luciferase refolding. J Cell Biol. 1997;139:1089–1095. doi: 10.1083/jcb.139.5.1089.0021-9525(1997)139[1089:THDHDF]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesic M, March JA, Cullinan SB, Gaber RF. Functional interactions between Hsp90 and the co-chaperones Cns1 and Cpr7 in Saccharomyces cerevisiae. J Biol Chem. 2003;278:32692–32701. doi: 10.1074/jbc.M304315200.0021-9258(2003)278[32692:FIBHAT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Tobaben S, Thakur P, Fernandez-Chacon R, Sudhof TC, Rettig J, Stahl B. A trimeric protein complex functions as a cynaptic chaperone machine. Neuron. 2001;31:987–999. doi: 10.1016/s0896-6273(01)00427-5.0896-6273(2001)031[0987:ATPCFA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, and Cagney G. et al. 2000 A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 403:623–627. [DOI] [PubMed] [Google Scholar]
- Wegele H, Haslbeck M, Reinstein J, Buchner J. Sti1 is a novel activator of the Ssa proteins. J Biol Chem. 2003;278:25970–25976. doi: 10.1074/jbc.M301548200.0021-9258(2003)278[25970:SIANAO]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Werner-Washburne M, Stone DE, Craig EA. Complex interactions among members of an essential subfamily of Hsp70 genes in yeast. Mol Cell Biol. 1987;7:2568–2577. doi: 10.1128/mcb.7.7.2568.1098-5549(1987)007[2568:CIAMOA]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnefeld M, Grewenig A, Schnolzer M, Spring H, Knock TA, Gan EC, Rommelaere J, Cziepuch C. Human SGT interacts with Bag-6/Bat-3/Synthe and cells with reduced levels of either protein display persistence of few misaligned chromosomes and mitotic arrest. Exp Cell Res. 2006;312:2500–2514. doi: 10.1016/j.yexcr.2006.04.020.0014-4827(2006)312[2500:HSIWSA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Wu S-J, Liu F-H, Hu S-M, Wang C. Different combinations of the heat-shock cognate protein 70 (hsc70) C-terminal functional groups are utilized to interact with distinct tetratricopeptide repeat-containing proteins. Biochem J. 2001;359:419–426. doi: 10.1042/0264-6021:3590419.0264-6021(2001)359[0419:DCOTHC]2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5:781–791. doi: 10.1038/nrm1492.1471-0080(2004)005[0781:POCPFI]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Young JC, Hoogenraad NJ, Hartl FU. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112:41–50.0092-8674(2003)112[0041:MCHAHD]2.0.CO;2 [Google Scholar]
- Zhao R, Davey M, and Hsu Y-C. et al. 2005 Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by Hsp90 chaperone. Cell. 120:715–727. [DOI] [PubMed] [Google Scholar]