Breast cancer is now the second leading cause of cancer death in women, with 181,600 new cases diagnosed in 1997. 1 Researchers in all disciplines of science are vigorously pursuing the identification of factors that might free women from the uncertainty of not knowing who will succumb to this disease. Some known risk factors, including early menarche, history of benign breast disease, and exposure to environmental factors such as radiation, have been shown to only moderately increase a woman’s risk for breast cancer. 2 More frustrating from a cancer prevention viewpoint is that two-thirds of women who develop this disease fit none of the currently identified at-risk groups. This has prompted scientists to look for clues at the level of the tumor genome. Although many genetic alterations involved in breast disease have been identified in recent years, clinical applications for this knowledge have for the most part not yet materialized. This article specifically examines the timing of chromosome 11p and 11q alterations in breast cancer progression pathways and their potential for clinical application. It is clear from this information that advancements with the greatest potential benefit to women already diagnosed with breast cancer would include characterization of preinvasive markers indicative of increased potential for invasion or, better yet, elucidation of genetic loci with predictive value for identifying women at high risk for developing this disease before clinical diagnosis. Both types of biomarkers could have an enormous impact on women’s health in general.
Why has the translational application of genetic information been so difficult in breast cancer? In part, the answer seems to be that breast cancer presents as a morphologically and biologically heterogeneous and progressive disease. Although this disease characteristic has been recognized at the histological level for many years, an equivalent multidimensional genetic profile has also emerged. The accepted paradigm that a serial, cumulative multistep progression of genetic events leads a normal cell down the pathway to becoming tumorigenic was first theorized by Nowell in 1976 3 and genetically deciphered for colon carcinoma by Fearon and Vogelstein in 1990. 4 This model allows for molecular heterogeneity as one of the characteristics of the tumor, this heterogeneity reflecting the successive and dynamic multistep genetic evolution of the disease. It also allows for the “selection” against genetic events that end in cell death, not cell transformation.
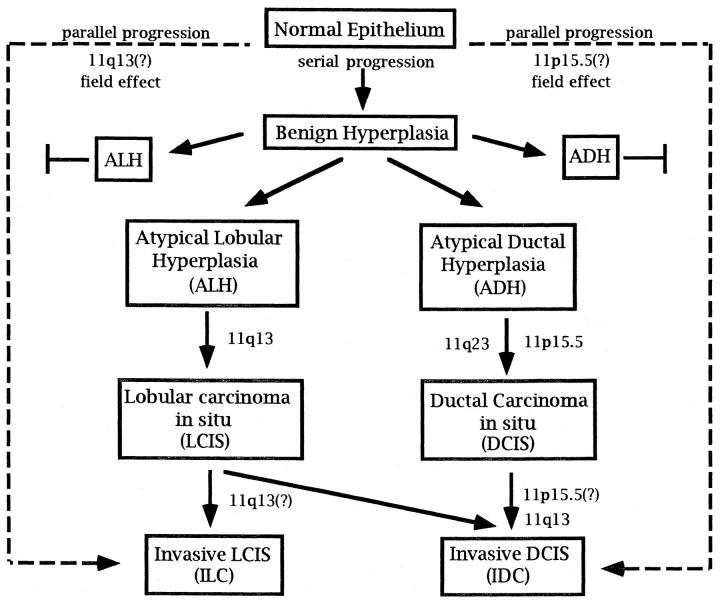
Current knowledge of the progression of breast cancer reveals that by analogy to colon and brain cancers, a subset of genetic aberrations may occur in a disease stage- or grade-specific manner suggestive of a serial progression (Figure 1) ▶ . Microsatellite marker alterations have recently provided evidence in support of a serial progressive pathway for the early stages of breast disease. Early changes, identified as benign proliferative breast lesions or atypical hyperplasias (AHs), are most often considered benign disorders, yet epidemiological studies indicate that these lesions are associated with a significantly increased risk (four- to fivefold) of developing breast cancer. 5,6 Clonal microsatellite alterations have been observed in histologically confirmed AHs and their concurrent ductal carcinoma in situ (DCIS) components, supporting the postulated model in which carcinomas may arise from proliferative lesions. 7 In contrast, other studies have also shown that microsatellite alterations identified in AHs from a patient were not shared by the concomitant carcinoma in situ (CIS) or invasive CISs examined. As theorized by Nowell, 3 these findings indicate that multiple independent clones can emerge that are not necessarily committed to a malignant phenotype. Such early molecular heterogeneity is further supported by the fact that 80% of patients with identified AH lesions do not develop malignancies. 8 Molecular heterogeneity in breast cancer is not restricted to the early stages of breast disease, however. Fujii et al 9 discovered, in addition to conserved alterations, that 40% of cases with synchronous DCIS and invasive ductal carcinoma (IDC) contained heterogeneous patterns of allelic loss at one or more loci in the DCIS components. This indicates genetic divergence can arise during the clonal evolution from the malignant to invasive phenotype as well.
Figure 1.
Serial and parallel progression pathways proposed for breast cancer. The potential placement of genetic events on chromosome 11p and 11q is shown within the context of these pathways.
The relationship of lobular neoplasia of the breast (LCIS) and atypical lobular hyperplasia (ALH) with the emerging genetic pathway for DCIS is less well defined. Although DCIS and LCIS can be distinguished histologically, both lesions arise in the terminal duct lobular unit and for both the cell of origin is unknown. Lobular neoplasia not only increases the incidence of invasive carcinoma in the same breast but is also a marker for increased risk of ductal and lobular carcinoma in the contralateral breast. 10,11 Few genetic analyses have been done on LCIS. In one such study, two of seven cases of multiple foci of LCIS from the same breast showed a different pattern of loss of heterozygosity (LOH), 12 suggesting this disease undergoes molecular heterogeneity comparable to DCIS. Whether LCIS and DCIS arise from the same or distinct genetic pathways requires further analysis.
In contrast to the single progressional pathway identified for colon cancer, some breast cells seem to be able to circumvent the prototypical serial accumulation of alterations to attain a malignant phenotype. A parallel progression from morphologically normal epithelium directly to advanced disease is supported by the discovery of many small invasive cancers that do not have accompanying atypical components. For such a progression to occur, a kind of “field effect” would be expected whereby at least some of the genetic alterations found in malignant tumors already exist in the corresponding morphologically normal-looking breast epithelium. Indeed, Deng et al 13 have identified a high frequency (60%) of DCIS cases with LOH at 3p24, in which the equivalent LOH was displayed in the adjacent normal breast tissue. These molecular observations clearly show that breast disease can potentially follow several tumorigenic pathways, creating a more histologically and genetically complex picture for this disease. Interestingly, a subset of malignant brain tumors, called de novo glioblastomas, do not possess the stage-related LOH events found in progressional glioblastomas, 14,15 another indication that both serial and parallel pathways exist in malignant tumor progression.
Cytogenetic and molecular analyses of breast tumors indicate that the functions of both oncogenes and tumor suppressor genes are altered during the tumorigenic process. 16,17 Loss of tumor suppressor gene function as measured by LOH has been investigated for a number of chromosomes in breast carcinoma, with the most frequently deleted loci at 3p, 6q, 11q, 16q, 17p, and 17q involving rates from 20 to 70%. 18,19 Both the long and short arms of chromosome 11 have received a great deal of attention in these LOH analyses. At least three separate regions of LOH have been consistently identified (11p15.5, 11q13, and 11q23), pointing to a potentially complicated role for this chromosome in breast tumorigenesis.
The presence of a gene or genes on chromosome 11p15.5 is of general interest given that LOH involving this band has been measured in many pediatric and adult neoplasms, including Wilms’ tumor, 20 embryonal rhabdomyosarcoma, 21 non-small-cell lung cancer, 22 and ovarian cancer. 23 For breast cancer, the predominant area harboring a putative tumor suppressor gene lies in a 2-Mb region between markers TH and D11S988. 24,25 The frequency with which allelic loss occurs at this site is less precisely measured. It has been reported at 3 to 35% in DCIS 24,26,27 and 10 to 41% in IDC. 28,29 In this issue, Lichy et al 27 use microdissection of concurrent intraductal, invasive, and metastatic tumor foci to readdress the issue of the frequency and timing for the 11p15.5 genetic event(s). Their studies show a frequency of allelic loss at 37.5%, 32%, and 44% in pure intraductal carcinoma, node-negative IDC, and node-positive IDC foci, respectively. It is likely, for several reasons, that these numbers represent the “best estimates” to date for breast tumors that undergo a serial-type progression. First, Lichy et al 27 used markers involving the well-defined 2-Mb LOH region for breast cancer on 11p15.5. In addition, microdissection of the individually analyzed components most likely eliminated a majority of the normal cell contamination that might have masked allelic deletions in previous studies using whole-tissue DNA samples. Not to be overlooked in this study is the small fraction of tumors that did not exhibit 11p15.5 allelic deletion until the later invasive stages of tumorigenesis. These tumors indicate that some heterogeneity exists in the appearance of 11p15.5 genetic alterations within the Nowell-like progression model for breast cancer (see Figure 1 ▶ ). Adding further complexity to this issue is the study on normal breast epithelium by Deng et al, 13 which revealed one in five tumors with LOH at 11p also showed LOH in the adjacent normal terminal duct lobular units. Their findings suggest that in some cases, loss of gene(s) at 11p15.5 could be a type of genetic shunt to invasive disease from morphologically normal breast epithelium. These tumors might be envisioned as following the alternate parallel pathway to tumorigenesis (Figure 1) ▶ , although clearly, a larger number of samples needs to be analyzed to demonstrate this.
Reaping clinical benefits from this genetic information will require both an increased understanding of what these alterations mean to the biology of the breast and a clearer picture of their place within tumorigenic progression. Lichy et al 27 and others have begun clarifying these issues by applying microdissection technologies to the examination of individual tumor components and by focusing their study on the now well-defined region of LOH on 11p15.5. In addition to this, future studies will need to be more rigid in grouping analyzed breast tumors into specific categories based on their histopathology (ductal versus lobular versus medullar versus intralobular, etc). Given the already identified heterogeneity and genetic complexity of this disease, it is quite possible that not all tumor subtypes will follow the same genetic progression to malignancy.
Maybe most important of all to understanding the consequence of the events on 11p15.5, and a component of breast cancer that is often studied in isolation of the 11p genetic events, are the allelic losses found on the long arm of chromosome 11. At present, up to three independent regions of LOH on 11q have been identified: two in 11q23 24,30,31 and one in 11q13. 26,32,33 The most common LOH region in 11q23 encompasses the recently isolated DNA repair-associated ataxia-telangiectasia gene (A-T), suggesting this may be the target of loss of function in these breast tumors. The fivefold increase in risk for breast cancer reported in A-T heterozygotes supports this hypothesis. 34 The third region of allelic loss on chromosome 11q centers around the INT2 locus in 11q13.
The frequency of LOH on chromosome 11q involving either one or all of the identified regions is consistently high, ranging from 40 to 50%. However, placement of 11q alterations in the tumorigenic pathway of breast disease has been less than adequately addressed. Chromosome band 11q has been demonstrated to be altered in a variety of other tumor types including cervical carcinoma, 35 ovarian cancer, 36 colorectal carcinoma, 37 and malignant melanoma. 38 By analogy to these studies, it has been inferred that events involving the long arm of chromosome 11 occur late in the tumor progression pathway. Furthermore, cytogenetic studies in metastatic breast carcinomas revealed frequent loss of chromosome 11q, supporting a later role for these sequences in breast disease. 39 However, little information as to the presence of 11q23 allelic deletion in DCIS or AHs is available to elaborate on this issue. Fujii et al 9 have studied 11q LOH in general in a sampling of DCIS and DCIS with synchronous IDC. They reported 23% LOH on 11q in DCIS with no synchronous IDC and 47% LOH in IDCs with synchronous DCIS lesions. These findings suggest, in contrast to current thinking, that 11q13–23 alterations occur early in the progression from normal or atypical hyperplastic tissue to DCIS. 9
Studies specifically analyzing 11q13 deletions and microdissection of DCIS and LCIS components have attempted to clarify this chromosomal region’s role in the pathway to breast cancer. In one such analysis of AH, DCIS, and IDC, LOH at 11q13 was found predominantly in the higher-grade DCIS tumors (35%), whereas a low frequency (0 to 9%) was measured in AHs and low-grade DCIS. 28,32 This places 11q13 alterations later between DCIS and IDC. However, another study of lobular lesions identified allelic deletion on 11q13 in both LCIS and invasive lobular carcinoma (ILCs), 33 with 11q13 LOH more frequent (50%) when LCIS and ILC components were found within the same tumor sample. This suggests 11q13 is altered at the earlier boundary to LCIS. In addition, as was described earlier for DCIS and 11p15.5, a small proportion of LCIS and ILC components showed 11q allelic deletion only in the more invasive component, and several ILCs and IDCs without other associated lesions also demonstrated 11q LOH. These ILC tumors may be the lobular equivalents to IDCs in which alterations of 11q13 place a cell on a parallel fast track to invasive carcinoma.
What is curious about the genetic events just described for the long and short arms of chromosome 11 in breast cancer is that they most often occur simultaneously. Monosomy of chromosome 11 is an infrequent occurrence in breast tumors. Studies by Winqvist et al 25 and Negrini et al 30 reveal that interstitial deletions involving both 11p and 11q regions in the same tumor sample are quite common, if not the predominant genetic event on chromosome 11. For the most part, tumors exhibiting LOH for both 11p15.5 and 11q23 display similar allelic ratios on densitometric analysis. These characteristics suggest both genetic events arose in the same chromosome homologue in the same tumor cell and further that they arose simultaneously through the same mechanism. However, examples are given in which these allelic ratios are discordant, implying independent events as an alternate mechanism. We can now add another level of complexity to the question of staging for these chromosome 11 alterations. If the alterations on chromosome 11p and 11q are synergistic either biologically or mechanistically, it may be difficult to assess their progressional and ultimately their clinical relevance independently. In much the same fashion as linkage analysis is performed to assess heritable breast cancer risk, haplotyping of the chromosomes 11 in breast tumors displaying both 11p and 11q allelic deletions could determine whether the events are linked by chromosome homologue or are truly independent. Such studies would, however, be hindered by the advanced age at diagnosis for a large proportion of cancer patients, most likely precluding access to the necessary parental blood samples.
Finally, what does all this mean for the prognostic value of 11p and 11q alterations? Grasping the levels at which molecular heterogeneity exists both in the breast tumor and in the genetic analyses performed to date, it is not surprising that the relevance of these observations to clinical disease remains controversial. Negrini et al 31 found no association between 11p15.5 or 11q23 regions of LOH and various clinical parameters including stage, lymph node metastasis, estrogen receptor, progesterone receptor, and mitotic index. However, an earlier study correlated chromosome 11p LOH with low estrogen receptor protein and tumor size in invasive breast cancer, both of which are indicators of poorer prognosis. 40 The study by Winqvist et al 24 determined that LOH at 11q23 (alone or in conjunction with 11p15) was predictive of postmetastatic disease course, with substantially reduced survival. Therefore, a potential synergy of genetic factors residing on 11q23 and 11p15.5, alone or in combination with other factors, might be playing a role in generating aggressive breast disease with extremely poor prognosis. Clearly, more subtype and region-specific studies need to be performed to clarify the prognostic value of chromosome 11p and 11q alterations in relation to each other as well as in the milieu of the many other genetic events that are taking place in progressively malignant breast tissue.
We are not yet at a stage in our understanding of breast disease at which genetic alterations can substitute for the histopathology of the lesion. Whether detection of LOH or other genetic abnormalities will ever allow for consistent and useful identification of preinvasive breast lesions at higher risk of progression is unclear at present. As studies on chromosome 11 and others continue, the goal should be discovering a molecular pathological role for providing pertinent clinical information regarding a woman’s risk for invasive or recurrent disease. What will be discovered by this process in the coming years could profoundly influence or change the treatment and/or follow-up for breast cancer patients. It seems that at the genetic level, the application of microdissection technologies to study the individual components of heterogeneous diseases such as breast cancer will be critical in predicting risk and clinical outcome on a patient-by-patient basis.
Footnotes
Address reprint requests to Irene Newsham, Ph.D., Department of Anatomy, MCV/VCU, P.O. Box 980709, Richmond, VA 23298. E-mail: inewsham@hsc.vcu.edu.
References
- 1.Parker SL, Tong T, Bolden S, Wingo PA: Cancer statistics, 1997. CA Cancer J Clin 1997, 47:5-27 [DOI] [PubMed] [Google Scholar]
- 2.Gusterson BA, Stratton M, Anbazhagan R: The importance of human breast development in mammary carcinogenesis. Li JJ Gustafasson JA Nandi S Sekely KO eds. Hormonal Carcinogenesis II. 1996, :pp 115-119 Springer, Berlin [Google Scholar]
- 3.Nowell PC: The clonal evolution of tumor cell populations. Science 1976, 94:23-28 [DOI] [PubMed] [Google Scholar]
- 4.Fearon ER, Vogelstein B: A genetic model for colorectal tumorigenesis. Cell 1990, 61:759-767 [DOI] [PubMed] [Google Scholar]
- 5.Tavassoli FA, Norris HJ: A comparison of the results of longterm follow-up for atypical intraductal hyperplasia and intraductal hyperplasia of the breast. Cancer 1990, 65:518-529 [DOI] [PubMed] [Google Scholar]
- 6.London SJ, Connolly JL, Schnitt SJ, et al: A prospective study of benign breast disease and the risk of breast cancer. J Am Med Assoc 1992, 267:941-944 [PubMed] [Google Scholar]
- 7.Rosenberg CL, Larson PS, Romo JD, de las Morenas A, Faller DV: Microsatellite alterations indicating monoclonality in atypical hyperplasias associated with breast cancer. Hum Pathol 1996, 28:214-219 [DOI] [PubMed] [Google Scholar]
- 8.Krieger N, Hiatt RA: Risk of breast cancer after benign breast diseases: variation by histologic type, degree of atypia, age at biopsy, and length of follow-up. Am J Epidemiol 1992, 135:619-631 [DOI] [PubMed] [Google Scholar]
- 9.Fujii H, Szumel R, Marsh C, Zhou W, Gabrielson E: Genetic progression, histological grade and allelic loss in ductal carcinoma in situ of the breast. Cancer Res 1996, 56:5260-5265 [PubMed] [Google Scholar]
- 10.Wheeler JE, Enterline JT: Lobular carcinoma of the breast: in situ and infiltrating. Pathol Annu 1976, 2:161-188 [PubMed] [Google Scholar]
- 11.Rosen PP, Kosloff C, Lieberman PH, Adair F, Braun DW: Lobular carcinoma in situ of the breast: detailed analysis of 99 patients with average follow-up of 24 years. Am J Surg Pathol 1978, 2:225-251 [DOI] [PubMed] [Google Scholar]
- 12.Lakhani SR, Collins N, Stratton MR: Loss of heterozygosity in lobular carcinoma in situ of the breast. J Clin Pathol Mol Pathol 1995, 48:M74-M78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng G, Lu Y, Zlotnikov G, Thor AD, Smith HS: Loss of heterozygosity in normal tissue adjacent to breast carcinomas. Science 1996, 274:2057-2059 [DOI] [PubMed] [Google Scholar]
- 14.Furnari FB, Huang H-JS, Cavenee WK: Genetics and malignant progression of human brain tumors. Cancer Surv 1995, 25:233-275 [PubMed] [Google Scholar]
- 15.von Deimling A, Louis DN, Wiestler OD: Molecular pathways in the formation of glioma. Glia 1995, 15:328-338 [DOI] [PubMed] [Google Scholar]
- 16.Devilee P, Cornelisse CJ: Somatic genetic changes in human breast cancer. Biochim Biophys Acta 1994, 1198:113-130 [DOI] [PubMed] [Google Scholar]
- 17.O’Connell P, Pekkel V, Fuqua S, Osborne CK, Allred DC: Molecular genetic studies of early breast cancer evolution. Breast Cancer Res Treat 1994, 32:5-12 [DOI] [PubMed] [Google Scholar]
- 18.Smith HS, Lu Y, Deng G, Martinez O, Krams S, Ljung B-M, Thor A, Lagios M: Molecular aspects of early stages of breast cancer progression. J Cell Biochem 1993, 17G:144-152 [DOI] [PubMed] [Google Scholar]
- 19.Beckman MW, Niederacher D, Schnurch H-G, Gusterson BA, Bender HG: Multistep carcinogenesis of breast cancer and tumour heterogeneity. J Mol Med 1997, 75:429-439 [DOI] [PubMed] [Google Scholar]
- 20.Coppes MJ, Bonetta L, Huang A, Hoban P, Chilton-MacNeill S, Campbell CE, Weksberg R, Yeger H, Reeve AE, Williams BRG: Loss of heterozygosity mapping in Wilms’ tumor indicates the involvement of three distinct regions and a limited role for non-disjunction or mitotic recombination. Genes Chromosomes Cancer 1992, 5:326-334 [DOI] [PubMed] [Google Scholar]
- 21.Scrable HJ, Witte DP, Lambkin BC, Cavenee WK: Chromosomal localization of the human rhabdomyosarcoma locus by mitotic recombination mapping. Nature 1989, 329:645-647 [DOI] [PubMed] [Google Scholar]
- 22.Tran YK, Newsham IF: High-density marker analysis of 11p15.5 in non-small cell lung carcinomas reveals allelic deletion of one shared and one distinct region when compared to breast carcinomas. Cancer Res 1996, 56:2916-2921 [PubMed] [Google Scholar]
- 23.Viel A, Giannini F, Tumiotto L, Sopracordevole F, Visentin C, Boiocchi M: Chromosomal localization of two putative 11p oncosuppressor genes involved in human ovarian tumours. Br J Cancer 1992, 66:1030-1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winqvist R, Hampton GM, Mannermaa A, Blanco G, Alavaikko M, Kiviniemi H, Taskinen PJ, Evans GA, Wright FA, Newsham I, Cavenee WK: Loss of heterozygosity for chromosome 11 in primary human breast tumors is associated with poor survival after metastasis. Cancer Res 1995, 55:2660-2664 [PubMed] [Google Scholar]
- 25.Winqvist R, Mannermaa A, Alavaikko M, Blanco G, Taskinen PJ, Kiviniemi H, Newsham I, Cavenee W: Refinement of regional loss of heterozygosity for chromosome 11p15.5 in human breast tumors. Cancer Res 1993, 53:4486-4488 [PubMed] [Google Scholar]
- 26.Radford DM, Phillips NJ, Fair KL, Ritter JH, Holt M, Donis-Keller H: Allelic loss and the progression of breast cancer. Cancer Res 1995, 55:5180-5183 [PubMed] [Google Scholar]
- 27.Lichy JH, Zavar M, Tsai MM, O’Leary TJ, Taubenberger JK: Loss of heterozygosity on chromosome 11p15 during histologic progression in microdissected ductal carcinoma of the breast. Am J Pathol 1998, 153:271-278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radford DM, Fair KL, Phillips NJ, Ritter JH, Steinbrueck T, Holt MS, Donis-Keller H: Allelotyping of ductal carcinoma in situ of the breast: deletion of loci on 8p, 13q, 16q, 17p and 17q. Cancer Res 1995, 55:3399-3405 [PubMed] [Google Scholar]
- 29.Devilee P, van Vliet M, van Sloun P, Luipers-Dijkshoorn N, Hermans J, Pearson PL, Cornelisse CJ: Allelotype of human breast carcinoma: a second major site for loss of heterozygosity is on chromosome 6q. Oncogene 1991, 6:1705-1711 [PubMed] [Google Scholar]
- 30.Hampton GM, Mannermaa A, Winqvist R, Alavaikko M, Blanco G, Taskinen PJ, Kiviniemi H, Newsham I, Cavenee WK, Evans GA: Loss of heterozygosity in sporadic human breast carcinoma: a common region between 11q22 and 11q23.3. Cancer Res 1994, 54:4586-4589 [PubMed] [Google Scholar]
- 31.Negrini M, Rasio D, Hampton GM, Sabbioni S, Rattan S, Carter SL, Rosenberg AL, Schwartz GF, Shiloh Y, Cavenee WK, Croce CM: Definition and refinement of chromosome 11 regions of loss of heterozygosity in breast cancer: identification of a new region at 11q23.3. Cancer Res 1995, 55:3003-3007 [PubMed] [Google Scholar]
- 32.Chuaqui RF, Zhuang Z, Emmert-Buck MR, Liotta LA, Merino MJ: Analysis of loss of heterozygosity on chromosome 11q13 in atypical ductal hyperplasia and in situ carcinoma of the breast. Am J Pathol 1997, 150:297-303 [PMC free article] [PubMed] [Google Scholar]
- 33.Nayar R, Zhuang Z, Merino MJ, Silverberg SG: Loss of heterozygosity on chromosome 11q13 in lobular lesions of the breast using tissue microdissection and polymerase chain reaction. Hum Pathol 1996, 28:277-282 [DOI] [PubMed] [Google Scholar]
- 34.Swift M, Morrell D, Massey RB, Chase CL: Incidence of cancer in 161 families affected by ataxia-telangiectasia. N Engl J Med 1991, 325:1831-1836 [DOI] [PubMed] [Google Scholar]
- 35.Hampton GM, Penny LA, Baergen LN, Larson A, Brewer C, Liao S, Busby-Earle RMC, Williams AWR, Steel CM, Bird CC, Stanbridge EJ, Evans GA: Loss of heterozygosity in cervical carcinoma: subchromosomal localization of a putative tumor suppressor gene to chromosome 11q22–q24. Proc Natl Acad Sci USA 1994, 91:6953-6957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foulkes WD, Campbell IG, Stamp GWH, Trowsdale J: Loss of heterozygosity and amplification on chromosome 11q in human ovarian cancer. Br J Cancer 1993, 67:268-273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keldysh PL, Dragani RA, Fleischman EW, Konstantinova LN, Perevoschikov AG, Pierotti MA, Della Porta G, Kopnin BP: 11q deletions in human colorectal carcinoma: cytogenetics and restriction fragment length polymorphism analysis. Genes Chromosomes Cancer 1993, 6:45-50 [DOI] [PubMed] [Google Scholar]
- 38.Tomlinson IPM, Gammack AJ, Strickland JE, Mann GJ, MacKie RM, Kefford RF, McGee JO: Loss of heterozygosity in malignant melanoma at loci on chromosome 11 and 17 implicated in the pathogenesis of other cancers. Genes Chromosomes Cancer 1993, 7:169-172 [DOI] [PubMed] [Google Scholar]
- 39.Thompson F, Emerson J, Dalton W, Yang J-M, McGee D, Massey K, Thompson F, Villar H: Clonal chromosome abnormalities in human breast carcinomas: twenty-eight cases with primary disease. Genes Chromosomes Cancer 1993, 7:185-193 [DOI] [PubMed] [Google Scholar]
- 40.Mackay J, Elder PA, Porteous DJ, Stell CM, Hawkins RA, Going JJ, Chetty U: Partial deletion of chromosome 11p in breast cancer correlates with size of primary tumour and oestrogen receptor level. Br J Cancer 1988, 58:710-714 [DOI] [PMC free article] [PubMed] [Google Scholar]