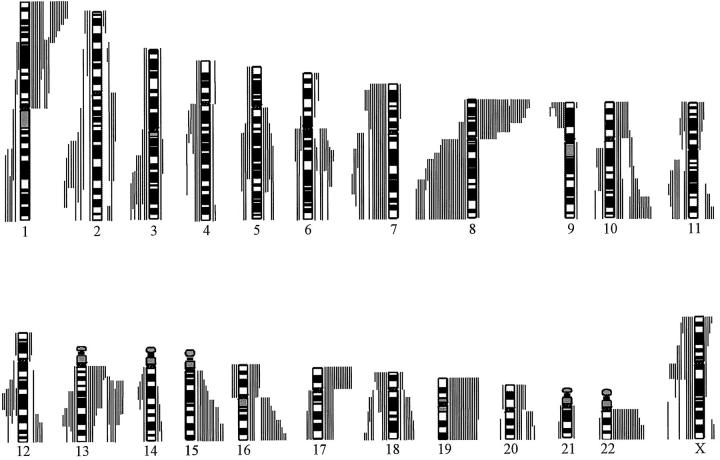Abstract
To study the genetic basis of tumor progression, we have screened 37 hormone-refractory prostate carcinomas for genetic changes by comparative genomic hybridization (CGH). All recurrent tumors showed genetic aberrations, with a mean total number of changes per tumor of 11.4 (range, 3 to 23). The most common genetic aberrations were losses of 8p (72.5%), 13q (50%), 1p (50%), 22 (45%), 19 (45%), 10q (42.5%), and 16q (42.5%) and gains of 8q (72.5%), 7q (40%), Xq (32.5%), and 18q (32.5%). The CGH results were further validated with fluorescence in situ hybridization (FISH) using probes for pericentromeric regions of chromosomes 7, 8, and 18 as well as probes for caveolin (7q31), c-myc (8q24), and bcl-2 (18q21.3). In addition, the samples had previously been analyzed for androgen receptor gene copy number. CGH and FISH results were concordant in 78% of cases. Seventeen of twenty-two tumors showed an increased copy number of c-myc by FISH. However, only 5 of 17 (29%) of the cases showed high-level (more than threefold) amplification. Both CGH and FISH findings suggested that in most of the cases 8q gain involves the whole q-arm of the chromosome. Four of seventeen (24%) cases showed increased copy number of bcl-2 by FISH; however, no high-level amplifications were found. To evaluate the clonal relationship of the primary and recurrent tumors, six primary-recurrent tumor pairs from the same patients were studied by CGH. In three of six cases (50%), the recurrent tumor had more than one-half of the aberrations found in the corresponding primary tumor, indicating a close clonal relationship. In the rest of the cases, such a linear clonal relationship was less evident. Altogether, these results suggest that recurrent prostate carcinomas are genetically unstable. The resulting heterogeneity may well underlie the poor responsiveness of hormone-refractory tumors to treatment.
Prostate cancer is the most common malignancy among men in many Western industrialized countries. Despite the improved early diagnosis of prostate cancer, approximately one-third of the patients are still diagnosed at a clinically advanced stage. 1 For these patients, androgen withdrawal remains the only effective treatment. Most of the prostate cancer patients initially respond to hormonal therapy, 2 but eventually the disease will progress. And there are no effective second-line treatments for such hormone-refractory tumors. 2
The mechanisms that lead to progression of prostate cancer during endocrine treatment are poorly understood. Several hypotheses on the molecular mechanisms of tumor recurrence have been suggested. These include overexpression of the bcl-2 oncogene, 3-5 activating mutations in the androgen receptor (AR) gene, 6,7 and amplification and overexpression of the AR gene. 8,9
Comparative genomic hybridization (CGH) is a fairly new molecular cytogenetic method that allows detection of DNA sequence copy number changes throughout the genome in a single hybridization. 10-12 CGH is helpful in defining chromosomal regions that may harbor amplified oncogenes or deleted tumor suppressor genes (TSGs). Thus, CGH provides a starting point for positional cloning of cancer-related genes. Several common malignancies, including prostate cancer, 13-16 have already been studied by CGH. These studies have also led to the identification of actual amplified genes in cancer. 8,17-19 As CGH detects only clonal genetic aberrations, it is also useful in the investigation of the clonal evolution of tumor progression. 20
To identify genetic aberrations that may underlie the progression of prostate cancer during endocrine therapy, we have now screened 37 hormone-refractory prostate tumors for genetic changes by CGH. In addition, we have studied the genetic relationship of six primary-recurrent tumor pairs. CGH findings were also further studied by fluorescence in situ hybridization (FISH) and locus-specific probes.
Materials and Methods
Tumor Specimens
The material consisted of 37 formalin-fixed, paraffin-embedded recurrent prostate carcinomas obtained from the Tampere University Hospital. According to histological grade, 21 there were 1 grade I, 15 grade II, and 21 grade III recurrent tumors. According to the TNM stage distribution of the recurrent prostate carcinomas, there were 1 T1NXMO, T2NXMX, T2NXM1; 5 T3NXMX; 6 T3NXMO, T3NXM1; 7 T4NXMO; 3 T4NXMX; and 5 T4NXM1. All samples were transurethral resection specimens taken from patients who had received only endocrine therapy (orchiectomy (28 cases), estrogen (3 cases), luteinizing hormone-releasing hormone agonist (1 case), and combinations of these therapies (5 cases)) and who had experienced local progression as evidenced by new onset of urethral obstruction. The choice of endocrine therapy instead of surgical treatment had been based either on clinical stage of the disease or general condition of the patients. An average time from the diagnosis (beginning of hormonal therapy) to progression was 44 (range, 8 to 113) months and from the progression to death was 29 (1 to 101) months. In addition, six primary-recurrent tumor pairs (five paraffin embedded, one freshly frozen) were available. The primary tumors (transurethral resection specimens) were taken before any treatment, and the recurrent tumors were from the same patients at the time of local relapse (urethral obstruction).
Five-micron sections were cut from tumor blocks and stained with hematoxylin and eosin to detect the histological representativeness of the malignant tissue. High molecular weight tumor DNAs for CGH and interphase nuclei for FISH were isolated from paraffin-embedded tumor blocks as described before. 22,23
CGH
CGH was done as described before. 12,22 Briefly, DNA samples from prostate tumors were labeled with fluorescein isothiocyanate (FITC)-dUTP (DuPont, Boston, MA) and normal reference male DNA with Texas Red-dUTP (DuPont) using nick translation. Labeled DNAs (400 ng) were hybridized to normal male lymphocyte metaphase slides (Vysis, Downers Grove, IL) together with unlabeled Cot-1 DNA (10 μg; Gibco BRL, Gaithersburg, MD). After hybridization, the slides were washed and counterstained with an anti-fade solution containing 4,6-diamidino 2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA).
Digital Image Analysis
Five high quality metaphases from each hybridization were captured using a Xillix CCD camera (Xillix Technologies Corp., Vancouver, British Columbia, Canada) mounted on an Olympus BX50 epifluorescence microscope (Tokyo, Japan) and interfaced to a Sun LX workstation (Sun Microsystems Computer Corp., Mountain View, CA). Relative DNA sequence copy number changes were detected by analyzing the fluorescence intensities of green (tumor) and red (normal) signals along the length of all chromosomes in the metaphase spreads using Quips CGH analysis program (Resource of Molecular Cytogenetics, Lawrence Berkeley National Laboratory, Berkeley, CA) based on the Scilimage program (TNO, Delft, The Netherlands). CGH results were plotted as a series of green-to-red ratio profiles and the interpretation of results followed previously described guidelines. 12 Hybridizations of FITC-labeled normal male DNA against Texas-Red-labeled normal female DNA, in each hybridization batch, were used as negative controls. The mean green-to-red ratio and corresponding SD for all autosomes remained between 0.85 and 1.15. Based on these control hybridizations, chromosomal regions with a mean ratio of 0.85 or less were considered lost, and those with a ratio 1.15 or more were considered gained in the prostate tumors. Chromosome Y was excluded from CGH analysis. The MCF-7 breast cancer cell line was used as a positive control in each hybridization batch.
FISH
Interphase FISH was performed with locus-specific probes for caveolin (obtained by screening the human PAC library with PCR using primers specific to caveolin) located at 7q31, c-myc (P1, c-myc, RMC08P001, Lawrence Berkeley National Laboratory) located at 8q24, and bcl-2 (obtained by screening the human P1 library with PCR using primers specific to bcl-2) at 18q21.3. Chromosomes 7 (p7αtet), 8 (pJM128), and 18 (p18r) pericentromeric alphoid probes were used as reference probes. Locus-specific probes were labeled with digoxigenin-11-dUTP (myc) (Gibco BRL) or biotin-14-dATP (caveolin, bcl-2; Gibco BRL) and centromeric probes with FITC-dUTP (chromosome 8; DuPont) or Texas-Red-dUTP (chromosomes 7 and 18; DuPont). FISH was performed as described in detail elsewhere. 23 Before FISH, the slides were pretreated by heating in 59% glycerol/0.1X standard saline citrate (SSC; pH 7.5) solution at 90°C for 3 minutes to improve hybridization efficiency. After hybridization, the slides were washed and counterstained with an anti-fade solution containing DAPI (Vector). In addition, 30 tumors had earlier been analyzed for chromosome X centromere and AR gene copy number by FISH. 9
The entire slide was first scanned through, and 120 to 150 randomly chosen individual nuclei were scored in detail to calculate signals for the locus-specific probes and centromeric probes. Control hybridizations to normal lymphocyte metaphase preparations were done to ascertain that the probes recognized a single-copy target and to evaluate the hybridization efficiencies of the probes.
Results
Genetic Changes in Recurrent Prostate Carcinomas
The mean total number of changes per tumor in recurrent prostate cancer was 11.4 (range, 3 to 23). The average number of gains per tumor was 4.5 (range, 0 to 13) and of losses was 6.6 (range, 1 to 15).
The most frequently lost chromosome arms were (Figure 1) ▶ 8p (73%), 1p (54%), 13q (51%), 22q (46%), 10q (46%), 16q (46%), 19 (43%, both arms), 15q (35%), 6q (27%), 18q (19%), 10p (22%), 17p (41%), and 20q (22%). The minimal commonly lost regions in these chromosome arms were 1p36-pter, 1p31, 5q15-q23, 6q16, 6q24-qter, 8p12-p22, 8p23, 10cen-q21, 10q26, 10p11, 13q12, 13q21, 15cen-q21, 15q25-qter, 16q24, 17p, 18q22-qter, 19pter-q13.1, 20cen-q22, and 22q13.
Figure 1.
Summary of all DNA sequence copy number changes in 37 recurrent prostate carcinomas detected by CGH. Gains are shown on the left side of the chromosome ideograms and losses on the right. One bar represents one tumor. Chromosome Y was excluded from the analysis.
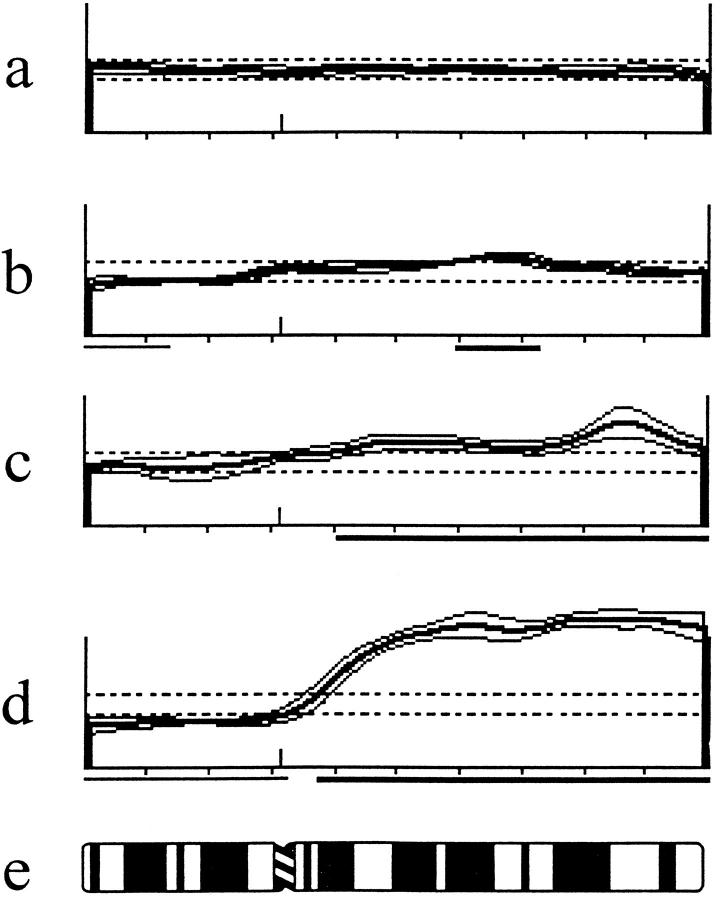
The most frequently gained chromosome arms were (Figure 1) ▶ 8q (73%), 7q (43%), Xq (35%), 7p (32%), 18q (30%), 2q (27%), 3q (24%), Xp (24%), 11q (22%), 12q (22%), 13q (19%), 4q (19%), and 5q (14%). And the minimal commonly gained regions were 1q25-q32, 2q33, 3q25-q26, 4q13-q23, 5q14-q31, 7p15-p21, 7q21, 7q31, 8q21, 8q23-qter, 11q22, 12q21, 13q31, 18q12, Xpter-Xp21, and Xcen-q13. Figure 2 ▶ shows examples of different types of CGH findings of chromosome 8 in hormone-refractory prostate carcinomas.
Figure 2.
Mean green-to-red ratio profiles with ±1 SD from pter to qter obtained from CGH analysis of four locally recurrent hormone-refractory paraffin-embedded prostate tumors. The dotted lines represent ratio values of 0.85 and 1.15. a: Normal; b: Loss of 8p22-pter, gain of 8q21-q22; c: Gain of 8q12-qter with higher-level amplification at 8q24; d: Loss of 8p and gain of 8q12-qter; e: Chromosome 8 idiogram.
CGH Analysis of Primary-Recurrent Tumor Pairs
Table 1 ▶ summarizes the results of CGH analysis of the primary-recurrent prostate tumor pairs. On average, 51% of the genetic alterations found in the primary tumors were also found in the recurrent tumors. Three recurrent tumors showed more than 50% of the genetic aberrations found in the corresponding primary tumor. However, there was, for example, a case in which only 8% of the aberrations were shared by both tumors (case 1).
Table 1.
Genetic Alterations in the Primary and Recurrent Tumor Pairs
| Case | Primary tumor | Recurrent tumor | Time to progression (months) |
|---|---|---|---|
| 1 | Gains: 22p, 22q | Gains: 3q, 7p, 7q, 8q, 13q, Xq | 15 |
| Losses: 1p, 1q, 2q, 4q, 5q, 6q, 8p, 12q, 13q, Xq | Losses: 8p, 19, 17, 20q | ||
| 2 | Gains: 7, 8q, 9p | Gains: 7, 8q | 36 |
| Losses: 8p, 13q | Losses: 8p, 13q | ||
| 3 | Gains: none | Gains: 3q, 8q, 17q, Xp, Xq | 59 |
| Losses: 1p, 6q, 11q, 12q, 13q, X | Losses: 6q, 8p, 10q, 11q, 13q, 15q, 18 | ||
| 4 | Gains: 8q, 20p | Gains: 8q, 4q, 5p | 46 |
| Losses: 16p, 17 | Losses: 6q | ||
| 5 | Gains: 2q, 3q, 5q, 7, 8q, 11q, 12q | Gains: 8q, 10q, 12q, 21q | 12 |
| Losses: 1p, 8p, 16p, 17p, 19, 22 | Losses: 1p, 8p, 13q, 15q, 16p, 17p, 18q, 19, 22 | ||
| 6 | Gains: 2p, 8q, 16p | Gains: 8q, 16p, Xq | 22 |
| Losses: 1p, 2q, 5p, 5q, 6q, 8p, 9p, 13q, 16q, 18q | Losses: 1p, 2q, 3, 5p, 5q, 6q, 8p, 9p, 13q, 16q, 18q, Xq |
The chromosome arms that contain either gains or losses are given. Genetic alterations that are shared by primary and recurrent tumors are in bold. The histological grades of the primary and the corresponding recurrent tumors were the same in each of the six sample pairs.
FISH Analyses
To validate the CGH findings, we chose a subset of tumors for FISH analysis of caveolin (7q31), c-myc (8q24), bcl-2 (18q21.3), and AR (Xq12). These were the regions that were most commonly gained by CGH. Table 2 ▶ summarizes the comparison of CGH and FISH findings. Altogether, 78% of cases showed similar results by CGH and FISH (κ value of 0.57). Figure 2 ▶ shows recurrent tumors studied by FISH for c-myc, caveolin, and bcl-2. Seventeen of twenty-two tumors showed increased copy number of c-myc. Of the seventeen tumors, five (29%) showed high-level (more than three times more signals of c-myc versus the chromosome 8 centromere) amplification of c-myc with a mean ± SD copy number of signals (excluding the nuclei with only two signals) of 6.2 ± 0.9. Occasional nuclei with more than 10 copies of c-myc were also found. Four of eight cases showed equally increased copy number of the chromosome 7 centromere and 7q31 (caveolin) with a mean copy number of signals of 3.9 ± 0.6. And four of seventeen cases showed equally increased copy number of the chromosome 18 centromere and bcl-2, with a mean copy number of signals of 4.4 ± 1.3.
Table 2.
Condordance between CGH and FISH Data for the 7q31 (Caveolin), 8q24 (c-myc), 18q21.3 (bcl-2), and Xq12 (AR) Regions
| CGH | FISH | |
|---|---|---|
| Aberrant | Normal | |
| Aberrant | 31 | 2 |
| Normal | 15 | 29 |
The κ value was 0.57.
Discussion
Here, we report results from the analyses of 37 recurrent hormone-refractory prostate carcinomas for genetic aberrations using CGH. The high number of alterations per tumors found by CGH emphasizes the genetic instability as an underlying mechanism of cancer development and progression. As compared with our previous CGH analysis of 31 unselected primary prostate carcinomas, 15 the recurrent tumors in this study contained almost four times more alterations than the primary tumors supporting our earlier findings that the clinical progression of prostate cancer is associated with genetic progression of the tumors. 8,24
We analyzed also six primary-recurrent tumor pairs. Three tumor pairs (50%) showed a close genetic relationship as evidenced by a high number of shared genetic alterations in primary and recurrent tumors. For example, in case 6, 13/14 of the alterations (93%) found in the primary tumor were also present in the recurrent tumor (Table 1) ▶ . The aberrations that were solely found in the recurrent tumors were losses of whole chromosome 3 and the telomeric part of Xq (Xq23-qter), as well as gain of Xcen-q13, indicating a relatively linear genetic progression of the tumor. The most likely target for the Xq gain in this tumor is the AR gene, which was found to be fivefold amplified in the specimen. 8 On the other hand, in case 1, only 1 of the 12 genetic aberrations found in the primary tumor were present in the recurrent counterpart. Prostate cancer is commonly considered to be heterogeneous disease. Studies on the whole-mount prostatectomy specimens have shown that prostate gland may contain several carcinoma foci, which appear not always to be physically in contact with each other. These foci may contain different genetic changes. 25-27 Jenkins and co-authors 27 have also shown that in some cases it is the minor, instead of the major, primary carcinoma focus that metastasizes. Still, the significance of such multifocality of prostate cancer is inadequately known. The results in this study suggest that the androgen ablation therapy acts as a strong selection force in a similar fashion as a metastases event. 20,27 In some cases, it may select for a clone that has acquired only a few additional genetic alterations, such as AR gene amplification. In other cases, the treatment may select for a genetically very different clone or a completely different clone than the primary tumor, suggesting that there are several different mechanisms that may underlie the progression of prostate cancer during endocrine treatment. The fact that recurrent tumors may share only a few genetic alterations with primary tumors indicates also that biomarkers measured exclusively from primary tumors give only a restricted view on the biological properties of hormone-refractory prostate cancer.
In this study, every chromosome arm showed genetic alterations in at least one tumor by CGH, probably as a result of random genetic instability. However, some of the chromosomal regions were often altered, suggesting that these regions contain oncogenes and tumor suppressor genes (TSGs) that are important in the development and progression of prostate cancer. The most commonly lost regions were 8p, 13q, 1p, 10q, 17q, 19, and 22, whereas the most commonly gained regions were 8q, 7q, Xq, 18q, and 7p. Most of these regions had been implicated in prostate cancer earlier by either CGH or loss of heterozygosity (LOH) studies. 13-16,28-30
Three chromosome arms that have most extensively been studied in prostate cancer, due to the high frequency of LOH found in these regions, are 8p, 10q, and 16q. Loss of 8p as detected either by LOH, CGH, or FISH may well be the most common genetic alterations in prostate cancer. 13-16,30-32 It also seems to be an early event in tumorigenesis, as prostate intraepithelial neoplasias have also shown LOH at 8p. 33 In this study, we found two minimal commonly deleted regions, 8p23 and 8p12-p22, suggesting that there are several still unknown target genes for the deletions in 8p.
Deletion of 10q was found in more than 40% of tumors. Chromosome 10q contains two candidate prostate cancer tumor suppressor genes: MXI1 at 10q25 34 and the recently cloned PTEN at 10q23. 35,36 Neither one of these genes map to the minimal commonly deleted regions (10cen-q21 and 10q26) found in this study, suggesting existence of yet another TSG in 10q. We have not commonly found losses at 10q in primary prostate carcinomas, 15 indicating that the inactivation of the TSGs in 10q may be a late event in the development of prostate cancer.
As in this study, LOH and deletions at 16q have been found in approximately 50% of prostate carcinomas. 15,16,28-30 One candidate TSG in chromosome 16 is E-cadherin (16q22.1), the decreased expression of which has been found in poorly differentiated and clinically aggressive prostate carcinomas. 37-39 However, no mutations have, so far, been identified in the coding region of the gene. We and others have also found that the minimal commonly lost region is 16q24, 16,40 suggesting that 16q may harbor TSGs other than E-cadherin.
Yet another frequently deleted chromosome region in prostate cancer is 13q. 15,16,30 Chromosome 13 contains at least three putative TSGs: Rb1 (13q14), BRCA2 (13q12-q13), and BRUSH-1 (13q12-q13). Although reduced expression of Rb has been found in prostate carcinomas, only few prostate tumors have shown mutations in the coding region of the gene. 41 Here, we found two minimal commonly deleted chromosomal regions: 13q12 and 13q21. Surprisingly, also gains of 13q, which has not previously been reported in prostate cancer, except in vitro, 42 was found in 20% of the tumors.
Two chromosomes that were frequently lost, but have not been previously implicated in prostate cancer by either LOH or CGH studies, were 1p and 19. Early studies suggested that these two regions may show artificial deletions by CGH as evidenced by alterations in 1p and 19 even in normal-normal control hybridization. However, these problems are believed to be caused by differential hybridization of biotin and digoxigenin-labeled DNAs. The use of directly fluorochrome-conjugated nucleotides, as in this study, has abolished this phenomenon. 12 Also, the negative controls (DNA from either paraffin-embedded blocks or blood lymphocytes) showed no variations in 1p or 19. Thus, we believe that the findings are true. Chromosome 19 contains some putative TSGs, such as BAX 43 and LKB1, 44 whereas the p53-related putative neuroblastoma TSG was recently mapped to 1p36. 45 However, the role of these TSGs in prostate cancer has not been studied so far.
Before CGH, there were no effective ways to screen the genome for gains of DNA sequences. Therefore, CGH has probably contributed most to the screening of tumors for amplifications. 10-12 CGH studies have already indicated that primary prostate cancers contain very few gains or high-level amplifications. 13,15 On the other hand, as in this study, hormone-refractory recurrent tumors commonly carry such gains. This is consistent with the notion that oncogene amplification is often a rather late event in tumor progression, affecting cells that are unstable enough to be able to amplify DNA. 46,47 In this study, the most common genetic alteration, together with the loss of 8p, was the gain of 8q. The 8q gain in prostate cancer was first described by Bova and co-workers 31 using Southern analysis. Subsequently, it has been shown that 8q gain is common in hormone-refractory 15 as well as in metastatic lesions of prostate cancer. 16 Recurrent tumors exhibit 8q gain >10 times more often than the primary tumors.15The presence of extra copies of chromosome 8 centromere is reported to be associated with a short progression-free interval, 48 and amplification of 8q24 region has been shown to be associated with the presence of lymph node metastases. 49 In addition to prostate cancer, the 8q gain has been found by CGH, for example, in breast and bladder cancer. 50,51 And, in breast cancer, the 8q gain seems to be associated with poor prognosis. 52
CGH analysis, here and earlier, 15,16 has identified two minimal commonly amplified regions: 8q21 and 8q23-q24. These findings suggest that there may well be several target genes for the gain of 8q. A natural candidate target gene for the 8q24 amplification is c-myc, which plays significant roles in the regulation of cellular proliferation, differentiation, and apoptosis. 53 Overexpression of c-myc has been detected in prostate cancer. 54,55 Still, until recently, amplification of c-myc had not been found in prostate carcinomas in vivo. 27 In this study, a subset of tumors with 8q gain showed high-level amplification of c-myc. However, based on these data, we cannot exclude the possibility that the real target gene for 8q24 amplification could also be some gene other than c-myc.
Other frequently gained chromosomal regions were 7q, 7p, Xq, and 18q. Increased copy number of chromosome 7 has been detected by FISH, 48,56,57 and trisomy 7 may be associated with progression of disease. 48,56 In several tumors, the entire chromosome 7 was gained, but we were able to narrow down three separate regions of minimal gains: 7p21-p15, 7q21, and 7q31. We applied also locus-specific (caveolin; 7q31) and centromeric FISH to evaluate the CGH findings. Equally increased copy number of centromere and caveolin was commonly found, whereas there were no high-level amplifications on the 7q31 region. Chromosome 7 contains several known genes, such as CLK2, MDR1, and elongation factor-1-γ, of which altered function could theoretically be involved in the progression of prostate cancer.
Gain of Xq was found in one-third of the cases. We have already earlier shown that the most likely target gene for Xq amplification is the AR gene. It is highly amplified and overexpressed in approximately 30% of hormone-refractory tumors. 8,9 Also, 18q was gained in approximately one-third of the tumors. Bcl-2 oncogene, which has commonly been found to be overexpressed in recurrent prostate cancers, 3-5 is located in 18q21.3. Although bcl-2 is located outside the minimal commonly gained regions (18q12) according to CGH, we decided to analyze the exact copy number of bcl-2 by FISH. Four cases showed increased copy number of bcl-2; however, no high-level amplifications were found. Thus, it seems that bcl-2 overexpression in prostate cancer is not due to the high-level amplification of the gene.
The ability to use DNA extracted from paraffin-embedded tumor blocks for CGH has made it possible to obtain samples from vast archives of routine pathology laboratories. It has been shown that the quality of CGH is almost equally good whether DNA from paraffin-embedded tumor blocks or from frozen tumor samples are used. 22 For example, the SDs of the mean fluorescence intensity ratio profiles, which are used to indicate the quality of CGH hybridizations, were practically equally as low in the CGH analysis of paraffin-embedded samples as we have found in the analysis of the frozen samples. Indeed, several studies using paraffin-embedded blocks for CGH have already been published. 58-60 We also used FISH to validate the CGH findings. In 78% of cases, FISH and CGH results were in agreement. However, some of the AR gene amplifications were undetected by CGH, whereas increased copy number of 7q31, 8q24, and 18q21.3 were found almost equally well by CGH and FISH (data not shown). This could be due to the small size of the AR gene amplicon as compared with the size of gained regions at 7q, 8q, and 18q.
In conclusion, the comparison of genetic alterations in the primary-recurrent tumor pairs revealed strong genetic heterogeneity and a nonlinear relationship of the tumors. The hormone-refractory recurrent prostate carcinomas contained a high number of both gains and losses of DNA sequences. Especially, the gain of 8q seemed to be associated with progression of the disease. Identification of target genes for the 8q gain would, therefore, most likely reveal important mechanisms of tumor progression.
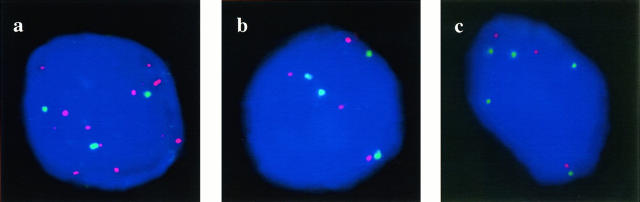
Figure 3.
Interphase FISH analyses of hormone-refractory prostate cancer with probes for chromosome 8 centromere (in green) and c-myc (in red) (a), chromosome 7 centromere (in red) and caveolin (in green) (b), and chromosome 18 centromere (in red) and bcl-2 (in green) (c). a: Multiple copies of c-myc and three copies of centromere are seen. b: Four copies of centromere and caveolin are seen. c: Three copies of centromere and five copies of bcl-2 are found. Nuclei are counterstained with DAPI.
Acknowledgments
We thank Mrs. Mariitta Vakkuri for technical assistance.
Footnotes
Address reprint requests to Dr. Tapio Visakorpi, Laboratory of Cancer Genetics, Institute of Medical Technology, University of Tampere, P.O. Box 607, FIN-33101 Tampere, Finland. E-mail: tapio.visakorpi@uta.fi.
Supported by grants from the Pirkanmaa Cancer Society, the Pirkanmaa Cultural Foundation, the Reino Lahtikari Foundation, the Yrjö Jahnsson Foundation, Academy of Finland, the Finnish Cancer Society, the Medical Research Fund of Tampere University Hospital, and CaPCURE.
References
- 1.Kosary CL, Ries LAG, Miller BA, Hankey BF, Harras A, Edwards BK (eds): SEER Cancer Statistics Review, 1973-1992, Tables and Graphs: National Institutes of Health Publication 96–2789. Bethesda, MD, National Cancer Institute, 1995
- 2.Stearns ME, McGarvey T: Prostate cancer: therapeutic, diagnostic, and basic studies. Lab Invest 1992, 67:540-552 [PubMed] [Google Scholar]
- 3.McDonnell TJ, Troncoso P, Brisbay SM, Logothetis C, Chung LW, Hsieh JT, Tu SM, Campbell ML: Expression of the protooncogene bcl-2 in the prostate and its association with emergence of androgen-independent prostate cancer. Cancer Res 1992, 52:6940-6944 [PubMed] [Google Scholar]
- 4.Colombel M, Symmans F, Gil S, O’Toole KM, Chopin D, Benson M, Olsson CA, Korsmeyer S, Buttyan R: Detection of the apoptosis-suppressing oncoprotein bcl-2 in hormone-refractory human prostate cancers. Am J Pathol 1993, 143:390-400 [PMC free article] [PubMed] [Google Scholar]
- 5.Krajewska M, Krajewski S, Epstein JI, Shabaik A, Sauvageot J, Song K, Kitada S, Reed JC: Immunohistochemical analysis of bcl-2, bax, bcl-X, and mcl-1 expression in prostate cancers. Am J Pathol 1996, 148:1567-1567 [PMC free article] [PubMed] [Google Scholar]
- 6.Taplin ME, Bubley GJ, Shuster TD, Frantz ME, Spooner AE, Ogata GK, Keer HN, Balk SP: Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. N Engl J Med 1995, 332:1393-1398 [DOI] [PubMed] [Google Scholar]
- 7.Tilley WD, Buchanan G, Hickey TE, Bentel JM: Mutations in the androgen receptor gene are associated with progression of human prostate cancer to androgen independence. Clin Cancer Res 1996, 2:277-285 [PubMed] [Google Scholar]
- 8.Visakorpi T, Hyytinen E, Koivisto P, Tanner M, Keinänen R, Palmberg C, Palotie A, Tammela T, Isola J, Kallioniemi O-P: In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nature Genet 1995, 9:401-406 [DOI] [PubMed] [Google Scholar]
- 9.Koivisto P, Kononen J, Palmberg C, Tammela T, Hyytinen E, Isola J, Trapman J, Cleutjens K, Noordzij A, Visakorpi T, Kallioniemi O-P: Androgen receptor gene amplification: A possible molecular mechanism for failure of androgen deprivation therapy in prostate cancer. Cancer Res 1997, 5:314-319 [PubMed] [Google Scholar]
- 10.Kallioniemi A, Kallioniemi O-P, Sudar D, Rutowitz D, Gray JW, Waldman F, Pinkel D: Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science 1992, 258:818-821 [DOI] [PubMed] [Google Scholar]
- 11.du Manoir S, Speicher MR, Joos S, Schröck E, Popp S, Döhner H, Kovacs G, Robert-Nicoud M, Lichter P, Cremer T: Detection of complete and partial chromosomal gains and losses by comparative genomic hybridization. Hum Genet 1993, 90:590-610 [DOI] [PubMed] [Google Scholar]
- 12.Kallioniemi O-P, Kallioniemi A, Piper J, Isola J, Waldman FM, Gray JW, Pinkel D: Optimizing comparative genomic hybridization for analysis of DNA sequence copy number changes in solid tumors. Genes Chromosomes & Cancer 1994, 10:231-243 [DOI] [PubMed] [Google Scholar]
- 13.Cher ML, MacGrogan D, Bookstein R, Brown JA, Jenkins RB, Jensen R: Comparative genomic hybridization, allelic imbalance, and fluorescence in situ hybridization on chromosome 8 in prostate cancer. Genes Chromosomes & Cancer 1994, 11:153-162 [DOI] [PubMed] [Google Scholar]
- 14.Joos S, Bergerheim U, Pan Y, Matsuyama H, Bentz M, du Manoir S, Lichter P: Mapping of chromosomal gains and losses in prostate cancer by comparative genomic hybridization. Genes Chromosomes & Cancer 1995, 14:267-276 [DOI] [PubMed] [Google Scholar]
- 15.Visakorpi T, Kallioniemi A, Syvänen A-C, Hyytinen ER, Karhu R, Tammela T, Isola JJ, Kallioniemi O-P: Genetic changes in primary and recurrent prostate cancer by comparative genomic hybridization. Cancer Res 1995, 55:342-347 [PubMed] [Google Scholar]
- 16.Cher ML, Bova GS, Moore DH, Small EJ, Carroll PR, Pin SS, Epstein JI, Isaacs WB, Jensen RH: Genetic alterations in untreated metastases and androgen-independent prostate cancer detected by comparative genomic hybridization and allelotyping. Cancer Res 1996, 56:3091-3102 [PubMed] [Google Scholar]
- 17.Houldsworth J, Mathew S, Rao PH, Dyomina K, Louie DC, Parsa N, Offit K, Changanti RS: REL proto-oncogene is frequently amplified in extranodal diffuse large cell lymphoma. Blood 1996, 87:25-29 [PubMed] [Google Scholar]
- 18.Monni O, Joensuu H, Franssila K, Klefstrom J, Alitalo K, Knuutila S: BCL2 overexpression associated with chromosomal amplification in diffuse large B-cell lymphoma. Blood 1997, 90:1168-1174 [PubMed] [Google Scholar]
- 19.Weber-Hall S, McManus A, Anderson J, Nojima T, Abe S, Pritchard-Jones K, Shipley J: Novel formation and amplification of the PAX7-FKHR fusion gene in a case of alveolar rhabdomyosarcoma. Genes Chromosomes & Cancer 1996, 17:7-13 [DOI] [PubMed] [Google Scholar]
- 20.Kuukasjärvi T, Karhu R, Tanner M, Kähkönen M, Schäffer A, Nupponen N, Pennanen S, Kallioniemi A, Kallioniemi O-P, Isola J: Genetic heterogeneity and clonal evolution underlying development of asynchronous metastasis in human breast cancer. Cancer Res 1997, 57:1597-1604 [PubMed] [Google Scholar]
- 21.Mostofi FK: Histological Typing of Prostate Tumours. 1980. World Health Organization, Geneva
- 22.Isola JJ, De Vries S, Chu LW, Ghazvini S, Waldman FM: Analysis of changes in DNA copy number by comparative genomic hybridization in archival paraffin-embedded tumor samples. Am J Pathol 1994, 145:1301-1398 [PMC free article] [PubMed] [Google Scholar]
- 23.Hyytinen E, Visakorpi T, Kallioniemi A, Kallioniemi O-P, Isola J: Improved technique for analysis of formalin-fixed paraffin-embedded tumors by fluorescence in situ hybridization. Cytometry 1994, 16:93-99 [DOI] [PubMed] [Google Scholar]
- 24.Koivisto P, Hyytinen E, Palmberg C, Tammela T, Visakorpi T, Isola J, Kallioniemi O-P: Analysis of genetic changes underlying local recurrence of prostate carcinoma during androgen deprivation therapy. Am J Pathol 1995, 147:624-630 [PMC free article] [PubMed] [Google Scholar]
- 25.Sakr WA, Macoska JA, Benson P, Grignon DJ, Wolman SR, Pontes JR, Grissman JD: Allelic loss in locally metastatic, multisampled prostate cancer. Cancer Res 1994, 54:3273-3277 [PubMed] [Google Scholar]
- 26.Greene DR, Taylor SR, Wheeler TM, Scardinoa PT: DNA ploidy by image analysis of individual foci of prostate cancer: a preliminary report. Cancer Res 1991, 51:4084-4089 [PubMed] [Google Scholar]
- 27.Jenkins RB, Qian J, Lieber MM, Bostwick DG: Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res 1997, 57:524-531 [PubMed] [Google Scholar]
- 28.Carter BS, Ewing CM, Ward WS, Treiger BF, Aalders TW, Schalken JA, Epstein JI, Isaacs WB: Allelic loss of chromosomes 16q and 10q in human prostate cancer. Proc Natl Acad Sci USA 1990, 22:8751-8755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunimi K, Bergerheim USP, Larsson I-L, Ekman P, Collins VP: Allelotyping of human prostatic adenocarcinoma. Genomics 1991, 11:530-536 [DOI] [PubMed] [Google Scholar]
- 30.Cunningham JM, Shan A, Wick MJ, McDonnel SK, Schaid DJ, Tester DJ, Qian J, Takahashi S, Jenkins RB, Bostwick DG, Thibodeau SN: Allelic imbalance and microsatellite instability in prostatic adenocarcinoma. Cancer Res 1996, 56:4475-4484 [PubMed] [Google Scholar]
- 31.Bova GS, Carter BS, Bussemakers MJG, Emi M, Fujiwara Y, Kyprianou N, Jacobs SC, Robinson JC, Epstein JI, Walsh PC, Isaacs WB: Homozygous deletion and frequent allelic loss of chromosome 8p22 loci in human prostate cancer. Cancer Res 1993, 53:3869-3873 [PubMed] [Google Scholar]
- 32.Macoska JA, Trybus TM, Benson PD, Sakr WA, Grignon DJ, Wojno KD, Pietruk T, Powell IJ: Evidence for three tumor suppressor gene loci on chromosome 8p in human prostate cancer. Cancer Res 1995, 55:5390-5395 [PubMed] [Google Scholar]
- 33.Emmert-Buck MR, Vocke CD, Pozzatti RO, Duray PH, Jennings SB, Florence CD, Zhuang Z, Bostwick DG, Liotta LA, Linehan WM: Allelic loss on chromosome 8p12–21 in microdissected prostatic intraepithelial neoplasia. Cancer Res 1995, 55:2959-2962 [PubMed] [Google Scholar]
- 34.Eagle LR, Yin X, Brothman AR, Williams BJ, Atkin NB, Prochownik EV: Mutation of the MXI1 gene in prostate cancer. Nature Genet 1995, 9:249-255 [DOI] [PubMed] [Google Scholar]
- 35.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner S, Giovanella BC, Ittman M, Tycko B, Hibshoosh H, Wigler MH, Parsons R: PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 1997, 275:1943-1947 [DOI] [PubMed] [Google Scholar]
- 36.Steck PA, Perhouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng DH, Tavtigan SV: Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nature Genet 1997, 15:293-299 [DOI] [PubMed] [Google Scholar]
- 37.Umbas R, Isaacs WB, Bringuier PP, Schaafma HE, Karthaus HFM, Oosterhof GON, Debruyne MJ, Schalken JA: Decreased E-cadherin expression is associated with poor prognosis in patients with prostate cancer. Cancer Res 1994, 54:3929-3933 [PubMed] [Google Scholar]
- 38.Richmond PJ, Karayiannakis AJ, Nagafuchi A, Kaisary AV, Pignatelli M: Aberrant E-cadherin and α-catenin expression in prostate cancer: correlation with patient survival. Cancer Res 1997, 57:3189-3193 [PubMed] [Google Scholar]
- 39.Umbas R, Schalken JA, Aalders TW, Carter BS, Karthaus HFM, Schaafma HE, Debruyne MJ, Isaacs WB: Expression of cellular adhesion molecule E-cadherin is reduced or absent in high-grade prostate cancer. Cancer Res 1992, 52:5104-5109 [PubMed] [Google Scholar]
- 40.Cher ML, Ito T, Weidner N, Carroll PR, Jensen RH: Mapping of regions of physical deletion on chromosome 16q in prostate cancer cells by fluorescence in situ hybridization (FISH). J Urol 1995, 153:249-254 [DOI] [PubMed] [Google Scholar]
- 41.Sarkar FH, Sakr W, Li Y-W, Macoska J, Ball DE, Crissman JD: Analysis of retinoblastoma (RB) gene deletion in human prostate carcinomas. Prostate 1992, 21:145-152 [DOI] [PubMed] [Google Scholar]
- 42.Hyytinen E-R, Thalmann GN, Zhau HE, Karhu R, Kallioniemi O-P, Chung LWK, Visakorpi T: Genetic changes associated with the acquisition of androgen-independent growth, tumorigenicity, and metastatic potential in a prostate cancer model. Br J Cancer 1997, 75:190-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oltvai ZN, Millman CL, Korsmeyer S: Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell 1993, 74:609-619 [DOI] [PubMed] [Google Scholar]
- 44.Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, Bignell G, Warren W, Aminoff M, Höglund P, Järvinen H, Kristo P, Pelin K, Ridanpää M, Salovaara R, Toro T, Bodmer W, Olschwang S, Olsen AS, Stratton MR, de la Chappelle A, Aaltonen LA: A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature 1998, 391:184-187 [DOI] [PubMed] [Google Scholar]
- 45.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, Ferrara P, McKeon F, Caput D: Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 1997, 90:809-819 [DOI] [PubMed] [Google Scholar]
- 46.Wintersberger E: DNA amplification: new insights into its mechanism. Chromosoma 1994, 103:73-81 [DOI] [PubMed] [Google Scholar]
- 47.Brison O: Gene amplification and tumor progression. Biochem Biophys Acta 1993, 1155:25-41 [DOI] [PubMed] [Google Scholar]
- 48.Takahashi S, Qian J, Brown JA, Alcaraz A, Bostwick DG, Lieber MM, Jenkins RB: Potential markers of prostate cancer aggressiveness detected by fluorescence in situ hybridization in needle biopsies. Cancer Res 1994, 54:3574-3579 [PubMed] [Google Scholar]
- 49.Van Den Berg C, Guan X-Y, Von Hoff D, Jenkins R, Bittner M, Griffin C, Kallioniemi O-P, Visakorpi T, McGill J, Herath J, Epstein J, Saroscy M, Meltzer P, Trent J: DNA sequence amplification in human prostate cancer identified by chromosome microdissection: potential prognostic implications. Clin Cancer Res 1995, 1:11-18 [PubMed] [Google Scholar]
- 50.Kallioniemi A, Kallioniemi O-P, Piper J, Tanner M, Stokke T, Chen L, Smith HS, Pinkel D, Gray JW, Waldman FM: Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci USA 1995, 91:2156-2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kallioniemi A, Kallioniemi O-P, Citro G, Sauter G, DeVries S, Kerschmann R, Carroll P, Waldman F: Identification of gains and losses of DNA sequences in primary bladder cancer by comparative genomic hybridization. Genes Chromosomes & Cancer 1995, 12:213-219 [DOI] [PubMed] [Google Scholar]
- 52.Isola JJ, Kallioniemi O-P, Chu LW, Fuqua SAW, Hilsenbeck SG, Osborne CK, Waldman FM: Genetic aberrations detected by comparative genomic hybridization predict outcome in node-negative breast cancer. Am J Pathol 1995, 147:905-911 [PMC free article] [PubMed] [Google Scholar]
- 53.Henriksson M, Lüsher B: Proteins of the Myc network: essential regulators of cell growth and differentiation. Adv Cancer Res 1996, 68:109-182 [DOI] [PubMed] [Google Scholar]
- 54.Fleming WH, Hamel A, MacDonald R, Ramsey E, Pettigrew NM, Johnston B, Dodd JG, Matusik RJ: Expression of the c-myc proto-oncogene in human prostatic carcinoma and benign prostatic hyperplasia. Cancer Res 1986, 46:1535-1538 [PubMed] [Google Scholar]
- 55.Buttyan R, Sawczuk IS, Benson MC, Siegal JD, Olsson CA: Enhanced expression of the c-myc protooncogene in high-grade human prostate cancers. Prostate 1987, 11:327-337 [DOI] [PubMed] [Google Scholar]
- 56.Alcaraz A, Takahashi S, Brown JA, Herath JF, Bergsralh EJ, Larso-Keller JJ, Lieber MM, Jenkins RB: Aneuploidy and aneusomy of chromosome 7 detected by fluorescence in situ hybridization are markers of poor prognosis in prostate cancer. Cancer Res 1994, 54:3998-4002 [PubMed] [Google Scholar]
- 57.Visakorpi T, Hyytinen E, Kallioniemi A, Isola J, Kallioniemi O-P: Sensitive detection of chromosome copy number aberrations in prostate cancer by fluorescence in situ hybridization. Am J Pathol 1994, 145:624-639 [PMC free article] [PubMed] [Google Scholar]
- 58.Boerman RH, Anderl K, Herath J, Borell T, Johnson N, Schaeffer-Klein J, Kirchhof A, Raap J, Scheithauer B, Jenkins RB: The glial and mesenchymal elements of gliosarcomas share similar genetic alterations. J Neuropathol Exp Neurol 1996, 9:973-981 [DOI] [PubMed] [Google Scholar]
- 59.Heselmeyer K, Macville M, Schröck E, Blegen H, Hellström A-C, Shah K, Auer G, Ried T: Advanced-stage cervical carcinomas are defined by a recurrent pattern of chromosomal aberrations revealing high genetic instability and a consistent gain of chromosome arm 3q. Genes Chromosomes & Cancer 1997, 19:233-240 [PubMed] [Google Scholar]
- 60.Kuukasjärvi T, Karhu R, Tanner M, Kähkönen M, Schäffer A, Nupponen N, Pennanen S, Kallioniemi A, Kallioniemi O-P, Isola J: Genetic heterogeneity and clonal evolution underlying development of asynchronous metastasis in human breast cancer. Cancer Res 1997, 57:1597-1604 [PubMed] [Google Scholar]