Abstract
The enriched polymerase chain reaction (PCR) assay has been used extensively in the detection of ras gene mutations in many types of human malignancies. Although it is very sensitive, it has a number of features that limit its use in the routine diagnostic laboratory. The aim of this study was to develop a novel enriched PCR strategy, in which the concurrent activity of the restriction enzyme BstNI and Taq polymerase allowed the amplification of mutant K-ras while inhibiting the formation of wild-type product. This restriction endonuclease-mediated selective PCR assay uses three sets of primers, together with BstNI, in the reaction mix, and the amplification products are analyzed by gel electrophoresis. The reliability of the restriction endonuclease-mediated selective PCR assay to detect activated K-ras was determined in a variety of clinical samples, including 139 fresh colorectal carcinomas and 113 paraffin-embedded blocks from 80 separate tumors of the colon and rectum, pancreas, breast, or kidney. Codon 12 mutations of the K-ras oncogene were identified in DNA from both fresh and paraffin-embedded tumors in a rapid, sensitive, and reproducible manner. Mutations were detected in 33 (24%) of the fresh colorectal cancers and 16 (20%) of the paraffin-embedded tumors. These results were 97% concordant in cases in which paraffin blocks and fresh specimens from the same tumor were available for analysis. We conclude that restriction endonuclease-mediated selective PCR is a sensitive, rapid, and robust assay for the detection of point mutations in a variety of clinical samples. Importantly, there is no need for manipulation of the sample once the PCR has been set up, and therefore, the chance of contamination is significantly reduced. In contrast to previous assays, restriction endonuclease-mediated selective PCR is not labor intensive, and its format is suitable for use in routine diagnostic laboratory.
Activating point mutations at codons 12, 13, or 61 of the ras proto-oncogenes occur frequently in human tumors. 1 In colon and pancreatic cancer, more than 90% of these mutations occur in the K-ras gene, and most of these are found in codon 12. 2,3 Many studies have suggested that detection of activated ras may have diagnostic or prognostic importance. Because these mutations are acquired early in tumor development, 4 the detection of activated K-ras in DNA from the stools of patients with colorectal cancer may allow diagnosis at a stage at which curative surgery is still possible. 5-7 Other studies have demonstrated that the presence of mutant K-ras in the regional lymph nodes or peripheral blood of patients with colorectal cancer identifies those individuals most likely to relapse. 8,9 Moreover, the recent development of specific treatments targeting the activated ras genes provides a further impetus for the development of new strategies for the detection of mutant ras in clinical samples.
A number of assays have been used for the detection of activated ras, and these protocols vary in their sensitivity and complexity. Typically, they rely on amplification of the ras gene by the polymerase chain reaction (PCR), followed by detection of the mutant product by electrophoresis, colorimetric analysis, or other means. Many of these PCR protocols have used mismatched bases within primer sequences, allowing the identification of mutant amplicons by the creation of restriction enzyme sites. 10 The sensitivity of this type of protocol has been significantly improved with the development of the enriched PCR assay, 11,12 which is based on an initial round of amplification followed by restriction enzyme digestion to cleave wild-type amplicons. Because only mutant amplicons remain as templates, a further round of amplification results in the “enrichment” of mutant ras product. This assay has been applied extensively to the analysis of ras gene mutations in many types of cancer, including colorectal tumors. 12,13 However, the assay remains a relatively long and labor-intensive procedure, with substantial risk of contamination.
We describe a novel enriched PCR strategy, known as restriction endonuclease-mediated selective PCR (REMS-PCR), in which the restriction enzyme BstNI is incorporated as part of a conventional PCR reaction. This approach exploits both the thermostability of the enzyme and its compatibility with standard PCR buffers. The concurrent activity of these enzymes in the one reaction allows simultaneous amplification of mutant signal and inhibition of amplification of wild-type K-ras. Further, we demonstrate the reliability and sensitivity of this assay for the detection of activated K-ras in a variety of clinical samples and its suitability for use in diagnostic laboratories.
Materials and Methods
Patient Samples
After informed consent was obtained, 137 individuals undergoing surgical resection of adenocarcinoma of the colon or rectum at St. Vincent’s Hospital (Sydney, NSW, Australia) were enrolled in this prospective study from 1993 to 1997. Fresh representative samples (500 μg) of all tumors were immediately frozen at −70°C. A total of 139 fresh tumor specimens were assayed from 80 males and 59 females, with ages ranging from 29 to 94 (mean, 67.3 ± 12.1). Of these tumors, 21% were modified Dukes’ stage A, 33% were stage B, 38% were stage C, and 7% were stage D. 14,15 In addition, paraffin-embedded blocks of 51 colorectal, 12 breast, 11 renal, and 6 pancreatic tumors were obtained from the Department of Anatomical Pathology, St. Vincent’s Hospital, after routine processing. The colorectal paraffin blocks were collected from 21 females and 30 males, ranging in age from 49 to 95 years. Of these tumors, 14% were Dukes’ stage A, 38% were stage B, 40% were stage C, and 8% were stage D. To determine the effect of paraffin block age on assay reliability, we selected tumors that had been processed at various time points over the last 10 years.
For all tumors, the histopathological diagnosis, stage and tumor size were determined independently by a histopathologist within the Department of Anatomical Pathology, St. Vincent’s Hospital.
DNA Preparation from Fresh Tissues and Paraffin Sections
For preparation of DNA from fresh tissues, the frozen tissue was macerated in a 500-μl solution of 10 mmol/L Tris-HCl, 1 mmol/L ethylenediamine tetra-acetic acid, 100 mmol/L NaCl, 1% sodium dodecyl sulfate, and 500 μg/ml proteinase K, using a sterile Eppendorf homogenizer at 4°C. The DNA was extracted with phenol/chloroform after incubation overnight with shaking at 50°C and precipitated with ethanol. It was then resuspended in water, and the concentration was determined by spectrophotometry.
For the analysis of paraffin-embedded tissues, three consecutive 10-μm sections were cut from paraffin blocks, and each section was placed in a separate sterile 2.0-ml screw-capped tube. To prevent cross contamination from tissue with flakes of paraffin, the blade was cleaned with a jet of compressed air after each section was cut. A sham tissue block, which did not contain tumor tissue, was cut after every 10 blocks. DNA was also extracted from this sample and used in subsequent PCR analysis. For each tumor block, an adjacent section was stained with hematoxylin and eosin and examined by light microscopy to determine the amount of tumor present.
For the extraction of DNA from paraffin-embedded tissues, each section was immersed in 300 μl of lysis buffer (50 mmol/L KCl, 3 mmol/L CaCl2, 0.4% Triton-X, and 10 mmol/L Tris-HCl, pH 8.0), together with 3 U of PreTaq (1 U/μl Boehringer Mannheim). The tubes were boiled for 5 minutes and then centrifuged at 14,000 rpm for 2 minutes. The supernatants were then transferred to tubes containing 100 μl of 0.25 mol/L ACES buffer (N-(2-acetamido)-2-aminoethanesulfonic acid, 0.125 mol/L NaOH, and 0.5% Tween 20, pH 6.8) and 25 μl of a cationic polymer (2.4% aqueous, pH 5, Ortho Diagnostics, Raritan, NJ), and the mixture was vortexed and then microfuged at 14,000 rpm for 2 minutes. The pellet was resuspended in 100 μl of 20 mmol/L NaOH and incubated at room temperature for 30 minutes. To detach the polymer, samples were boiled for 5 minutes, and the solution containing the DNA was stored at −20°C.
REMS-PCR Mutational Analysis
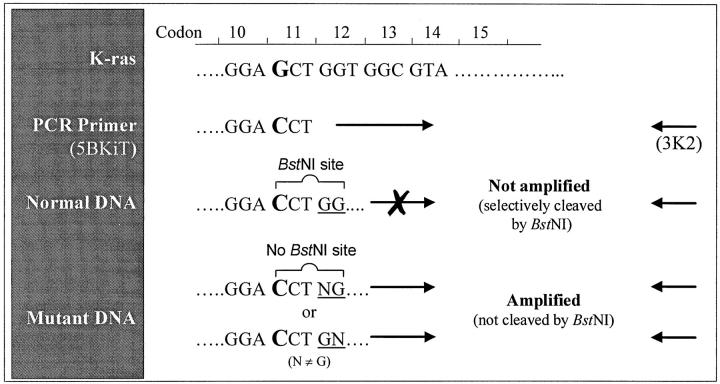
Mutations at the first and second bases of codon 12 of the K-ras gene were detected using REMS-PCR. Each PCR reaction contained three sets of primers (Table 1 ▶ , Figure 1 ▶ ).
Table 1.
Oligonucleotides Used in the REMS-PCR Reaction
| Primers | Sequence | Significance of amplicon |
|---|---|---|
| Diagnostic | 5BKIT 5′-TAT AAA CTT GTG GTA GTT GGA CCT | Presence of this amplicon indicates that the template contains mutant K-ras |
| 3K2 5′-CGT CCA CAA AAT GAT TCT GA | ||
| PCR control | 5BK38 5′-GTA CAC ATG AAG CCA TCG TAT A | Absence of this amplicon indicates PCR failure |
| 3K39 5′-CCA CTT GTA CTA GTA TGC CTT AAG | ||
| Restriction enzyme control | 5BK36 5′-CTA GAA CAG TAG ACA CAA ACC A | Presence of this amplicon indicates BstN1 failure |
| 3K37 5′-GAT TTT GCA GAA AAC AGA TC |
The underscored C in 5BKIT and 5BK36 indicates a mismatch with the K-ras gene.
Figure 1.
Schematic representation of diagnostic primers (5BKIT and 3K2) used in REMS-PCR amplification of K-ras.
The diagnostic primers 5BKIT and 3K2 induce a cleavage site for BstNI in wild-type K-ras amplicons. Because BstNI is included in the REMS-PCR reaction, the amplification of such a wild-type template is prevented. However, when a template also contains K-ras gene with mutations at bases 1 and 2 of codon 12, no BstNI cleavage site is induced by the diagnostic primers, and the mutant template is thus selectively amplified. An 81-bp amplicon therefore indicates the presence of mutant K-ras template in the test sample. The template control primers 5BK38 and 3K39 amplify a 214-bp DNA sequence in exon 4B of the K-ras gene. An amplicon of this size indicates that all conditions for amplification are satisfactory, including the amount and integrity of the DNA in the reaction. Amplicons incorporating these primers must be present for the unambiguous interpretation of negative results. The enzyme control primers 5BK36 and 3K37 amplify a 130-bp fragment of exon 3 of the K-ras gene and always induce a BstNI cleavage site in this amplicon. This 130-bp product is seen only if BstNI is inactive in the REMS-PCR reaction. Therefore, a test sample is considered mutant only when this enzyme control band is absent and BstNI has therefore been active throughout the PCR reaction.
The REMS-PCR was performed using 0.1 to 0.6 μg of DNA extracted from fresh tissue or 5–20 μl of DNA from the paraffin-embedded tissues. Reaction mixtures also contained the following: 40 pmol of each diagnostic primer (5BKIT and 3K2), 2 pmol of each enzyme control primer (5BK36 and 3K37), 20 pmoles of each PCR control primer (5BK38 and 3K39), 1 mmol/L dithiothreitol, 50 μmol/L deoxynucleotide triphosphate, 4 mmol/L Mg2+, and 40 U of BstN1 (Biolabs, Northbrook, IL). Taq DNA polymerase (Perkin-Elmer, Norwalk, CT) was preincubated with 0.26 μg of TaqStart antibody (Clontech, Palo Alto, CA) in buffer (50 mmol/L KCl and 10 mmol/L Tris-HCl, pH 7.0) for 15 minutes at room temperature, and 6 U of the enzyme was added to the reaction mix. The final reaction volume was brought to 50 μl in PCR buffer (50 mmol/L Tris-HCl and 100 mmol/L NaCl, pH 8.3).
The reactions were cycled in a Perkin-Elmer 9600 PCR cycler as follows: 94°C for 2 minutes (one cycle) and 58°C for 60 seconds followed by 92°C for 20 seconds (30 cycles); the reactions were then held at 4°C before analysis.
Samples were analyzed by electrophoresis on a 5% NuSieve agarose gel (FMC Bioproducts, Rockland, ME) and photographed using a Stratagene (La Jolla, CA) Eagle Eye II video system. In all assays, DNA from the human K562 cell line, known to be homozygous wild type for K-ras, was amplified both with and without BstNI in the reaction.
For the analysis of all paraffin blocks, the three cut sections were analyzed independently by three operators who were masked with respect to both the tissue of origin of each sample and the results of the other operators. In the case of fresh samples, extracted DNA was analyzed independently by two operators. REMS-PCR was performed at least twice on all samples, and the mutational status of each tumor or block was interpreted from analytical gels by two investigators independently.
Samples were considered mutant when the diagnostic and template control bands were present, and the enzyme control band was absent. They were considered wild type when the template control band was present and the diagnostic band was absent. In all other cases, the K-ras status of the sample was considered indeterminate.
The presence or absence of mutations in the K-ras gene was confirmed by DNA sequencing of PCR products in eight selected samples (four mutant, four wild type). Sequencing reactions were performed with the fmol DNA sequencing system (Promega) using the 3K2 primer labeled with [γ-33P]ATP according to the manufacturer’s instructions.
Statistical Analysis
The relationship between K-ras mutational status and pathological variables including Dukes’ stage, differentiation, site, and tumor size was investigated using logistic regression analysis. All data were analyzed using the SPSS statistical software (SPSS Inc., Chicago, IL).
Results
Assay Validation
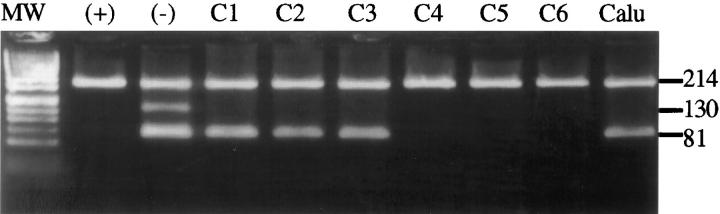
The ability of the REMS-PCR assay to detect mutations of K-ras codon 12 was determined in cell lines of known K-ras status. When performed on DNA from the heterozygous mutant cell line Calu I, the assay resulted in template control and diagnostic amplicons, the latter indicating the presence of a K-ras codon 12 mutation (Figure 2) ▶ . Only the template control fragment was present in amplification of DNA from the homozygous wild-type cell line K562. When BstN1 was omitted from this reaction, three amplicons were seen. Absence of the restriction enzyme control band in reactions containing BstN1 indicated that the enzyme was sufficiently active during these reactions (Figure 2) ▶ . The limit of detection of this assay has previously been determined by serial dilution of Calu I DNA in K562 DNA at ratios from 1:10 to 1:10,000 (A. V. Todd, manuscript in preparation). Under these conditions, a ratio of 1 mutant gene to 1000 wild-type genes was sufficient to give a diagnostic amplicon (data not shown).
Figure 2.
REMS-PCR detection of mutant K-ras in DNA from fresh colorectal tumor tissue. Results from six representative colorectal cancers are shown (C1 to C6). DNA from the wild-type cell line K562 analyzed with (+) and without (−) BstN1 in the reaction, and DNA from the heterozygous mutant cell line Calu 1 (Calu) are also shown. Mutant K-ras (81-bp amplicon) is present in three tumors (C1 to C3). All samples contain the template control band (214 bp). Only the K562 DNA analyzed without BstN1 contains the enzyme control band. Absence of this band in the other lanes indicates that the restriction enzyme was active in all other reactions. Products are shown on a 5% NuSieve gel stained with ethidium bromide. MW, phiX174/Hinf.
The reliability of the assay in the detection of K-ras mutations in both fresh and paraffin-embedded tumor tissue was analyzed by repeated testing of four samples known to be mutant or wild type at codon 12. When tested on six occasions more than 1 day apart, each of these samples consistently showed the expected result. Similarly, no intra-assay variation was noted when aliquots of the same sample were tested simultaneously.
Analysis of Fresh Tumor Tissues
The K-ras status of DNA isolated from 139 fresh colorectal tumor samples was determined by REMS-PCR. All samples contained a 214-bp template control product, indicating that the DNA was of sufficient integrity for the amplification of the K-ras gene. Twenty-four percent (33 of 139) of the samples contained an additional band at 81 bp, indicating the presence of mutant K-ras (Figure 2) ▶ . The remaining samples contained only wild-type K-ras, and there were no samples of indeterminate status. No statistical differences were found between mutant and nonmutant tumors in terms of tumor volume, anatomical site, Dukes’ stage, or histological differentiation.
Analysis of Paraffin-Embedded Tumor Tissues
REMS-PCR was used to analyze the DNA from 113 paraffin blocks of 80 carcinomas, including those of the pancreas, colon and rectum, breast, and kidney (Figure 3 ▶ , Table 2 ▶ ). For the three consecutive paraffin sections cut from each tumor block, the REMS-PCR results were always concordant. The processing of sham tissue blocks in parallel with test samples indicated that there was no cross-contamination of samples during sectioning or analysis. In general, the signal intensity of amplicons of DNA extracted from paraffin-embedded tissue were less than that observed with comparable amounts of DNA from fresh tissue (Figures 2 and 3 ▶ ▶ , samples C2 to C6). Further, there was greater variability in the amount of PCR product from different paraffin-embedded tumors, when compared with either fresh tumor tissues or with cell lines.
Figure 3.
REMS-PCR detection of mutant K-ras in DNA from paraffin-embedded tumor tissue. DNA from paraffin-embedded sections of the colorectal tumors analyzed in Figure 2 ▶ are shown (C1 to C6). Control samples (+), (−), and Calu are as for Figure 2 ▶ . Mutant K-ras is present in tumors C1 to C3, whereas tumors C4 and C5 are wild type. Although a weak control amplicon is present in C6, the sample was designated as indeterminate because of insufficient amplification. Also shown is analysis of DNA from two pancreatic tumors (P1 and P2) that contain mutant K-ras. Products are shown on a 5% NuSieve gel stained with ethidium bromide. MW, phiX174/Hinf.
Table 2.
Analysis of K-ras Status of DNA Extracted from Paraffin Tissues Using REMS-PCR
| Site | Wild type | Mutant | Indeterminate |
|---|---|---|---|
| Colorectal | 32 | 12 | 7 |
| Breast | 11 | Nil | 1 |
| Renal | 10 | Nil | 1 |
| Pancreas | 1 | 4 | 1 |
Sequence analysis of representative mutant and wild-type PCR products from both fresh and paraffin-embedded tissues confirmed the results obtained with REMS-PCR. Of the four mutant samples sequenced, three showed A/G substitutions at the second position of codon 12, and the fourth showed a C/G substitution at the same position.
Overall, 16 (20%) of these tumors were found to contain mutant K-ras, and 54 contained only wild type (Table 2) ▶ . In 10 samples (12.5%), the PCR profile was indeterminate, either because of poor amplification (Figure 3 ▶ , sample C6) or because of insufficient activity of BstNI, as evidenced by an enzyme control band.
The age of the paraffin block was the only factor that appeared to contribute to the failure of REMS-PCR in indeterminate samples, and this was particularly the case in those blocks stored for more than 6 years (Table 3) ▶ . No other factors were associated with assay failure. Histological examination of adjacent sections showed no clear differences with other samples with respect to the proportion of tumor to normal tissue or to other histological features, such as the amount of necrosis, fibrosis, mucin, or adipose tissue in the section. Importantly, four of the smallest samples were biopsies of pancreatic tumors, yet mutations of K-ras were reproducibly detected from these blocks. Assay failure was also not related to the amount or extent of degradation of DNA in the paraffin samples, as determined by gel electrophoresis. In fact, most samples contained only high-molecular weight DNA (>4000 bp), whereas REMS-PCR was often conclusive in those samples with clear evidence of some DNA degradation (data not shown).
Table 3.
Effect of Tumor Age on REMS-PCR Reliability
| Tumor age | Paraffin | Fresh | ||||
|---|---|---|---|---|---|---|
| Wild type | Mutant | Indeterminate | Wild type | Mutant | Indeterminate | |
| <1 year | 11 | 0 | 0 | 27 | 7 | 0 |
| 1–3 years | 22 | 4 | 2 | 60 | 15 | 0 |
| 3–6 years | 18 | 5 | 1 | 19 | 11 | 0 |
| 6–10 years | 1 | 7 | 3 | NA | NA | NA |
| >10 years | 2 | 0 | 4 | NA | NA | NA |
Results of REMS-PCR analysis are shown for both paraffin-embedded and fresh (frozen) tissue, according to the age of the tumor at the time of analysis. Paraffin sections were from colorectal, pancreas, renal, and breast cancers, whereas fresh tissues are from only colorectal tumors. Numbers represent the number of tumors in each category.
NA, not available.
Correlation with the Subset of Tissues with Fresh Results
Of the 33 colorectal tumors in which both fresh and paraffin-embedded tissues were analyzed, 27 were wild type in both assays, 5 were mutant, and in 1 case there was a discordant result. In this case, the tumor was mutant by analysis of the frozen tissue, but wild type in DNA from the paraffin sections, despite the fact that it was shown by histological examination to contain tumor. DNA sequence analysis of this case confirmed the REMS-PCR results from fresh and paraffin-embedded tissues.
To assess the possibility that tumors are heterogeneous for K-ras mutations, we examined 4 to 6 tissue blocks taken from each of 10 colorectal tumors. In 2 of these 10 tumors, REMS-PCR showed mutations of K-ras in all 9 blocks examined. Likewise, 32 blocks from the other 8 tumors contained only wild-type K-ras. These results show that there is no clear evidence to support tumor heterogeneity for mutations of K-ras in colorectal tumors.
Discussion
The REMS-PCR assay exploits the thermostable properties of BstNI and Taq polymerase, as well as their ability to function effectively in a common buffer system, thereby allowing simultaneous amplification of mutant signal and inhibition of amplification of wild-type K-ras. This strategy represents a significant advance over existing enriched PCR approaches, producing a robust assay suitable for use in the diagnostic laboratory. The principal advantage of the REMS procedure comes from the greatly reduced handling of amplified product. Unlike conventional enriched PCR, in which the two rounds of PCR are separated by an enzyme digestion step, all reactions in the REMS-PCR occur concurrently in the one tube. This reduced handling has two important implications for the applicability of the assay as a diagnostic procedure. Firstly, there is greatly reduced risk of contamination by PCR product, a major problem in most PCR assays and a common cause of false positive results. Secondly, reduced handling means considerable savings in both time and labor costs and makes the process more amenable to automation.
A further significant advantage of the REMS-PCR assay over current enriched PCR methods is that each reaction includes internal controls for both template integrity and BstN1 enzyme activity. The control for template integrity allows discrimination between those reactions that are negative because of complete digestion of wild-type product and those in which amplification of either wild-type or mutant product has failed for any reason, including insufficient quantity or quality of the template. This proved to be particularly important in the analysis of the paraffin-embedded samples. The template control primers have been designed to amplify a longer DNA template than the diagnostic primers. Thus, a reduction in the integrity of the template is readily apparent in the proportionally greater loss of the template control product.
Incomplete BstNI digestion is a potential problem in REMS-PCR, particularly because restriction enzyme activity is influenced by a number of factors including formalin fixation of the tissue. 16 The inclusion of a restriction enzyme control primer set within the PCR reaction allows ready identification of this problem. If BstNI has remained active throughout the reaction, the enzyme control amplicon will not be seen after REMS-PCR.
By incorporating these features into the REMS-PCR procedure, we were able to detect mutations in codon 12 of K-ras, with minimal manipulation of samples, in a time of less than 3 hours, including DNA extraction and gel electrophoresis. Despite this, the sensitivity of the assay remained at a level of 1 mutant per 1000 wild-type genes, a level comparable with that achieved in previous enriched PCR strategies. 17
To be applicable in routine laboratories, it is important that the assays be reliable as well as rapid and sensitive. The reliability of the REMS-PCR assay was evaluated in a rigorous manner, with assessment of coded samples by two independent operators on several occasions. There was no inter- or intra-assay variation when the technique was applied to fresh specimens. Moreover, the assay allowed the rapid, unambiguous, and reproducible determination of K-ras status in all 139 fresh colorectal specimens examined.
When assayed with methods of appropriate sensitivity, the frequency of K-ras codon 12 mutations in colorectal cancer has been reported to vary between 20 and 50%. 18,19 In part, this may represent geographical variation in the frequency of mutations. 20,21 Two recent studies in the Australian population have reported that, in samples of 233 and 103 individuals, respectively, 24% of colorectal tumors contained mutations at codon 12 of the K-ras gene. 22,23 The results with REMS-PCR are clearly in agreement with these reports and suggest that the assay reflects the true incidence in this population.
Large archives of frozen tissues are rarely available for retrospective analysis, and it can be difficult to confirm the presence of tumor cells within these tissues. We therefore also sought to evaluate the utility of REMS-PCR in the analysis of K-ras mutations in paraffin-embedded tumor blocks. A number of factors are known to influence the success of PCR analysis of paraffin-embedded tissues, including the type and amount of tumor and the methods used for fixation and embedding. 16,24-26 The effect of variation in these factors was of particular concern in the REMS-PCR assay, in which simultaneous activity of both Taq polymerase and restriction enzymes is required. We therefore used the assay to analyze randomly selected blocks from a variety of tumors collected at different time points over the previous 10 years. The REMS-PCR assay proved robust in the analysis of DNA from paraffin tissue collected up to 6 years previously and proved effective on a range of tissues containing varying amounts of tumor. The analysis of paraffin tissue did not appear to generate false positive results, because no amplicons were seen in sham blocks, no diagnostic amplicons were seen in multiple blocks from wild-type tumors, and there was a 97% concordance with the assay on fresh colorectal tumors. Furthermore, activated K-ras was not detected in either the breast or renal cancers, tumors that have rarely been reported to contain mutations in this gene. 27,28 Although REMS-PCR was often less efficient using paraffin blocks, the K-ras status of the tumor could still be determined in more than 90% of cases in which the block age was less than 6 years.
The factors responsible for the suboptimal amplification in some tissues were not easily identified. Neither the fragment size, the amount of DNA extracted from paraffin blocks, nor the method of extraction, appeared to influence the success of REMS-PCR. It is possible that the fixation process itself may have contributed to the failure of amplification or inactivation of BstN1 seen in some paraffin samples. Structural modifications induced in DNA by formaldehyde may not only reduce the effective amount of PCR template, but may also interfere with the activity of restriction enzymes. 16 This possibility is supported by our observation that the inclusion of dithiothreitol in the assay markedly reduced the number of indeterminate results.
There are conflicting data concerning the effect of block age on successful PCR amplification. 29,30 Our study indicated that, in the case of REMS-PCR, the frequency of indeterminate results increased with increasing block age, and we would therefore not recommend its use for blocks collected more than 6 years previously.
In conclusion, REMS-PCR provides a sensitive, fast, and robust assay for the detection of point mutations in a variety of clinical samples. Importantly, there is no need for manipulation of the sample once the PCR has been set up, and thus the chance of contamination is significantly reduced. In contrast to previous assays, the REMS-PCR is not labor intensive, and its format would be suitable for use in a routine diagnostic laboratory.
Footnotes
Address reprint requests to Dr. Robyn Ward, Department of Medical Oncology, St. Vincent’s Hospital, Victoria Street, Darlinghurst, 2010 New South Wales, Australia. E-mail: r.ward@garvan.unsw.edu.au.
Supported by the Leo and Jenny Leukaemia and Cancer Foundation and the St. Vincent’s Clinic Foundation.
References
- 1.Barbacid M: ras genes. Annu Rev Biochem 1987, 56:779-827 [DOI] [PubMed] [Google Scholar]
- 2.Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M: Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell 1988, 53:549-554 [DOI] [PubMed] [Google Scholar]
- 3.Bos JL: ras oncogenes in human cancer: a review. Cancer Res 1989, 49:4682-4689 [PubMed] [Google Scholar]
- 4.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AMM, Bos JI: Genetic alterations during colorectal-tumor development. N Engl J Med 1988, 319:525-532 [DOI] [PubMed] [Google Scholar]
- 5.Sidransky D, Tokino Y, Hamilton SR, Kinzler KW, Levin B, Frost P, Vogelstein B: Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science 1992, 256:102-105 [DOI] [PubMed] [Google Scholar]
- 6.Losi L, Benhattar J, Costa J: Stability of K-ras mutations throughout the natural history of human colorectal cancer. Eur J Cancer 1992, 28A:1115-1120 [DOI] [PubMed] [Google Scholar]
- 7.Ratto C, Flamini G, Sofo L, Nucera P, Ippoliti M, Curigliano G, Ferretti G, Sgambato A, Merico M, Doglietto GB, Cittadini A, Crucitti F: Detection of oncogene mutation from neoplastic colonic cells exfoliated in feces. Dis Colon Rectum 1996, 39:1238-1244 [DOI] [PubMed] [Google Scholar]
- 8.Hayashi N, Ito I, Yanagisawa A, Kato Y, Nakamori S, Imaoka S, Watanabe H, Ogawa M, Nakamura Y: Genetic diagnosis of lymph-node metastasis in colorectal cancer. Lancet 1995, 345:1257-1259 [DOI] [PubMed] [Google Scholar]
- 9.Hardingham JE, Kotasek D, Sage RE, Eaton MC, Pascoe VH, Dobrovic A: Detection of circulating tumor cells in colorectal cancer by immunobead-PCR is a sensitive prognostic marker for relapse of disease. Mol Med 1995, 1:789-794 [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen JB, Levinson AD: A point mutation in the last intron responsible for increased expression and transforming activity of the c-Ha-ras oncogene. Nature 1988, 334:119-124 [DOI] [PubMed] [Google Scholar]
- 11.Todd AV, Iland HJ: Allele-specific enrichment: a method for the detection of low level N-ras gene mutations in acute myeloid leukemia. Leukemia 1991, 5:160-161 [PubMed] [Google Scholar]
- 12.Levi S, Urbano-Ispizua A, Gill R, Thomas DM, Gilbertson J, Foster C, Marshall CJ: Multiple K-ras codon 12 mutations in cholangiocarcinomas demonstrated with a sensitive polymerase chain reaction technique. Cancer Res 1991, 51:3497-3502 [PubMed] [Google Scholar]
- 13.Singh J, Rao CV, Kulkarni N, Simi B, Reddy BS: Molecular markers as intermediate end-points in chemoprevention of colon cancer: modulation of ras activation by sulindac and phenylhexylisothiocyanate during colon carcinogenesis. Int J Oncol 1994, 5:1009-1018 [DOI] [PubMed] [Google Scholar]
- 14.Dukes CE, Bussey HJR: The spread of rectal cancer and its effect on prognosis. Br J Cancer 1958, 12:309-320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lineweaver W: Staging of colon cancer. Contemp Surg 1984, 25:19 [Google Scholar]
- 16.Hamazaki S, Koshiba M, Habuchi T, Takahashi R, Sugiyama T: The effect of formalin fixation on restriction endonuclease digestion of DNA and PCR amplification. Pathol Res Pract 1993, 189:553-557 [DOI] [PubMed] [Google Scholar]
- 17.Ward RL, Santiago F, Hawkins NJ, Coomber D, Oconnor T, Todd AV: A rapid PCR ELISA for the detection of activated K-ras in colorectal cancer. J Clin Pathol 1995, 48:M273-M277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasegawa H, Ueda M, Watanabe M, Teramoto T, Mukai M, Kitajima M: K-ras gene mutations in early colorectal cancer: flat elevated vs. polyp-forming cancer. Oncogene 1995, 10:1413-1416 [PubMed] [Google Scholar]
- 19.Minamoto T, Ronai Z, Yamashita N, Ochiai A, Sugimura T, Mai M, Esumi H: Detection of Ki-ras mutation in non-neoplastic mucosa of Japanese patients with colorectal cancers. Int J Oncol 1994, 4:397-401 [DOI] [PubMed] [Google Scholar]
- 20.Sasaki H, Nishii H, Takahashi H, Tada A, Furusato M, Terashima Y, Siegal GP, Parker SL, Kohler MF, Berchuck A: Mutation of the Ki-ras protooncogene in human endometrial hyperplasia and carcinoma. Cancer Res 1993, 53:1906-1910 [PubMed] [Google Scholar]
- 21.Scarpa A, Capelli P, Villanueva A, Zamboni G, Lluìs F, Accolla R, Mariuzzi G, Capella G: Pancreatic cancer in Europe: Ki-ras gene mutation pattern shows geographical differences. Int J Cancer 1994, 57:167-171 [DOI] [PubMed] [Google Scholar]
- 22.Fung C, Bragg T, Newland R, Dent O, Nicholson G, Bokey L, Chapuis P: K-ras mutation, and loss of heterozygosity of chromosome 17p, and survival in colorectal cancer. Aust NZ J Surg 1997, 67:239-244 [DOI] [PubMed] [Google Scholar]
- 23.Thomas RJS, Liu YS, St Clair F, Norris PM, Valentine R, Phillips WA: Frequency and clinico-pathological associations of ras mutations in colorectal cancer in the Victorian population. Aust NZ J Surg 1997, 67:233-238 [DOI] [PubMed] [Google Scholar]
- 24.Greer CE, Wheeler CM, Manos MM: Sample preparation and PCR amplification from paraffin-embedded tissues. PCR Methods Appl 1994, 3:S113-S122 [DOI] [PubMed] [Google Scholar]
- 25.Alaibac M, Filotico R, Giannella C, Paradiso A, Labriola A, Marzullo F: The effect of fixation type on DNA extracted from paraffin-embedded tissue for PCR studies in dermatopathology. Dermatology 1997, 195:105-107 [DOI] [PubMed] [Google Scholar]
- 26.Merkelbach S, Gehlen J, Handt S, Fuzesi L: Novel enzyme immunoassay and optimized DNA extraction for the detection of polymerase chain reaction-amplified viral DNA from paraffin-embedded tissue. Am J Pathol 1997, 150:1537-1546 [PMC free article] [PubMed] [Google Scholar]
- 27.Koffa M, Malamou-Mitsi V, Agnantis NJ, Spandidos DA: Mutational activation of K-ras oncogene in human breast tumors. Int J Oncol 1994, 4:573-576 [DOI] [PubMed] [Google Scholar]
- 28.Nagata Y, Abe M, Kobayashi K, Saiki S, Kotake T, Yoshikawa K, Ueda R, Nakayama E, Shiku H: Point mutations of c-ras genes in human bladder cancer and kidney cancer. Jpn J Cancer Res 1990, 81:22-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shibata D, Martin WJ, Arnheim N: Analysis of DNA sequences in forty-year-old paraffin-embedded thin-tissue sections: a bridge between molecular biology and classical histology. Cancer Res 1988, 48:4564-4566 [PubMed] [Google Scholar]
- 30.Goelz SE, Hamilton SR, Vogelstein B: Purification of DNA from formaldehyde fixed and paraffin embedded human tissue. Biochem Biophys Res Commun 1985, 130:118-126 [DOI] [PubMed] [Google Scholar]