Abstract
Peroxisome proliferator-activated receptors (PPARs) regulate genes involved in lipid metabolism and adipocyte differentiation. Steroid receptor coactivator-1 (SRC-1) and PPAR-binding protein (PBP) interact with PPARγ and act as coactivators to enhance ligand-dependent transcription. We report here that PPARγ, SRC-1, and PBP are differentially expressed in the brown fat, transitional epithelium of the urinary bladder, colonic mucosa, and mammary epithelium of the adult mouse. PPARγ and PBP are expressed in the transitional epithelium of urinary bladder and in brown adipose tissue, but not SRC-1. In the colonic mucosa, PPARγ expression occurs throughout the villi, whereas the expression of both SRC-1 and PBP is confined mostly to the crypts. The expression of both SRC-1 and PBP is prominent in the breast epithelium of nonpregnant, pregnant, and lactating mice, whereas PPARγ expression appeared prominent during lactation. During early embryonic development, PPARγ, SRC-1, and PBP are differentially expressed, with only limited cell types displaying overlapping expression. PPARγ and PBP expression overlapped in the brown fat and urogenital sinus at stage E15.5 of embryogenesis, whereas SRC-1 expression occurred mostly in neuroepithelium and cartilage between stages E9.5 and E13.5 of embryogenesis.
Peroxisomes are single membrane-bound organelles present in most eukaryotic cells. These organelles participate in a variety of metabolic functions such as lipid metabolism; synthesis of cholesterol; production of bile acids; and catabolism of purines, polyamines, and amino acids. 1 Peroxisomes in liver parenchymal cells can be stimulated to proliferate by the administration of nonmutagenic chemicals designated as peroxisome proliferators. 2 The induction of peroxisome proliferation is mediated by peroxisome proliferator-activated receptors (PPARs) that regulate the expression of genes associated with lipid metabolism and adipocyte differentiation. 3-7 Three isotypes of this subfamily of nuclear receptor superfamily, namely PPARα, PPARδ, and PPARγ have been identified as products of separate genes from Xenopus, rodents, and humans. 8-12 PPARs form heterodimers with 9-cis-retinoic acid receptor (RXR), and this heterodimer complex binds to peroxisome proliferator response elements on the target gene promoter as PPAR-RXR heterodimers to initiate transcriptional activity. 13 All three PPAR isotypes are generally co-expressed in most cell types, but their relative levels of expression appear to vary from one cell type to the other. 14-16
Recent investigations have revealed that nuclear receptors also interact with other nuclear proteins designated as coactivators and co-repressors and form macromolecular complexes that appear to modulate transcription. 17-21 It is highly feasible that these coactivators and co-repressors may act in an intricate way to modulate the extent of tissue/cell and species-specific physiological processes controlled by various nuclear receptors. In an effort to understand possible tissue- and species-specific differences in the transcriptional activity of PPAR isotypes, we initiated studies to identify cofactors that modulate PPAR transcriptional activity and identified mouse SRC-1 22 and PBP 23 as PPAR coactivators. Analysis of spatiotemporal expression patterns of these accessory molecules by in situ hybridization and immunohistochemical approaches will be necessary to gain insights into the PPAR-controlled lipid metabolism and adipocyte differentiation. In this report, we describe the expression patterns of PPARγ and its two activators SRC-1 and PBP in the mouse brown adipose tissue, urinary bladder, colon, and breast.
Materials and Methods
Animals
Male C57BL/6J mice were used in these experiments. Embryos were from timed mating, with stage E0.5 defined as noon of the day when the copulatory plug was observed. Mouse embryos or tissues from the adult mice were fixed in 4% paraformaldehyde for 16 to 20 hours at 4°C and paraffin embedded.
Probes
The sense and antisense riboprobes for in situ hybridization were derived from cDNAs as follows: PPAR (nucleotides 870 to 1390), 10 SRC-1 (nucleotides 1 to 530), 22 and PBP (nucleotides 1461 to 2217). 23
In Situ Hybridization
Sections 5 μm thick were cut under ribonuclease-free conditions. 24 Every 6th sagittal section for the embryos and every 10th section for the adult tissues were stained with hematoxylin and eosin. Each plasmid was linearized with appropriate restriction enzymes in the polylinker to generate sense and antisense riboprobes in the presence of α-S35-labeled UTP, (80 μCi, >1000 Ci/mmol; Amersham Corp., Arlington Heights, IL). Hybridization, washing, and developing conditions were as described by Wilkinson and Nieto. 25 The sections were exposed in NTB2 emulsion (Eastman Kodak, Rochester, NY) at 4°C for 10 days. Heochst 33258 (Boehringer Mannheim, Mannheim, Germany) was used to counterstain the nuclei. Visualization and photography were performed using epifluorescence and dark-field microscopy on a Zeiss Axiophot microscope or simple dark-field microscopy. Identical conditions were used for sense and antisense sections.
Results
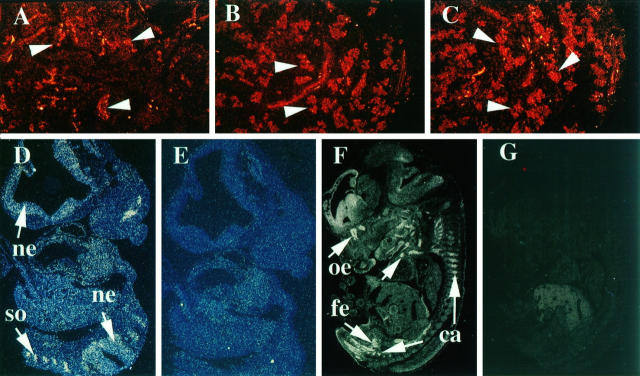
In the adult mouse, PPARγ was expressed abundantly in all cells of the transitional epithelium of the urinary bladder (Figure 1 ▶ , A and B). PBP expression was also noted in the urinary bladder, but this expression appeared to be prominent in the basal cells as compared with the differentiated umbrella cell layer (Figure 1 ▶ , C and F). SRC-1 expression in the urinary bladder was not appreciable. Transitional urothelium lining the ureter and renal pelvis also showed intense labeling for PPARγ. During the embryonic development, PPARγ expression appeared prominent in the urogenital sinus at E15.5 of embryogenesis (Figure 1 ▶ , D and E). Brown adipose tissue expressed PPARγ abundantly at stage E15.5 of embryogenesis along with PBP (Figure 1 ▶ , G to I), and this level of expression was also maintained in the adult brown fat (not shown). SRC-1 expression was not detected either in the embryonic or adult brown fat.
Figure 1.
Differential expression of PPARγ, PBP, and mSRC-1 mRNAs in bladder, brown fat, and colon by in situ hybridization. A to F: Expression of PPARγ and PBP in bladder. A and B: Expression of PPARγ in adult mouse bladder transitional epithelium at low (×5) and high (×20) power, respectively. C: Expression of PBP in the adult bladder transitional epithelium. D and E: antisense and sense panels, respectively, for PPARγ expression in the urogenital sinus of a stage E15.5 mouse. F: Hematoxylin and eosin-stained adult urinary bladder for histological reference. A to F, arrows: Transitional epithelium; lu, lumen of the bladder. G and H: High PPARγ and PBP expression in the E15.5 brown fat (arrows), respectively. I: Sense control for PPARγ showing background signal levels compared with G. J to O: Expression of PPARγ, PBP, and SRC-1 in the adult colon. J to L: Photographs taken at low power (×5). M to O: Photographs taken at high power (×20). J and M denote PPARγ expression (J, arrows, colonic villi; M, arrows, entire villus), K and N denote PBP expression, and L and O denote SRC-1 expression. The arrows in K, L, N, and O point to the crypts of the villus. In J to O, lu denotes the lumen of the colon.
In the colonic mucosa, PPARγ, SRC-1, and PBP displayed a limited extent of overlapping expression (Figure 1 ▶ , J to O). PPARγ expression appeared dynamic throughout the colonic mucosa, with intense labeling of all cells lining the villi (Figure 1 ▶ , J and M). In contrast, the expression of both PBP (Figure 1 ▶ , K and N) and SRC-1 (Figure 1 ▶ , L and O) appeared restricted almost exclusively to cells lining the crypts.
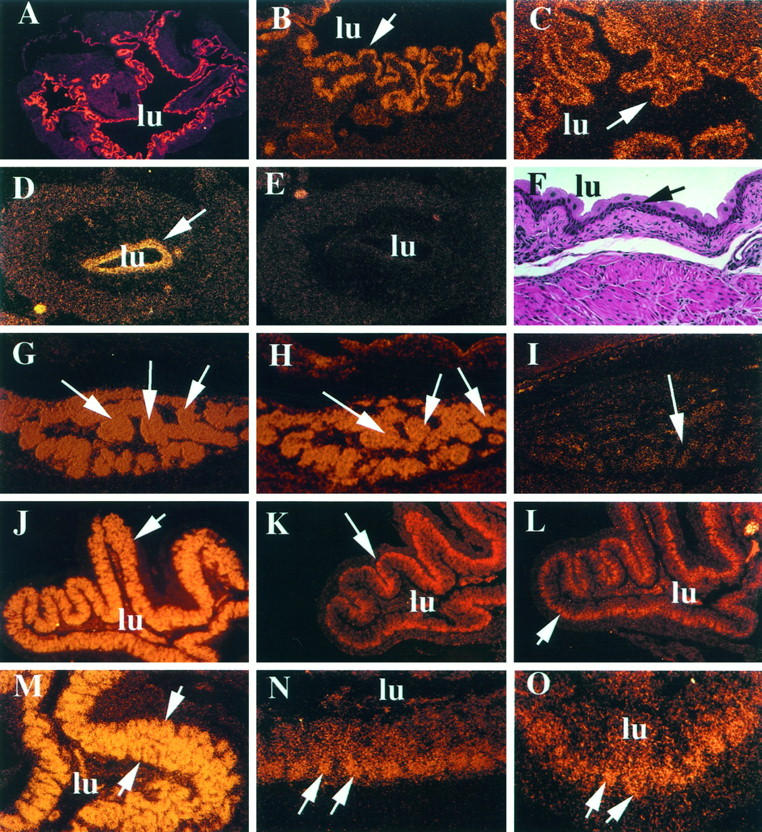
Another site of differential expression of PPARγ and its two coactivators was the mammary gland, in which PBP and SRC-1 transcripts were detectable in the virgin, pregnant, and lactating mouse mammary epithelium (Figure 2 ▶ , A to C). In the mammary gland of virgin mice, few ducts and acinar elements were present among the white adipose tissue, and these revealed the SRC-1 and PBP transcripts, but very little or no expression of PPARγ (Figure 2A ▶ and data not shown). During pregnancy, there was a remarkable proliferation of ductular and acinar structures, and these showed a prominent expression of both PBP and SRC-1 (Figure 2 ▶ , B and C). Lactating mammary epithelium also expressed both coactivators, and during this phase a modest level of expression of PPARγ was observed (not shown).
Figure 2.
Differential expression of PBP and mSRC-1 mRNAs in murine breast and SRC-1 expression during embryogenesis by in situ hybridization. A and B: PBP in normal and pregnant mouse breast, respectively. C: SRC-1 expression in pregnant breast. A to C, arrowheads, mammary gland epithelium. D and F: SRC-1 expression in E9.5 and E13.5 mouse embryos, respectively. ne, neuroepithelium; so, somites; oe, olfactory epithelium; fe, femur head; ca, cartilage primordium of the vertebral column. F, arrowhead, a developing rib. E and G are the corresponding sense controls showing background hybridization levels.
During murine embryogenesis, SRC-1 was detectable as early as stage E9.5 of development and displayed high expression in the somites and the neuroepithelium (Figure 2D) ▶ . This specific expression continued in stage E13.5 embryo (Figure 2F) ▶ with primordia of the developing vertebrae, femur head, and ribs showing high SRC-1 expression. Interestingly, olfactory epithelium also displayed high SRC-1 expression. The sense controls for E9.5 and E13.5 (Figure 2 ▶ , E and G, respectively) showed no significant signal. We were unable to detect any significant expression of PPARγ at these E9.5 and E13.5 embryogenic developmental time points (data not shown).
Discussion
PPARγ is an important member of the nuclear receptor superfamily. 7,9,10 It has been shown to regulate adipocyte differentiation and thus has been implicated as a key protein that may be involved in regulating biological responses, such as thermogenesis and fat metabolism. 7 Evidence also suggests that ligands for PPARγ are highly effective in reducing levels of inflammatory cytokines and other mediators of inflammatory responses generated by activated macrophages. 26,27 Furthermore, recent studies also demonstrate that PPARγ and its heterodimeric partner act synergistically in the conversion of monocytes into foam cells. 28,29 In the present study, we determined the mRNA expression patterns of PPARγ, SRC-1, and PBP in several mouse tissues by in situ hybridization to gain additional insights into the possible functional role of PPARγ at biological sites other than the adipose and lymphoid tissues. In addition, we are also interested in whether, at such biological sites, the PPARγ expression is coincident with the two coactivators, ie, SRC-1 and PBP. This information is essential in understanding which coactivators are biologically relevant in mediating PPARγ signaling. Our results demonstrate that, in the urinary bladder and brown fat, SRC-1 is not co-expressed with PPARγ in the adult and during embryogenesis, whereas PBP is spatiotemporally co-expressed with PPARγ in these two tissues (Figure 1 ▶ , A to I, and data not shown), thus suggesting that SRC-1 is perhaps not biologically relevant in PPARγ signaling at these sites. In addition, the expression of PPARγ and PBP in brown adipose tissue and urothelium during embryogenesis implies a possible role for these proteins in the differentiation of brown adipose tissue and transitional epithelium of the urinary bladder. Another important possibility is that PPARγ, which is activated by 15-deoxy-Δ 12,14 prostaglandin J2 which is present abundantly in urine, may serve as a regulator of inflammatory response in the urinary bladder. Because chronic inflammation appears to predispose the transitional epithelium of the urinary bladder mucosa to neoplastic conversion, it is reasonable to speculate that PPARγ, by virtue of its ability to inhibit inflammatory response and induce differentiation, may be important in the pathogenesis of bladder cancer.
An important finding of the current study is the differential expression of PPARγ, SRC-1, and PBP in the colonic mucosa. The expression of all three molecules appeared abundant in the cells lining the colonic crypts. PPARγ expression appeared robust in the differentiated enterocytes of the colon, whereas the expression of the two coactivators is confined mostly to the cells within the crypts of these villi (Figure 1 ▶ , J to O). It is possible that the co-expression of both coactivators in cells lining the crypts may be synergistically regulating the PPARγ activity in determining a differentiation commitment of the immature enterocytes. Thus, PPARγ may play a basal function in the undifferentiated enterocytes but possibly requires additional coactivators for its optimal functioning in mature intestinal epithelium. The relative paucity of SRC-1 and PBP in the differentiated epithelium lining the intestinal villi suggests the presence of additional coactivators of PPARγ in the mature epithelium of the villi. It will be of interest to determine whether PGC-1, 30 a recently discovered PPARγ coactivator that is cold-inducible in brown adipose tissue, is also expressed in the differentiated epithelium of the intestinal villi along with PPARγ. Strong RXRα expression has been reported in the intestinal mucosa, 31 which appears similar to PPARγ distribution described in the present study, suggesting a biological role for these two heterodimerization partners in this cell type.
On a similar note, both PBP and SRC-1 were also identified in the ductular epithelium of the nonlactating and pregnant breast tissue, in which PPARγ was only found at low levels (Figure 2 ▶ , A to C, and data not shown). Nonetheless, during lactation, PPARγ expression in the breast appeared prominent. From these findings, it is tempting to speculate that PBP and SRC-1 regulate ductular proliferation but require PPARγ for differentiation. Recently, SRC-1-null mice were shown to have defects in ductular branching of the breast epithelium. 32 SRC-1, and to some extent PBP, modulate the activity of other members of the steroid receptor family besides PPARγ. Thus, an understanding of their biological role is essential, especially during early development, because embryogenesis is under the tight control of different nuclear receptors.
We have previously shown that PBP is ubiquitously expressed in several mouse tissues as early as stage E9.5 of mouse gestation. Because PBP and SRC-1 both interact with several common nuclear receptors, we also investigated the developmental regulation of SRC-1 in the present study. Unlike PBP, SRC-1 expression appeared to be limited to neural tissue and the somites in the stage E9.5 embryo, thus suggesting that SRC-1 may be important in the differentiation of somites into cartilage and muscle. In this regard, its expression is coincident to RXRγ. 31 We were unable to detect significant expression of PPARγ in the somites. It will be of interest to ascertain whether PPARα and -δ isotypes are co-expressed with SRC-1. If this is true, then it is tempting to speculate that RXRγ and PPARα and PPARδ along with SRC-1 are the relevant players in regulating target genes in the somites. SRC-1 is also a member of the basic helix-loop-helix-PAS family of transcription factors, 33 and to date, its putative heterodimerization partner has not been identified. Thus, our results regarding its expression pattern allude to putative biological sites for this purpose.
Detailed analysis of the relative levels of co-expression of the three PPAR isotypes in a variety of tissues will be necessary in view of the increasing functional implications of these three receptors. It has been reported that in the rat, PPARα and PPARγ are co-expressed in the same cell types in the intestinal mucosa. 15 In the rabbit urinary tract, PPARβ and PPARγ are co-expressed in the transitional epithelium. 16 A limitation of our study is that it does not provide information regarding the protein expression pattern of these nuclear proteins. As appropriate reagents become available, such characterization would be valuable. Nevertheless, mRNA expression profiles are an important step toward beginning to understand the biology of transcription factors and co-factors. Although the PPARγ probe we used for in situ hybridization in this study was not designed to distinguish between PPARγ1 from PPARγ2, 10 these results provide important preliminary understanding of the differential expression of PPARγ and its two coactivators. Our study focused on selected tissues that have either been shown to be biologically regulated by PPARγ (eg, fat) or that demonstrated novel expression profiles for these transcription factors. The knowledge of the interplay between PPARs and accessory proteins becomes important in understanding the control of certain biological processes, especially during development, differentiation, and carcinogenesis. These results confirm and extend some of the previous findings on the expression of PPARγ 14-16 and provide novel expression profiles for its coactivators, SRC-1 and PBP.
Footnotes
Address reprint requests to Dr. Janardan K. Reddy, Department of Pathology, Northwestern University Medical School, 303 East Chicago Avenue, Chicago, IL 60611. E-mail: jkreddy@nwu.edu.
Supported by National Institutes of Health Grant GM-23750 (to JKR) and a Department of Veterans Affairs Merit Review Grant (to AVY and MSR).
References
- 1.Tolbert NE: Metabolic pathways in peroxisomes and glyoxisomes. Annu Rev Biochem 1981, 50:133-157 [DOI] [PubMed] [Google Scholar]
- 2.Reddy JK, Krishnakantha TP: Hepatic peroxisome proliferation: induction by two novel compounds structurally unrelated to clofibrate. Science 1975, 200:787-789 [DOI] [PubMed] [Google Scholar]
- 3.Reddy JK, Lalwani ND: Carcinogenesis by hepatic peroxisome proliferators: evaluation of the risk of hypolipidemic drugs and plasticizers to human. CRC Crit Rev Toxicol 1983, 12:1-58 [DOI] [PubMed] [Google Scholar]
- 4.Reddy JK, Goel SK, Nemali MR, Carrino JJ, Laffler TG, Reddy MK, Sperbeck SJ, Osumi T, Hashimoto T, Lalwani ND, Rao MS: Transcriptional regulation of peroxisomal fatty acyl-CoA oxidase and enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase in rat liver by peroxisome proliferators. Proc Natl Acad Sci USA 1986, 83:1747-1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Issemann I, Green S: Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature 1990, 347:645-650 [DOI] [PubMed] [Google Scholar]
- 6.Reddy JK, Chu R: Peroxisome proliferator-induced pleiotropic responses: pursuit of a phenomenon. Ann NY Acad Sci 1996, 384:176-201 [DOI] [PubMed] [Google Scholar]
- 7.Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM: mPPAR 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev 1994, 8:1224-1234 [DOI] [PubMed] [Google Scholar]
- 8.Dreyer C, Krey G, Keller H, Givel F, Helftenbein G, Wahli W: Control of the expression of peroxisomal β-oxidation pathway by a novel family of nuclear hormone receptors. Cell 1992, 68:879-887 [DOI] [PubMed] [Google Scholar]
- 9.Zhu Y, Alvares K, Huang Q, Rao MS, Reddy JK: Cloning of a new member of the peroxisome proliferator-activated receptor gene family from mouse liver. J Biol Chem 1993, 268:26817-26820 [PubMed] [Google Scholar]
- 10.Zhu Y, Qi C, Korenberg JR, Chen ZN, Noya D, Rao MS, Reddy JK: Structural organization of mouse peroxisome proliferator-activated receptor (mPPARγ) gene: alternative promoter use and different splicing yield two PPARγ isoforms. Proc Natl Acad Sci USA 1995, 92:4318-4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kliewer SA, Forman BM, Blumberg B, Ong ES, Borgmeyer U, Mangelsdorf DJ, Umesono K, Evans RM: Differential expression and activation of a family of murine peroxisome proliferator activated receptors. Proc Natl Acad Sci USA 1994, 91:7355-7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sher T, Yi HF, McBride OW, Gonzalez F: cDNA cloning, chromosomal mapping, and functional characterization of the human peroxisome proliferator activated receptor. Biochemistry 1993, 32:5598-5604 [DOI] [PubMed] [Google Scholar]
- 13.Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM: Convergence of 9-cis retinoic acid and peroxisome proliferator-activated signaling pathways through heterodimer formation of their receptors. Nature 1992, 358:771-774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang Q, Yeldandi AV, Alvares K, Ide H, Reddy JK, Rao MS: Localization of peroxisome proliferator activated receptor in mouse and rat tissues and demonstration of its nuclear translocation in transfected CV-1 cells. Int J Oncol 1995, 6:307-312 [DOI] [PubMed] [Google Scholar]
- 15.Braissant O, Foufelle F, Scotto C, Dauca M, Wahli W: Differential expression of peroxisome proliferator activated receptors (PPARs): tissue distribution of PPAR-α, -β, and -γ in the adult rat. Endocrinology 1996, 137:354-366 [DOI] [PubMed] [Google Scholar]
- 16.Guan Y, Zhang Y, Davis L, Breyer MD: Expression of peroxisome proliferator-activated receptors in urinary tract of rabbits and humans. Am J Physiol 1997, 273:F1013-F1022 [DOI] [PubMed] [Google Scholar]
- 17.Onate SA, Tsai SY, Tsai MJ, O’Malley BM: Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science 1995, 270:1354-1357 [DOI] [PubMed] [Google Scholar]
- 18.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin SC, Heyman RA, Rose DW, Glass CK, Rosenfeld MG: A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 1996, 85:403-414 [DOI] [PubMed] [Google Scholar]
- 19.Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, Rosenfeld MG: Ligand-dependent repression by the thyroid hormone receptor mediated by a nuclear receptor corepressor. Nature 1995, 377:397-404 [DOI] [PubMed] [Google Scholar]
- 20.Voegel JJ, Heine MJS, Zechel C, Chambon P, Gronemeyer H: TIF2, a 160 kd transcriptional mediator for the ligand-dependent activation function Af-2 of nuclear receptors. EMBO J 1996, 15:3667-3675 [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H, Lin RJ, Schlitz RL, Chakrabarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM: Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell 1997, 90:569-580 [DOI] [PubMed] [Google Scholar]
- 22.Zhu Y, Qi C, Calandra C, Rao MS, Reddy JK: Cloning and identification of mouse steroid receptor coactivator-1 (mSRC-1), as a coactivator of peroxisome proliferator activated receptorγ. Gene Exp 1996, 6:185-195 [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Y, Qi C, Jain S, Rao MS, Reddy JK: Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J Biol Chem 1997, 72:25500-25506 [DOI] [PubMed] [Google Scholar]
- 24.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K: Current Protocols in Molecular Biology. 1990, Vol 1. Green Publishing Associates and Wiley-Interscience New York
- 25.Wilkinson DG, Nieto MA: Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol 1993, 225:361-373 [DOI] [PubMed] [Google Scholar]
- 26.Jiang C, Ting AT, Seed B: PPAR-γ agonists inhibit production of monocyte inflammatory cytokines. Nature 1998, 391:82-86 [DOI] [PubMed] [Google Scholar]
- 27.Ricote M, Li AC, Willson TM, Kelly CJ, Glass CK: The peroxisome proliferator-activated receptor is a negative regulator of macrophage activation. Nature 1998, 391:79-82 [DOI] [PubMed] [Google Scholar]
- 28.Tontonoz P, Nagy L, Alvarez JGA, Thomazy VA, Evans RM: PPARγ promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell 1998, 93:241-252 [DOI] [PubMed] [Google Scholar]
- 29.Nagy L, Tontonoz P, Alvarez JGA, Chen H, Evans RM: Oxidized LDL regulates macrophage gene expression through ligand activation of PPARγ. Cell 1998, 93:229-240 [DOI] [PubMed] [Google Scholar]
- 30.Puigserver P, Wu Z, Cheol-Won P, Graves R, Wright M, Spiegelman BM: A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 1998, 92:829-839 [DOI] [PubMed] [Google Scholar]
- 31.Mangelsdorf DJ, Borgmeyer U, Heyman RA, Yang-Zhou J, Ong ES, Oro AE, Kakizuka A, Evans RM: Characterization of three genes that mediate the action of 9-cis retinoic acid. Genes Dev 1992, 6:329-344 [DOI] [PubMed] [Google Scholar]
- 32.Xu J, Qiu Y, DeMayo FJ, Tsai SY, Tsai M, O’Malley BW: Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science 1998, 279:1922-1925 [DOI] [PubMed] [Google Scholar]
- 33.Schmidt JV, Bradfield CA: Ah receptor signaling pathways. Annu Rev Cell Dev Biol 1996, 12:55-89 [DOI] [PubMed] [Google Scholar]