Abstract
The enzymatic breakdown of sphingomyelin by sphingomyelinases is considered the major source of the second messenger ceramide. Studies on the contribution of the various described acidic and neutral sphingomyelinases to the signaling pool of ceramide have been hampered by the lack of molecular data on the neutral sphingomyelinases (nSMases). We recently identified a mammalian nSMase, an integral membrane protein with remote similarity to bacterial sphingomyelinases. However, its ubiquitous expression pattern is in contrast to previous findings that sphingomyelinase activity is found mainly in brain tissues. By using an improved database search method, combined with phylogenetic analysis, we identified a second mammalian nSMase (nSMase2) with predominant expression in the brain. The sphingomyelinase activity of nSMase2 has a neutral pH optimum, depends on Mg2+ ions, and is activated by unsaturated fatty acids and phosphatidylserine. Immunofluorescence reveals a neuron-specific punctate perinuclear staining, which colocalizes with a Golgi marker in a number of cell lines. The likely identity of nSMase2 with cca1, a rat protein involved in contact inhibition of 3Y1 fibroblasts, suggests a role for this enzyme in cell cycle arrest. Both mammalian nSMases are members of a superfamily of Mg2+-dependent phosphohydrolases, which also contains nucleases, inositol phosphatases, and bacterial toxins.
Keywords: sequence analysis, generalized profiles, Ccr4, cytolethal distending toxin, three-dimensional structure
Regulated sphingomyelin hydrolysis, triggered by various inducers of cellular stress, is thought to be the major pathway for the generation of the lipid second messenger molecules ceramide, sphingosine, and sphingosine 1-phosphate (1, 2). A number of different sphingomyelinase activities have been described in mammalian tissues (1). These isoenzymes are thought to transmit different signals and to give rise to different pools of ceramide, eliciting cellular responses ranging from apoptosis and cell cycle arrest to cell survival and cell proliferation (3–5).
Several neutral sphingomyelinase activities have been described in mammalian tissues and cell lines (1, 2). The most prominent one, the membrane-bound Mg2+-dependent sphingomyelinase activity, is found predominantly in the brain and, to a lesser extent, in other tissues (6–9). Because this activity is held responsible for the stress-induced ceramide generation and for relaying anti-apoptotic signals from cell surface receptors (2, 10), molecular data on the corresponding protein(s) are highly desirable. Studies addressing the role of neutral sphingomyelinase (nSMase)-generated ceramide in the stress response and in apoptosis had to rely on bacterial sphingomyelinases or on externally applied ceramide mimetics. The recent debate on the interpretation of those experiments (4, 11, 12) underscores the need to work with the genuine mammalian nSMase in its native environment.
By using a bioinformatics-based gene discovery approach, we recently identified a mammalian nSMase, an integral membrane protein with remote similarity to the secreted bacterial sphingomyelinases (9). However, its ubiquitous expression pattern was in contrast to the restricted localization of the bulk of mammalian nSMase activity. This discrepancy prompted us to search for alternative nSMases. By using an improved database search method combined with phylogenetic analysis, we identified a brain-specific nSMase with a different domain structure and only marginal sequence similarity to other SMases.
Materials and Methods
Sequence Analysis.
Profile searches and template-guided multiple alignments were done with the pftools v2.1 package (P. Bucher, available by ftp from ftp.isrec.isb-sib.ch) as described previously (13). Dendrogram analysis was done with the phylip package, by using the neighbor-joining algorithm (14). For prediction of membrane-spanning helices, the tmpred service was used (http://www.ch.embnet.org/software/TMPRED_form.html).
Cloning and Expression of Mouse and Human nSMase2 cDNA.
Poly(A)+ RNA was extracted from intestine of CD1 mice by using affinity purification on oligo(dT)-cellulose, according to the manufacturer's protocol (Roche Molecular Biochemicals). The coding region of mouse nSMase2 cDNA was amplified from the poly(A)+ mRNA with reverse transcription (RT)–PCR using the primer pair 5′-gcg gcc gcg caa tgg ttt tgt aca cga ccc cct ttc-3′/5′-tct aga cta cgc ctc ctc ttc ccc tgc aga cac c-3′. Restriction sites appended to the oligonucleotides are depicted in italics. The coding region of human nSMase2 cDNA was amplified from a human cDNA plasmid library (pCMV Sport; GIBCO). nSMase2 cDNAs were cloned into the eukaryotic expression vector pRc/CMV (Stratagene) using the restriction sites introduced by the PCR primers (NotI/XbaI). Stable transfection of HEK 293 was performed as described (15).
Measurement of SMase Activity.
nSMase activity was determined as described (15) with [N-methyl-14C]sphingomyelin (SPM) as substrate. Putative effectors of nSMase2 activity were added directly to the mixture before starting the assay. Phospholipids and fatty acids were purchased from Sigma.
Cell Culture.
Human embryonic kidney cells (HEK 293), rat adrenal pheochromocytoma cells (PC-12), and human neuroblastoma cells (SH-SY5Y) were purchased from the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany) and cultivated as described by the supplier.
Generation of Polyclonal Antibodies.
A truncated murine nSMase (mnSMase)2-cDNA construct coding for amino acids 310–655 was fused to a C-terminal 6xHis-tag derived from the plasmid vector pcDNA3.1/Myc-His (Invitrogen) and expressed in Escherichia coli [BL21(DE3)pLysS] using the pET-expression system (Novagen). Before immunization of rabbits, recombinant protein was extracted from the bacterial inclusion bodies and purified using metal chelating Talon columns (CLONTECH) following the manufacturer's instructions. Recombinant proteins were coupled to CNBr-activated Sepharose FF (Amersham Pharmacia) and used for affinity purification of nSMase2-directed antibodies, according to the manufacturer's instructions.
Immunoblotting.
Crude membrane extracts from wild-type, stably mock-, and mnSMase2-transfected cells, as well as from different tissues of 3-mo-old CD1 mice, were separated by SDS/PAGE and transferred onto a nitrocellulose membrane (Schleicher & Schuell) by semidry blotting. After blocking, the membrane was incubated with purified anti-mnSMase2 antibody (1 μg/ml) for 1 h. After various washing steps, the blot was incubated with an anti-rabbit IgG/horseradish peroxidase conjugate (Sigma) and then washed several times. Bands were visualized by chemiluminescence staining using Lumi-Light Western Blotting Substrate (Roche Molecular Biochemicals).
Northern Blot Analysis.
mRNA expression analysis was performed by using a commercially available human multitissue Northern blot (CLONTECH). As hybridization probe, a fragment corresponding to the complete coding region of the human nSMase2 cDNA was used and labeled by the random-primed labeling method using [α-32P]dATP nucleotides. Signal intensities were quantified with a phosphorimager (Packard).
Immunofluorescence Microscopy.
Cells were cultured on collagen-covered glass chamber slides and fixed with 4% paraformaldehyde in PBS or with ice-cold methanol/acetone (1:1, vol/vol) and lysed with PBS/0.5% Triton X-100. After blocking with PBS/3% BSA, cells were treated with the purified anti-mnSMase2 antibody (1 μg/ml) and different organelle-specific antibodies: mouse monoclonal anti-Golgi 58-kDa protein (clone 58K9; Sigma), mouse monoclonal anti-grp78 (BiP; StressGen Biotechnologies, Victoria, Canada), goat polyclonal anti-caveolin-2 (N-20; Santa Cruz Biotechnology) or mouse monoclonal anti-Na+/K+-ATPase (β1-subunit, IEC 1–48; kindly provided by A. Quaroni, Cornell University, Ithaca, NY) at 4°C overnight. After washing with PBS/0.5% Triton X-100, cells were incubated with a Cy3-conjugated anti-rabbit IgG antibody (Jackson ImmunoResearch) and a Cy2-conjugated anti-mouse or anti-goat IgG antibody (Jackson ImmunoResearch) for 1 h at 37°C. After washing with PBS/0.5% Triton X-100, cells were analyzed with a Leica confocal microscope.
After removal of the paraffin and rehydration, paraformaldehyde-fixed coronal slices of total brain from 1-mo-old CD1 mice were lysed by incubation in 0.5% Triton X-100/PBS and blocked with 3% goat serum/PBS. The staining procedure was the same as described above, except the concentration of the purified anti-mnSMase2 antibody was 4 μg/ml. Slices were analyzed with a fluorescence microscope (Zeiss) or a confocal microscope (Leica).
Results
nSMase2 Is a Member of the Superfamily of Mg2+-Dependent Phosphohydrolases.
Because all biochemical attempts to isolate nSMases from mammalian brain tissues failed to yield a sufficient amount of pure protein, we used an approach based on highly sensitive sequence comparisons. Profile-based sequence database searches offer increased sensitivity, as compared with sequence-based searches, because the query profiles exploit the additional information contained in multiple alignments of related sequences (16, 17). The generalized profile method (18), an extension of the original sequence profile method (19), allows the reliable determination of statistical significance (20).
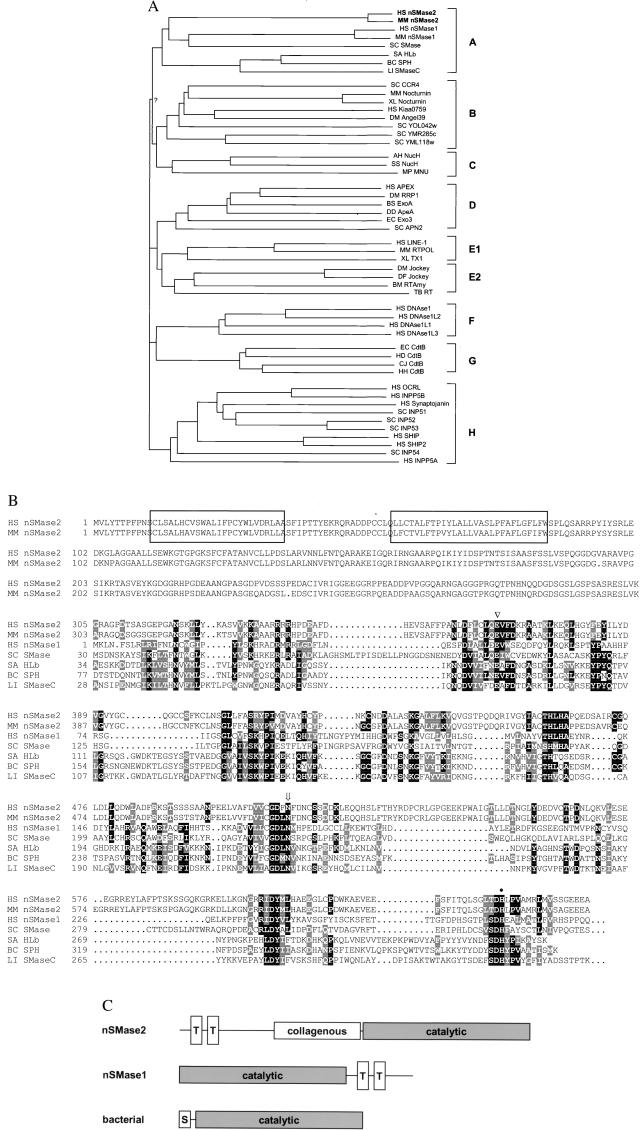
The ubiquitously expressed nSMase1 had originally been found as the best-matching eukaryotic sequence to a profile derived from the catalytic domain of bacterial SMases. Iterative refinement of the original profile by including newly found sequences into the profile generation process allowed us to identify a large enzyme superfamily. Although the enzymatic reactions present in the superfamily are heterogeneous (nSMases, exonucleases, endonucleases, inositol polyphosphate 5-phosphatases), all enzymes hydrolyze phosphate bonds and require Mg2+ or other divalent cations. A “Neighbor-Joining” dendrogram (14) shows the subfamily relationships within the superfamily (Fig. 1A).
Figure 1.
Sequence analysis of nSMase2. (A) Neighbor-joining dendrogram for representative members of the phosphohydrolase superfamily. All bifurcations are supported by more than 400 of 1,000 bootstrap samples, except for the one labeled “?”. The letters on the right represent the subfamilies, as described in Discussion. Species abbreviations are as follows: HS, human; MM, mouse; SC, Saccharomyces cerevisiae; SA, Staphylococcus aureus; BC, Bacillus cereus; LI, Leptospira interrogans; XL, Xenopus laevis; DM, Drosophila melanogaster; AH, Aeromonas hydrophila; SS, Synechocystis sp.; MP, Mycoplasma pulmonis; BS, Bacillus subtilis; DD, Dictyostelium discoideum; EC, Escherichia coli; BM, Bombyx mori; TB, Trypanosoma brucei; HD, Haemophilus ducreyi; and CJ, Campylobacter jejuni. In cases where no systematic gene nomenclature is available, the following abbreviations were used: SC SMase, yeast sphingomyelinase Yer019w; HLb, β-hemolysin; RRP1, recombination repair protein 1; Exo3, exonuclease III; LINE-1, LINE-1 reverse transcriptase; TX1, transposon TX1 ORF2; RTAmy, reverse transcriptase of Amy transposon; NucH, extracellular nuclease H; MNU, membrane nuclease A; CtdB, cytolethal distending toxin B-subunit. (B) Alignment of human and murine nSMase2 with representative other SMases. Sequence numbering is relative to unprocessed protein. The Mg2+-complexing glutamic acid (▿), the asparagine involved in substrate binding (arrow), and the general base histidine (●) are indicated above the sequence. The two transmembrane regions of nSMase2 are boxed. (C) Schematic diagram of SMase domain structure. Open boxes labeled “T” or “S” represent putative transmembrane or signal sequences, respectively.
We reasoned that the missing brain-specific isoform of Mg2+-dependent nSMase might be a member of this superfamily, which, in addition to the enzymes listed above, contains many sequences with unknown functionality. Although the dendrogram (Fig. 1A) shows a robust clustering of bacterial and eukaryotic nSMases, no additional uncharacterized protein appears in this subfamily (labeled “A” in Fig. 1A). Expressed sequence tag (EST) data (21), a rich source for sequence-based gene discovery, frequently contain truncated coding regions, which are riddled by frameshift-inducing sequencing errors. As a consequence, not all of the highly conserved motifs defining a sequence superfamily are present in the same reading frame of a target sequence. To overcome this restriction, we applied two additional methods: (i) the estscan program corrects frameshift errors by detecting irregularities in the coding potential (22), and (ii) the 2ft program of the pftools package allows frameshift-tolerant general profile searches. Both methods independently yielded a number of additional superfamily members, which had been missed previously because important sequence motifs are found spread over different reading frames. One sequence, represented by a mRNA from rat and several ESTs, was of particular interest because the “corrected” ORF clustered with the nSMase subfamily (nSMase2 in Fig. 1A).
We cloned the cDNA coding for the human and murine proteins and verified that, in both sequences, all necessary sequence motifs were present in one reading frame. An alignment of the two nSMase2 sequences with nSMase1 and representative bacterial sphingomyelinases is shown in Fig. 1B. Both the human and murine ORFs encode proteins of 655 aa, resulting in a predicted molecular mass of 71 kDa. The N terminus contains two hydrophobic regions that are predicted to span the membrane, whereas the C terminus contains the putative catalytic domain matched by the superfamily profile. Situated between the membrane-anchoring domain and the catalytic domain is a 200-residue linker region with the potential to form collagen-like triple helices. The domain arrangement of nSMase2 is different from that found in nSMase1, which contains two C-terminal transmembrane helices, or the bacterial SMases, which are not membrane-anchored, but contain a signal sequence for secretion (Fig. 1C). The sequence similarity to other SMases is restricted to the catalytically important motifs. Overall sequence identity of the catalytic domain to human nSMase1 is 23%, whereas identity to bacterial SMases is in the range of 18–21%. Nevertheless, the statistical significance of belonging to the Mg2+-dependent phosphohydrolase superfamily is P < 0.001, underscoring the superior sensitivity of profile searches.
Characterization of nSMase2 Enzymatic Activity.
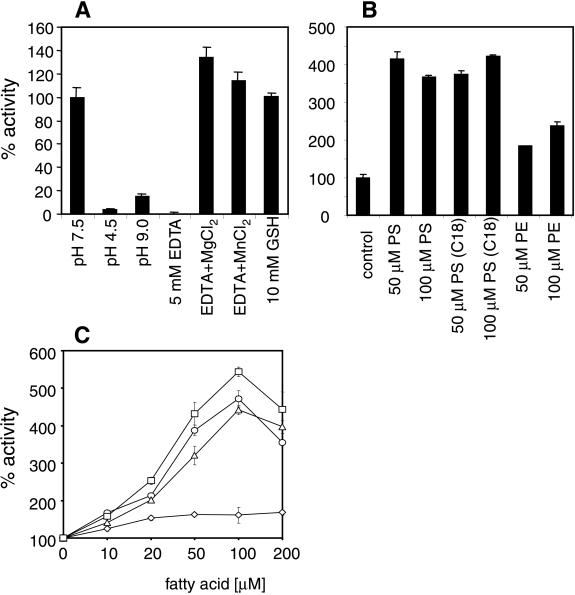
Enzymatic characterization of the nSMase2 protein was performed by transient expression in HEK 293 cells and in stable nSMase2-overexpressing HEK 293 cell lines. Enzymatic assays of membrane extracts showed a strong increase of the nSMase activity compared with mock-transfected cells. Various cell lines exhibited specific nSMase activities between 50 and 200 nmol of SPM per mg of protein per h corresponding to a 3- to 40-fold overexpression compared with the endogenous HEK 293 cell nSMase activity (5–15 nmol of SPM per mg of protein per h). nSMase2 specifically hydrolyzed SPM and showed no detectable phospholipase C-like activity against phosphatidylcholine, lysophosphatidylcholine, platelet-activating factor (PAF), or lyso-PAF in the micellar assay system (data not shown). Its enzymatic activity revealed a neutral pH optimum with no detectable substrate cleavage at acidic or alkaline conditions (Fig. 2A). Murine nSMase2 activity was inhibited by the metal-chelating reagent EDTA and completely restored by the subsequent addition of the divalent cations Mg2+ and Mn2+ (Fig. 2A). The addition of reducing agents such as DTT stabilized the enzymatic activity during preparation. Interestingly, nSMase2 showed a significant stimulation by unsaturated fatty acids (Fig. 2C) similar to previous observations of arachidonic acid stimulation of nSMase activity in HL60 cells (23). The phospholipid phosphatidylserine (PS) has been described to stimulate purified brain-specific Mg2+-dependent nSMase specifically up to 20-fold at PS concentrations of 15 mol% (8). SMase2 activity in crude membrane extracts of stably transfected HEK 293 cells was stimulated 4-fold at concentrations of 50 μM PS, corresponding to 6.5 mol% (Fig. 2B). Higher concentrations of the lipid or the use of dioleoyl-PS instead of bovine brain PS did not cause a further increase in SPM hydrolysis. The structurally related phospholipid phosphatidylethanolamine exhibited a minor stimulatory effect on the enzyme activity (Fig. 2B). Thus, the enzymatic properties of nSMase2 are similar to those of nSMase1, except for its stimulation by PS. They also match the characteristics of the membrane-bound, Mg2+-dependent “signaling sphingomyelinase” activity found mainly in the brain but also in other tissues and cell lines (1).
Figure 2.
Enzymatic properties of murine nSMase2. (A) pH optimum and ion requirement of murine nSMase2 from membrane extracts of stably transfected HEK 293 cells. Control assay mixtures contain 5 mM MgCl2, 0.05% Triton X-100, 5 mM DTT, 100 mM Tris⋅Cl at pH 7.4, and a protease inhibitor mixture (1× Complete without EDTA; Roche Molecular Biochemicals). Measurements with EDTA and metal ions were done with 5 mM EDTA and 10 mM of the respective metal ion. (B) Influence of phospholipids on nSMase2 activity. Phospholipids were dissolved in 0.05% Triton X-100 by sonication and premixed with the assay components before adding the enzyme preparation. Results represent mean values (±SD) from three independent measurements. PS, phosphatidylserine; PS(C18), dioleoyl-PS; PE, phosphatidylethanolamine. (C) Activation of nSMase2 activity by unsaturated fatty acids. Fatty acids, dissolved in EtOH at various concentrations, were added directly to the assay mixture. ⋄, Stearic acid; ▵, linolenic acid; ○, linoleic acid; and □, arachidonic acid.
Tissue Specificity and Subcellular Localization of nSMase2.
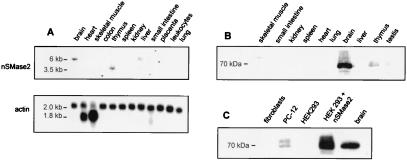
To address the question of whether nSMase2 is the elusive nSMase of mammalian brain tissue, we analyzed the localization of nSMase2 message and protein product both on the tissue and subcellular level. Northern blot analysis using poly(A)+ RNA from multiple tissues and a probe from the coding region revealed a moderate 6-kb signal in brain, and a weak signal in liver (Fig. 3A). A weak signal at the smaller size of 3.5 kb was observed in thymus RNA. The enrichment of nSMase2 in brain was even more pronounced on the protein level (Fig. 3B). In Western blot experiments, a polyclonal antibody against the cytoplasmic portion of murine nSMase2 yielded a strong brain-specific signal at 70 kDa, the same size as observed in cell lines overexpressing recombinant nSMase2 (Fig. 3C). Untransfected murine embryonal fibroblasts or human HEK 293 cells did not show a Western blot signal, whereas the rat pheochromocytoma cell line PC-12 exhibits a double band at 70 kDa (Fig. 3C).
Figure 3.
Expression of nSMase2 mRNA and protein. (A) Northern blot analysis of poly(A)+ mRNA derived from various human tissues, using a fragment containing the coding region of the murine nSMase2 cDNA as hybridization probe. The membrane was rehybridized with a β-actin probe for standardization. (B) Western blot analysis with protein extracts from different murine tissues, using the purified anti-mouse nSMase2 antibody in a concentration of 1 μg/ml. (C) Western blot analysis of murine embryonal fibroblasts, rat PC-12 cells, mock-transfected and nSMase2-transfected HEK 293 cells, and mouse brain extract, using the anti-mouse nSMase2 antibody at 1 μg/ml. The only specific signal is a single or double band at 70 kDa.
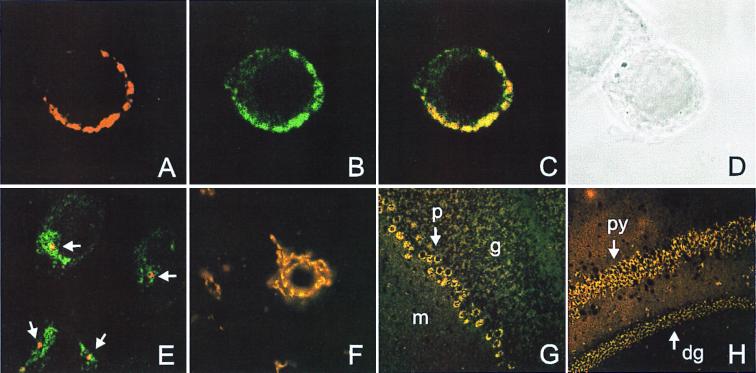
To address the question which cells in the brain express nSMase2, we performed immunofluorescence microscopy with sections from murine brain regions and confocal microscopy with human neuroblastoma (SH-SY5Y) and undifferentiated rat pheochromocytoma (PC-12) cells. (Fig. 4). The latter show an inhomogeneous intensive nSMase2 immunofluorescence overlapping with the Golgi-marker 58-kDa protein (Fig. 4 A–D). No colocalization with markers for plasma membrane (Na+/K+-ATPase), endoplasmic reticulum (BiP), or caveolae (caveolin2) was observed. Interestingly, the nSMase2 staining of SH-SY5Y and HEK 293 (not shown) cells revealed a single discrete focus of nSMase2 staining within, but only partially overlapping with, the area stained by the Golgi marker (Fig. 4E). The staining intensity correlates well with the endogenous nSMase activity of these cell lines, being 5–6 times higher in PC-12 cells as compared with HEK 293 and SH-SY5Y cells.
Figure 4.
Immunofluorescence analysis. (A–D) Localization of nSMase2 protein in the Golgi apparatus of rat PC-12 cells. Cells were costained with anti-mnSMase2 and anti-Golgi 58-kDa protein antibodies and the corresponding Cy2- and Cy3-conjugated secondary antibodies and analyzed by confocal microscopy. (A) Anti-mnSMase2-Cy3. (B) Anti-58-kDa-Cy2. (C) Superposition of the Cy2 and Cy3 channels. Yellow indicates colocalization. (D) Corresponding light-microscopic image. (E) Confocal microscopic image of SH-SY5Y cells costained with anti-mnSMase2 (red) and anti-58-kDa (green) antibodies. Note the distinct punctate staining of the nSMase2 protein (indicated by arrows) within the Golgi apparatus. (F–H) Immunofluorescence of nSMase2 in coronal slices of total mouse brain is restricted to neurons. (F) Cerebellar neurons stained with anti-mnSMase2 antibody. (G) nSMase2 staining pattern in the cerebellum. Labels: p, Purkinje cell layer; g, granular layer; m, molecular layer. (H) nSMase2 staining of pyramidal cells (py) and dentate gyrus granular layer (dg).
In brain sections, immunofluorescence is restricted to neurons and especially prominent in large cells, including Purkinje cells (Fig. 4G), pyramidal cells, neurons of the dentate gyrus granular layer (Fig. 4H), and neurons in the pontine nuclei. Other stained regions include the hypothalamic nuclei (Fig. 4 B and C), neurons in the piriform cortex, and nuclei of the brainstem (not shown). All nSMase2-positive cells were negative for astrocyte- or oligodendrocyte-specific markers such as glial fibrillary acidic protein and myelin basic protein, respectively (not shown). nSMase2 staining in neurons appears as a perinuclear punctate staining of internal membranes (Fig. 4F). In all immunofluorescence studies, the nucleus is excluded from nSMase2 immunoreactivity, demonstrating that nSMase2 is unlikely to be responsible for the nSMase activity observed in chromatin (24).
Discussion
Although the enzymatic properties of the previously reported nSMase1 closely matched those of the Mg2+-dependent nSMase activity of mammalian brain, three factors make it unlikely that the activities are identical: (i) nSMase1 is ubiquitously expressed, (ii) it is not stimulated by PS, (iii) in immunofluorescence experiments, nSMase1 mainly colocalizes with markers of the endoplasmic reticulum, and (iv) brain-derived nSMase activity is resistant to precipitation by an anti-nSMase1 antibody that precipitates nSMase activity from other tissues (15). We report here the identification, cloning, and molecular characterization of mouse and human brain-specific neutral sphingomyelinase nSMase2, an enzyme belonging to a large family of Mg2+-dependent phosphohydrolases with neutral pH optimum.
Several features of nSMase2 suggest that this protein is responsible for the high Mg2+-dependent nSMase activity of mammalian brain. The nSMase2 protein is present in the brain in significant amounts (Fig. 3) and contains membrane-spanning regions. The nSMase2 enzymatic activity has a neutral pH optimum, depends on Mg2+ or Mn2+ ions, and is activated by unsaturated fatty acids and PS (Fig. 2). The hydrolyzing activity is specific for SPM: no effect on structurally related lipids was observed. These properties are hallmarks of the neutral brain SMase, as described earlier (7, 8, 25). The molecular mass of nSMase2 is 71 kDa, which is compatible with the estimated molecular mass of 150 kDa in Triton-X100 micellar solution, given that the Triton micelles contribute approximately 90 kDa to the apparent mass (8). The nSMase activity observed in other tissues is probably because of a combination of the ubiquitous nSMase1, the brain-specific nSMase2, and possibly other as yet undefined nSMases.
The question of whether nSMase2 also accounts for the nSMase activity localized at the plasma membrane is equivocal. Localization of the nSMase activity to the plasma membrane has been done in liver (26), a tissue where nSMase2 is expressed at very low levels. Localization studies in cultivated cells indicate a localization in a subcompartment of the Golgi apparatus. PC-12 cells show an essential overlap of nSMase2 staining with a Golgi marker. By contrast, HEK 293 cells and SY5Y cells show a single discrete focus of nSMase2 staining contained within the area stained by the Golgi marker. Further experiments with markers specific for Golgi subcompartments will be necessary to fully elucidate the localization of nSMase2 in those cells.
The nSMase2 protein, as well as nSMase1 and the bacterial SMases, belong to a large superfamily of phosphohydrolases, most likely sharing a common three-dimensional fold and a common reaction mechanism. The enzymatic reactions catalyzed by members of this superfamily are diverse. The dendrogram analysis (Fig. 1A) shows that the superfamily can be subdivided into eight major subfamilies, where members within each subfamily (A–H) show considerable sequence similarity and catalyze similar reactions.
Subfamily A comprises both prokaryotic and eukaryotic SMases. Subfamily B is an interesting group of proteins acting as transcriptional repressors, with the budding yeast protein Ccr4p being the most prominent member. The mode of action of these proteins is not known at present, but conservation of the active site residues and position in the dendrogram suggest a role as nucleases. Subfamily C is formed by a small family of bacterial-secreted nucleases. Subfamily D comprises the AP-endonucleases, a family of repair enzymes cutting DNA at abasic sites, as well as some related bacterial nucleases, including exonuclease III. Subfamily E contains endonucleases found in retrotransposons, typically in combination with reverse transcriptases. This subfamily can be further subdivided into the vertebrate LINE-1 type retrotransposons and the insect jockey-type retrotransposons, which share only little sequence similarity. Subfamily F is formed by the mammalian DNase 1-type secreted nucleases. Subfamily G contains the B subunits of the bacterial cytolethal distending toxin (cdt) family. cdt is an AB-type toxin with a lectin-like A chain distantly related to ricin (K.H., unpublished observation) mediating the cellular entry of cdtB. Intracellular cdtB causes a G2 arrest of the cell cycle by an unknown mechanism. Again, the position in the dendrogram suggests a phosphohydrolase, most likely a nuclease activity. Subfamily H comprises a large family of inositol-polyphosphate 5-phosphatases. Among the well-characterized members of this family are synaptojanin and the OCRL gene product.
For exonuclease III, DNase 1, and the AP-nuclease HAP1, three representative members of the superfamily, the three-dimensional structure has been determined by x-ray crystallography (27–29). Similar to the nSMase activity, these enzymes require Mg2+ or Mn2+ ions, a property most likely shared by other members of the superfamily as well. The residues required for the coordination of the divalent ions and for the hydrolysis reaction are totally invariant throughout the superfamily.
An interesting aspect is the probable identity of nSMase2 with the rat Cca1 protein (30), which had been identified in a screen for genes involved in contact inhibition of rat 3Y1 fibroblasts (31). The Cca1 mRNA was found to be undetectable in growing 3Y1 cells, but was significantly up-regulated on growth arrest triggered by confluence. This up-regulation was not found in variant 3Y1 cells, which had lost contact inhibition. By ectopic expression of Cca1, the confluence-induced growth arrest could be restored in those cells (31). The published sequence for Cca1 (accession no. AB000215) differs from nSMase2 in the C-terminal region, which lacks the highly conserved “T/S-D-H” motif found in all members of the superfamily. However, our frameshift-tolerant database search methods picked up the database sequence, because the missing motif is present in the DNA sequence on a different reading frame. We amplified the corresponding region using rat brain RNA as a template and obtained a fragment with the T/S-D-H motif being in-frame. The most likely explanation is a sequencing error in the original Cca1 sequence, although the presence of a frameshifted, nonfunctional version of Cca1 in fibroblasts cannot be excluded. The biological significance of a putative role of nSMase2 in cell cycle arrest remains to be established. Our preliminary experiments in murine 3T3 fibroblasts did not show an increased nSMase expression on confluence (data not shown). It is possible that the reported effect is specific for 3Y1 cells, or that the 3T3 cells used lack a ceramide-responsive factor important for cell cycle arrest. A role of ceramide in cell cycle regulation by causing a G1 arrest has been proposed before (32, 33). Further experiments are needed to address this interesting perspective.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft, Project Sto32/36-1 (to W.S.) and by the Federal Ministry of Education, Science, Research and Technology (BMBF), Project 0311-686 and Project 01 KS 9502 in the Interdisciplinary Center for Molecular Medicine, Cologne (ZMMK), Project 24.
Abbreviations
- SPM
sphingomyelin
- SMase
sphingomyelinase
- nSMase
neutral SMase
- mnSMase
murine nSMase
- PAF
platelet-activating factor
- EST
expressed sequence tag
- RT
reverse transcription
- PS
phosphatidylserine
- cdt
cytolethal distending toxin
Footnotes
References
- 1.Levade T, Jaffrezou J P. Biochim Biophys Acta. 1999;1438:1–17. doi: 10.1016/s1388-1981(99)00038-4. [DOI] [PubMed] [Google Scholar]
- 2.Liu B, Obeid L M, Hannun Y A. Semin Cell Dev Biol. 1997;8:311–322. doi: 10.1006/scdb.1997.0153. [DOI] [PubMed] [Google Scholar]
- 3.Hannun Y A. Science. 1996;274:1855–1859. doi: 10.1126/science.274.5294.1855. [DOI] [PubMed] [Google Scholar]
- 4.Hofmann K, Dixit V M. Trends Biochem Sci. 1998;23:374–377. doi: 10.1016/s0968-0004(98)01289-4. [DOI] [PubMed] [Google Scholar]
- 5.Kolesnick R N, Krönke M. Annu Rev Physiol. 1998;60:643–665. doi: 10.1146/annurev.physiol.60.1.643. [DOI] [PubMed] [Google Scholar]
- 6.Das D V, Cook H W, Spence M W. Biochim Biophys Acta. 1984;777:339–342. doi: 10.1016/0005-2736(84)90437-1. [DOI] [PubMed] [Google Scholar]
- 7.Spence M W, Burgess J K. J Neurochem. 1978;30:917–919. doi: 10.1111/j.1471-4159.1978.tb10804.x. [DOI] [PubMed] [Google Scholar]
- 8.Liu B, Hassler D F, Smith G K, Weaver K, Hannun Y A. J Biol Chem. 1998;273:34472–34479. doi: 10.1074/jbc.273.51.34472. [DOI] [PubMed] [Google Scholar]
- 9.Tomiuk S, Hofmann K, Nix M, Zumbansen M, Stoffel W. Proc Natl Acad Sci USA. 1998;95:3638–3643. doi: 10.1073/pnas.95.7.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiegmann K, Schütze S, Machleidt T, Witte D, Krönke M. Cell. 1994;78:1005–1015. doi: 10.1016/0092-8674(94)90275-5. [DOI] [PubMed] [Google Scholar]
- 11.Kolesnick R, Hannun Y A. Trends Biochem Sci. 1999;24:224–225. doi: 10.1016/s0968-0004(99)01408-5. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann K, Dixit V M. Trends Biochem Sci. 1999;24:227. doi: 10.1016/s0968-0004(99)01410-3. (lett.). [DOI] [PubMed] [Google Scholar]
- 13.Weimbs T, Low S H, Chapin S J, Mostov K E, Bucher P, Hofmann K. Proc Natl Acad Sci USA. 1997;94:3046–3051. doi: 10.1073/pnas.94.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 15.Tomiuk S, Zumbansen M, Stoffel W. J Biol Chem. 2000;275:5710–5717. doi: 10.1074/jbc.275.8.5710. [DOI] [PubMed] [Google Scholar]
- 16.Gribskov M. Methods Mol Biol. 1994;25:247–266. doi: 10.1385/0-89603-276-0:247. [DOI] [PubMed] [Google Scholar]
- 17.Luthy R, Xenarios I, Bucher P. Protein Sci. 1994;3:139–146. doi: 10.1002/pro.5560030118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bucher P, Karplus K, Moeri N, Hofmann K. Comput Chem. 1996;20:3–23. doi: 10.1016/s0097-8485(96)80003-9. [DOI] [PubMed] [Google Scholar]
- 19.Gribskov M, McLachlan A D, Eisenberg D. Proc Natl Acad Sci USA. 1987;84:4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hofmann K, Bucher P. Trends Biochem Sci. 1995;20:347–349. doi: 10.1016/s0968-0004(00)89072-6. [DOI] [PubMed] [Google Scholar]
- 21.Lennon G, Auffray C, Polymeropoulos M, Soares M B. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 22.Iseli C, Jongeneel C V, Bucher P. In: Seventh International Conference on Intelligent Systems for Molecular Biology. Lengauer T, Schneider R, Bork P, Brutlag D, Glasgow J, Mewes H-W, Zimmer R, editors. Menlo Park, CA: AAAI Press; 1999. pp. 138–148. [Google Scholar]
- 23.Jayadev S, Linardic C M, Hannun Y A. J Biol Chem. 1994;269:5757–5763. [PubMed] [Google Scholar]
- 24.Albi E, Magni M P. Biochem Biophys Res Commun. 1997;236:29–33. doi: 10.1006/bbrc.1997.6803. [DOI] [PubMed] [Google Scholar]
- 25.Maruyama E N, Arima M. J Neurochem. 1989;52:611–618. doi: 10.1111/j.1471-4159.1989.tb09163.x. [DOI] [PubMed] [Google Scholar]
- 26.Hostetler K Y, Yazaki P J. J Lipid Res. 1979;20:456–463. [PubMed] [Google Scholar]
- 27.Mol C D, Kuo C F, Thayer M M, Cunningham R P, Tainer J A. Nature (London) 1995;374:381–386. doi: 10.1038/374381a0. [DOI] [PubMed] [Google Scholar]
- 28.Gorman M A, Morera S, Rothwell D G, de La Fortelle E, Mol C D, Tainer J A, Hickson I D, Freemont P S. EMBO J. 1997;16:6548–6558. doi: 10.1093/emboj/16.21.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weston S A, Lahm A, Suck D. J Mol Biol. 1992;226:1237–1256. doi: 10.1016/0022-2836(92)91064-v. [DOI] [PubMed] [Google Scholar]
- 30.Hayashi Y, Kiyono T, Fujita M, Ishibashi M. J Biol Chem. 1997;272:18082–18086. doi: 10.1074/jbc.272.29.18082. [DOI] [PubMed] [Google Scholar]
- 31.Hayashi Y, Kiyono T, Fujita M, Ishibashi M. Biochim Biophys Acta. 1997;1352:145–150. doi: 10.1016/s0167-4781(97)00051-1. [DOI] [PubMed] [Google Scholar]
- 32.Jayadev S, Liu B, Bielawska A E, Lee J Y, Nazaire F, Pushkareva M, Obeid L M, Hannun Y A. J Biol Chem. 1995;270:2047–2052. doi: 10.1074/jbc.270.5.2047. [DOI] [PubMed] [Google Scholar]
- 33.Nickels J T, Broach J R. Genes Dev. 1996;10:382–394. doi: 10.1101/gad.10.4.382. [DOI] [PubMed] [Google Scholar]