Abstract
MRP-1/CD9, KAI1/CD82, and ME491/CD63, have been reported to be associated with the metastatic potential of solid tumors. The aim of this study was to determine whether their expression in tumor tissues is a useful indicator for prognosis in breast cancer patients. We studied 109 breast cancer patients who underwent surgery. Quantitative reverse transcription-polymerase chain reaction analysis was performed to evaluate the expression of these genes. The results were confirmed with immunohistochemistry. All of the carcinomas were ME491/CD63 positive. Thirty-six tumors were MRP-1/CD9 negative. The disease-free survival rate and the 5-year survival rate of patients with MRP-1/CD9-negative tumors were both significantly lower than that in patients with MRP-1/CD9-positive tumors (P = 0.0005 and P = 0.0380, respectively). Sixty-five tumors were KAI1/CD82 negative. The disease-free survival rate of patients with KAI1/CD82-negative tumors was significantly lower than that of patients with KAI1/CD82-positive tumors (P = 0.0065). Cox regression analysis demonstrated that MRP-1/CD9 status (P = 0.0016) and KAI1/CD82 status (P = 0.0234) were useful indicators for the disease-free survival of breast cancer patients. The disease-free survival rate and 5-year survival rate of patients with either MRP-1/CD9-negative or KAI1/CD82-negative tumors were both significantly lower than patients who were positive for both genes (P = 0.0003 and P = 0.0292, respectively). The expression of MRP-1/CD9 and KAI1/CD82 genes are useful indicators of a poor prognosis in breast cancer patients.
When compared with other types of solid human cancers, breast cancer is interesting because of its high sensitivity to hormonal therapy and chemotherapy. 1 However, the prognosis varies according to the extent of the disease and its biological behavior. Although the histopathological presence of axillary lymph node metastases is considered to be the most informative parameter for predicting the occurrence of relapses and the prognosis in breast cancer patients, there is a possibility of recurrence after resection even in patients with an early stage of node-negative breast cancer. 2 Therefore, it is important to understand the biological behavior of each individual tumor and to determine which types of tumor will have a more malignant course and thus need more intensive adjuvant therapy. It is widely accepted that several kinds of cancers are caused by the accumulation of genetic alterations. 3-6 Recent investigations have revealed that some of these genetic changes can be used as prognostic factors for predicting a poor prognosis. For example, the amplification of some oncogenes such as c-erbB-2, 3,4 myc, 5 ras, 6 and c-fos 6 have been found to be associated with a poor prognosis in patients with breast cancers. Mutations of p53, a well known tumor suppressor gene, also may be important for the prognosis of breast cancer patients. 5,7 In addition, assessing a combination of these genetic alterations may enable the more precise prediction of the outcomes of patients with breast cancers. 6
Recently, three members of the transmembrane 4 superfamily MRP-1/CD9,8,9KAI1/CD82, 10 and ME491/CD6311 have been reported to be associated with the biological behavior of solid tumors, especially with their metastatic potential. Initially, we found that MRP-1/CD9 was recognized by the murine MAb M31–15, which inhibited cell motility. 12 After the transfection of MRP-1/CD9 into human lung cancer cell lines, we demonstrated that cell motility was suppressed in the MRP-1/CD9-expressing cells. 8 In addition, we showed that reduced MRP-1/CD9 protein expression was associated with metastasis and a poor prognosis in breast cancer patients 13 and that reduced MRP-1/CD9 gene expression was also correlated with a poor prognosis in non-small cell lung cancer patients.14 KAI1/CD82 gene is located on human chromosome 11p11.2–13, and encodes a protein of 267 amino acids. 10 Initially, it was identified by cDNA cloning as the R2 antigen, which was strongly up-regulated in mitogen-activated human T cells.15 KAI1/CD82 gene was also found to suppress tumor metastasis in a prostate cancer cell line 10 and a breast cancer cell line, 16,17 and thus may function as a metastatic suppressor gene. 10 A clinical analysis of patients with non-small cell lung cancers also revealed that reduced KAI1/CD82 gene expression was associated with the metastasis of these tumors. 18 In addition, it has been reported that ME491/CD63 is strongly expressed on the cell surface during the early stage of malignant melanoma but becomes weaker in the advanced stages. 19 These findings are similar to those observed for MRP-1/CD9 and KAI1/CD82. Thus, of the many genetic markers available for evaluating the prognosis in breast cancers, MRP-1/CD9, KAI1/CD82, and ME491/CD63 gene expression may have significant value as prognostic indicators in breast cancer patients. In the present study, we investigated whether the levels of MRP-1/CD9, KAI1/CD82, and ME491/CD63 gene expression in tumor tissues are of value as prognostic factors in predicting the clinical behavior of breast cancer. Therefore, we performed a reverse transcription-polymerase chain reaction (RT-PCR) analysis to quantify the expression of these genes in tumor tissues from 109 patients with breast cancer. Immunohistochemical assays were also performed to confirm the results of the RT-PCR.
Materials and Methods
Clinical Characteristics of the Patients
From February 1987 to December 1995, 109 patients who underwent surgery at the Department of Thoracic Surgery of Kitano Hospital, Medical Research Institute of Osaka in Japan, were studied. The complete clinical records of all patients were available, and their histopathological diagnoses were fully documented. The postsurgical stage of each tumor was classified according to the Union International Contre Cancer TNM system. 20 In total, 109 patients with breast cancer up to stage IIIB were investigated.
Ninety-five patients had undergone a mastectomy, and 14 patients had undergone a quadrantectomy followed by immediate radiotherapy. Adjuvant systemic chemotherapy was given according to the patients’ estrogen receptor (ER) status. Patients who were node positive or premenopausal (n = 73) underwent chemotherapy with oral 5-fluorouracil (200 mg/day) for 2 years, and eight patients with N2 disease were also treated with six cycles of cyclophosphamide/Adriamycin. Fifty-two ER-positive patients were treated with tamoxifen (20 mg/day) for 2 years or before recurrence. Sixteen postmenopausal patients of node-negative and receptor-negative status did not have any further adjuvant treatment. Thirty-three patients had recurrences during the observation period. After the recurrence, the locoregional tumor or lymph nodes were principally resected, followed by radiotherapy. Patients with distant metastases were treated with more effective adjuvant chemotherapies, including cisplatin and pirarubicin. This report includes follow-up data as of May 1, 1997. The median follow-up period was 48.5 months.
Tumor Specimens
To ascertain the presence of cancer cells, one-half of each fresh tumor tissue specimen was immediately embedded in optimum cutting temperature compound (Miles, Kankakee, IL), and frozen sections were then cut on the cryostat to a thickness of 6 μm and immediately stained with hematoxylin and eosin. After the connective tissues were trimmed off, the other half of the tumor specimen containing greater than 80% cancer cells of all tissue cells was selected for the RT-PCR analysis.
Quantitative RT-PCR Analysis
Total cellular RNA was extracted from the frozen tumor tissues by the acid guanidinium thiocyanate procedure. 21 First-strand cDNA synthesis was performed with 5 μg of total RNA using a cDNA synthesis kit (Pharmacia, Piscataway, NJ) according to the manufacturer’s protocol. All of the subsequent assays were then carried out using the same procedures as described previously. 14,18 The generated cDNAs were amplified using primers for MRP-1/CD9 (5′-TGCATCTGTATCCAGCGCCA-3′ and 5′-CTCAGGGATGTAAGCTGACT-3′), KAI1/CD82 (5′-AGTCCTCCCTGCTGCTGTGTG-3′ and 5′-TCAGTCAGGGTGGGCAAGAGG-3′) and ME491/CD63 (5′-CCCGAAAAACAACCACACTGC-3′ and 5′-GATGAGGAGGCTGAGGAGACC-3′). The internal control was β-actin (5′-GAGAAGATGACCCAGATCATGT-3′ and 5′-ACTCCATGCCCAGGAAGGAAGG-3′). 22 All of the subsequent assays were then carried out under conditions that yielded amplifications of MRP-1/CD9, KAI1/CD82, ME491/CD63, and β-actin within a linear range. Twenty-six cycles of PCR amplification were performed as follows: denaturation at 94°C for 40 seconds, annealing at 60°C for 40 seconds, and extension at 72°C for 90 seconds, followed by the final extension at 72°C for 7 minutes. The same PCR conditions were used to amplify the β-actin DNA. Tubes containing all of the ingredients except templates were included in all runs and served as negative controls. The human endothelial cell line ECV304 was used as a positive control, which has positive expression of MRP-1/CD9, KAI1/CD82, and ME491/CD63. 23 The amplified PCR products were electrophoresed on a 1% agarose gel containing ethidium bromide, and the bands were visualized under ultraviolet light followed by densitometric analysis (Figure 1) ▶ .
Figure 1.
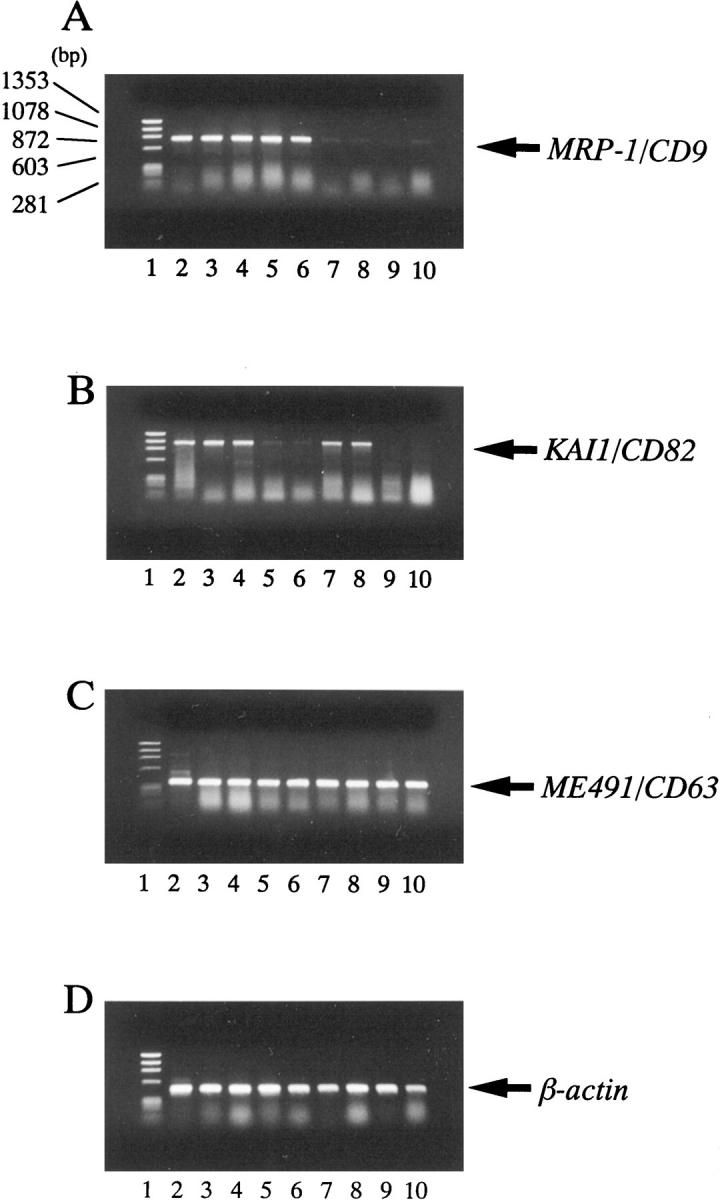
Agarose gel electrophoresis of RT-PCR-amplified MRP-1/CD9 KAI1/CD82 and ME491/CD63 cDNA. A: MRP-1/CD9 cDNA. B: KAI1/CD82 cDNA. C: ME491/CD63 cDNA. D: β-actin cDNA (internal PCR control). Lane 1, size marker; lane 2, human endothelial cell line ECV304 (positive control); lanes 3 and 4, breast cancers with both MRP-1/CD9- and KAI1/CD82-positive expression; lanes 5 and 6, breast cancers with MRP-1/CD9-positive but KAI1/CD82-reduced expression; lanes 7 and 8, breast cancers with MRP-1/CD9-reduced but KAI1/CD82-positive expression; lanes 9 and 10, breast cancers with both MRP-1/CD9- and KAI1/CD82-reduced expression.
Because it has been difficult to quantitate the absolute amount of specific mRNA without an internal standard of known concentration, the adjustment with a housekeeping gene has been used for the precise quantitation of mRNA of a specific gene in Northern blotting. Recently, quantitative RT-PCR has been developed using this method. 24 The densitometric values obtained for MRP-1/CD9, KAI1/CD82, and ME491/CD63 bands in a given tumor tissue sample were divided by the corresponding value of β-actin for normalization, and the ratio was referred to as the gene expression ratio for each gene. The expression ratio of the tumor was divided by the expression ratio of the human endothelial cell line ECV304 to obtain the gene conservation rates. When the conservation rate of a given specimen was ≥1.0, it was considered to indicate conserved (positive) gene expression. If the value was <1.0, this denoted nonconserved (reduced) gene expression.
Immunohistochemical Assays
To confirm the results of MRP-1/CD9 and KAI1/CD82 gene expression on RT-PCR, immunohistochemical studies were performed as described previously. 25 Because MRP-1/CD9 and KAI1/CD82 are not well preserved in formalin-fixed, paraffin-embedded tissues, frozen sections were used instead. After quenching the endogenous peroxidase activity with 0.3% H2O2 (in absolute methanol) for 30 minutes, the sections were blocked for 2 hours at room temperature with 5% bovine serum albumin. Subsequently, duplicate sections were incubated for 2 hours with the anti-MRP-1/CD9 monoclonal antibody M31-15 12 and the anti-KAI1/CD82 monoclonal antibody C33, 26 respectively, and were then incubated for 1 hour with biotinylated horse anti-mouse immunoglobulin G (Vector Laboratories Inc., Burlingame, CA). The sections were incubated with the avidin-biotin-peroxidase complex (Vector) for 1 hour, and the antibody binding was visualized with 3,3′-diaminobenzidine tetrahydrochloride. Finally, the sections were lightly counterstained with Mayer’s hematoxylin (Figure 2) ▶ . Specimens of fibroadenoma of the breast were used as positive controls.
Figure 2.
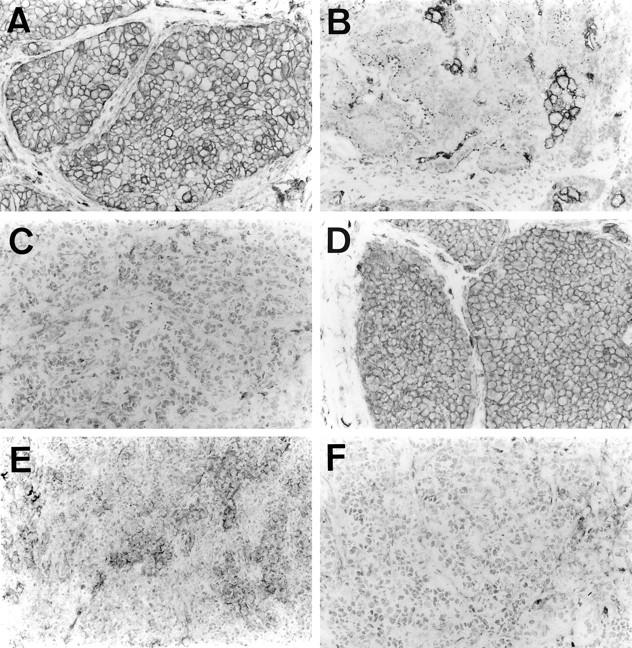
Immunohistochemical staining of human breast cancer tissues using the avidin-biotin-peroxidase complex procedure (original magnification, ×100). A: Invasive ductal carcinoma with positive MRP-1/CD9 expression. B: Invasive ductal carcinoma with lower MRP-1/CD9 expression classified as negative MRP-1/CD9 expression. C: Invasive ductal carcinoma with negative MRP-1/CD9 expression. D: Invasive ductal carcinoma with positive KAI1/CD82 expression. E: Invasive ductal carcinoma with lower KAI1/CD82 expression classified as negative KAI1/CD82 expression. F: Invasive ductal carcinoma with negative KAI1/CD82 expression.
All of the immunostained sections were reviewed by two pathologists who had no knowledge of the patients’ clinical status. Slides were examined under low power (×4 objective) to identify regions containing low-staining invasive tumors cells. In cases of multiple areas of low intensity, five areas selected at random were scored, and in sections where all of the staining appeared intense, one random field was selected. The proportion of high- and low-staining tumor cells in each selected field was determined by counting individual tumor cells at high magnification. At least 200 tumor cells were scored per ×40 field. Positive tumor cells were stained equivalent to normal breast glands and benign fibroadenoma tumor cells. All sections were scored in a semiquantitative fashion according to the method described previously, 27 which considers both the intensity and percentage of cells staining at each intensity. Intensities were classified as 0 (no staining), +1 (weak staining), +2 (distinct staining), and +3 (very strong staining), whereas 10% groupings were used for the percentage of cells that stained positive. For each slide, a value designated HSCORE was obtained by application of the following algorithm: HSCORE = Σ(I × PC), where I and PC represent intensity and percentage cells that stain at each intensity, respectively, and corresponding HSCOREs were calculated separately. Specimens with an HSCORE of ≥50 were classified as MRP-1/CD9 or KAI1/CD82-positive (+), and when HSCORE was <50, specimens were classified as reduced (−).
Statistical Analyses
The statistical significance of differences between MRP-1/CD9 or KAI1/CD82 gene expression and several other clinical pathological parameters was assessed by the χ 2 test. The disease-free survival and the overall survival curves were constructed according to the Kaplan-Meier method, 28 and differences in the survival of subgroups of patients were compared using Mantel’s log-rank test. 29 Multivariate analyses were performed using the Cox regression model to study the effects of different variables on survival, 30 and six factors (MRP-1/CD9 status, KAI1/CD82 status, ER status, age at surgery, T status, and N status) were studied. Scores were assigned to each variable for the regression analysis. All P values were based on two-tailed statistical analyses, and a P value < 0.05 was considered to indicate statistical significance.
Results
MRP-1/CD9, KAI1/CD82 and ME491/CD63 Gene Expression in Breast Cancer Tissues Analyzed by RT-PCR
Of all 109 breast cancers studied, ME491/CD63 gene expression was preserved and no reduced levels of ME491/CD63 DNA were detected (Figure 1) ▶ . All of the carcinomas were evaluated to be ME491/CD63 positive, and no statistically significant relationships were found between ME491/CD63 gene expression and other known prognostic factors (Figure 1C) ▶ . Thus, ME491/CD63 might play a different role from the other two transmembrane 4 superfamily members in breast cancer. On the other hand, of the 109 breast cancer patients, 73 tumors (67.0%) were evaluated as MRP-1/CD9 positive, and 36 tumors (33.0%) were MRP-1/CD9 negative (Figure 1A) ▶ . Forty-four tumors (40.4%) were evaluated as KAI1/CD82 positive, and 65 tumors (59.6%) were KAI1/CD82 negative (Figure 1B) ▶ . However, no relationship was found between MRP-1/CD9 expression and KAI1/CD82 expression (r = 0.138, P = 0.3526, data not shown), and the expression of these genes were independent of each other.
MRP-1/CD9 and KAI1/CD82 Protein Expression Analyzed by Immunohistochemistry
Of the 109 breast cancers studied using the immunohistochemical method, 72 (66.0%) were classified as MRP-1/CD9 positive. In these cases, the MRP-1/CD9 expression resembled that of benign fibroadenomas, and the immunostaining was intense and uniform on the cell surface membrane (Figure 2A) ▶ . There were 37 cases (34.0%) with reduced MRP-1/CD9 expression (Figure 2, B and C) ▶ , and the immunostaining from most of these tumors was heterogeneous. The MRP-1/CD9 gene expression was readily evident in those primary tumors that were classified as positive in the immunohistochemical assays. In contrast, the MRP-1/CD9 gene expression was weak or entirely absent in those breast cancers that had reduced immunohistochemically detectable MRP-1/CD9. The MRP-1/CD9 gene expression evaluated by RT-PCR was highly associated with MRP-1/CD9 protein expression, as determined by immunohistochemical staining (r = 0.755, P < 0.0001) (Figure 3A) ▶ . Overall, the immunohistochemical results agreed well with those from the RT-PCR assays, and 89.9% of the samples coincided exactly.
Figure 3.
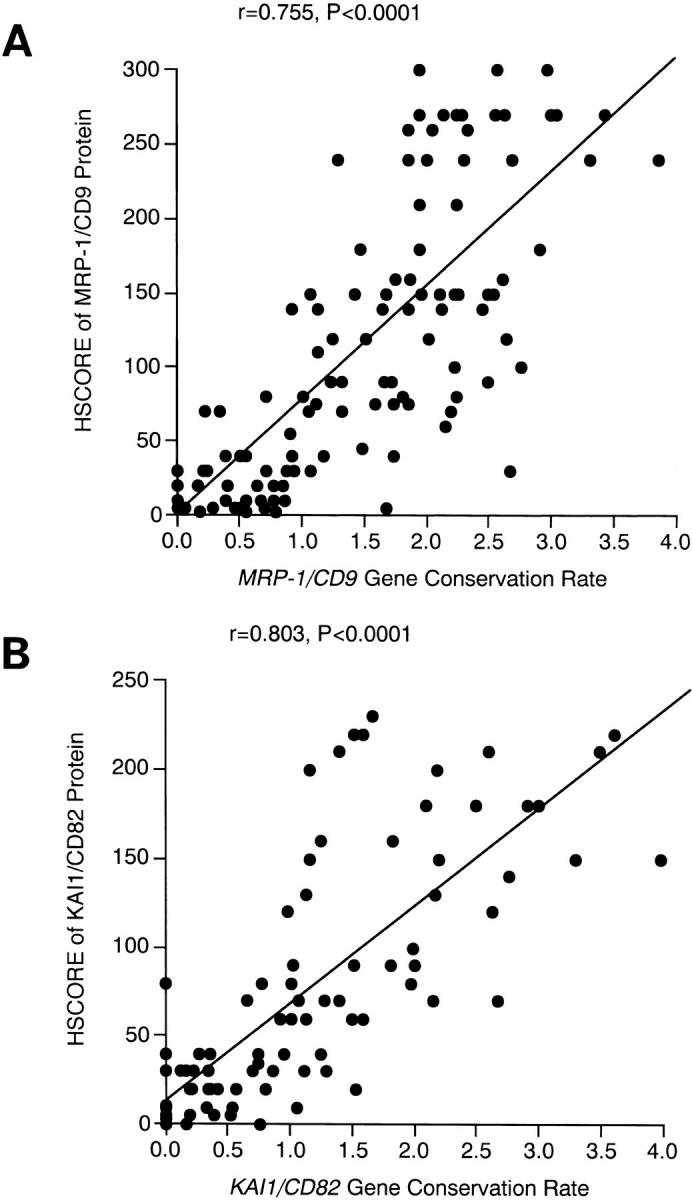
Pearson’s correlation coefficient between gene conservation rate by RT-PCR and HSCORE by immunohistochemistry. A: MRP-1/CD9. B: KAI1/CD82.
On the other hand, there were 44 cases (40.4%) with positive KAI1/CD82 expression and 65 cases (59.6%) with reduced KAI1/CD82 expression (Figure 2, D to F) ▶ . The KAI1/CD82 gene expression evaluated by RT-PCR was also associated with KAI1/CD82 protein expression, as determined by immunohistochemical staining (r = 0.803, P < 0.0001) (Figure 3B) ▶ . These results also agreed well with those from the RT-PCR assays, and 90.8% of the samples coincided exactly. In cases of discrepancy, the results from the RT-PCR analysis were used in the specimen classification.
Association of Tumor MRP-1/CD9 Status with Disease-Free and 5-Year Survival of Breast Cancer Patients
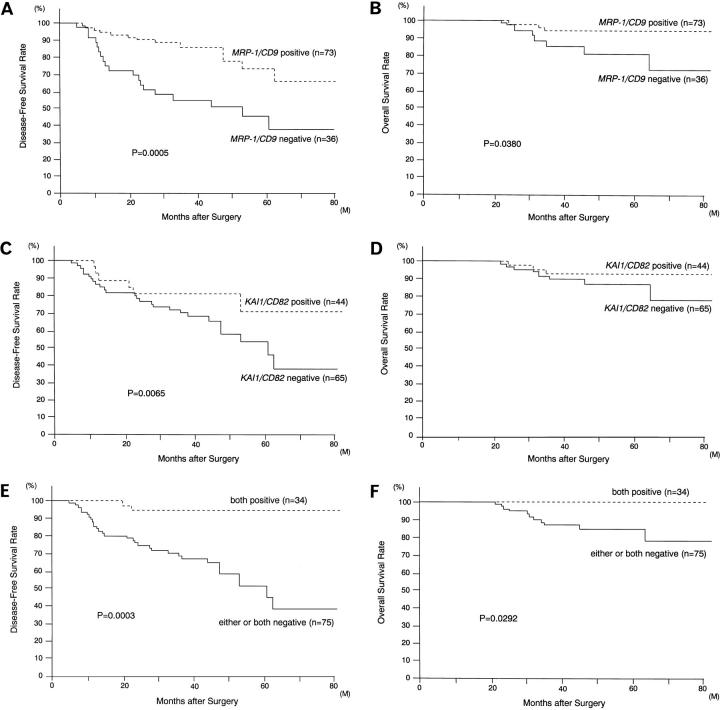
As described in our results from the Western blotting analysis, comparing survival of 109 patients with breast cancer demonstrated that the disease-free survival rate of patients with MRP-1/CD9-negative tumors was significantly lower than that of patients with MRP-1/CD9-positive tumors (37.7% versus 65.7%, P = 0.0005) (Table 1 ▶ and Figure 4A ▶ ). In particular, MRP-1/CD9 was an effective indicator of patients with early-stage tumors such as T1, N0, stage I, and stage II (P = 0.0005, P = 0.0007, P = 0.0168, and P = 0.0249, respectively). In addition, the 5-year survival rate of patients with MRP-1/CD9-negative tumors was significantly lower than that of patients with MRP-1/CD9-positive tumors (80.8% versus 94.0%, P = 0.0380) (Table 1 ▶ and Figure 4B ▶ ).
Table 1.
Disease-Free Survival Rate and 5-Year Survival Rate of 109 Patients with Breast Cancer According to Their Clinicopathological Characteristics and MRP-1/CD9 Gene Status
| Characteristics | Disease-free survival rate (%) | 5-year survival rate (%) | ||||
|---|---|---|---|---|---|---|
| MRP-1/CD9 + | MRP-1/CD9 − | P value | MRP-1/CD9 + | MRP-1/CD9 − | P value | |
| Age at surgery (years) | ||||||
| ≤50 | 68.8 | 45.9 | 0.0409 | 96.2 | 83.7 | 0.1855 |
| >50 | 63.9 | 28.6 | 0.0013 | 92.8 | 76.2 | 0.0815 |
| ER status | ||||||
| + | 66.2 | 45.0 | 0.0244 | 100.0 | 90.0 | 0.4000 |
| − | 64.4 | 34.3 | 0.0081 | 88.7 | 70.3 | 0.0587 |
| Tumor status | ||||||
| T1 | 95.2 | 44.4 | 0.0005 | 100.0 | 100.0 | >0.9999 |
| T2 | 52.0 | 52.9 | 0.4539 | 90.3 | 83.7 | 0.4179 |
| T3 | 80.0 | 25.0 | 0.0570 | 100.0 | 75.0 | >0.9999 |
| T4 | 100.0 | 0.0 | >0.9999 | 100.0 | 33.3 | >0.9999 |
| Nodal status | ||||||
| N0 | 93.0 | 45.0 | 0.0007 | 100.0 | 85.6 | 0.2000 |
| N1 | 52.5 | 33.9 | 0.0438 | 90.1 | 85.1 | 0.6659 |
| N2 | 40.0 | 0.0 | 0.2536 | 66.7 | 0.0 | 0.0445 |
| Pathological stage | ||||||
| I | 93.8 | 37.5 | 0.0168 | 100.0 | 100.0 | >0.9999 |
| II | 69.5 | 46.3 | 0.0249 | 93.2 | 81.5 | 0.1302 |
| III | 55.6 | 14.3 | 0.0887 | 85.7 | 68.6 | 0.3759 |
| Total number of patients | 65.7 | 37.7 | 0.0005 | 94.0 | 80.8 | 0.0380 |
Figure 4.
A: Disease-free survival of 109 breast cancer patients according to their tumor MRP-1/CD9 gene status. B: Overall survival of 109 breast cancer patients according to their tumor MRP-1/CD9 gene status. C: Disease-free survival of 109 breast cancer patients according to their tumor KAI1/CD82 gene status. D: Overall survival of 109 breast cancer patients according to their tumor KAI1/CD82 gene status. E: Disease-free survival of 109 breast cancer patients in relation to their classification (both MRP-1/CD9-positive and KAI1/CD82-positive subgroup versus either MRP-1/CD9-negative or KAI1/CD82-negative subgroup). F: Overall survival of 109 breast cancer patients in relation to their classification (both MRP-1/CD9-positive and KAI1/CD82-positive subgroup versus either MRP-1/CD9-negative or KAI1/CD82-negative subgroup).
Association of Tumor KAI1/CD82 Status with Disease-Free and 5-Year Survival of Breast Cancer Patients
The disease-free survival rate of patients with KAI1/CD82-negative tumors was significantly lower than that of patients with KAI1/CD82-positive tumors (38.2% versus 80.2%, P = 0.0065) (Table 2 ▶ and Figure 4C ▶ ). In particular, the disease-free survival rate of patients with KAI1/CD82-negative, early-stage tumors (ie, T1, T2, N0, stage I, and stage II) was significantly lower than that of patients with KAI1/CD82-positive, early-stage tumors (P = 0.0226, P = 0.0105, P = 0.0243, P = 0.0429, and P = 0.0108, respectively). However, there was no significant difference between the 5-year survival rates of patients with KAI1/CD82-negative tumors and patients with KAI1/CD82-positive tumors (Table 2 ▶ and Figure 4D ▶ ).
Table 2.
Disease-Free Survival Rate and 5-Year Survival Rate of 109 Patients with Breast Cancer According to Their Clinicopathological Characteristics and KAI1/CD82 Gene Status
| Characteristics | Disease-free survival rate (%) | 5-year survival rate (%) | ||||
|---|---|---|---|---|---|---|
| KAI1/CD82 + | KAI1/CD82 − | P value | KAI1/CD82 + | KAI1/CD82 − | P value | |
| Age at surgery (years) | ||||||
| ≤50 | 64.3 | 54.6 | 0.8587 | 87.1 | 93.2 | 0.5449 |
| >50 | 92.9 | 25.7 | 0.0005 | 96.2 | 81.4 | 0.0860 |
| ER status | ||||||
| + | 94.4 | 39.6 | 0.0207 | 100.0 | 94.4 | >0.9999 |
| − | 70.7 | 40.8 | 0.0719 | 87.8 | 78.2 | 0.2174 |
| Tumor status | ||||||
| T1 | 75.0 | 67.3 | 0.0226 | 100.0 | 100.0 | >0.9999 |
| T2 | 90.9 | 35.7 | 0.0105 | 95.0 | 84.4 | 0.2012 |
| T3 | 57.1 | 50.0 | 0.6415 | 83.3 | 66.7 | 0.5637 |
| T4 | 0.0 | 33.3 | 0.5151 | 0.0 | 66.7 | 0.0833 |
| Nodal status | ||||||
| N0 | 95.7 | 54.1 | 0.0243 | 95.2 | 93.3 | 0.8570 |
| N1 | 72.9 | 27.5 | 0.0729 | 94.4 | 85.5 | 0.3488 |
| N2 | 33.3 | 20.0 | 0.3727 | 66.7 | 33.3 | 0.9883 |
| Pathological stage | ||||||
| I | 100.0 | 70.1 | 0.0429 | 100.0 | 100.0 | >0.9999 |
| II | 83.6 | 36.6 | 0.0108 | 92.3 | 86.8 | 0.3911 |
| III | 42.9 | 33.3 | 0.5373 | 85.7 | 71.4 | 0.6945 |
| Total number of patients | 80.2 | 38.2 | 0.0065 | 92.8 | 87.0 | 0.3080 |
Prognostic Value of MRP-1/CD9 and KAI1/CD82 Status
The Cox regression model was used to evaluate the disease-free survival and overall survival as shown in Table 3 ▶ . The MRP-1/CD9 status (hazard ratio, 3.332; P = 0.0016), KAI1/CD82 status (hazard ratio, 2.778; P = 0.0234), ER status (hazard ratio, 2.242; P = 0.0336), and nodal status (hazard ratio, 3.089; P = 0.0003) were found to be useful indicators for the disease-free survival of breast cancer patients. On the other hand, only two variables, ER status (hazard ratio, 6.358; P = 0.0230) and nodal status (hazard ratio, 3.376; P = 0.0221), were significant factors for predicting the overall survival of breast cancer patients.
Table 3.
Multivariate Regression Analysis in Predicting the Disease-Free Survival and Overall Survival of 109 Patients with Breast Cancer
| Variables | Assigned score | Disease-free survival | Overall survival | ||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| MRP-1/CD9 status | |||||
| + | 0 | 3.332 (1.579–7.032) | 0.0016 | 3.092 (0.732–13.052) | 0.1245 |
| − | 1 | ||||
| KAI1/CD82 status | |||||
| + | 0 | 2.778 (1.149–6.721) | 0.0234 | 2.260 (0.559–9.142) | 0.2528 |
| − | 1 | ||||
| Age at Surgery (years) | |||||
| ≤50 | 0 | 1.996 (0.972–4.101) | 0.0598 | 1.983 (0.549–7.158) | 0.2959 |
| >50 | 1 | ||||
| ER status | |||||
| + | 0 | 2.242 (1.065–4.723) | 0.0336 | 6.358 (1.291–31.316) | 0.0230 |
| − | 1 | ||||
| Tumor status | |||||
| T1 | 1 | 1.239 (0.721–2.128) | 0.4374 | 1.353 (0.552–3.316) | 0.5080 |
| T2 | 2 | ||||
| T3 | 3 | ||||
| T4 | 4 | ||||
| Nodal status | |||||
| N0 | 0 | 3.089 (1.688–5.654) | 0.0003 | 3.376 (1.190–9.573) | 0.0221 |
| N1 | 1 | ||||
| N2 | 2 |
CI, confidence interval.
Classification of Breast Cancer According to MRP-1/CD9 and KAI1/CD82 Gene Expression
Initially, the 109 breast cancer patients were divided into four groups according to their MRP-1/CD9 and KAI1/CD82 gene status; 34 patients had both MRP-1/CD9- and KAI1/CD82-positive tumors, 10 patients had MRP-1/CD9-negative but KAI1/CD82-positive tumors, 39 patients had MRP-1/CD9-positive but KAI1/CD82-negative tumors, and 26 patients had both MRP-1/CD9- and KAI1/CD82-negative tumors. The disease-free survival rates of these patients were 94.1%, 40.0%, 26.9%, and 37.6%, respectively. The disease-free survival rate of patients with tumors positive for both genes was significantly higher than that of patients with the other three types of tumors. On the other hand, there were no significant differences among the latter three groups. Therefore, the 109 breast cancer patients were reclassified into two subgroups: one subgroup with both MRP-1/CD9- and KAI1/CD82-positive tumors and the other subgroup with either or both negative tumors. The disease-free survival rate of the former double-positive subgroup was significantly higher than the latter subgroup (94.1% versus 38.4%, P = 0.0003) (Table 4 ▶ and Figure 4E ▶ ). In addition, the former subgroup also had a higher 5-year survival rate than the latter subgroup (100.0% versus 84.4%, P = 0.0292) (Table 4 ▶ and Figure 4F ▶ ). The Cox multivariate regression analysis of disease-free survival is shown in Table 5 ▶ . This simultaneous evaluation for both MRP-1/CD9 and KAI1/CD82 expression was found to be a significant indicator of a poor prognosis (P = 0.0006), and nodal status is also a significant indicator of a poor prognosis. The other variables (age at surgery, ER status, and T status) did not correlate with the MRP-1/CD9 and KAI1/CD82 gene status or its prognostic value. However, because no patients in the double-positive subgroup have died, it is impossible to perform a Cox multivariate regression analysis for their 5-year survival.
Table 4.
Disease-Free Survival Rate and 5-Year Survival Rate of 109 Patients with Breast Cancer according to Their MRP-1/CD9 and KAI-1/CD82 Gene Status
| Characteristics | Disease-free survival rate (%) | 5-year survival rate (%) | ||||
|---|---|---|---|---|---|---|
| Both + | Either − or both − | P value | Both + | Either − or both − | P value | |
| Age at surgery (years) | ||||||
| ≤50 | 80.0 | 51.9 | 0.3928 | 100.0 | 88.4 | 0.5242 |
| >50 | 100.0 | 25.4 | 0.0001 | 100.0 | 80.4 | 0.0609 |
| ER status | ||||||
| + | 92.9 | 46.1 | 0.0782 | 100.0 | 94.7 | >0.9999 |
| − | 95.0 | 35.6 | 0.0010 | 100.0 | 73.0 | 0.0571 |
| Tumor status | ||||||
| T1 | 100.0 | 72.0 | 0.0360 | 100.0 | 100.0 | >0.9999 |
| T2 | 94.1 | 40.3 | 0.0178 | 100.0 | 83.7 | 0.2782 |
| T3 | 80.0 | 25.0 | 0.0570 | 100.0 | 75.0 | >0.9999 |
| T4 | 25.0 | 50.0 | ||||
| Nodal status | ||||||
| N0 | 100.0 | 55.1 | 0.0147 | 100.0 | 90.9 | 0.5238 |
| N1 | 92.9 | 28.3 | 0.0090 | 100.0 | 83.9 | 0.2621 |
| N2 | 50.0 | 16.7 | 0.2125 | 100.0 | 26.7 | >0.9999 |
| Pathological stage | ||||||
| I | 100.0 | 72.9 | 0.2157 | 100.0 | 100.0 | >0.9999 |
| II | 100.0 | 38.3 | 0.0080 | 100.0 | 84.4 | 0.1725 |
| III | 60.0 | 27.3 | 0.1465 | 100.0 | 67.5 | 0.4965 |
| Total number of patients | 94.1 | 38.4 | 0.0003 | 100.0 | 84.4 | 0.0292 |
Table 5.
Multivariate Regression Analysis in Predicting the Disease-Free Survival of 109 Patients with Breast Cancer
| Variables | Assigned score | Hazard ratio (95% CI) | P value |
|---|---|---|---|
| MRP-1/CD9 and KAI1/CD82 status | |||
| Both+ | 0 | 13.375 (3.068–58.312) | 0.0006 |
| Either− or both− | 1 | ||
| Age at surgery (years) | |||
| ≤50 | 0 | 2.468 (1.175–5.183) | 0.0170 |
| >50 | 1 | ||
| ER status | |||
| + | 0 | 2.299 (1.112–4.752) | 0.0247 |
| − | 1 | ||
| Tumor status | |||
| T1 | 1 | 1.347 (0.791–2.293) | 0.2727 |
| T2 | 2 | ||
| T3 | 3 | ||
| T4 | 4 | ||
| Nodal status | |||
| N0 | 0 | 2.870 (1.572–5.241) | 0.0006 |
| N1 | 1 | ||
| N2 | 2 |
CI, confidence interval.
Discussion
Previously, we have shown that MRP-1/CD9 protein expression was a good predictive factor for poor prognosis in breast cancer patients. 13 As part of our evaluation of the members of the transmembrane 4 superfamily as possible prognostic predictors for breast cancer, we have extended our study to other genes, such as ME491/CD63 and KAI1/CD82. It has been reported that ME491/CD63 is strongly expressed on the cell surface during the early stage of malignant melanoma, but is weaker or absent in the more malignant stages and in the normal melanocyte. 19 However, the ME491/CD63 mRNA levels in almost all of the breast cancers were well preserved, and no cases with a reduction in ME491/CD63 mRNA levels were detected. This suggests that ME491/CD63 may limit the progression only in malignant melanomas and that its expression might have no effect on the characteristics of breast cancers. On the other hand, our present study demonstrated that a reduction in not only MRP-1/CD9 gene expression but also KAI1/CD82 gene expression in breast tumors correlated with a poor prognosis. In addition, a simultaneous classification according to both MRP-1/CD9 and KAI1/CD82 gene expression was a very useful indicator for predicting the recurrence of breast cancer. Of patients with N0 status or stage I diseases, those with both MRP-1/CD9- and KAI1/CD82-positive tumors had no recurrences. However, among patients with either MRP-1/CD9- or KAI1/CD82-negative tumors, the disease-free survival rates were only 55.1% for patients with N0 status and 72.9% for patients with stage I. Therefore, when breast cancer patients have either MRP-1/CD9- or KAI1/CD82-negative tumors, they should be carefully observed after surgery even in early pathological stages. Furthermore, these patients might need additional adjuvant chemotherapy and/or hormonal therapy. In contrast, of the 34 patients with both MRP-1/CD9- and KAI1/CD82-positive tumors, only 2 patients (T2N2 and T3N1) had recurrences, and all of the patients were alive for 5 years after their surgery. These results suggested that the tumors positive for both MRP-1/CD9 and KAI1/CD82 may have a lower-grade malignancy and that adjuvant chemotherapy might not be necessary before recurrence.
Both MRP-1/CD9 and KAI1/CD82 are cell surface membrane glycoproteins that are widely expressed in various normal epithelia. 25 Members of the transmembrane 4 superfamily include MRP-1/CD9, 8 KAI1/CD82, 10 ME491/CD63, 19 TAPA-1/CD81, 31 CD53, 32 CD37, 33 and others. The members of this family have four hydrophobic transmembrane domains divided by two extracellular loops, with cytoplasmic N and C termini. 34 Their extracellular loops usually have some N-glycosylation sites. This type of structure suggests that these cell surface glycoproteins might play an important role in signal transduction and may regulate cell growth, cell differentiation, cell adhesion, and cell motility. 34,35 Although the precise functions of this family still remain unclear, many studies using immunoprecipitation have demonstrated the possible existence of a “tetraspan network.” 36 By connecting with other molecules such as integrins 35,37,38 and human lymphocyte antigens, 39-41 this family may organize the positioning of cell surface proteins and thus play a role in signal transduction. In addition, MRP-1/CD9, KAI1/CD82, CD63, and CD81 are considered to form complexes with each other. 36 These results suggest that MRP-1/CD9 or KAI1/CD82 gene expression might be declining as the malignant tumors advance and could disrupt the tetraspan network and enable the malignant cells to acquire their metastatic potential. In fact, MRP-1/CD9 gene has been reported to be more highly expressed in a primary colon cancer as compared with its corresponding metastatic tumor using differential display cloning. 42 These studies also indicated that MRP-1/CD9 and KAI1/CD82 might function as metastatic suppressor genes and are useful indicators of a poor outcome in patients with some solid tumors. 13,14,18
On the other hand, the down-regulation of KAI1/CD82 gene during the progression of human prostate cancer infrequently involves a gene mutation or allelic loss. 43 We also have found no mutations of MRP-1/CD9 gene in 143 resected lung tumor specimens (data not shown). To date, we believe that the mechanism underlying the reduction in the expression of these genes in tumor tissues might be an abnormal gene promoter or an abnormal down-regulation somewhere upstream in their signal pathway. Various oncogenes and oncosuppressor genes have their actions relatively upstream of the signal pathway. During the progression of these tumors, the accumulations of abnormalities might act as triggers inducing the tumors to become more aggressive.
Another interesting aspect of the current research is that our study showed that the results of MRP-1/CD9 and KAI1/CD82 gene expression as evaluated by RT-PCR agreed well quantitatively with their protein expression as evaluated by immunohistochemistry. In addition, because the results in larger-scale studies might also depend on the quality of the mRNA, it may be relatively easy to quantitate MRP-1/CD9 and KAI1/CD82 protein expression in an actual clinical setting. Our present study demonstrated that MRP-1/CD9 and KAI1/CD82 gene expression in breast cancers are important factors for predicting recurrence. The classification of breast cancers according to both MRP-1/CD9 and KAI-1/CD82 gene expression will prove to be useful for the clinical treatment of breast cancers patients.
Acknowledgments
We thank Tatsuya Hirai for his technical assistance and Drs. Tadashi Obayashi and Sadayuki Matsumoto for the pathological examination.
Footnotes
Address reprint requests to Dr. Masayuki Miyake, Department of Thoracic Surgery and Department V of Oncology, Kitano Hospital, Tazuke Kofukai Medical Research Institute, 13-3, Kamiyama-cho, Kita-ku, Osaka 530-0026, Japan.
Supported in part by Grants-in-Aid from the Ministry of Education, Science and Culture of Japan (No 07557253 and 08407040) to M. Miyake.
References
- 1.Early Breast Cancer Trialists’ Collaborative Group: Systemic treatment of early breast cancer by hormonal, cytotoxic, or immune therapy: 133 randomized trials involving 31 000 recurrences and 24 000 deaths among 75 000 women. Lancet 1992, 339:1–15,71–85 [PubMed]
- 2.Allred DC, Clark GM, Elledge R, Fuqua SA, Brown RW, Chamness GC, Osborne CK, McGuire WL: Association of p53 protein expression with tumor cell proliferation rate and clinical outcome in node-negative breast cancer. J Natl Cancer Inst 1993, 85:200-206 [DOI] [PubMed] [Google Scholar]
- 3.Hartmann LC, Ingle JN, Wold LE, Farr GH, Grill JP, Su JQ, Maihle NJ, Krook JE, Witzig TE, Roche PC: Prognostic value of c-erbB2 overexpression in axillary lymph node positive breast cancer. Cancer 1994, 74:2956-2963 [DOI] [PubMed] [Google Scholar]
- 4.Giai M, Roagna R, Ponzone R, Bortoli MD, Dati C, Sismondi P: Prognostic and predictive relevance of c-erbB-2 and ras expression in node positive and negative breast cancer. Anticancer Res 1994, 14:1441-1450 [PubMed] [Google Scholar]
- 5.Berns EMJJ, Klijn JGM, Smid M, Staveren IL, Look MP, Putten WLJ, Foekens JA: TP53, and MYC gene alterations independently predict poor prognosis in breast cancer patients. Genes Chromosomes Cancer 1996, 16:170-179 [DOI] [PubMed] [Google Scholar]
- 6.Bland KI, Konstadoulakis MM, Vezeridis MP, Wanebo HJ: Oncogene protein co-expression: value of Ha-ras, c-myc, c-fos, and p53 as prognostic discriminants for breast carcinoma. Ann Surg 1995, 221:706-720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kovach JS, Hartmann A, Blaszyk H, Cunningham J, Schaid D, Sommer SS: Mutation detection by highly sensitive methods indicates that p53 gene mutations in breast cancer can have important prognostic value. Proc Natl Acad Sci USA 1996, 93:1093-1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikeyama S, Koyama M, Yamaoko M, Sasada R, Miyake M: Suppression of cell motility and metastasis by transfection with human motility-related protein (MRP-1/CD9) DNA. J Exp Med 1993, 177:1231-1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyake M, Nakano K, Ieki Y, Adachi M, Huang C, Itoi S, Koh T, Taki T: Motility related protein 1 (MRP-1/CD9) expression: inverse correlation with metastases in breast cancer. Cancer Res 1995, 55:4127-4131 [PubMed] [Google Scholar]
- 10.Dong JT, Lamb PW, Rinker-Schaeffer CW, Vukanovic J, Ichikawa T, Isaacs JT, Barrett JC: KAI1, a metastasis suppressor gene for prostate cancer on human chromosome 11p11.2. Science 1995, 268:884-886 [DOI] [PubMed] [Google Scholar]
- 11.Radford KJ, Mallesch J, Hersey P: Suppression of human melanoma cell growth and metastasis by the melanoma-associated antigen CD63 (ME491). Int J Cancer 1995, 62:631-635 [DOI] [PubMed] [Google Scholar]
- 12.Miyake M, Koyama M, Seno M, Ikeyama S: Identification of the motility-related protein (MRP-1), recognized by monoclonal antibody M31–15, which inhibits cell motility. J Exp Med 1991, 174:1347-1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyake M, Nakano K, Itoi S, Koh T, Taki T: Motility-related protein-1 (MRP-1/CD9) reduction as a factor of poor prognosis in breast cancer. Cancer Res 1996, 56:1244-1249 [PubMed] [Google Scholar]
- 14.Higashiyama M, Taki T, Ieki Y, Adachi M, Huang C, Koh T, Kodama K, Doi O, Miyake M: Reduced motility related protein-1 (MRP-1/CD9) gene expression as a factor of poor prognosis in non-small cell lung cancer. Cancer Res 1995, 55:6040-6044 [PubMed] [Google Scholar]
- 15.Gaugitsch HW, Hofer E, Huber NE, Schnabl E, Baumruker T: A new superfamily of lymphoid and melanoma cell proteins with extensive homology to Schistosoma mansoni antigen Sm23. Eur J Immunol 1991, 21:377-383 [DOI] [PubMed] [Google Scholar]
- 16.Yang X, Welch DR, Phillips KK, Weissman BE, Wei LL: KAI1, a putative marker for metastatic potential in human breast cancer. Cancer Lett 1997, 119:149-155 [DOI] [PubMed] [Google Scholar]
- 17.Phillips KK, White AE, Hicks DJ, Welch DR, Barrett JC, Wei LL, Weissman BE: Correlation between reduction of metastasis in the MDA-MB-435 model system and increased expression of the Kai-1 protein. Mol Carcinog 1998, 21:111-120 [PubMed] [Google Scholar]
- 18.Adachi M, Taki T, Ieki Y, Huang C, Higashiyama M, Miyake M: Correlation of KAI1/CD82 gene expression with good prognosis in patients with non-small cell lung cancer. Cancer Res 1996, 56:1751-1755 [PubMed] [Google Scholar]
- 19.Hotta H, Ross AH, Huebner K, Isobe M, Wendeborn S, Chao MV, Ricciardi RP, Tsujimoto Y, Croce CM, Koprowski H: Molecular cloning and characterization of an antigen associated with early stages of melanoma tumor progression. Cancer Res 1988, 48:2955-2962 [PubMed] [Google Scholar]
- 20.Hermanek P, Sobin LH: TNM Classification of Malignant Tumors. ed 4 Hermanek P Sobin LH eds. 1987, :pp 103-109 Springer-Verlag, Berlin [Google Scholar]
- 21.Chomczynski P, Sacchi N: Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 1987, 162:156-159 [DOI] [PubMed] [Google Scholar]
- 22.Nakajima-Iijima S, Hamada H, Reddy P, Kakunaga T: Molecular structure of the human cytoplasmic β-actin gene: interspecies homology of sequences in the introns. Proc Natl Acad Sci USA 1985, 82:6133-6137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahasi K, Sawasaki Y: Rare spontaneously transformed human endothelial cell line provides useful research tool. In Vitro Cell Dev Biol 1992, 28A:380-382 [DOI] [PubMed] [Google Scholar]
- 24.Coen DM: Quantitation of rare DNAs by PCR. Current Protocols in Molecular Biology. Edited by Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. New York, John Wiley & Sons, 1995, pp 15.3.1–15.3.8
- 25.Huang C, Taki T, Adachi M, Yagita M, Sawada S, Takabayashi A, Inufusa H, Yoshie O, Miyake M: MRP-1/CD9 and KAI1/CD82 expression in normal and various cancer tissues. Int J Oncol 1997, 11:1045-1051 [DOI] [PubMed] [Google Scholar]
- 26.Imai T, Fukudome K, Takagi S, Nagira M, Furuse M, Fukuhara N, Nishimura M, Hinuma Y, Yoshie O: C33 antigen recognized by monoclonal antibodies inhibitory to human T cell leukemia virus type 1-induced syncytium formation is a member of a new family of transmembrane proteins including CD9, CD37, CD53, and CD63. J Immunol 1992, 149:2879-2886 [PubMed] [Google Scholar]
- 27.McCarty KS, Jr, Szabo E, Flowers JL, Cox EB, Leight GS, Miller L, Konrath J, Soper JT, Budwit DA, Creasman WT, Seigler HF, McCarty KS, Sr: Use of a monoclonal anti-estrogen receptor antibody in the immunohistochemical evaluation of human tumors. Cancer Res 1986, 46:4244s-4248s [PubMed] [Google Scholar]
- 28.Kaplan EL, Meier P: Nonparametric estimation from incomplete observations. J Am Stat Assoc 1958, 53:457-481 [Google Scholar]
- 29.Mantel N: Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep 1966, 50:163-170 [PubMed] [Google Scholar]
- 30.Cox DR: Regression models and life-tables. J R Stat Soc B 1972, 34:187-220 [Google Scholar]
- 31.Oren R, Takahashi S, Doss C, Levy R, Levy S: TAPA-1, the target of an antiproliferative antibody, defines a new family of transmembrane proteins. Mol Cell Biol 1990, 10:4007-4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amiot M: Identification and analysis of cDNA clones encoding CD53: a pan-leukocyte antigen related to membrane transport proteins. J Immunol 1990, 145:4322-4325 [PubMed] [Google Scholar]
- 33.Classon BJ, Williams AF, Willis AC, Seed B, Stamenkovic I: The primary structure of the human leukocyte antigen CD37, a species homologue of the rat MRC OX-44 antigen. J Exp Med 1989, 169:1497-1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright MD, Tomlinson MG: The ins and outs of the transmembrane 4 superfamily. Immunol Today 1994, 15:588-594 [DOI] [PubMed] [Google Scholar]
- 35.Hemler ME, Mannion BA, Berditchevski F: Association of TM4SF proteins with integrins: relevance to cancer. Biochim Biophys Acta 1996, 1287:67-71 [DOI] [PubMed] [Google Scholar]
- 36.Rubinstein E, Naour FL, Lagaudriere-Gesbert C, Billard M, Conjeaud H, Boucheix C: CD9, CD63, CD81, and CD82 are components of a surface tetraspan network connected to HLA-DR, and VLA integrins. Eur J Immunol 1996, 26:2657-2665 [DOI] [PubMed] [Google Scholar]
- 37.Berditchevski F, Zutter MM, Hemler ME: Characterization of novel complexes on the cell surface between integrins and proteins with 4 transmembrane domains (TM4 proteins). Mol Biol Cell 1996, 7:193-207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mannion BA, Berditchevski F, Kraeft SK, Chen LB, Hemler ME: Transmembrane-4 superfamily proteins CD81 (TAPA-1), CD82, CD63, and CD53 specifically associate with integrin α4β1 (CD49d/CD29). J Immunol 1996, 157:2039-2047 [PubMed] [Google Scholar]
- 39.Angelisova P, Hilgert I, Horejsi V: Association of four antigens of the tetraspans family (CD37, CD53, TAPA-1, and R2/C33) with MHC class II glycoproteins. Immunogenetics 1994, 39:249-256 [DOI] [PubMed] [Google Scholar]
- 40.Szollosi J, Horejsi V, Bene L, Angelisova P, Damjanovich S: Supramolecular complexes of MHC class I, MHC class II, CD20, and tetraspan molecules (CD53, CD81, and CD82) at the surface of a B cell line JY. J Immunol 1996, 157:2939-2946 [PubMed] [Google Scholar]
- 41.Lagaudriere-Gesbert C, Lebel-Binay S, Wiertz E, Ploegh HL, Fradelizi D, Conjeaud H: The tetraspanin protein CD82 associates with both free HLA class I heavy chain and heterodimeric β2-microglobulin complexes. J Immunol 1997, 158:2790-2797 [PubMed] [Google Scholar]
- 42.Cajot JF, Sordat I, Silvestre T, Sordat B: Differential display cloning identifies motility-related protein (MRP1/CD9) as highly expressed in primary compared to metastatic human colon carcinoma cells. Cancer Res 1997, 57:2593-2597 [PubMed] [Google Scholar]
- 43.Dong JT, Suzuki H, Pin SS, Bova GS, Schalken JA, Isaacs WB, Barrett JC, Isaacs JT: Down-regulation of the KAI1 metastasis suppressor gene during the progression of human prostatic cancer infrequently involves gene mutation or allelic loss. Cancer Res 1996, 56:4387-4390 [PubMed] [Google Scholar]