Abstract
DNA copy number changes were investigated in 29 leiomyosarcomas by comparative genomic hybridization. The most frequent losses were detected in 10q (20 cases, 69%) and 13q (17 cases, 59%). The most frequent gains were detected in 17p (16 cases, 55%). The most frequent high-level amplifications were detected in 17p (7 cases, 24%) and 8q (6 cases, 21%). A total of 137 losses and 204 gains were detected. Small tumors (less than 5 cm in diameter) displayed fewer changes per sample (3 to 11; mean, 7) than the other tumors (4 to 22; mean, 13). There was an increase in the number of gains from small tumors (mean, 4) to very large tumors (>20 cm; mean, 10). However, the number of losses was similar in small, large, and very large tumors (mean, 4.5). Tumor size-related aberrations were observed. Gains in 16p were detected in all small tumors but were infrequent in large and very large tumors (27% and 11%, respectively). Similarly, gains and high-level amplifications in 17p were more common in small (80%) than in very large tumors (33%). Gains in 1q, 5p, 6q, and 8q were not seen in any of the small tumors but were detected in large and very large tumors. Gains in 6q and 8q occurred in 8 of 9 cases (89%) of very large tumors, 5 of them with a high-level amplification in 8q.
Leiomyosarcoma (LMS) is the designation for malignant mesenchymal neoplasms that display phenotypic features of smooth muscle differentiation. These tumors typically show spindle cells with blunt-ended nuclei and eosinophilic cytoplasm and are immunoreactive for α-smooth muscle-actin and often, but not always, for desmin. LMS occurs in a wide variety of body sites, especially in the retroperitoneum, skin, and superficial soft tissues and in the deep compartments of the extremities. 1 Gastrointestinal spindle cell sarcomas and their benign counterparts appear to differ from smooth muscle tumors histologically, ultrastructurally, and immunophenotypically, and recently they have been classified as gastrointestinal stromal tumors (GISTs) separate from leiomyomas and LMSs. 2
Standard karyotyping and fluorescent in situ hybridization techniques have shown that the genetic changes in LMS are complex. 3-7 Numerical aberrations as well as structural rearrangements in chromosomes 1, 7, 10, 13, 14, 16, and 17 have been reported in LMS. 3-9 Molecular studies on LMS have suggested that p53 gene mutations and p53 protein overexpression may be associated with the development of LMS. 10-13
Comparative genomic hybridization (CGH) enables the screening of entire tumor genomes for gains and losses of DNA copy number and consequent mapping of aberrations to chromosomal subregions. 14-16 To identify genomic areas that may be involved in the oncogenesis of LMS and the areas that may be related to tumor size and progression, we used CGH to screen whole tumor genomes for DNA copy number changes.
Materials and Methods
Characterization of Tumors
All soft tissue LMS cases included in this study were extensively characterized histologically and immunohistochemically. All superficial tumors were primary tumors, and the possibility of metastatic LMS from a deeper primary was ruled out in each case. All tumors were classified from intermediate to high grade, except one case (case 1) that showed a low mitotic count. According to the size, the tumors were classified as small (less than 5 cm; n = 5), large (more than 5 cm but less than 20 cm; n = 15), and very large (more than 20 cm; n = 9). All but two LMSs (cases 21 and 22) were histologically spindle cell tumors that typically showed intersecting fascicles of spindle cells. The tumor cells had blunt-ended nuclei and variably eosinophilic cytoplasm that showed strong α-smooth muscle actin immunoreactivity and that was also desmin positive at least focally, except in two desmin-negative cases. All tumors were negative for CD34. Sixteen of the tumors were from the retroperitoneum (none were located in the mesentery or associated with the small intestine), one originated from the inferior vena cava, seven from the leg, and single tumors from the arm, buttock, and perineum. Malignant GISTs, typically displaying rudimentary, if any, smooth muscle differentiation, were excluded from this study. No mesenteric tumors were included.
Two LMSs (cases 21 and 22) were classified as epitheloid LMS. Both of these showed focal α-smooth muscle actin and desmin immunoreactivity. Both cases were very large retroperitoneal tumors exceeding 20 cm in diameter. None of the patients had received chemotherapy before surgery.
CGH
DNA was extracted from paraffin-embedded tumors as described elsewhere. 17 CGH was performed according to standard procedures, 18 with a modification using fluorochromes conjugated to a mixture of dCTP and dUTP for standard nick translation. 19 Briefly, the tumor DNA was labeled with a mixture of fluorescein isothiocyanate-dCTP and fluorescein isothiocyanate-dUTP (DuPont, Boston, MA), whereas reference genomic DNA was labeled with a mixture of Texas red-dCTP and Texas red-dUTP (DuPont) by nick translation to obtain DNA fragments ranging from 600 to 2000 bp. The hybridization mixture consisted of 800 ng labeled tumor DNA, 800 ng labeled reference genomic DNA, and 20 μg unlabeled Cot-1 DNA dissolved in 10 μl of hybridization buffer (50% formamide, 10% dextran sulfate, and 2× standard saline citrate (SSC)). The hybridization mixture was denatured at 75°C for 5 minutes and hybridized to a slide preparation with normal metaphase spreads denatured in 70% formamide/2× SSC at 68°C for 2 minutes. Hybridization was performed at 37°C for 48 hours. The slides were washed three times in 50% formamide/2× SSC (pH 7), twice in 2× SSC, and once in 0.1× SSC at 45°C, followed by 2× SSC, 0.1 mol/L NaH2PO4/0.1 mol/L Na2HPO4/0.1% Nonidet P40 (pH 8), and distilled water at room temperature for 10 minutes each. After air drying, the slides were counterstained with 4′-6-diamidino-2-phenylindole-dihydrochloride (Sigma Chemical Co., St. Louis, MO) and then mounted with an antifading medium (Vectashield; Vector Laboratories Inc., Burlingame, CA).
Digital Image Analysis
The hybridizations were analyzed using an Olympus fluorescence microscope and the ISIS digital image analysis system (Metasystems GmbH, Altlussheim, Germany) based on an integrated high-sensitivity monochrome charge-coupled device camera and automated CGH analysis software. Three-color images (red for reference DNA, green for tumor DNA, and blue for counterstaining) were acquired from 8 to 10 metaphases per sample. Only metaphases of good quality with strong uniform hybridization were included in the analysis. Chromosomes not suitable for CGH analysis were excluded (ie, chromosomes heavily bent, overlapping, or with overlying artifacts). Chromosomal regions were interpreted as overrepresented when the corresponding ratio exceeded 1.17 (gains) or 1.5 (high-level amplification) and as underrepresented (losses) when the ratio was less than 0.85. All of the results were confirmed using a 99% confidence interval. Briefly, intraexperiment standard deviations for all positions in the CGH ratio profiles were calculated from the variation of the ratio values of all homologous chromosomes within the experiment. Confidence intervals for the ratio profiles were then computed by combining them with an empirical interexperiment standard deviation and by estimating error probabilities based on the t-distribution.
Controls
In each CGH experiment, a negative control (peripheral blood DNA from a healthy donor) and a positive control were included. The positive control was a tumor with known changes in DNA copy numbers.
Results
The results are summarized in Table 1 ▶ and Figure 1 ▶ . Complex changes in DNA copy numbers were detected in all cases of LMS. A total of 137 losses and 204 gains were detected. The most consistent findings were seen as losses in DNA copy numbers with minimal common overlapping regions at 10q11–q21.2 (20 cases, 69%) and 13q14.3–q21.1 (17 cases, 59%) and as gains with a minimal common overlapping region at 17p11–p12 (16 cases, 59%). In 16 of 20 cases with 10q loss, another common overlapping region was observed at 10q11–q24. The most frequent high-level amplifications were detected at 17p11–p12 (7 cases, 27%) and 8q (6 cases, 21%). The number of aberrations was related to the tumor size. Small tumors displayed fewer changes per sample (3 to 11; mean, 7) than other tumors (4 to 22; mean, 13). One low-grade case (case 1) showed only three aberrations. The number of gains per sample increased from small tumors (mean, 4) to very large tumors (mean, 10), whereas the number of losses was similar in small tumors (mean, 4) and in large and very large tumors (mean, 5).
Table 1.
Clinical, Histological, and CGH Karyotype Findings in Leiomyosarcoma
| No./age/sex | Tumor site* (clinical outcome†) | CGH karyotype | ||
|---|---|---|---|---|
| No. of aberrations | Losses | Gains and high-level amplifications‡ | ||
| Small tumors (less than 5 cm) | ||||
| 1/67/F | Subcutis of upper thigh (alive at 1 year, NED) | 3 | 13q14–qter | 15, 16p |
| 2/25/F | Inferior vena cava (recurrence after 13 years, died after 14 years) | 6 | 10q, 13q14.2–qter, 16q | 16p, 17p, X |
| 3/54/F | Perineum, subcutaneous (alive at 1.5 years, NED) | 7 | 7p, 10, 11q14–qter, 17p | 4, 15, 16p |
| 4/55/M | Inguinal, subcutaneous (alive at 2 years, NED) | 10 | 3p22–pter, 4p15–pter, 11q23–qter, 13q21–q22, 16q, 17q | 10p12–pter, 13q31–qter, 16p, 17p (17p12) |
| 5/62/F | Skin of upper arm (alive 22 years, NED) | 11 | 2p14–pter, Xp, 10, 13q14.2–qter | 2q11–q14.3, 3q13.3–qter, 12q23–qter, 15, 16p, 17p, 20q |
| Large tumors (more than 5 cm less than 20 cm) | ||||
| 6/41/M | Leg, deep intramuscular | 4 | 10, 11p, 13 | X |
| 7/50/F | RP | 6 | 1p33–pter, 13q | 1p32–qter, 4q, 5q23–qter, X |
| 8/67/F | RP | 7 | 2p, 7p, 10q11–q21, 14q13–qter | 1q, 3q, 16p |
| 9/40/F | RP | 9 | 10q, 11p, 13q13–qter, 14q22–qter, 16q, 18q22–qter | 1q, 16p11–p12, 17 |
| 10/63/F | RP | 10 | 2p, 6p, 10q, 11, 13, 16q, 22 | 15, 17p (17p11–p12), Xp |
| 11/88/F | Leg, deep intramuscular | 10 | 4q31–qter, 6p21.2–pter10, 18q | 2q11–q23, 5p, 8q24, 9, 14q11–q23, 17p, 18p |
| 12/85/F | RP | 10 | 6p21.2–pter, 9, 10q22–qter, 16q | 3q23–q25, 4pter–q31.3, 5, 6q21–q24, 18q12–q21, X |
| 13/51/M | RP | 11 | 6p, 10, 11p, 13q | 1q, 4q, 5, 6q, 8q21–qter, 17p, Xp |
| 14/78/F | RP | 12 | 1p35–pter, 10q11–q21, 11q22–qter, 17, 18p, 19 | 2q22–q33, 4q22–q31.3, 5q13.2–q32, 6q14–q24, Xp11.3–p22.2 |
| 15/71/M | Buttock, subcutis | 14 | 2q24–qter, 6p, 7p, 8p21–pter, 10q, 13q21–qter, 19q, 20p | 7q, 10p, 15q11–q23, 17p, 19p, Xp |
| 16/80/M | Thigh, intramuscular | 15 | 1p34.3–pter, 6, 10, 11p14–pter, 13q11–q21.2, 22 | 2q11–q14.3, 5, 7, 8, 11q14–q22, 12p, 13q21.3–qter, 15, 17p |
| 17/68/F | RP | 16 | 4q11–q26, 6q21–qter, 7p, 10, 11, 13, 16q | 1q, 3p, 5, 7q, 8, 9, 15q14–qter, 17, X (Xp) |
| 18/75/M | Popliteal, intramuscular | 16 | 2p, 4q31.2–qter, 6p, 9p, 10q, 16q | 1q, 2q11–q22, 3, 5q21–qter, 7p11–p14, 8, 9q, 11q13–q23.1, 13q21–qter, 16p |
| 19/70/F | RP | 18 | 9p, 10pter–q21.2, 11q, 13q11–q14.2, 16q, 18q22–qter | 1q22–qter, 3, 4pter–q31.3, 5, 6p, 7p13–p15, 8, 9q, 16p, 17, 19q11–q13.2, 20 |
| 20/49/F | Perirenal | 20 | 8p22–pter | 1pter–q31 (1q23–q25), 2q, 3q, 5p, 6, 7p, 8q22–qter (8q23), 9q, 10p, 11p, 12, 14q, 15, 17 (17p), 18q11–q21, 20, 21, Xp |
| Very large tumors (more than 20 cm) | ||||
| 21/46/F | RP | 7 | 1p, 3q, 7q, 14 | 3p, 7p, 8 |
| 22/81/F | RP | 9 | 13q31–qter, 14 | 1q24–qter, 2, 4, 6q, 8q, 9, 18q |
| 23/86/F | RP | 9 | 3p, 9p, 10q, 12p, 22 | 2q33–q35, 4q25–q31, 6q (6q21–qter), 12q14–q24 |
| 24/84/F | Thigh, subcutaneous | 11 | 1p32–pter, 8p, 10q11–q22, 13, 18p | 2pter–q14.3, 4p13–q25, 6, 7, 8q, 17q12–qter |
| 25/72/M | RP | 13 | 18q, X | 1q21–qter, 2q13–qter, 3q13.3–qter, 5, 6, 7p13–p15, 8q (8q11–q13), 12, 17q, 19, 20p |
| 26/60/F | RP | 15 | 4q31.3–qter, 11p, 16q, 19p, 22 | 1p21–p31, 2p21–pter, 4pter–q31.2, 5pter–q31, 6q22–q25.2, 7p14–qter, 8 (8q21–q23), 16p, 17p12pter, 19q |
| 27/37/F | RP | 17 | 9p, 11q23–qter, 19p | 1 (1q), 3, 4, 5 (5p), 6q14–qter, 8, 9q, 11q14–q22, 14q13–qter, 15q13–qter, 17p11–p12, 19q11–q13.2, 21, Xp11–p21 |
| 28/46/M | RP | 22 | 2p, 5q31.3–qter, 7p, 10q11–q21.2, 11, 12p, 13q11–q14.2, 17q22–qter, 18q21–qter, 20p | 1p21–pter, 3p21–pter, 3q, 4pter–q28, 5pter–q31.2, 6, 8 (8q21.2–qter), 10p, 13q21–qter, 14q21–qter, 15q21–qter, 20q |
| 29/48/F | RP | 22 | 1p35–pter, 2, 6p22–pter, 7p, 10q, 12q24–qter, 13q11–q14, 19p | 1p34–qter, 3, 4q, 5q12–qter, 6q14–qter, 7q, 8q21.3–q24.1, 11p14–p12, 11q14–qter, 13q21–qter, 14q21–qter, 15q21–qter, 18q12–q21, X (Xq) |
*NED, no evidence of disease; RP, retroperitoneal.
†Small tumors only.
‡High-level amplifications appear in bold type.
Figure 1.
Summary of DNA copy number gains and losses detected by CGH in LMS. Each bar represents one tumor sample. Gains are on the right side, losses on the left. Dotted lines represent small tumors, broken lines large tumors, and continuous lines very large tumors.
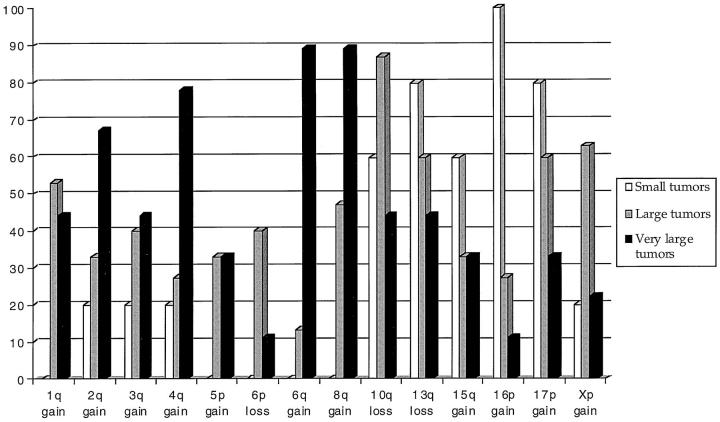
Figure 2 ▶ shows the tumor size-related aberrations. Gains in 16p were detected in all small tumors but were infrequent in other tumors (27% and 11%, respectively). Similarly, gains and high-level amplifications in 17p were more common in small and large tumors (80% and 60%, respectively) than in very large tumors (33%). Gains in 1q, 5p, 6q, and 8q were only seen in large and very large tumors. Gains in 1q with a minimal common overlapping region at 1q23–q31 were detected in eight large tumors (44%) and six very large tumors (67%). Gains of 5p were seen in five large tumors (33%) and three very large tumors (33%). Gains in 6q with a minimal common overlapping region at 6q22–q26 were seen in two cases of large tumors (13%) and eight cases of very large tumors (89%). Gains in 8q with a minimal common overlapping region at 8q24.1 were frequently seen in large tumors (eight cases, 47%) and very large tumors (eight cases, 89%), with a high-level amplification in six of them. Other gains that were associated with large and very large tumors included minimal common overlapping regions at 2q11–q14 (33% and 67%, respectively), 3q25–q26 (40%), and 4q27–q28 (27% and 78%, respectively). Gains in Xp were frequently detected in large tumors (63%) but less frequently in small and very large tumors (20%). Losses in 6p were detected in six large tumors but in none of the small tumors and only in one very large tumor.
Figure 2.
Comparative frequencies of common losses and gains detected in small, large, and very large LMS tumors.
Losses in 14q were seen in both epitheloid LMS cases but were rarely observed in the other tumors (7%). In 19 cases, 34 aberrations were seen as a gain accompanied by a loss in the same chromosome. These were either a loss of one chromosome arm with a gain of the other arm, suggesting the presence of an isochromosome or a breakpoint with a gain and a loss in the same chromosome arm indicating unbalanced translocations in these cases.
In addition, several other DNA copy number changes were detected, as shown in Table 1 ▶ and Figure 1 ▶ .
Discussion
We studied the cytogenetic changes in typical narrowly defined LMSs of soft tissues. All of these tumors were histologically typical and immunohistochemically showed phenotypic features of smooth muscle cells. All cases showed α-smooth muscle actin immunoreactivity, and most cases were at least focally positive for desmin. The GISTs, typically negative for desmin and showing only sporadic, if any, actin reactivity, were excluded from the study. Previously, these tumors have often been classified as smooth muscle tumors. The narrow definition for LMS was used in this study to better highlight the changes typical of LMS, considering that previous cytogenetic studies have shown marked heterogeneity within this group of tumors. Although genetic heterogeneity remained, regardless of the narrow definition used for LMS, some typical and nearly consistent DNA copy number changes were seen. The most striking finding in this study was the high frequency of losses in DNA copy numbers in 10q and 13q, gains in 17p, and the presence of tumor size-related aberrations. These findings suggest that the changes in 10q, 13q, and 17p may be early changes during the tumorigenesis of LMS. However, more cases need to be studied to confirm the finding. Of these changes, gains in chromosome 17 were also seen in two well-differentiated esophageal leiomyomas in our earlier study, 20 further supporting the idea that such a change might be related to an early stage in the development of smooth muscle tumors.
The DNA copy number changes in LMS differed from those in malignant GIST, suggesting different pathogenesis. Losses in 14q, typically observed in our earlier study of GIST, were only rarely seen in LMS, and losses in 10q and 13q and gains in 17p, seen frequently in LMS, were rare among GISTs. 15 The differences in DNA copy number changes in LMS and malignant GIST further support separate classification of these tumors. Losses in chromosome 14 were detected in both epitheloid LMS cases. Epitheloid LMS constitutes a morphologically distinctive group of tumors. This finding may suggest that epitheloid LMS is related to GIST more than to ordinary LMS.
Rearrangements in 10q have rarely been reported in LMS studied by standard cytogenetic techniques. 7,8 However, losses of DNA copy numbers in 10q have frequently been observed by CGH and in molecular studies of prostate carcinoma and gliomas. 21-24 Recent studies of the 10q23 region have led to the isolation of a candidate tumor suppressor gene, PTEN, that appears to be mutated at a considerable frequency in human cancers. In preliminary screenings, mutations of PTEN have been detected in glioblastoma, prostate cancer, breast cancer, and endometrial carcinoma. 25-27 Our minimal common overlapping region was 10q11–q24 in 16 of 20 cases with 10q loss. Moreover, the 10q region is known to contain the MXI1 gene located at 10q24–q25, which overlaps with the minimal common overlapping region (10q11–q24) in our cases. The MXI1 gene may negatively regulate c-myc oncogene activity, and it may have a tumor suppressor function. Altered MXI1 function as such might contribute to tumorigenesis. 21,28
Deletions in 13q have been found in several tumors, and the region contains the tumor suppressor genes RB1 at 13q14 and BRCA2 at 13q12. From 17 cases with losses in 13q, 12 (71%) had losses in 10q, which could imply a cumulative effect of deletions of several tumor suppressor genes in LMS.
Gains and high-level amplifications in 17p were strikingly present in most of our cases. A translocation involving 10q and 17p has been reported in an LMS case. 5 Gains in 17p11–p12 were frequently detected using CGH in osteosarcomas, chondrosarcoma, and malignant fibrous histiocytoma. 29-31 Although p53 mutations and p53 protein overexpression have been described in LMS, the 17p11–p12 region seems to be frequently involved in sarcomas, indicating that it may harbor other genes relevant for the development of these tumors.
Gains in 1q, 6q, and 8q were found in large tumors and more frequently in very large tumors, but not in any of the small tumors. Gains and high-level amplifications in 1q have been found in sarcomas as well as in other malignancies. 22,29,32-34 The 6q24 region contains two oncogenes, MYB and AAS1. The DNA copy gains and high-level amplifications seen at 8q involved the 8q24 region that contains the proto-oncogene c-myc, which is known to be amplified in several tumors. 35 These, in addition to some unknown oncogenes, may be related to tumor size and progression in LMS.
Gains and high-level amplifications in 3q have been detected in several tumors by CGH and they have been found to indicate tumor progression in mantle cell lymphoma, cervical carcinoma, and ovarian tumors. 15,23,36-41 Gains detected in the X chromosome had the minimal overlapping region at Xp which, according to the Genome Data Base, contains several putative target genes that have been implicated in recurrent prostate cancer, small cell lung cancer, and malignant stromal tumors of the gastrointestinal tract. 15,42-44 These regions seem to contain potential oncogenes relevant in several tumors.
In conclusion, LMSs, even when narrowly defined, show multiple and complex DNA copy number changes. The number of such changes correlates with the tumor size. DNA losses in 10q and 13q and gains in 16p and 17p may be important for tumor development, as they are seen preferentially in smaller tumors, whereas amplifications, especially at 1q, 6q, and 8q, appear to be associated with tumor progression. Further studies are needed to more accurately delineate the changes in specific genes and their possible value as clinical and prognostic markers in LMS.
Footnotes
Address reprint requests to Dr. Sakari Knuutila, Laboratory of Medical Genetics, Helsinki University Central Hospital, PO Box 404 (Haartmanink. 3, 4th floor), FIN-00029 HUCH, Helsinki, Finland. E-mail: sakari.knuutila@helsinki.fi
Supported by the Sigrid Jusélius Foundation and the Finnish Cancer Society.
References
- 1.Enzinger FM, Weiss SW: Soft Tissue Tumors, ed 3 1995, :pp 491-510 Mosby, St. Louis [Google Scholar]
- 2.Miettinen M, Virolainen M, Sarlomo-Rikala M: Gastrointestinal stromal tumors: value of CD34 antigen in their identification and separation from true leiomyomas and schwannomas. Am J Surg Pathol 1995, 19:207-216 [DOI] [PubMed] [Google Scholar]
- 3.Boghosian L, Dal Cin P, Turc-Carel C, Rao U, Karakousis C, Sait SJ, Sandberg AA: Three possible cytogenetic subgroups of leiomyosarcoma. Cancer Genet Cytogenet 1989, 43:39-49 [DOI] [PubMed] [Google Scholar]
- 4.Bardi G, Johansson B, Pandis N, Heim S, Mandahl N, Bak-Jensen E, Frederiksen H, Andren-Sandberg A, Mitelman F: Recurrent chromosome aberrations in abdominal smooth muscle tumors. Cancer Genet Cytogenet 1992, 62:43-46 [DOI] [PubMed] [Google Scholar]
- 5.Dal Cin P, Boghosian L, Crickard K, Sandberg AA: t(10;17) as the sole chromosome change in a uterine leiomyosarcoma. Cancer Genet Cytogenet 1988, 32:263-266 [DOI] [PubMed] [Google Scholar]
- 6.Han K, Lee W, Harris CP, Simsiman RC, Lee K, Kang C, Meisner LF: Comparison of chromosome aberrations in leiomyoma and leiomyosarcoma using FISH on archival tissues. Cancer Genet Cytogenet 1994, 74:19-24 [DOI] [PubMed] [Google Scholar]
- 7.Sreekantaiah C, Davis JR, Sandberg AA: Chromosomal abnormalities in leiomyosarcomas. Am J Pathol 1993, 142:293-305 [PMC free article] [PubMed] [Google Scholar]
- 8.Kiechle-Schwarz M, Berger CS, Surti U, Sandberg AA: Rearrangement of band 10q22 in leiomyoma and leiomyosarcoma of the uterus. Cancer Genet Cytogenet 1990, 47:95-100 [DOI] [PubMed] [Google Scholar]
- 9.Saunders AL, Meloni AM, Chen Z, Sandberg AA, Lauwers GY: Two cases of low-grade gastric leiomyosarcoma with monosomy 14 as the only change. Cancer Genet Cytogenet 1996, 90:184-185 [DOI] [PubMed] [Google Scholar]
- 10.Dei Tos AP, Maestro R, Doglioni C, Piccinin S, Libera DD, Boiocchi M, Fletcher CD: Tumor suppressor genes and related molecules in leiomyosarcoma. Am J Pathol 1996, 148:1037-1045 [PMC free article] [PubMed] [Google Scholar]
- 11.Hall KL, Teneriello MG, Taylor RR, Lemon S, Ebina M, Linnoila RI, Norris JH, Park RC, Birrer MJ: Analysis of Ki-ras, p53, and MDM2 genes in uterine leiomyomas and leiomyosarcomas. Gynecol Oncol 1997, 65:330-335 [DOI] [PubMed] [Google Scholar]
- 12.Patterson H, Gill S, Fisher C, Law MG, Jayatilake H, Fletcher CD, Thomas M, Grimer R, Gusterson BA, Cooper CS: Abnormalities of the p53, MDM2 and DCC genes in human leiomyosarcomas. Br J Cancer 1994, 69:1052-1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoo J, Lee HK, Kang CS, Park WS, Lee JY, Shim SI: p53 gene mutations and p53 protein expression in human soft tissue sarcomas. Arch Pathol Lab Med 1997, 121:395-399 [PubMed] [Google Scholar]
- 14.Kallioniemi A, Kallioniemi OP, Piper J, Tanner M, Stokke T, Chen L, Smith HS, Pinkel D, Gray JW, Waldman FM: Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci USA 1994, 91:2156-2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El-Rifai W, Sarlomo-Rikala M, Miettinen M, Knuutila S, Andersson LC: DNA copy number losses in chromosome 14: an early change in gastrointestinal stromal tumors. Cancer Res 1996, 56:3230-3233 [PubMed] [Google Scholar]
- 16.Ried T, Knutzen R, Steinbeck R, Blegen H, Schrock E, Heselmeyer K, du Manoir S, Auer G: Comparative genomic hybridization reveals a specific pattern of chromosomal gains and losses during the genesis of colorectal tumors. Genes Chromosomes Cancer 1996, 15:234-245 [DOI] [PubMed] [Google Scholar]
- 17.Miller SA, Dykes DD, Polesky HF: A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988, 16:1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kallioniemi OP, Kallioniemi A, Piper J, Isola J, Waldman FM, Gray JW, Pinkel D: Optimizing comparative genomic hybridization for analysis of DNA sequence copy number changes in solid tumors. Genes Chromosomes Cancer 1994, 10:231-243 [DOI] [PubMed] [Google Scholar]
- 19.El-Rifai W, Larramendy ML, Björkqvist A-M, Hemmer S, Knuutila S: Optimization of comparative genomic hybridization using fluorochrome conjugated to dCTP and dUTP nucleotides. Lab Invest 1997, 77:699-700 [PubMed] [Google Scholar]
- 20.Sarlomo-Rikala M, El-Rifai W, Lahtinen T, Andersson L, Miettinen M, Knuutila S: Different patterns of DNA copy number changes in gastrointestinal stromal tumors, leiomyomas, and schwannomas. Hum Pathol 1998, 29:476-481 [DOI] [PubMed] [Google Scholar]
- 21.Gray IC, Phillips SM, Lee SJ, Neoptolemos JP, Weissenbach J, Spurr NK: Loss of the chromosomal region 10q23–25 in prostate cancer. Cancer Res 1995, 55:4800-4803 [PubMed] [Google Scholar]
- 22.Weber RG, Sommer C, Albert FK, Kiessling M, Cremer T: Clinically distinct subgroups of glioblastoma multiforme studied by comparative genomic hybridization. Lab Invest 1996, 74:108-119 [PubMed] [Google Scholar]
- 23.Cher ML, Bova GS, Moore DH, Small EJ, Carroll PR, Pin SS, Epstein JI, Isaacs WB, Jensen RH: Genetic alterations in untreated metastases and androgen-independent prostate cancer detected by comparative genomic hybridization and allelotyping. Cancer Res 1996, 56:3091-3102 [PubMed] [Google Scholar]
- 24.Weber RG, Sabel M, Reifenberger J, Sommer C, Oberstrass J, Reifenberger G, Kiessling M, Cremer T: Characterization of genomic alterations associated with glioma progression by comparative genomic hybridization. Oncogene 1996, 13:983-994 [PubMed] [Google Scholar]
- 25.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, Parsons R: PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 1997, 275:1943-1947 [DOI] [PubMed] [Google Scholar]
- 26.Tashiro H, Blazes MS, Wu R, Cho KR, Bose S, Wang SI, Li J, Parsons R, Ellenson LH: Mutations in PTEN are frequent in endometrial carcinoma but rare in other common gynecological malignancies. Cancer Res 1997, 57:3935-3940 [PubMed] [Google Scholar]
- 27.Rhei E, Kang L, Bogomolniy F, Federici MG, Borgen PI, Boyd J: Mutation analysis of the putative tumor suppressor gene PTEN/MMAC1 in primary breast carcinomas. Cancer Res 1997, 57:3657-3659 [PubMed] [Google Scholar]
- 28.Wechsler DS, Shelly CA, Dang CV: Genomic organization of human MXI1, a putative tumor suppressor gene. Genomics 1996, 32:466-470 [DOI] [PubMed] [Google Scholar]
- 29.Forus A, Weghuis DO, Smeets D, Fodstad O, Myklebost O, van Kessel AG: Comparative genomic hybridization analysis of human sarcomas. II. Identification of novel amplicons at 6p and 17p in osteosarcomas. Genes Chromosomes Cancer 1995, 14:15-21 [DOI] [PubMed] [Google Scholar]
- 30.Larramendy ML, Tarkkanen M, Valle J, Kivioja AH, Ervasti H, Karaharju E, Salmivalli T, Elomaa I, Knuutila S: Gains, losses, and amplifications of DNA sequences evaluated by comparative genomic hybridization in chondrosarcomas. Am J Pathol 1997, 150:685-691 [PMC free article] [PubMed] [Google Scholar]
- 31.Larramendy ML, Tarkkanen M, Blomqvist C, Virolainen M, Wiklund T, Asko-Seljavaara S, Elomaa I, Knuutila S: Comparative genomic hybridization of malignant fibrous histiocytoma reveals a novel prognostic marker. Am J Pathol 1997, 151:1153-1161 [PMC free article] [PubMed] [Google Scholar]
- 32.Gronwald J, Storkel S, Holtgreve-Grez H, Hadaczek P, Brinkschmidt C, Jauch A, Lubinski J, Cremer T: Comparison of DNA gains and losses in primary renal clear cell carcinomas and metastatic sites: importance of 1q and 3p copy number changes in metastatic events. Cancer Res 1997, 57:481-487 [PubMed] [Google Scholar]
- 33.Björkqvist AM, Tammilehto L, Anttila S, Mattson K, Knuutila S: Recurrent DNA copy number changes in 1q, 4q, 6q, 9p, 13q, 14q, and 22q detected by comparative genomic hybridization in malignant mesothelioma. Br J Cancer 1997, 75:523-527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forus A, Weghuis DO, Smeets D, Fodstad O, Myklebost O, van Kessel AG: Comparative genomic hybridization analysis of human sarcomas. I. Occurrence of genomic imbalances and identification of a novel major amplicon at 1q21–q22 in soft tissue sarcomas. Genes Chromosomes Cancer 1995, 14:8-14 [DOI] [PubMed] [Google Scholar]
- 35.Brison O: Gene amplification and tumor progression. Biochim Biophys Acta 1993, 1165:25-41 [DOI] [PubMed] [Google Scholar]
- 36.Heselmeyer K, Schrock E, du Manoir S, Blegen H, Shah K, Steinbeck R, Auer G, Ried T: Gain of chromosome 3q defines the transition from severe dysplasia to invasive carcinoma of the uterine cervix. Proc Natl Acad Sci USA 1996, 93:479-484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iwabuchi H, Sakamoto M, Sakunaga H, Ma YY, Carcangiu ML, Pinkel D, Yang Feng TL, Gray JW: Genetic analysis of benign, low-grade, and high-grade ovarian tumors. Cancer Res 1995, 55:6172-6180 [PubMed] [Google Scholar]
- 38.Balsara BR, Sonoda G, du Manoir S, Siegfried JM, Gabrielson E, Testa JR: Comparative genomic hybridization analysis detects frequent, often high-level, overrepresentation of DNA sequences at 3q, 5p, 7p, and 8q in human non-small cell lung carcinomas. Cancer Res 1997, 57:2116-2120 [PubMed] [Google Scholar]
- 39.Marchio A, Meddeb M, Pineau P, Danglot G, Tiollais P, Bernheim A, Dejean A: Recurrent chromosomal abnormalities in hepatocellular carcinoma detected by comparative genomic hybridization. Genes Chromosomes Cancer 1997, 18:59-65 [PubMed] [Google Scholar]
- 40.Monni O, Oinonen R, Elonen E, Franssila K, Teerenhovi L, Joensuu H, Knuutila S: Gains of 3q and deletion of 11q22 are frequent aberrations in mantle cell lymphoma. Genes Chromosomes Cancer 1998, 21:298-307 [DOI] [PubMed] [Google Scholar]
- 41.Björkqvist A-M, Husgafvel-Pursiainen K, Anttila S, Karjalainen A, Tammilehto L, Mattson K, Vainio H, Knuutila S: DNA gains in 3q occur frequently in squamous cell carcinoma of the lung, but not in adenocarcinoma. Genes Chromosomes Cancer 1998, 22:79-82 [PubMed] [Google Scholar]
- 42.Ried T, Petersen I, Holtgreve Grez H, Speicher MR, Schrock E, du Manoir S, Cremer T: Mapping of multiple DNA gains and losses in primary small cell lung carcinomas by comparative genomic hybridization. Cancer Res 1994, 54:1801-1806 [PubMed] [Google Scholar]
- 43.Visakorpi T, Kallioniemi AH, Syvänen AC, Hyytinen ER, Karhu R, Tammela T, Isola JJ, Kallioniemi OP: Genetic changes in primary and recurrent prostate cancer by comparative genomic hybridization. Cancer Res 1995, 55:342-347 [PubMed] [Google Scholar]
- 44.Visakorpi T, Hyytinen E, Koivisto P, Tanner M, Keinanen R, Palmberg C, Palotie A, Tammela T, Isola J, Kallioniemi OP: In vivo amplification of the androgen receptor gene, and progression of human prostate cancer. Nat Genet 1995, 9:401-406 [DOI] [PubMed] [Google Scholar]